Abstract
Background
In a world of chemical cues, smell and taste are essential senses for survival. Here we focused on Heliconius, a diverse group of butterflies that exhibit variation in pre- and post-zygotic isolation and chemically-mediated behaviors across their phylogeny. Our study examined the ionotropic receptors, a recently discovered class of receptors that are some of the most ancient chemical receptors.
Results
We found more ionotropic receptors in Heliconius (31) than in Bombyx mori (25) or in Danaus plexippus (27). Sixteen genes in Lepidoptera were not present in Diptera. Only IR7d4 was exclusively found in butterflies and two expansions of IR60a were exclusive to Heliconius. A genome-wide comparison between 11 Heliconius species revealed instances of pseudogenization, gene gain, and signatures of positive selection across the phylogeny. IR60a2b and IR60a2d are unique to the H. melpomene, H. cydno, and H. timareta clade, a group where chemosensing is likely involved in pre-zygotic isolation. IR60a2b also displayed copy number variations (CNVs) in distinct populations of H. melpomene and was the only gene significantly higher expressed in legs and mouthparts than in antennae, which suggests a gustatory function. dN/dS analysis suggests more frequent positive selection in some intronless IR genes and in particular in the sara/sapho and melpomene/cydno/timareta clades. IR60a1 was the only gene with an elevated dN/dS along a major phylogenetic branch associated with pupal mating. Only IR93a was differentially expressed between sexes.
Conclusions
All together these data make Heliconius butterflies one of the very few insects outside Drosophila where IRs have been characterized in detail. Our work outlines a dynamic pattern of IR gene evolution throughout the Heliconius radiation which could be the result of selective pressure to find potential mates or host-plants.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2572-y) contains supplementary material, which is available to authorized users.
Keywords: Chemosensory receptors, Speciation, Butterfly, Lepidoptera, Smell, Taste, Olfaction, Gustation
Background
Animals utilize a variety of cues to make key decisions over a range of behaviors, including navigating [1, 2], foraging [3], avoiding predators [4], kin recognition [5], and mate choice [6, 7]. Among the different signals, all organisms from bacteria to mammals utilize chemical cues. Chemosensory receptors (CRs) are the molecular tools that animals have evolved to distinguish a myriad of odors and tastes. This complexity in chemical sensing is manifested by the high degree of inter- and intraspecific variation at CR loci which is partly explained by the adaptation of organisms to different environments. Some of this variation is also due to genetic drift, and the random processes of gene duplication and deletion, which can generate pseudogenes and copy number variations (CNVs). Thus, given the co-founding effect of selection and genetic drift, explaining the mechanisms that underlie differences in CRs between and within species is not a simple task. To date, only a few species have been studied and our understanding of the biochemical basis of chemo-sensation is limited.
Only with massive parallel sequencing have a variety of genomic projects begun to investigate the evolution of chemosensory genes across different levels of taxonomic organization [8–18]. These studies have shown that chemosensory receptor genes and pseudogenes vary enormously among different animal species. For example, in tetrapods the olfactory receptor (OR) genes range from 400 to 2,100 depending on the species, with 20–50 % described as pseudogenes [19]. These receptors are the most studied chemosensory genes, especially in mammals where ORs have been estimated to represent around 3 % of all the genes in the genome and are thus the largest mammalian gene family known to date [20]. However, compared to mammals, insects display a more compact and simpler gene repertoire but with similar chemo-sensing functions [21]. The insect chemosensory receptors, which have evolved independently from mammals, consist mainly of gustatory receptors (GRs), olfactory receptors (ORs), and ionotropic receptors (IRs). GRs are involved in tasting sweet and bitter compounds [22] but also act as CO2 sensors [23] and are thought to be involved in heat avoidance [24]. The expression of GRs occurs mostly in tissues directly in contact with food or other objects being tasted. ORs are the starting point for odor coding and are expressed in olfactory organs such as antennae. IRs are the most primitive class of receptors and are expressed both in olfactory and gustatory tissues [16, 25]. Their ability to detect a wide array of solid and volatile molecules probably has to do with their ancestral role in the early protostomes [26], and the necessity to cover functions that during the evolution of chemo-sensation were reassigned to GRs and ORs.
The recent discovery and annotation of the ionotropic receptors in Drosophila [16, 27] has provided the opportunity to gain novel insight into the genetic and molecular basis of smell and taste in insects. Unlike mammals, insects possess IRs, which have most likely evolved from ionotropic glutamate receptors (iGluRs), a conserved family of synaptic ligand-gated ion channels [26]. IRs have been implicated in detection of phagostimulants [25], pheromones [25], salt [28], volatiles [16, 26, 27] and also seem to be involved in hearing [29]. IRs have mostly been studied in Drosophila, in which they form two groups, the olfaction oriented “antennal” IRs and the gustatory, mostly intronless, “divergent” IRs [25, 26]. The “antennal” IRs display higher sequence conservation, lower dN/dS ratios, fewer duplications, and fewer pseudogenes than the “divergent” intronless IRs. The divergent IR genes, also display a pattern of rapid gain and loss between species [26]. However, to understand whether the patterns of evolution in Drosophila IRs are similar to the patterns in other insects, additional data are needed across the insect phylogeny. The increasing availability of insect genomes offers the opportunity to better understand patterns of chemosensory gene evolution at a broader scale.
Heliconius butterflies are a group of insects where chemical cues have likely played a critical role in their evolution, adaptation, and speciation [30–32]. Heliconius butterflies have extraordinary phenotypic diversity and complex behaviors. Composed of almost 50 species, which represent a continuum of taxa across the stages of speciation, Heliconius includes distantly-related species with identical wing color patterns that are sexually incompatible and also closely-related interbreeding species with different wing color patterns. These incomplete species boundaries are best represented by H. melpomene, H. cydno, and H. timareta, which are closely related species that occasionally exchange genes while showing strong assortative mating [33–35]. It is known that strong mating preference is partially linked to the same genes controlling wing color pattern variation [36]. However, chemosensory genes likely play an important role in pre- and post-zygotic isolation. Indeed, smell and taste are strongly involved in insect prezygotic isolation [6, 7], host plant choice [37], and food recognition [38]. Heliconius butterflies, for example, show different degrees of host plant specialization, with species that are generalists and species that are specialists on one or a few species of Passiflora [39].
Previous studies of chemosensory receptors in Heliconius butterflies have revealed an unexpected diversity in ORs and GRs [34, 37], and suggested a link between the evolution of GRs and female oviposition behavior [37]. However, to date Heliconius IRs have not yet been examined. Bombyx mori [16, 40, 41] and Danaus plexippus [1, 16] are the only two representatives of 180,000 species of Lepidoptera for which the GR, OR and IR genes have been annotated. Here, we characterize the IR genes in Heliconius and conduct a comparative analysis with B. mori and D. plexippus.
Methods
Annotation of ionotropic receptors in H. melpomene and comparative analysis with Lepidoptera and Diptera
IR gene annotation was performed by TBLASTX [42] searches of IR and related iGluR genes of Bombyx mori [26], Drosophila melanogaster [26] and Danaus plexippus [1] against a Trinity [43] assembled RNA-Seq library of Heliconius melpomene rosina males (n = 3) and females (n = 3). The RNA-Seq data is deposited in the European nucleotide archive (ERP002272) [37]. The contigs from the Trinity assembly were aligned against the H. melpomene genome (v1.1) [34] in Mega 6.0 [44] using MUSCLE [45]. This resulted in almost complete genes including exon-intron boundaries and a physical location in the genome. When RNA-Seq data did not recover the whole IR gene, missing parts were identified by aligning homologues of reference species against the H. melpomene genome. All genes identified in H. melpomene as IRs were clear homologs of genes identified by Croset et al. [26] as ionotropic receptors in Danaus plexippus and Bombyx mori. For 15 H. melpomene genes, when using TBLASTX, the closest homolog had an E-value of 0, while the lowest of the remaining 16 was a Heliconius-specific duplication, IR60a2c, with an E-value of 1E-97. We used the gene models constructed from H. melpomene to improve IR annotations in B. mori (v2.0) and D. plexippus (v3) and discover unannotated genes in those genomes. The H. melpomene IR genes were aligned against the genomes of D. plexippus or B. mori, and if applicable the IR sequences of Croset et al. [26]. As a result of our deep RNA sequencing and annotation in H. melpomene we were able to improve the IR gene models significantly in all the three butterflies genomes.
H. melpomene IRs were named after their closest homologs described in B. mori or D. plexippus [26]. Gene phylogenies were constructed using MAFFT [46] and RAxML [47] (best of 200 trees, 500 bootstraps). We used the H. melpomene genome 1.1 [34] to precisely locate each IR gene across the 21 linkage groups (see Additional file 1).
