Abstract
Coagulase-negative staphylococci, particularly Staphylococcus epidermidis, can be regarded as potential reservoirs of resistance genes for pathogenic strains, e.g., Staphylococcus aureus. The aim of this study was to assess the prevalence of different resistance phenotypes to macrolide, lincosamide, and streptogramins B (MLSB) antibiotics among erythromycin-resistant S. epidermidis, together with the evaluation of genes promoting the following different types of MLSB resistance:ermA, ermB, ermC,msrA, mphC, and linA/A’. Susceptibility to spiramycin was also examined. Among 75 erythromycin-resistantS. epidermidis isolates, the most frequent phenotypes were macrolides and streptogramins B (MSB) and constitutive MLSB (cMLSB). Moreover, all strains with the cMLSB phenotype and the majority of inducible MLSB (iMLSB) isolates were resistant to spiramycin, whereas strains with the MSB phenotype were sensitive to this antibiotic. The D-shape zone of inhibition around the clindamycin disc near the spiramycin disc was found for some spiramycin-resistant strains with the iMLSB phenotype, suggesting an induction of resistance to clindamycin by this 16-membered macrolide. The most frequently isolated gene was ermC, irrespective of the MLSB resistance phenotype, whereas the most often noted gene combination wasermC, mphC, linA/A’. The results obtained showed that the genes responsible for different mechanisms of MLSB resistance in S. epidermidis generally coexist, often without the phenotypic expression of each of them.
Keywords: Staphylococcus epidermidis, MLSB antibiotics, resistance, genotypes, spiramycin
Coagulase-negative staphylococci (CoNS), particularly Staphylococcus epidermidis, belong to the microbiota of human skin and the mucosal membrane of the upper respiratory tract, and they express low pathogenic potential as commensals in healthy people (Voung & Otto 2002, Otto 2009). However, they can be responsible for several serious infections in immunocompromised patients, particularly those associated with biomaterials (e.g., catheters, prosthetics etc.), leading to bacteraemia and sepsis (Ziebuhr et al. 2006,Caesy et al. 2007, Schoenfelder et al. 2010, Castro-Alarcón et al. 2011). On the other hand, as a natural part of the microflora, drug resistant strains may be selected during antibiotic therapy, which is a potential source of the resistance genes for pathogenic strains, e.g.,Staphylococcus aureus (Reyes et al. 2007, Otto 2013, Vitali et al. 2014).
Resistance to macrolide, lincosamide, and streptogramins B (MLSB antibiotics) in staphylococci is associated with the following three mechanisms: (i) target modification, (ii) efflux pumps, and (iii) enzymatic modification of antibiotics. The first macrolide-resistant staphylococcal strains were identified in the 1950s (Roberts 2004). Currently, a large number of strains exhibit resistance to these antibiotics via different mechanisms. It is known that macrolide-resistant strains often exhibit co-resistance to other MLSB antibiotics. The most common mechanism is the modification of ribosomes as a result of methylation of adenine within 23S rRNA ribosomal subunits by a methylase encoded by the erm genes (predominantlyermC). Conformational changes in the ribosome result in the reduced binding of all MLSB antibiotics; these strains are resistant to all MLSB antibiotics (the combination of quinupristin/dalfopristin loses bactericidal activity as the result of the development of resistance to quinupristin). The phenotypic expression of MLSB resistance can be either inducible (iMLSB) (generally induced by 14 and 15-membered macrolides) or constitutive (cMLSB) (Weisblum 1995). The active efflux of antibiotics is mediated by msr genes (mainlymsrA) and is responsible for resistance only to 14 and 15-membered macrolides and streptogramins B (MSB) phenotype (Reynolds et al. 2003). The third mechanism of resistance is based on the production of antibiotic-inactivating enzymes (e. g., phosphorylase encoded bymph or lin, the gene responsible for inactivation of lincosamides) (Chesneau et al. 2007, Achard et al. 2008).
The aim of this study was to assess the prevalence of different MLSBresistance phenotypes among S. epidermidis, together with the evaluation of genes responsible for target modification (ermA,ermB, ermC), antibiotic efflux (msrA) or antibiotic inactivation (mphC,linA/A’). The evaluation of susceptibility to the 16-membered macrolide spiramycin was also performed.
This paper was developed using the equipment purchased within agreement POPW.01.03.00-06-010/09-00 Operational Program Development of Eastern Poland 2007-2013, Priority Axis I, Modern Economy, Operations 1.3. Innovations Promotion.
SUBJECTS, MATERIALS AND METHODS
Bacterial strains - A total of 197 strains of S. epidermidis were obtained from the mucosal membranes of the upper respiratory tracts of patients with nonsmall cell lung cancer who underwent hospitalisation. Nasal and pharyngeal swabs were obtained on the second day of the patients’ stays at the hospital. Among the strains, resistance to erythromycin was detected in 75 isolates.
Isolation and identification - Isolation and identification of bacterial strains were performed using routine microbiological tests. The following tests were used in the identification of CoNS: the coagulase test tube using rabbit plasma (Biomed, Poland) and API Staph strips (bioMérieux, France).
Identification of resistance to MLS B antibiotics - Susceptibility to MLSB antibiotics, including the detection of resistance mechanisms, was based on the D-test according to European Centre for Disease Prevention and Control (EUCAST) recommendations. In addition, disks containing lincomycin (15 mg) were used to identify the L-phenotype. Moreover, for detection of the effects of spiramycin on clindamycin susceptibility, discs containing spiramycin (100 mg) were applied next to clindamycin (2 mg).
Determination of minimal inhibitory concentrations (MICs) to spiramycin - Detection of MICs to spiramycin was based on EUCAST recommendations using the double broth dilution method. In the absence of breakpoints for spiramycin in EUCAST, only the MICs were evaluated without grouping the strains as susceptible or resistant.
Isolation of bacterial DNA - The DNA Genomic Mini Kit (A&A Biotechnology, Poland) was used to isolate S. epidermidis DNA according to the manufacturer’s guidelines.
Identification of genes by polymerase chain reaction (PCR) - The sequences of the primers and the conditions of the PCR reactions are presented inTable I. For the PCR reactions, PCR REDTaq® Ready MixTM PCR Mix with MgCl2(Sigma-Aldrich, USA) was used. The final volume of each PCR reaction was 25 ml and contained 12.5 ml of REDTaq Ready Mix, 1 ml of each forward and reverse primer (concentration between 0.1-1.0 mM), 1 ml of DNA (50-200 ng), and 9 ml of water. The reactions were performed using a Whatman Biometra thermocycler, whereas the PCR products were subjected to agarose gel electrophoresis (2% agarose, 1xTRIS-acetate-EDTA, 120 mV, 40 min). The gels were stained with ethidium bromide and the PCR products were visualised using a Wilbert Lambert transilluminator and compared with molecular size markers [Gene RulerTM 100 bp DNA Ladder (Fermentas, Thermo Scientific, USA)].
TABLE I. Primers sequence, thermal cycling profile, and size of amplified polymerase chain reaction (PCR) fragment in each PCR reaction in the detection of genes of Staphylococcus epidermidisresistant to erythromycina .
| Gene | Primers sequence | PCR conditions | PCR fragment size (bp) |
|---|---|---|---|
| ermA | 5’-TCTAAAAAGCATGTAAAAGAA-3’ 5’-CTTCGATAGTTTATTAATATTAGT-3’ | 35 (30 s at 94ºC, 1 min at 48ºC, 2 min at 72ºC) | 645 |
| ermB | 5’-GAAAAGGTACTCAACCAAATA-3’ 5’-AGTAACGGTACTTAAATTGTTTAC-3’ | 35 (30 s at 94ºC, 30 s at 50ºC, 2 min at 72ºC) | 639 |
| ermC | 5’-AGTACAGAGGTGTAATTTCG-3’ 5’-AATTCCTGCATGTTTTAAGG-3’ | 35 (55 s at 94ºC, 1 min at 53ºC, 1 min at 72ºC) | 642 |
| msrA | 5’-GGCACAATAAGAGTGTTTAAAGG-3’ 5’-AAGTTATATCATGAATAGATTGTCCTGTT-3’ | 25 (1 min at 94ºC, 1 min at 50ºC, 90 s at 72ºC) | 399 |
| mphC | 5’-GAGACTACCAGACCTGACG-3’ 5’-CATACGCCGATTCTCCTGAT-3’ | 35 (1 min at 94ºC, 1 min at 59ºC, 1 min at 72ºC) | 530 |
| linA/A’ | 5’-GGTGGCTGGGGGGTAGATGTATTAACTGG-3’ 5’-GCTTCTTTTGAAATACATGGTATTTTTCGATC-3’ | 30 (30 s at 94ºC, 30 s at 57ºC, 1 min at 72ºC) | 323 |
a: Sutcliffe et al. (1996) and Lina et al. (1999).