Ionotropic receptor evolution within the genus Heliconius
We assembled 10 Heliconius genomes (H. cydno chioneus, H. timareta timareta, H. wallacei flavescens, H. hecuba, H. doris doris, H. clysonymus tabaconas, H. telesiphe sotericus, H. erato petiverana, H. sara magdalena and H. sapho sapho) using ABySS v1.5.2 [48] (command: abyss-pe n = 5 k = 31 c = 2) as previously described by Briscoe et al. [37]. We used TBLASTX and Tophat [49] to identify the homolgous H. melpomene IRs in each species. Depending on the species we were able to obtain complete or nearly complete IR gene sequences (n = 282, mean of 97.9 % of bases complete).
Detecting positive selection in IRs across the Heliconius adaptive radiation
The IRs annotated in the 11 Heliconius species and the out-group D. plexippus were stripped of their terminal stop codon and aligned in the correct reading frame using MEGA 6 [44] with the aligner MUSCLE [45]. Alignments were analyzed in Datamonkey [50] using the HyPhy branch-site Random Effects Likelihood (REL) model [51] to test if a branch shows signs of positive diversifying selection. In branch-site REL, Ω (the dN/dS ratio) is modeled as three variables, Ω1, Ω2 and Ω3. While Ω1 and Ω2 remain between zero and one, with Ω2 bigger than Ω1, the value of Ω3 ≥1. Ω3 is the estimate of positive selection. All Ω parameters are estimated by maximum likelihood. The likelihood ratio test (LRT) test determines if Ω3 is bigger than one with a p-value corrected for multiple testing. The HyPhy branch-site random effects likelihood method [51] produces its own neighbor joining trees, and when the neighbor joining gene tree and the species tree from Kozak et al. [52] were not identical, the resulting branch under positive selection does not exist on the species tree of Kozak et al. [52] and is shown as a dotted line. In the case of a gene with many duplications, such as IR60a2 (a, b, c, d and e), only the closest copy to the orthologous gene in the out-group species D. plexippus was utilized.
Copy number variation in natural populations of H. melpomene and H. cydno
We tested IR Copy Number Variations (CNVs) by utilizing twenty previously published whole genome sequences [37, 53], from distinct natural population of H. melpomene and H. cydno. More specifically, we analyzed four individuals for the following populations: H. melpomene amaryllis, H. melpomene aglaope, H. melpomene rosina, H. cydno chioneus, and H. melpomene melpomene. We first aligned the Illumina re-sequenced genomes against the H. melpomene reference genome (v1.1) [34] and then analyzed the read depth for each IR gene using CNVnator (100 bp sliding window) [54]. The output of CNVnator was used to determine candidate variable insertions and deletions. We considered estimated copy number of >2 as a potential duplication and <0.5 as a potential deletion. To control for possible artifacts in the H. melpomene genome assembly (v1.1) we also ran CNVnator on the raw reads used to create the reference genome and identified false positives then discarded those from further analyses.
Ionotropic receptor expression in sensory tissues of male and female individuals of H. melpomene
As mentioned above, we used Illumina RNA-sequencing data (European nucleotide archive ERP002272) [37], collected from legs, mouthparts and antennae of three males and three females (two paired-end runs for every sample and three additional single-end 100 bp run for one male and one female on a HiSeq 2000 for a total of 730,617,415 reads, ~33900× coverage). Raw data was trimmed (adapter sequence, distorted 10 bp at beginning and sliding window:4:20) using Trimmomatic [55] and assembled using Trinity [43]. A differential expression analyses was done on the entire transcriptome using Trinity scripts built around the edgeR package from Bioconductor, which uses the trimmed mean of M values (TMM) normalization method [56–59]. Genes were called differentially expressed when the FDR <0.05. We calculated tissue- and sex-specific expression, and used FPKM value to generate a combined heat map for each biological replicate as well as for individual specimens. Because a Trinity assembly is de novo, only genes expressed high enough can be assembled and analyzed for expression. IR60a1a and IR60a2b were expressed sufficiently but IR60a1b and IR60a2 (a, c, d and e) were not and were therefore omitted from analysis.
Results
Annotation of ionotropic receptors in the Heliconius melpomene genome
Overall, with our annotation (Additional files 1 & 2) we have generated a complete description of 31 IR genes in H. melpomene. This is more than B. mori (25) and D. plexippus (27), but around half the number found in Drosophila melanogaster (66) and other Diptera [26] (Table 1, Fig. 1). We also significantly improved the gene models of most of the previously described ionotropic receptors genes in B. mori (IR68a, IR7d1, IR7d2, IR7d3, IR8a, IR21a, IR40a, IR41a, IR75d, IR75p2, IR75q1, IR75q2, IR76b, IR87a, IR93a and IR143a) and D. plexippus (IR8a, IR21a, IR25a, IR31a, IR40a, IR41a, IR60a2, IR64a, IR68a, IR75p1, IR75p2, IR75q1, IR75q2, IR76b, IR87a and IR93a). Moreover we identified new putative IRs in B. mori and in D. plexippus: seven in B. mori (IR1, IR31a, IR60a2, IR60a1, IR75p1, IR75p2pseudo, and IR85a) (Table 1), and eleven in D. plexippus (IR7d1, IR7d2, IR7d3, IR7d4, IR60a, IR85a and IR143a) including four genes (IR75p1, IR75p2, IR75q1 and IR75q2) which were reported as two gene models by Zhan et al. [1] (Table 1, & Additional file 2).
Table 1.
IR gene diversity in H. melpomene compared to B. mori or D. plexippus
| H. melpomene IR | B. mori | D. plexippus |
|---|---|---|
| HmIR1 | discovered | unchanged |
| HmIR7d1 | improved | discovered |
| HmIR7d2 | improved | discovered |
| HmIR7d3 | improved | discovered |
| HmIR7d4 | absent | discovered |
| HmIR8a | improved | improved |
| HmIR21a | improved | improved |
| HmIR25a | unchanged | improved |
| HmIR31a | discovered | improved |
| HmIR40a | improved | improved |
| HmIR41a | improved | improved |
| HmIR60a | absent | discovered |
| HmIR60a1a | discovered | unchanged |
| HmIR60a1b | absent | absent |
| HmIR60a2a | discovered | improved |
| HmIR60a2b | absent | absent |
| HmIR60a2c | absent | absent |
| HmIR60a2d | absent | absent |
| HmIR60a2e | absent | absent |
| HmIR64a | unchanged | improved |
| HmIR68a | improved | improved |
| HmIR75d | improved | unchanged |
| HmIR75p1 | discovered | improved |
| HmIR75p2 | improved & discovered pseudogene | improved |
| HmIR75q1 | improved | improved |
| HmIR75q2 | improved | improved |
| HmIR76b | improved | improved |
| HmIR85a | discovered | discovered |
| HmIR87a | improved | improved |
| HmIR93a | improved | improved |
| HmIR143a | improved | discovered |
The list of homologous IR genes between the three Lepidoptera genomes is reported relative to the IRs in H. melpomene. Information is also provided for each gene if its sequence was either left unchanged, improved, or discovered in our study when compared to data from Croset et al. [26]
Fig. 1.