Ethics - The study design and protocols were approved by the Ethical Committee of the Medical University of Lublin (KE-0254/75/2011).
RESULTS
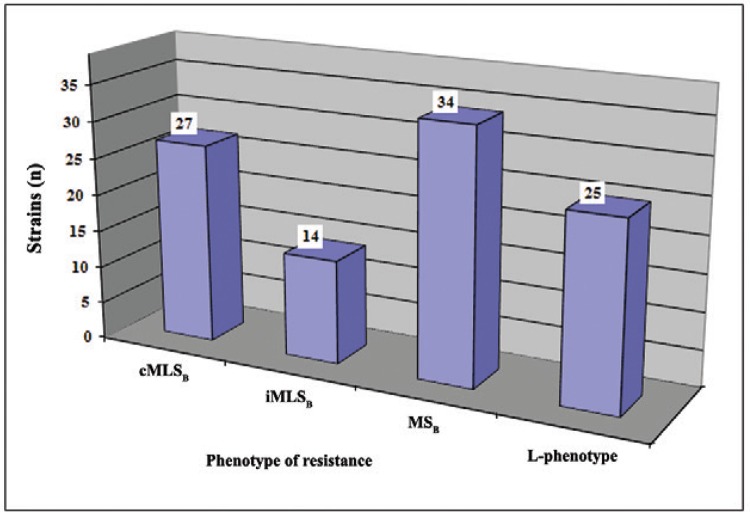
The 75 S. epidermidis isolates expressed resistance to erythromycin with the following mechanisms of resistance: 27 (36%) strains exhibited cMLSB resistance, 14 (18.7%) strains exhibited iMLSBresistance, and 34 (45.3%) strains exhibited MSB resistance (Figure). Twenty-five isolates exhibited L-phenotypes and were determined to be either resistant to only lincomycin (24 strains) or resistant to lincomycin and clindamycin (1 strain).
The prevalence of different mechanisms of resistance to macrolide, lincosamide, and streptogramins B (MLSB) antibiotics among erythromycin-resistant Staphylococcus epidermidis. cMLSB: constitutive resistance to MLSB antibiotics; iMLSB: inducible resistance to MLSB antibiotics; MSB: resistance of MSB type.
The MICs of spiramycin among erythromycin-resistant S. epidermidiswere evaluated as follows: > 128 mg/L for all cMLSB strains, from 4-> 128 mg/L for iMLSB strains, and from 1-4 mg/L for strains exhibiting the MSB phenotype. The MIC50 and MIC90values were also calculated. Strains with cMLSB and iMLSBphenotypes exhibited MIC50 and MIC90 values > 128 mg/L, whereas the MIC50 and MIC90 values for the MSBstrains were determined to 4 mg/L (Table II). Moreover, for the 11 (78.6%) strains exhibiting iMLSB phenotypes, the noninhibition zone around the spiramycin disc was found together with a D-shaped zone around the clindamycin disk.
TABLE II. The minimal inhibitory concentrations (MICs) to spiramycin among erythromycin-resistant Staphylococcus epidermidis .
| mg/L | iMLSB | cMLSB | MSB |
|---|---|---|---|
| MIC range | 4-> 128 | > 128 | 1-4 |
| MIC50 | > 128 | > 128 | 4 |
| MIC90 | > 128 | > 128 | 4 |
cMLSB: constitutive resistance to macrolide, lincosamide, and streptogramins B (MLSB) antibiotics; iMLSB: inducible resistance to MLSBantibiotics; MSB: resistance of MSB type.
As shown in Table III, among the strains with cMLSB resistance, the predominant genes were ermCand mphC in 23 (85.2%) and 24 (88.9%) strains, respectively.linA/A’ was found to occur in 14 (51.8%) strains. The presence of other genes (e.g., ermA and ermB) was detected in a few strains; two strains did not possess any of the erm genes. The isolates with iMLSB possessed the following genes:ermC - 14 (100%) strains, msrA - 7 (50%) strains, mphC - 13 (92.9%) strains, and linA/A’ - 10 (71.4%) strains; ermA and ermB were not detected. The strains exhibiting MSB resistance were found to possess the following genes: ermC in 20 (58.8%) strains, msrAin 32 (94.1%) strains, mphC in 33 (97.1%) strains, andlinA/A’ in 24 (70.6%) strains; these strains did not carryermA or ermB. The strains exhibiting L-phenotypes contained linA/A’ in 24 (96%) strains,mphC in 23 (92%) strains, and ermC in 24 (96%) strains. ermA, ermB, and msrAwere not detected in the isolates with L-phenotypes. One strain did not carry any of the evaluated genes.
TABLE III. The prevalence of genes responsible for resistance to macrolide, lincosamide, and streptogramins B (MLSB) antibiotics among erythromycin-resistant Staphylococcus epidermidis .
| Gene | Phenotypes n (%) | |||
|---|---|---|---|---|
|
| ||||
| cMLSB (n = 27) | iMLSB (n = 14) | MSB (n = 34) | L-phenotype (n = 25) | |
| ermA | 4 (14.8) | 0 (0) | 0 (0) | 0 (0) |
| ermB | 1 (3.7) | 0 (0) | 0 (0) | 0 (0) |
| ermC | 23 (85.2) | 14 (100) | 20 (58.8) | 24 (96) |
| msrA | 5 (18.5) | 7 (50) | 32 (94.1) | 0 (0) |
| mphC | 24 (88.9) | 13 (92.9) | 33 (97.1) | 23 (92) |
| linA/A’ | 14 (51.8) | 10 (71.4) | 24 (70.6) | 24 (96) |
cMLSB: constitutive resistance to MLSBantibiotics; iMLSB: inducible resistance to MLSB antibiotics; MSB: resistance of MSB type.
Table IV shows the combination of genes responsible for resistance to MLSB antibiotics among staphylococci. In isolates exhibiting cMLSB resistance, 11 different combinations were detected. The most frequent gene combination was ermC,mphC, and linA/A’, which was found in 10 (37%) strains. Among the strains exhibiting iMLSB resistance, four gene combinations were evaluated. The most frequent combinations contained the following genes: ermC, mphC, and linA/A’ in five (35.7%) isolates and ermC, msrA,mphC, and linA/A’, also in five (35.7%) isolates. The MSB-positive strains contained six different gene combinations in three major groups: ermC, msrA,mphC, and linA/A’ in 14 (41.2%) strains;msrA, mphC, and linA/A’ in nine (26.5%) strains, and ermC, msrA, andmphC in six (17.6%) strains. In the isolates with L-phenotypes, the most significant three-gene combination was ermC,mphC, and linA/A’ in 21 (84%) strains.