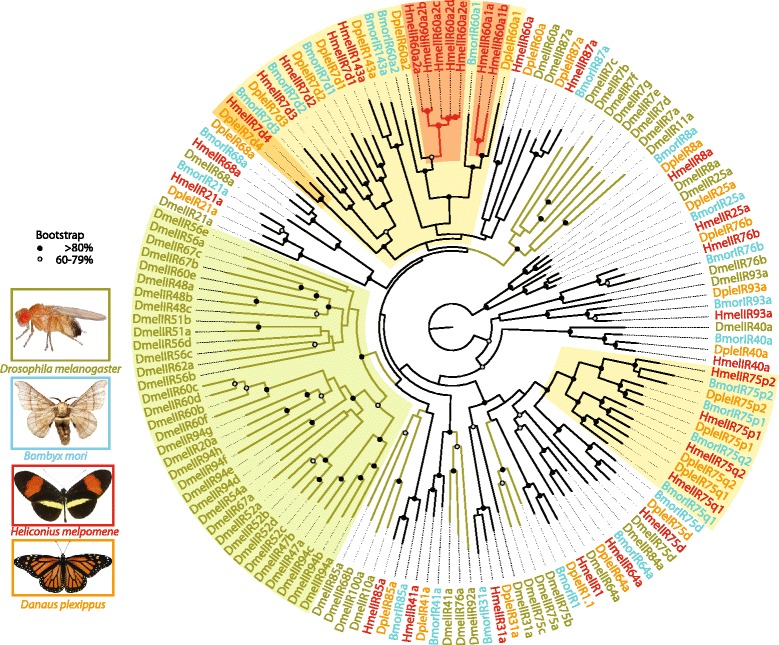
Phylogenetic relationships of ionotropic receptors in Lepidoptera and Diptera. Black dots indicate >80 % bootstrap support, grey dots indicated 60–79 % bootstrap support. Shaded colors represent taxon-specific IR genes: Diptera-specific (green lines), IR20a clade (green shade), Lepidoptera-specific (yellow), butterfly-specific (orange), and Heliconius-specific (red). While a large number of genes are conserved between Lepidoptera and Diptera, instances of lepidopteran-, butterfly- and Heliconius-specific IRs emerged from our analysis
Our IR gene models in H. melpomene consisted of one to 19 exons with 14 of 31 genes being intronless and only four with an elevated number of introns (≥15) (Fig. 2, Additional file 1). The IR gene models were on average 647 amino acids (AAs) in length with individual gene models ranging from 546 to 899 AAs. A genomic location for four of the 31 IRs (IR75d, IR40a, IR8a, IR7d3) could not be identified and thus these genes were unmapped. The remaining 27 IRs are located on 24 scaffolds, representing 13 chromosomes (Fig. 2, Additional file 1). From our H. melpomene genomic and transcriptomic dataset we did not find any evidence of possible pseudogenes or alternative splicing. However, we found evidence of four recent gene duplication events in the H. melpomene genome. These genes are IR60a1a and IR60a1b on Chromosome 19, and IR60a2a, IR60a2b, IR60a2c, IR60a2d, and IR60a2e on Chromosome 20 (Fig. 2). We also found evidence of older duplications shared with B. mori and D. plexippus, but not with Diptera; therefore they happened between 100 and 270 MY ago [60]. These lepidopteran duplications are IR7d1 and IR7d2 on Chromosome 1, IR75p1 and IR75p2 on Chromosome 14, and IR75q1 and IR75q2 on Chromosome 15 of the H. melpomene reference genome. While some IRs map to chromosomes containing major wing color pattern genes, none of these IR genes are tightly linked to color pattern loci (Fig. 2).
Fig. 2.
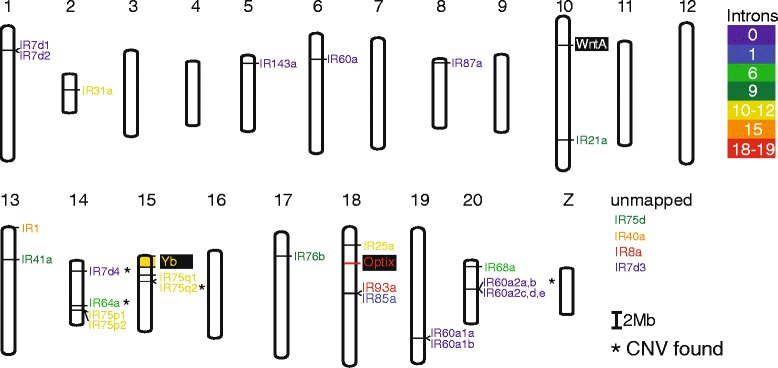
Chromosomal locations of H. melpomene IRs. Chromosomal location of IR genes that map to the H. melpomene reference genome and IRs for which the position is unknown are reported in the right corner. Different colors are used to represent the number of introns in each IR gene. Black boxes indicate the genomic location of known color pattern genes controlling black (WntA), yellow (Yb) and red (Optix) wing color patterns [78]. The asterisks indicate genes that display copy number variations in H. melpomene and/or H. cydno
Evolution of ionotropic receptors in Lepidoptera
Our comparative analysis between two butterflies’ (H. melpomene, D. plexippus), a moth’s (B. mori) and a fly’s (D. melanogaster) genomes allowed us to identify dipteran-, lepidopteran-, butterfly-, and Heliconius-specific IRs (Fig. 1). D. melanogaster demonstrated higher diversity, with 35 more IR genes than H. melpomene, mainly due to the IR20a clade [25], a large group with no orthologues in the three Lepidopteran species (Fig. 1, green background). Ten IR genes were Lepidoptera-specific (yellow background), namely: IR7d1, IR7d2, IR7d3, IR143a, IR60a2, IR60a1, IR75p1, IR75p2, IR75q1and IR75q2 (Fig. 1). IR7d4 was found in H. melpomene, and D. plexippus but not in B. mori, thus representing a butterfly-specific receptor (Fig. 1, orange background). Intriguingly, we identified two H. melpomene-specific IR gene expansions (IR60a1 and IR60a2) (Fig. 1, red background). IR60a1 has two copies and IR60a2 has five copies which represent Heliconius-specific expansions.
Ionotropic receptor diversity and evolution across the Heliconius genus
Comparision of 11 Heliconius species allowed us to study the diversity and evolution of the IRs in this genus (Fig. 3 & Additional file 3). We identified 28 out of the 31 genes in almost all 11 species. The level of conservation of these 28 genes is higher than the other two chemosensory gene families (GRs and ORs) previously analyzed by Briscoe et al. [37]. Overall, our analysis revealed a strong level of conservation in the IRs, suggesting an ancestral function.
Fig. 3.
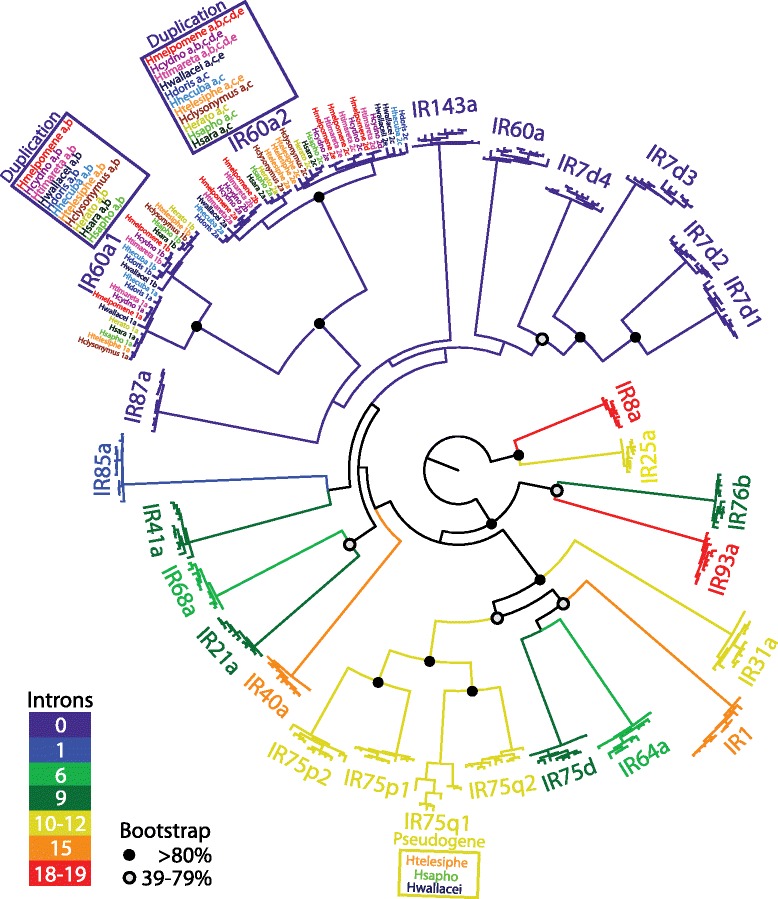
IR phylogeny in Heliconius. A phylogenetic relationship of the IRs across 11 Heliconius species is presented. Weak (50–79 %) and strong (80 %) bootstrap values are shown with solid and empty black dots respectively, but only for major branches. Support for branches of homologues of the 11 species that group into one gene are left out. When a gene is present in one copy for all 11 species, full gene names were left out. While the majority of the IRs are conserved between the 11 species analyzed we observed Heliconius-specific expansions at IR60a1, IR60a2, and the divergent IR75q1 gene. Colors of each branch represent the number of exons in each specific IR gene
Despite the broad IR gene conservation across Heliconius, we identified several examples of gene duplication and pseudogenization. We found that the two H. melpomene-specific duplications (IR60a1 and IR60a2, see Fig. 1) were also duplicated in several other Heliconius species (Fig. 3). We found IR60a1a and IR60a1b in all 11 Heliconius species. While IR60a2 had five copies IR60a2(a, b, c, d and e) in H. melpomene, H. cydno, and H. timareta, only IR60a2 (a, c and e) were found in H. wallacei and H. telesiphe (Figs. 3 and 4b). All the other species displayed only one duplication, IR60a2a and IR60a2c (Fig. 3). The most interesting pattern emerging from the data is the presence of the two extra copies IR60a2b and IR60a2d unique to H. melpomene, H. cydno, and H. timareta clade (Figs. 4 and 5b). Finally, IR75q1 was the only receptor with pseudogenes, and showed lower sequence conservation as shown by longer branch lengths in Fig. 3. Conservation among H. wallacei, H. sapho, and H. telesiphe was so low that in some cases the complete sequence was not traceable in the genome (Fig. 4b).