TABLE IV. The prevalence of gene combinations responsible for resistance to macrolide, lincosamide, and streptogramins B (MLSB) antibiotics among erythromycin-resistant Staphylococcus epidermidis .
| Gene combinations | Phenotypes n (%) | |||
|---|---|---|---|---|
|
| ||||
| cMLSB (n = 27) | iMLSB (n = 14) | MSB (n = 34) | L-phenotype (n = 25) | |
| ermC | 1 (3.7) | 1 (7.1) | 0 (0) | 0 (0) |
| mphC | 0 (0) | 0 (0) | 2 (5.9) | 0 (0) |
| ermC, mphC | 4 (14.8) | 3 (21.4) | 0 (0) | 0 (0) |
| ermB, mphC | 1 (3.7) | 0 (0) | 0 (0) | 0 (0) |
| ermC, linA/A’ | 1 (3.7) | 0 (0) | 0 (0) | 2 (8) |
| ermA, mphC | 1 (3.7) | 0 (0) | 0 (0) | 0 (0) |
| msrA, mphC | 0 (0) | 0 (0) | 2 (5.9) | 0 (0) |
| msrA, linA/A’ | 0 (0) | 0 (0) | 1 (2.9) | 0 (0) |
| mphC, linA/A’ | 0 (0) | 0 (0) | 0 (0) | 1 (4) |
| ermC, msrA,mphC | 3 (11.1) | 0 (0) | 6 (17.6) | 0 (0) |
| ermC, mphC,linA/A’ | 10 (37) | 5 (35.7) | 0 (0) | 21 (84) |
| msrA, mphC,linA/A’ | 1 (3.7) | 0 (0) | 9 (26.5) | 0 (0) |
| ermA, ermC,mphC | 2 (7.4) | 0 (0) | 0 (0) | 0 (0) |
| ermC, msrA,mphC, linA/A’ | 1 (3.7) | 5 (35.7) | 14 (41.2) | 0 (0) |
| ermA, ermC,mphC, linA/A’ | 1 (3.7) | 0 (0) | 0 (0) | 0 (0) |
| Without genes | 1 (3.7) | 0 (0) | 0 (0) | 1 (4) |
cMLSB: constitutive resistance to MLSBantibiotics; iMLSB: inducible resistance to MLSB antibiotics; MSB: resistance of MSB type.
DISCUSSION
CoNS are potential reservoirs of antibiotic resistance genes, which can be transferred to S. aureus not only in vitro but also in vivo (Reyes et al. 2007, Otto 2013). Erythromycin resistance among CoNS was previously reported to result from a methylase encoded by different erm family genes that can be horizontally transferred to recipient strains (Zmantar et al. 2011, Vitali et al. 2014). Hence, surveillance of erythromycin resistance and MLSB resistance in CoNS at phenotypic and genetic levels can provide important information regarding their current epidemiology.
Among the S. epidermidis strains studied, the most frequently identified gene in strains exhibiting both cMLSB and iMLSBphenotypes was ermC, which is consistent with previous reports (Reyes et al. 2007, Gherardi et al. 2009, Coutinho et al. 2010, Bouchami et al. 2011,Brzychczy-Wloch et al. 2013, Heb & Gallert 2014). Only a few S. epidermidis exhibiting cMLSB phenotypes possessedermA and/or ermB. Similar data have been previously reported (Bouchami et al. 2011,Teodoro et al. 2012, Szczuka et al. 2016). Moreover, the presence of other erm genes (e.g., ermF) has been rarely detected in Staphylococcus spp (Roberts 2004). Notably, the distribution of erm genes depends on the bacterial species. For example, ermA is more characteristic of S. aureus, whereas ermB is more characteristic of beta-haemolytic streptococci (Roberts 2004, Buter et al. 2010,Meehan et al. 2014, Vitali et al. 2014). Moreover, among CoNS, the type oferm gene also depends on the geographical region of their isolation. For example, ermC was previously detected in 50% of the strains exhibiting MLSB resistance in Great Britain, whereas it was detected 90% of those in Denmark (Lim et al. 2002, Gatermann et al. 2007, Cetin et al. 2010, Bouchami et al. 2011) and in Mexico, ermA was reported as predominant in S. epidermidis (Castro-Alarcón et al. 2011).
The MSB S. epidermidis isolates examined contained anmsrA gene encoding an ATP-dependent efflux pump, which actively removes 14-,15-membered MSB. The MSB phenotype observed inmsrA-negative S. epidermidis strains may be the result of the presence of mphC, which encodes for a macrolide-modifying enzyme (Gatermann et al. 2007), thereby resulting in a “false-positive” MSBphenotype.