Fig. 4.
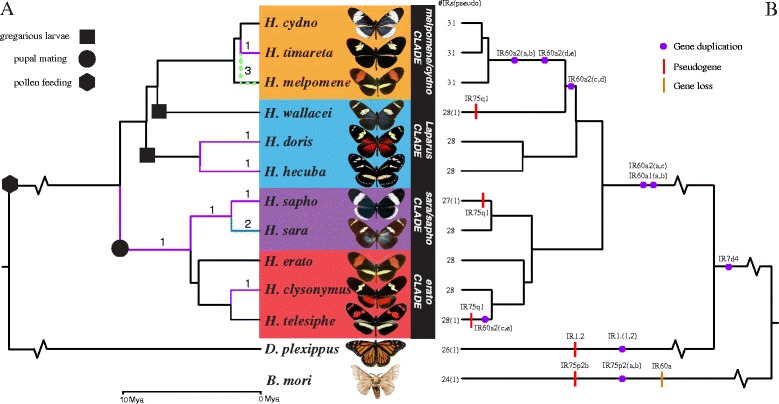
IR evolution in Heliconius butterflies. a Number of genes per branch under positive diversifying selection as determined using HyPhy’s branch-site random effects likelihood model [51]. Most positive selection maps onto the H.melpomene/H. cydno clade and H.sara/H.sapho clade. The phylogeny taken from Kozak et al. [52]. b Overview of gene duplication, pseudogenization and gene loss in Heliconius, D. plexippus and B. mori. Duplications of IR60a1 and IR60a2 map within the Heliconius genus. IR7d4 is shared by the butterflies but not with the moth
Fig. 5.
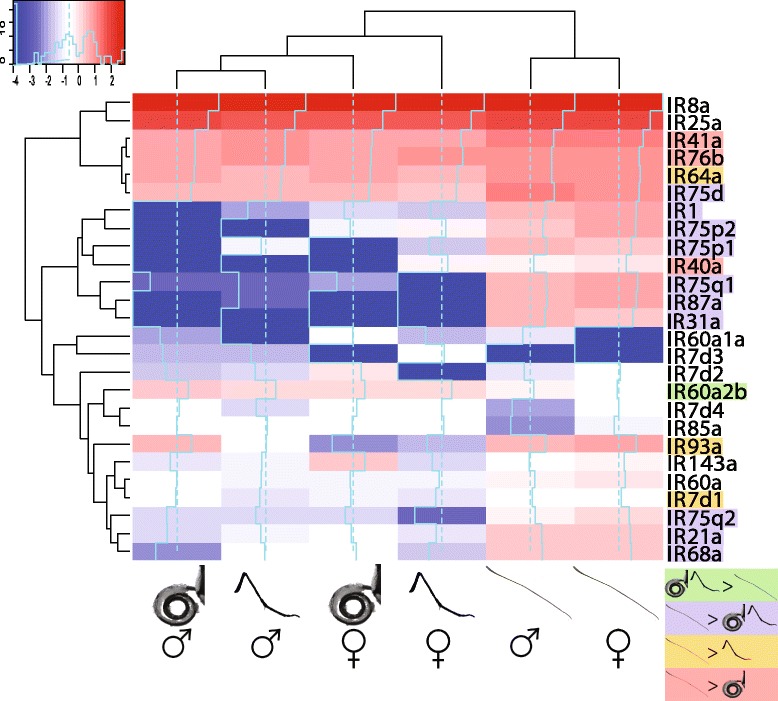
H. melpomene IR gene expression heatmap for sensory organs and sexes. Names of IR genes significantly differentially expressed (FDR <0.05) between mouthparts, legs and antenna are highlighted with different background colors. Most of the differentially expressed genes display higher expression in antennae than in mouthparts and legs (purple background). Only IR60a2b displayed higher expression in mouthparts and legs compared to antenna (green background). The remaining mRNAs that are significantly DE were more abundant in antennae than in legs (yellow background) or mouthparts (red background). Note the similarity in overall expression of legs and mouthparts compared to antennae. FPKM values are represented with a color gradient from blue to red. Exact FPKM values can be found in Table 4 and Additional file 4. Individual heatmaps are presented in the Additional file 5
Evidence for positive diversifying selection of ionotropic receptors within Heliconius
Using HyPhy [51] in Datamonkey [50] we found 12 instances of positive diversifying selection (Fig. 4a). A total of 8 IRs (IR7d1, IR7d4, IR60a2a, IR60a1a, IR143a, IR68a, IR75d, IR93a) had branches which show signs of positive selection (Table 2). Out of the 12 instance of positive diversifying selection, the majority was found in the H. sapho/sara clade (four out of 12), and the H. melpomene/timareta clade (four out of 12) (Fig. 4a and Table. 2). Only IR60a1a was under positive selection on a major branch of the phylogeny, during the formation of the H. erato/sapho/sara clade (Fig. 4a and Table 2). Interestingly three genes, IR143a, IR60a2a, and IR75d showed signs of positive selection in H. melpomene and H. timareta but not in the species H. cydno (Fig. 4a and Table. 2). This resulted in a gene tree that grouped H. timareta with its mimic H. melpomene instead of H. cydno (Fig. 4. green dotted line). Finally, the majority of the genes (50 %) showing sign of positive selection were intronless. Only 19 % of genes with introns had at least one branch with a dN/dS >1.
Table 2.
Branches of gene trees under positive selection as determined using HyPhy branch site random effects likelihood model [51]
| Gene | Branch | Mean Ω | Ω1 | p1 | Ω2 | p2 | Ω3 | p3 | LRT | p-value | Corrected p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IR143a | H. timareta*H. melpomene | 0.151 | 0 | 0.963 | 0 | 0.035 | 10000 | 0.002 | 11.309 | 0.000 | 0.008 |
| IR60a1a | H. erato*H. telesiphe*H. clysonymus*H. sapho*H. sara | 0.196 | 0.165 | 0.994 | 0.167 | 0.001 | 3333 | 0.005 | 11.042 | 0.000 | 0.009 |
| IR60a1a | H. sapho*H. sara | 0.178 | 0.151 | 0.983 | 0.149 | 0.013 | 10000 | 0.004 | 8.989 | 0.001 | 0.027 |
| IR60a2a | H. sapho | 0.434 | 0.055 | 0.942 | 0.163 | 0.026 | 22 | 0.032 | 19.785 | 0.000 | 0.000 |
| IR60a2a | H. sara | 0.839 | 0.762 | 0.975 | 0.763 | 0.015 | 199 | 0.010 | 14.949 | 0.000 | 0.001 |
| IR60a2a | H. timareta*H. melpomene | 0.501 | 0.402 | 0.984 | 0.401 | 0.000 | 5197 | 0.016 | 36.954 | 0.000 | 0.000 |
| IR68a | H. hecuba | 0.244 | 0.206 | 0.992 | 0.204 | 0.005 | 263 | 0.003 | 8.198 | 0.002 | 0.044 |
| IR75d | H. timareta*H. melpomene | 0.155 | 0.091 | 0.971 | 0.151 | 0.025 | 2181 | 0.004 | 14.184 | 0.000 | 0.002 |
| IR7d1 | H. sara | 0.498 | 0.463 | 0.973 | 0.462 | 0.025 | 10000 | 0.002 | 9.936 | 0.001 | 0.017 |
| IR7d4 | H. timareta | 0.079 | 0.033 | 0.994 | 0.032 | 0.002 | 5447 | 0.004 | 13.677 | 0.000 | 0.002 |
| IR93a | H. clysonymus | 0.131 | 0.079 | 0.991 | 0.080 | 0.005 | 80 | 0.004 | 12.607 | 0.000 | 0.004 |
| IR93a | H. doris | 0.287 | 0 | 0.950 | 0 | 0.001 | 8 | 0.049 | 10.858 | 0.000 | 0.010 |
IR gene name and phylogenetic branch showing positive selection is reported in the first two columns. Values obtained from HyPhy for positive selection are shown only for the IR genes supported by a significant p-value obtained from the likelihood ratio test (LRT) and corrected for multiple testing with the Holm formula
Copy number variations in H. melpomene subspecies and H. cydno
We observed significant copy number variation only in seven out of 20 individuals analyzed (Table 3). Overall, the percentages of IRs for which CNVs are found in H. melpomene are similar to ORs (12.9 % vs 18.5 %, Fisher’s exact test, two-tailed, P = 0.57) but lower than GRs (12.9 % vs 54.4 %, Fisher’s exact test, two-tailed, P = 0.0001). We found evidence of CNVs for IR75q1, IR64a, IR60a2b and IR7d4 in H. melpomene, and IR75q2 and IR60a2a in H. cydno (Table 3). Population-specific copy number variation was found in IR60a2b (2 H. m. aglaope, and 2 H. m. amaryllis), and in IR7d4 (2 H. m. melpomene) (Table 3). All together these results suggest the possible existence of individual differences in the number of IR genes across Heliconius populations.