All S. epidermidis isolates with L-phenotypes generally contained the linA/A’ gene. Data from Novotna et al. (2005, 2007) also indicated a connection between the presence of the linA/A’ gene and resistance to only lincomycin among staphylococci. The S. epidermidis strains studied exhibited resistance to lincomycin, but susceptibility to clindamycin as a result of increased enzyme affinity for lincomycin (Achard et al. 2005). Resistance both to lincomycin and clindamycin may be a consequence of the presence of otherlin family genes orvga(A) LC, which encodes a “new” variant of the SgA protein that is responsible for cross-resistance to streptogramins A and all lincosamides (Novotna & Janata 2006).
Among the iMLSB and cMLSB S. epidermidisstrains, the erm genes do not exist separately, but in combination with others (predominantly with mphC). Notably, othererm genes (e.g., ermF), which are rarely detected in Sta- phylococcus spp, may encode both the inducible or constitutive MLSB phenotypes (Roberts 2004). In MSB-positive S. epidermidisstrains, the msrA genes predominantly coexist withermC, mphC, and linA/A’, and the coexistence of msrA and ermC has also been previously reported (Roberts 2004, Novotna et al. 2007, Wang et al. 2008, Teodoro et al. 2012). Moreover, the presence of the linA/A’ gene inmsrA-positive strains results in resistance to lincomycin. TheS. epidermidis strains exhibiting L-phenotypes correlated with the presence of the linA/A’ gene in most of the strains that also contained the ermC and mphC genes, whereas those strains did not contain the msrA gene. Notably, theermC genes were also detected in both of the MSB and L-phenotype S. epidermidis strains - but without its expression - suggesting a defect in ermC expression.
Previous studies have reported (Leclercq 2002,Coutinho et al. 2010) that 16-membered macrolides (e.g., spiramycin) are not inducers of MLSB resistance in staphylococci. According to our data, spiramycin is able to induce resistance to clindamycin among the iMLSB S. epidermidis isolates examined. Moreover, iMLSB S. epidermidis strains, which contain ermC, exhibited resistance to spiramycin in vitro. These observations contradict previous reports that 16-membered macrolides remain active against staphylococci that exhibit iMLSB phenotypes (Leclercq 2002, Szczuka et al. 2016). Notably, resistance to spiramycin appears to be characteristic of iMLSB streptococci containing ermB(Leclercq 2002, Acikgoz et al. 2003).
The diversity of genes involved in different mechanisms that are responsible for the resistance of S. epidermidis to MLSB antibiotics suggests that the insensitivity of CoNS strains to these antibacterial drugs is not necessarily a unidirectional process and that the coexistence of various genes may influence the nature of their resistance.
REFERENCES
- Achard A, Guérin-Faublée V, Pichereau V, Villers C, Leclercq R. Emergence of macrolide resistance gene mph(B) in Streptococcus uberis and cooperative effects with rdmC-like gene. Antimicrob Agents Chemother. 2008;52:2767–2770. doi: 10.1128/AAC.00481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard A, Villers C, Pichereau V, Leclercq R. New lnuC gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob Agents Chemother. 2005;49:2716–2719. doi: 10.1128/AAC.49.7.2716-2719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acikgoz ZC, Gocer S, Tuncer S. Macrolide resistance determinants of group A streptococci in Ankara, Turkey. J Antimicrob Chemother. 2003;52:110–112. doi: 10.1093/jac/dkg300. [DOI] [PubMed] [Google Scholar]