Table 3.
Copy Number Variations (CNVs) in populations of H. melpomene and H. cydno
| Species | Butterfly | Del/dup | Gene | Chromosome | Scaffold size | Begin gene | End gene | Begin CNV | End CNV | Length CNV | Norm. RD | P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H. c. chioneus | 553 | dup | IR75q2 | Chr15 | 190001 | 51255 | 62232 | 41501 | 62300 | 20800 | 2.64 | 3.87E-08 |
| H. m. amaryllis | 11-160 | del | IR75q1 | Chr15 | 190001 | 67972 | 71764 | 66901 | 72200 | 5300 | 0.44 | 3.01E-11 |
| H. m. amaryllis | 11-160 | del | IR64a | Chr14 | 16065 | 3076 | 6448 | 2301 | 5400 | 3100 | 0.5 | 3.50E-06 |
| H. m. melpomene | 9315 | dup | IR7d4 | Chr14 | 89318 | 65125 | 66960 | 55501 | 68600 | 13100 | 6.52 | 0 |
| H. m. melpomene | 13435 | dup | IR7d4 | Chr14 | 89318 | 65125 | 66960 | 55001 | 68600 | 13600 | 3.75 | 0 |
| H. m. aglaope | 09-122 | del | IR60a2b | Chr20 | 72055 | 37488 | 39341 | 38201 | 39400 | 1200 | 0.23 | 0.0174 |
| H. m. aglaope | 11-569 | del | IR60a2b | Chr20 | 72055 | 37488 | 39341 | 38201 | 39400 | 1200 | 0.16 | 0.0023 |
| H. m. amaryllis | 09-216 | del | IR60a2b | Chr20 | 72055 | 37488 | 39341 | 37401 | 39400 | 2000 | 0.37 | 0.0472 |
| H. m. amaryllis | 11-160 | del | IR60a2b | Chr20 | 72055 | 37488 | 39341 | 38101 | 39400 | 1300 | 0.2 | 0.0127 |
| H. c. chioneus | 553 | dup | IR60a2a | Chr20 | 72055 | 47451 | 49370 | 41401 | 49500 | 8100 | 2.15 | 0 |
Output of CNVnator is reported here. Butterfly ID, deletion or insertion, IR gene name, chromosomal position, scaffold size, coordinates of the IR gene, coordinates and length of the CNV, normalization and P-value are shown. Only insertion and deletion variants with a read depth >2 (duplications) and <0.5 (deletions) are reported
H. melpomene RNA-Seq expression profile of ionotropic receptor genes in mouthparts, legs, and antennae of males and females
Several IR genes showed differential expression between mouthparts, legs and antennae in males and females of H. melpomene (Fig. 5, Additional files 4 & 5). A total of 26 out of the 31 genes were expressed in at least one tissue type, but most genes were expressed in both sexes and all three tissues (Table 4). We did not detect expression for some Heliconius specific duplications, IR60a2a, IR60a2c, IR60a2d, IR60a2e and IR60a1b, which is likely due to their expression being below our detection threshold. The expression profile of the 26 detected IR genes showed strong similarity between mouthparts and legs, and a distinct expression pattern in the antennae (Fig. 5). However, within each tissue type we also observed a sex-specific expression profile (Fig. 5).
Table 4.
H. melpomene IR gene expression levels between sensory tissues and sexes. Average FPKM values obtained from three biological replicates are reported for each IR gene. The false discovery rate (FDR) is provided only for the gene that is significantly differentially expressed. Only for these genes do we reported the direction of the significant comparison and the FDR value. More detailed information on FPKM values can be found in Additional files 4 & 5
| IR | FPKM antenna | FPKM legs | FPKM mouthparts | FPKM average | Tissue specificity | FDR antennae vs. legs | FDR antennae vs. mouthparts | FDR legs vs. mouthparts | FDR Male vs Female |
|---|---|---|---|---|---|---|---|---|---|
| IR60a1a | 0.02 | 0 | 0.08 | 0.04 | |||||
| IR7d3 | 0 | 0.26 | 0.01 | 0.09 | |||||
| IR85a | 0.05 | 0.41 | 0.21 | 0.22 | |||||
| IR7d1 | 0.4 | 0.09 | 0.17 | 0.22 | ant>leg | 9.86E-03 | |||
| IR7d4 | 0.11 | 0.12 | 0.43 | 0.22 | |||||
| IR7d2 | 0.17 | 0.02 | 0.63 | 0.27 | |||||
| IR60a | 0.79 | 0.14 | 0.24 | 0.39 | |||||
| IR40a | 0.77 | 0.47 | 0 | 0.41 | ant>mouth | 5.28E-03 | |||
| IR75q2 | 1.22 | 0.03 | 0.05 | 0.43 | ant>leg&mouth | 1.41E-12 | 6.16E-10 | ||
| IR143a | 0.39 | 0.08 | 1.23 | 0.57 | |||||
| IR21a | 2.2 | 0.08 | 0.07 | 0.78 | ant>leg&mouth | 2.49E-15 | 1.75E-08 | ||
| IR68a | 2.57 | 0.08 | 0.14 | 0.93 | ant>leg&mouth | 1.13E-04 | 1.43E-03 | ||
| IR31a | 3.82 | 0 | 0 | 1.27 | ant>leg&mouth | 6.65E-28 | 4.41E-28 | ||
| IR75p1 | 3 | 0.06 | 0 | 1.02 | ant>leg&mouth | 5.87E-09 | 6.11E-15 | ||
| IR60a2b | 0.26 | 1.51 | 2.02 | 1.26 | leg&mouth>ant | 3.51E-04 | 1.06E-04 | ||
| IR75p2 | 4.06 | 0.17 | 0.05 | 1.43 | ant>leg&mouth | 8.98E-04 | 4.67E-10 | ||
| IR1 | 4.68 | 0.01 | 0.02 | 1.57 | ant>leg&mouth | 5.14E-30 | 2.59E-29 | ||
| IR87a | 5.89 | 0 | 0 | 1.96 | ant>leg&mouth | 8.58E-37 | 4.86E-37 | ||
| IR75q1 | 6.93 | 0 | 0.01 | 2.31 | ant>leg&mouth | 5.84E-30 | 1.79E-30 | ||
| IR93a | 5.4 | 0.15 | 2.23 | 2.59 | ant>leg | 7.26E-09 | M>F, legs FDR = 0.032, mouthparts FDR = 1.67E-05 | ||
| IR64a | 12.44 | 4.72 | 5.89 | 7.68 | ant>leg | 1.17E-02 | 8.99E-02 | ||
| IR75d | 14.77 | 3.16 | 3.81 | 7.24 | ant>leg&mouth | 9.09E-04 | 1.28E-04 | ||
| IR76b | 10.77 | 9 | 6.86 | 8.88 | ant>mouth | 3.62E-03 | |||
| IR41a | 19.5 | 8.46 | 6.47 | 11.47 | ant>mouth | 2.31E-03 | |||
| IR25a | 75.39 | 44.89 | 57.54 | 59.28 | |||||
| IR8a | 734.67 | 462.02 | 474.75 | 557.15 |
From our RNA-Seq data we identified a total of six genes (IR8a, IR25a, IR41a, IR76b, IR64a and IR75d) highly expressed across all tissues and sexes. Overall, the gene expression profile indicated higher expression in the antennae than in mouthparts and legs (Fig. 5). Sixteen IRs were significantly more highly expressed in the antennae (Table 4): antennae > mouthparts and legs (IR1, IR21a, IR31a, IR68a, IR75d, IR75p1, IR75p2, IR75q1, IR75q2 and IR87a) (Fig. 5, purple shade); antennae > mouthparts (IR41a, IR76b, IR40a) (Fig. 5, red shade); and antennae > legs (IR64a, IR93a, IR7d1)(Fig. 5, yellow shade). IR60a2b was the only gene with a gustatory expression profile, being significantly higher expressed in mouthparts and legs than antennae (Fig. 5, green shade). No gene showed a significant difference in expression between legs and mouthparts. Only one gene, IR93a was differentially expressed between sexes, with males showing higher expression in the legs and mouthparts than females (Table 4, Fig. 6). The remaining genes had a similar expression between the sexes, at a very low level (IR7d2, IR7d3, IR7d4, IR60a1b, IR60a, IR85a and IR143a), or a very high level (IR8a, IR25a). The very high expression for IR8a and IR25a was expected because they function as co-receptors [27] with IR8a expression about 8 times higher than IR25a.
Fig. 6.
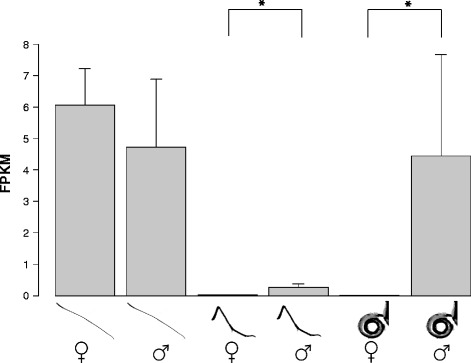
Histogram showing sex-specific gene expression of IR93a. Average level of IR93a expression (FPKM) and standard deviation for each tissue type and sex is reported. Asterisks indicate significant differential expression between male and female legs and mouthparts
Discussion
A complete annotation of ionotropic receptors in Heliconius melpomene provides novel information on IR evolution in Lepidoptera
With 31 IRs in Heliconius and a significantly increased number of IR genes in Danaus and Bombyx, our work suggests that Lepidoptera in general may have more IR genes than previously recognized (Table 1). Our analyses increased the number of IRs in Danaus and Bombyx [26], and significantly improved many of their gene models (Table 1). We revealed instances of taxon-specific IR duplications in H. melpomene, D. plexippus, B. mori, and D. melanogaster. Drosophila displayed by far the highest number of IRs and lineage-specific receptors. However, our data suggested that Lepidoptera have also evolved specific ionotropic receptors (Fig. 1, yellow shade). The majority of these Lepidoptera specific genes, which encompasses four large sub-families (IR7d, IR60a, IR75p, and IR75q) and a single IR locus (IR143a), were either described here for the first time or their annotation significantly improved with our gene models (see Table 1). We know from Drosophila that members of the IR7 group are involved in gustation [26], while expansion in the IR75 sub-family has been associated with the increased need to discriminate among very closely related chemicals [16]. However, it is not yet known to what extent such functions are conserved between Diptera and Lepidoptera.
On a broader scale, our data is generally in agreement with patterns observed in Drosophila [26] where “antennal” IRs are more conserved than “divergent” intronless IRs. In Lepidoptera however, the amount of gain and loss is less profound than in Drosophila, which have approximately four times as many “divergent” IRs. This pattern of diversification between “divergent” and “antennal” IRs is similar to that observed in Heliconius between GRs and ORs, with GRs being more diverse than ORs [37].
Candidate IR genes of evolutionary significance across the Heliconius adaptive radiation
To the best of our knowledge Heliconius is the only other genus after Drosophila where the evolution of chemosensing has been explored in several species [37]. The H. melpomene clade displayed the largest IR repertoire (31) across the 11 Heliconius species analyzed (Fig. 4b). Only one pseudogene was found, IR75q1in H. wallacei, H. sapho, and H. telesiphe (Figs. 3 and 4b). However, IR75q1 is still a potentially functional gene in other species. The reason why IR75q1, which is shared between butterflies and moths, is less conserved than any other IR gene (see its long branch in Fig. 3) in the Heliconius lineage is unknown.
Possibly the most intriguing observation that emerged from our analyses is the repeated gene duplication and evolution of IR60a, especially for H. melpomene, H. cydno, and H. timareta, which have two extra copies IR60a2b and IR60a2d. Interestingly, H. melpomene, H. cydno, and H. timareta are incompletely reproductively isolated, but with strong assortative mating. Moreover, H. melpomene and H. timareta are sometimes near-perfect mimics of each other [61–63]. It is likely that these species utilize specialized pheromones to distinguish among themselves [32]. The complex mating behaviors observed in Heliconius butterflies supports this interpretation. While the initial approach to females is strongly influenced by visual cues, close contact during courtship offers multiple opportunities for volatile and non-volatile chemical communications to occur. Thus, it is highly possible that chemical and behavioral cues may also contribute to prezygotic barriers [64]. The two extra copies, IR60a2b and IR60a2d, might be used by the melpomene/cydno/timareta complex to distinguish chemical cues during mate choice.
Evidence for positive selection of ionotropic receptors within Heliconius
The ratio of substitution at non-synonymous (dN) versus synonymous (dS) sites has been widely utilized to quantify evolutionary pressures on proteins [65]. Despite limitations and controversies over the model used to evaluate gene evolution [66, 67], the dN/dS ratio is a widely used approach to quantify selection pressures acting on protein-coding regions, owing in part to its simplicity and robustness. Our results are based on only one sequence per species, because dN/dS is not designed for within population comparison [67]. Our analysis did not take in consideration instances of population and species-specific nucleotide variation.
We found the majority of the genes with elevated dN/dS in the two lineages represented by H. sara and H. sapho (IR60a1a, IR60a2a, and IR7d1) and H. melpomene and H.timareta (IR143a, IR60a2a, and IR75d) (Fig. 4a). Interestingly, H. sapho and H. sara are unusual in that they do not synthesize, but instead sequester most of their toxic cyanide compounds from their host plants and therefore specialize on only a few species of Passiflora [19]. It is therefore possible that these receptors represent candidate genes for host-plants recognition. Conversely, H. melpomene and H. timareta are more generalists, but represent a clade of very young species which are still not reproductively isolated [16, 17, 31]. Thus, the three receptors under positive selection in these two species (IR143a, IR60a2a, and IR75d) might be part of a toolkit of chemosensory genes that contributes to the establishment of prezygotic isolation based on chemical cues. In contrast, we found very few IRs with elevated dN/dS in the erato clade (H. erato, H. clysonymus, and H. telesiphe), and the Laparus clade (H. wallacei, H. doris, and H. hecuba). Lastly, only IR60a1a had a significantly elevated dN/dS at an older phylogenetic node comprising several distinct clades (Fig. 3 and Table 2). This phylogenetic node represents the ancestor of the pupal-maters, a group of butterflies that share a unique behavior, which consists of males being able to localize and mate with uneclosed or with freshly emerged females [68].
Copy number variation in natural populations of H. melpomene and H. cydno
Copy number variations (CNVs) are now recognized as important components of genomic diversity, alongside single nucleotide polymorphisms (SNPs) [69]. However, few studies have explored CNVs in chemosensory gene families of non-human populations [70, 71]. To date, only one study has explored CNVs in chemical receptor families across a butterfly genome [37]. This study of H. melpomene found widespread CNVs in GRs and ORs, with CNVs more frequent among GRs than ORs [37]. Our data suggest a modest number of H. melpomene IR genes with CNVs, comparable to their ORs [37]. Intriguingly, half of the CNVs occur in IR60a2b, one of two receptors uniquely found in H. melpomene, H. cydno and H. timareta and the only gene with higher expression in legs and mouthparts than in antennae (Figs. 3, 4 and 5). IR60a2b showed reduced read depth in the populations of H. m. aglaope and H. m. amaryllis (Table 3). Although these CNVs might be involved in the evolution of ecologically relevant traits, further work will be needed to elucidate their functional significance, if any.
RNA-Seq provides insights into tissue- and sex-specific IR expression in the sensory organs of H. melpomene
Very little is known about the expression patterns of ionotropic receptors. In D. melanogaster IR genes are not only expressed in antennae [16] but also in the labellum, legs, pharynx, and anterior wing margin [25], thus suggesting a more complex function for this receptor family than previously envisioned. The majority of the H. melpomene IRs were more highly expressed in antennae compared to mouth parts and legs (Fig. 5, Additional files 4 & 5). In accordance with previous studies, the two co-receptors, IR8a and IR25a, have the highest and most homogeneous expression level across all tissues and sexes [72, 73]. However, IR8a is expressed ~eight times more than IR25a, which contrasts with Drosophila, where the two genes have similar expression levels [16], and with Aedes aegypti where IR25a is expressed tenfold more than IR8a [72]. Only in Culex quinquefasciatus is the expression of IR8a and IR25a similar to that in Heliconius [73]. These differences in gene expression at IR25a and IR8a could be driven by selection for expression levels of other chemical-specific receptors,for example, the IR20a clade is mostly expressed together with IR25a [16, 74], which implies that chemical-specific IRs differ significantly between species. In addition to IR8a and IR25a, we also identified four additional genes (IR41a, IR76b, IR64a and IR75d) with elevated expression across tissue types and sexes. All these genes display a strong sequence homology between Diptera and Lepidoptera. Although no specific function is known in Heliconius, studies in Drosophila have shown the involvement of IR41a and IR64a in amino-sensing: specifically IR41a is sensitive to 1,4-diaminobutane [75], while IR64a is sensitive to acetate, propionate and butyrate [76]. Moreover, in D. melanogaster IR76b has been shown to be co-expressed with other IRs suggesting that it might act as a second type of co-receptor [16, 74]. The ubiquitous and high expression of IR76b in H. melpomene seems to support this theory.