- Bouchami O, Achour W, Hassen AB. Prevalence of resistance phenotypes and genotypes to macrolide, lincosamide, and streptogramin in Gram-positive cocci isolated in Tunisian Bone Marrow Transplant Center. Pathol Biol. 2011;59:199–206. doi: 10.1016/j.patbio.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Brzychczy-Wloch M, Borszewska-Kornacka M, Gulczynska E, Wojkowska-Mach J, Sulik M, Grzebyk M, Luchter M, Heczko PB, Bulanda M. Prevalence of antibiotic resistance in multi-drug resistant coagulase-negative staphylococci isolated from invasive infection in very low birth weight neonates in two Polish NICUs. Ann Clin Microbiol Antimicrob. 2013;12(41) doi: 10.1186/1476-0711-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buter CCVL, Mouton JW, Klaassen CHW, Handgraaf CMA, Sunnen S, Melchers WJG, Sturm PDJ. Prevalence and molecular mechanism of macrolide resistance in b-haemolytic streptococci in The Netherlands. Int J Antimicrob Agents. 2010;35:590–592. doi: 10.1016/j.ijantimicag.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Caesy AL, Lambert PA, Elliott TSJ. Staphylococci. Int J Antimicrob Agents. 2007;29(Suppl. 3):S23–S32. doi: 10.1016/S0924-8579(07)72175-1. [DOI] [PubMed] [Google Scholar]
- Castro-Alarcón N, Ribas-Aparicio RM, Silva-Sánchez J, Calderón-Navarro A, Sánchez-Pérez A, Parra-Rojas I, Aparicio-Ozores G. J Med Microbiol. Vol. 60. 730-736: 2011. Molecular typing and characterization of macrolide, lincosamide, and streptogramin resistance in Staphylococcus epidermidis strains isolated in a Mexican hospital. [DOI] [PubMed] [Google Scholar]
- Cetin ES, Gunes H, Kaya S, Aridogan BC, Demirci M. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among clinical staphylococcal isolates in a Turkish university hospital. J Microbiol Immunol Infect. 2010;43:524–529. doi: 10.1016/S1684-1182(10)60081-3. [DOI] [PubMed] [Google Scholar]
- Chesneau O, Tsvetkova K, Courvalin P. Resistance phenotypes conferred by macrolide phosphotransferases. FEMS Microbiol Lett. 2007;269:317–322. doi: 10.1111/j.1574-6968.2007.00643.x. [DOI] [PubMed] [Google Scholar]
- Coutinho VLS, Paiva RM, Reiter KC, de-Paris F, Barth AL, Mombach AB, Machado P. Distribution of erm genes and low prevalence of inducible resistance to clindamycin among staphylococci isolates. Braz J Infect Dis. 2010;14:564–568. doi: 10.1016/s1413-8670(10)70113-6. [DOI] [PubMed] [Google Scholar]
- Gatermann SG, Koschinski T, Friedrich S. Distribution and expression of macrolide resistance genes in coagulase-negative staphylococci. Clin Microbiol Infect. 2007;13:777–781. doi: 10.1111/j.1469-0691.2007.01749.x. [DOI] [PubMed] [Google Scholar]
- Gherardi G, Florio L, Lorino G, Fico L, Dicuonzo G. Macrolide resistance genotypes and phenotypes among erythromycin-resistant clinical isolates of Staphylococcus aureus and coagulase-negative staphylococci, Italy. FEMS Immunol Med Microbiol. 2009;55:62–67. doi: 10.1111/j.1574-695X.2008.00499.x. [DOI] [PubMed] [Google Scholar]
- Heb S, Gallert C. Resistance behavior of inducible clindamycin-resistant staphylococci from clinical samples and aquatic environments. J Med Microbiol. 2014;63:1446–1453. doi: 10.1099/jmm.0.077081-0. [DOI] [PubMed] [Google Scholar]
- Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- Lim JA, Kwon AR, Kim SK, Chong Y, Lee K, Choi AC. Prevalence of resistance to macrolide, lincosamide, and streptogramin antibiotics in Gram-positive isolated in a Korean hospital. J Antimicrob Chemother. 2002;49:489–495. doi: 10.1093/jac/49.3.489. [DOI] [PubMed] [Google Scholar]
- Lina G, Quaglia A, Reverdy ME, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43:1062–1066. doi: 10.1128/aac.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan M, Cunney R, Cafferkey M. Molecular epidemiology of group B streptococci in Ireland reveals a diverse population with evidence of capsular switching. Eur J Clin Microbiol Infect Dis. 2014;33:1155–1162. doi: 10.1007/s10096-014-2055-5. [DOI] [PubMed] [Google Scholar]