Like D. melanogaster [27], our analysis identified several genes that are significantly higher expressed in antennae than in legs or mouthparts (Fig. 5, Table 4, Additional files 4 & 5). This translates into the antennae differing from legs and mouthparts in their overall expression profile (Fig. 5). Such clustering likely represents the different chemosensing functions of these tissues, with mouthparts and legs being utilized in tasting while antennae are more attuned for smelling [16, 25]. Only one gene, IR60a2b, was significantly higher expressed in legs and mouthparts compared to antennae (Fig. 5, Table 4, Additional files 4 & 5), suggesting a gustatory function, possibly related to host plant recognition or mate choice. Our heat map groups male legs with male mouthparts and vice versa for females, which suggests a distinct ability of males and females to perceive compounds (Fig. 5, Additional file 5). However, only IR93a was differentially expressed between sexes but only in legs and mouth parts (Table 4, Figs. 5 and 6). Unfortunately nothing is known of the function of these two promising candidate genes, IR60a2b and IR93a.
Conclusions
Although Heliconius are best known for their stunning visual signals and vision far into the ultraviolet [77], our work and the recent study of Briscoe et al. [37] have shown an elaborate chemosensory system consisting of ~165 chemosensory receptors. Our work characterized the least studied chemosensory gene family in Lepidoptera, the ionotropic receptors (IRs). We identified instances of IR duplication and pseudogenization in Lepidoptera, butterflies, and within Heliconius. The most notable butterfly-specific IR gene duplications are of IR7d4, and of IR60a1 and IR60a2, which are Heliconius-specific. Among these Heliconius duplications, IR60a2b was uniquely found in the incompletely reproductively isolated species, H. melpomene, H. cydno, and H. timareta, and was the only gene significantly more expressed in legs and mouthparts than in antennae. Moreover, two additional genes that should be mentioned for their unique characteristics are: IR60a1, which displayed an elevated dN/dS in a major phylogenetic branch encompassing several species associated with the evolution of pupal mating, and IR93a that was the only gene with sex specific expression. Overall our work has generated a list of Heliconius candidate IR genes of evolutionary significance, which could have important implications for their chemical-mediated behaviors and ecological adaptations leading to speciation. Unfortunately functional studies of chemosensory genes are still absent in Lepidoptera, thus hampering our understanding of the specific roles of these genes. As we continue to learn more about their function, our understanding of the link between these receptors, butterfly behaviors and the evolution of relevant ecological traits will greatly improve.
Availability of supporting data
Raw data for Heliconius cydno (ERS235661), Heliconius doris (ERS977668), Heliconius hecuba (ERS977683), Heliconius timareta (ERS977723), H. sara (ERP002444), H. sapho (ERP002444), Heliconius clysonymus (ERS1061651) and Heliconius telesiphe (ERS1061652) are available at the European nucleotide archive.
All H. melpomene IR genes (KU702609-KU702639), and the intronless genes for the other 10 Heliconius species (KU756934 - KU757038) are available on genbank.
Acknowledgements
We would like to thank Brian X. Leon Ricardo, Gilbert Smith and Krzysztof Kozak for advice, and Aide Macias-Muñoz for preparing the RNA-Seq libraries. We thank the SGF at the UPR,RP supported by the INBRE (NIH/NIGMS-award number P20 GM103475), and the NSF/IFN (EPSCoR-award number 1002410), for continued assistance in the project development, bioinformatic advice and computational support.
Abbreviations
- IR
ionotropic receptor
- GR
gustatory receptor
- OR
olfactory receptor
Additional files
Excel spreadsheet with H. melpomene IRs with their genomic location and intron- and exon- boundaries. (XLSX 37 kb)
Fasta file for all manually curated IRs in the three lepidopterans: Bombyx mori, Danaus plexippus and Heliconius melpomene. (TXT 155 kb)
Fasta file for all manually curated IRs in the 11 Heliconius species utilized in this study. (TXT 610 kb)
Excel spreadsheet with all detailed FPKM expression values. (XLSX 13 kb)
Heatmap of IR gene expression for each H. melpomene biological replicate utilized in this study. Exact FPKM numbers can be found in Table 4 and Additional file 4. (EPS 8200 kb)
Footnotes
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
All authors have read and approved the manuscript. BvS Design, acquisition of data, analysis and interpretation of data, drafting the manuscript and revising. CDJ Acquisition of data, interpretation of data, revising the manuscript. ADB Acquisition of data, interpretation of data, revising the manuscript. RP Design, acquisition of data, interpretation of data, drafting the manuscript and revising.
Contributor Information
Bas van Schooten, Email: basvanschooten@gmail.com.
Chris D. Jiggins, Email: c.jiggins@zoo.cam.ac.uk
Adriana D. Briscoe, Email: abriscoe@uci.edu
Riccardo Papa, Email: rpapa.lab@gmail.com.
References
- 1.Zhan S, Merlin C, Boore JL, Reppert SM. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147:1171–85. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 3.Andersson S, Dobson HEM. Antennal responses to floral scents in the butterfly Heliconius melpomene. J Chem Ecol. 2003;29:2319–30. doi: 10.1023/A:1026278531806. [DOI] [PubMed] [Google Scholar]
- 4.Kats LB, Dill LM. The scent of death: chemosensory assessment of predation risk by prey animals. Ecoscience. 1998;5(3):361–394. [Google Scholar]
- 5.Breed MD, Diaz PH, Lucero KD. Olfactory information processing in honeybee, Apis mellifera, nestmate recognition. Anim Behav. 2004;68:921–928. doi: 10.1016/j.anbehav.2003.10.033. [DOI] [Google Scholar]
- 6.Smadja C, Butlin RK. On the scent of speciation: the chemosensory system and its role in premating isolation. Heredity. 2009;102:77–97. doi: 10.1038/hdy.2008.55. [DOI] [PubMed] [Google Scholar]
- 7.Horth L. Sensory genes and mate choice: evidence that duplications, mutations, and adaptive evolution alter variation in mating cue genes and their receptors. Genomics. 2007;90:159–75. doi: 10.1016/j.ygeno.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 8.Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 10.Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006;16:1395–403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohbot JD. Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect Mol Biol. 2007;16:525–537. doi: 10.1111/j.1365-2583.2007.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Beeman RW, Brown SJ, Bucher G, Friedrich M, Grimmelikhuijzen CJP, Klingler M, Lorenzen M, Richards S, Roth S, Schröder R, Tautz D, Zdobnov EM, Muzny D, Gibbs RA, Weinstock GM, Attaway T, Bell S, Buhay CJ, Chandrabose MN, Chavez D, Clerk-Blankenburg KP, Cree A, Dao M, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–55. [DOI] [PubMed]
- 13.Kent LB, Walden KKO, Robertson HM. The Gr family of candidate gustatory and olfactory receptors in the yellow-fever mosquito Aedes aegypti. Chem Senses. 2008;33:79–93. doi: 10.1093/chemse/bjm067. [DOI] [PubMed] [Google Scholar]
- 14.Smith CD, Zimin A, Holt C, Abouheif E, Benton R, Cash E, Croset V, Currie CR, Elhaik E, Elsik CG, Fave M-J, Fernandes V, Gadau J, Gibson JD, Graur D, Grubbs KJ, Hagen DE, Helmkampf M, Holley J-A, Hu H, Viniegra ASI, Johnson BR, Johnson RM, Khila A, Kim JW, Laird J, Mathis KA, Moeller JA, Munoz-Torres MC, Murphy MC, et al. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc Natl Acad Sci. 2011;108:5673–8. [DOI] [PMC free article] [PubMed]
- 15.Smith CR, Smith CD, Robertson HM, Helmkampf M, Zimin A, Yandell M, Holt C, Hu H, Abouheif E, Benton R, Cash E, Croset V, Currie CR, Elhaik E, Elsik CG, Fave M-J, Fernandes V, Gibson JD, Graur D, Gronenberg W, Grubbs KJ, Hagen DE, Viniegra ASI, Johnson BR, Johnson RM, Khila A, Kim JW, Mathis KA, Munoz-Torres MC, Murphy MC, et al. Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc Natl Acad Sci. 2011;108:5667–72. [DOI] [PMC free article] [PubMed]