- Novotna G, Adamkova V, Janat J, Melter O, Spižek J. Prevalence of resistance mechanisms against macrolides and lincosamides in methicillin-resistant coagulase-negative staphylococci in the Czech Republic and occurrence of undefined mechanism of resistance to lincosamides. Antimicrob Agents Chemother. 2005;49:3586–3589. doi: 10.1128/AAC.49.8.3586-3589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotna G, Janata J. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob Agents Chemother. 2006;50:4070–4076. doi: 10.1128/AAC.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotna G, Spižek J, Janata J. In vitro activity of telithromycin and quinupristin/dalfopristin against methicillin-resistant coagulase-negative staphylococci with defined resistance genotypes. Folia Microbiol. 2007;52:593–599. doi: 10.1007/BF02932188. [DOI] [PubMed] [Google Scholar]
- Otto M. Staphylococcus epidermidis - the “accidental” pathogen. Nat Rev Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays. 2013;35:4–11. doi: 10.1002/bies.201200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J, Hidalgo M, Díaz L, Rincón S, Moreno J, Vanegas N, Castañeda E. Characterization of macrolide resistance in Gram-positive cocci from Colombian hospitals: a countrywide surveillance. Int J Infect Dis. 2007;11:329–336. doi: 10.1016/j.ijid.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Reynolds E, Ross JI, Cove JH. msr(A) and related macrolide-streptogramin resistance determinants: incomplete transporters? Int J Antimicrob Agents. 2003;22:228–236. doi: 10.1016/s0924-8579(03)00218-8. [DOI] [PubMed] [Google Scholar]
- Roberts MC. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett. 2004;282:147–159. doi: 10.1111/j.1574-6968.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- Schoenfelder SM, Lange C, Eckart M, Hennig S, Kozytska S, Ziebuhr W. Success through diversity - how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int J Med Microbiol. 2010;28:380–386. doi: 10.1016/j.ijmm.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczuka E, Makowska N, Bosacka K, Słotwińska A, Kaznowski A. Molecular basis of resistance to macrolides, lincosamides, and streptogramins in Staphylococcus hominis strains isolated from clinical specimens. Folia Microbiol (Praha) 2016;61:143–147. doi: 10.1007/s12223-015-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro CRS, Mattos CS, Cavalcante FS, Pereira EM, Santos KRN. Characterization of MLSb resistance among Staphylococcus aureus and Staphylococcus epidermidis isolates carrying different SCCmec types. Microbiol Immunol. 2012;56:647–650. doi: 10.1111/j.1348-0421.2012.00481.x. [DOI] [PubMed] [Google Scholar]
- Vitali LA, Petrelli D, Lamikanra A, Prenna M, Ainkunmi EO. Diversity of antibiotic resistance genes and staphylococcal cassette chromosome mec elements in faecal isolates of coagulase-negative staphylococci from Nigeria. BMC Microbiol. 2014;14(106) doi: 10.1186/1471-2180-14-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voung C, Otto M. Staphylococcus epidermidis infections. Microbes Infect. 2002;4:481–489. doi: 10.1016/s1286-4579(02)01563-0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu CM, Lu LM, Ren GWN, Cao XY, Shen JZ. Macrolide-lincosamide-resistant phenotypes and genotypes of Staphylococcus aureus isolated from bovine clinical mastitis. Vet Microbiol. 2008;130:118–125. doi: 10.1016/j.vetmic.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr W, Henning S, Eckart M, Kränzler H, Batzilla C, Kozitskaya S. Nosocomial infections by Staphylococcus epidermidis: how a commensal bacterium turns into a pathogen. Int J Antmicrob Agents. 2006;28(Suppl. 1):S14–S20. doi: 10.1016/j.ijantimicag.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Zmantar T, Kouidhi B, Miladi H, Bakhrouf A. Detection of macrolide and disinfectant resistance genes in clinical Staphylococcus aureus and coagulase-negative staphylococci. BMC Res Notes. 2011;4(453) doi: 10.1186/1756-0500-4-453. [DOI] [PMC free article] [PubMed] [Google Scholar]