- 16.Rytz R, Croset V, Benton R. Ionotropic Receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol Biol. 2013;43:888–97. doi: 10.1016/j.ibmb.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Corey E a, Bobkov Y, Ukhanov K, Ache BW. Ionotropic crustacean olfactory receptors. PLoS One. 2013;8:e60551. doi: 10.1371/journal.pone.0060551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chipman AD, Ferrier DEK, Brena C, Qu J, Hughes DST, Schröder R, Torres-Oliva M, Znassi N, Jiang H, Almeida FC, Alonso CR, Apostolou Z, Aqrawi P, Arthur W, Barna JCJ, Blankenburg KP, Brites D, Capella-Gutiérrez S, Coyle M, Dearden PK, Du Pasquier L, Duncan EJ, Ebert D, Eibner C, Erikson G, Evans PD, Extavour CG, Francisco L, Gabaldón T, Gillis WJ, et al. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 2014;12:e1002005. [DOI] [PMC free article] [PubMed]
- 19.Keller A. Different noses for different mice and men. BMC Biol. 2012;10:75. doi: 10.1186/1741-7007-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young JM, Friedman C, Williams EM, Ross J a, Tonnes-Priddy L, Trask BJ. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum Mol Genet. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- 21.Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–33. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- 22.Montell C. A taste of the Drosophila gustatory receptors. Curr Opin Neurobiol. 2010;19:345–353. doi: 10.1016/j.conb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon JY, Dahanukar A, Weiss L a, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci U S A. 2007;104:3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni L, Bronk P, Chang EC, Lowell AM, Flam JO, Panzano C, Theobald DL, Griffith LC, Garrity PA. A gustatory receptor paralog controls rapid warmth avoidance in Drosophila. Nature. 2013;500:580–4. [DOI] [PMC free article] [PubMed]
- 25.Koh T-W, He Z, Gorur-Shandilya S, Menuz K, Larter NK, Stewart S, Carlson JR. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron. 2014;83:850–65. [DOI] [PMC free article] [PubMed]
- 26.Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. [DOI] [PMC free article] [PubMed]
- 27.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–62. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang YV, Ni J, Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340:1334–8. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senthilan PR, Piepenbrock D, Ovezmyradov G, Nadrowski B, Bechstedt S, Pauls S, Winkler M, Möbius W, Howard J, Göpfert MC. Drosophila auditory organ genes and genetic hearing defects. Cell. 2012;150:1042–54. [DOI] [PubMed]
- 30.Estrada C, Yildizhan S, Schulz S, Gilbert LE. Sex-specific chemical cues from immatures facilitate the evolution of mate guarding in Heliconius butterflies. Proc Biol Sci. 2010;277:407–13. doi: 10.1098/rspb.2009.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Estrada C, Schulz S, Yildizhan S, Gilbert LE. Sexual selection drives the evolution of antiaphrodisiac pheromones in butterflies. Evolution. 2011;65:2843–54. doi: 10.1111/j.1558-5646.2011.01352.x. [DOI] [PubMed] [Google Scholar]
- 32.Mérot C, Frérot B, Leppik E, Joron M. Beyond magic traits: multimodal mating cues in Heliconius butterflies. Evolution. 2015;69:2891–2904. doi: 10.1111/evo.12789. [DOI] [PubMed] [Google Scholar]
- 33.Nadeau NJ, Martin SH, Kozak KM, Salazar C, Dasmahapatra KK, Davey JW, Baxter SW, Blaxter ML, Mallet J, Jiggins CD. Genome-wide patterns of divergence and gene flow across a butterfly radiation. Mol Ecol. 2012;22:814–26. [DOI] [PubMed]
- 34.The Heliconius Genome Consortium Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–8. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kronforst MR. Gene flow persists millions of years after speciation in Heliconius butterflies. BMC Evol Biol. 2008;8:98. doi: 10.1186/1471-2148-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merrill RM, Van Schooten B, Scott J a, Jiggins CD. Pervasive genetic associations between traits causing reproductive isolation in Heliconius butterflies. Proc Biol Sci. 2011;278:511–8. doi: 10.1098/rspb.2010.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briscoe AD, Macias-Muñoz A, Kozak KM, Walters JR, Yuan F, Jamie G a, Martin SH, Dasmahapatra KK, Ferguson LC, Mallet J, Jacquin-Joly E, Jiggins CD. Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet. 2013;9:e1003620. [DOI] [PMC free article] [PubMed]
- 38.Sánchez-Gracia a, Vieira FG, Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity. 2009;103:208–16. doi: 10.1038/hdy.2009.55. [DOI] [PubMed] [Google Scholar]
- 39.Engler-Chaouat HS, Gilbert LE. De novo synthesis vs. sequestration: negatively correlated metabolic traits and the evolution of host plant specialization in cyanogenic butterflies. J Chem Ecol. 2007;33:25–42. doi: 10.1007/s10886-006-9207-8. [DOI] [PubMed] [Google Scholar]
- 40.Wanner KW, Robertson HM. The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol Biol. 2008;17:621–629. doi: 10.1111/j.1365-2583.2008.00836.x. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, Yamaoka R, Touhara K. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol. 2009;19:881–90. [DOI] [PubMed]
- 42.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, Macmanes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat Protoc. 2013;8:1494–512. [DOI] [PMC free article] [PubMed]
- 44.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–23. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delport W, Poon AF, Frost SDW, Kosakovsky Pond SLK. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kosakovsky Pond SL, Murrell B, Fourment M, Frost SD, Delport W, Scheffler K. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol. 2011;28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozak KM, Wahlberg N, Neild A, Dasmahapatra KK, Mallet J, Jiggins CD. Multilocus species trees show the recent adaptive radiation of the mimetic Heliconius butterflies. Syst Biol. 2015;64(3):505–24. doi: 10.1093/sysbio/syv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin SH, Dasmahapatra KK, Nadeau NJ, Salazar C, Walters JR, Simpson F, Blaxter M, Manica A, Mallet J, Jiggins CD. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 2013;23:1817–28. [DOI] [PMC free article] [PubMed]
- 54.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–84. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–32. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 57.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–97. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–7. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 60.Wheat C, Wahlberg N. Critiquing blind dating: the dangers of over-confident date estimates in comparative genomics. Trends Ecol Evol. 2013;28:636–42. doi: 10.1016/j.tree.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 61.Brower AVZ. A new mimetic speices of Heliconius (Lepidoptera: Nymphalidae), from southeastern Colombia, revealed by cladistic analysis of mitochondrial DNA sequences. Zool J Linn Soc. 1996;116:317–332. doi: 10.1111/j.1096-3642.1996.tb00126.x. [DOI] [Google Scholar]
- 62.Giraldo N, Salazar C, Jiggins CD, Bermingham E, Linares M. Two sisters in the same dress: Heliconius cryptic species. BMC Evol Biol. 2008;8:324. doi: 10.1186/1471-2148-8-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mallet J. Speciat patterns Divers. 2009. Rapid speciation, hybridization and adaptive radiation in the Heliconius melpomene group; pp. 177–194. [Google Scholar]
- 64.Mérot C, Mavárez J, Evin A, Dasmahapatra KK, Mallet J, Lamas G, Joron M. Genetic differentiation without mimicry shift in a pair of hybridizing Heliconius species (Lepidoptera: Nymphalidae). Biol J Linn Soc. 2013;109:830–47.
- 65.Yang Z, Bielawski JR. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spielman SJ, Wilke CO. The relationship between dN/dS and scaled selection coefficients. Mol Biol Evol. 2015;32(4):1097–108. doi: 10.1093/molbev/msv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gilbert L, Lewinsohn T, Fernandes T, Benson W. Plant–animal Interact Evol Ecol Trop Temp Reg. 1991. Biodiversity of a Central American Heliconius community: pattern, process, and problems; pp. 403–427. [Google Scholar]
- 69.Freeman JL, Perry GH, Feuk L, Redon R, Mccarroll S a, Altshuler DM, Aburatani H, Jones KW, Tyler-smith C, Hurles ME, Carter NP, Scherer SW, Lee C. Copy number variation : new insights in genome diversity. Genome Res. 2006;16:949–61. [DOI] [PubMed]
- 70.Duvaux L, Geissmann Q, Gharbi K, Zhou J-J, Ferrari J, Smadja CM, Butlin RK. Dynamics of copy number variation in host races of the pea aphid. Mol Biol Evol. 2014;32:63–80. [DOI] [PMC free article] [PubMed]
- 71.Paudel Y, Madsen O, Megens H-J, Frantz L a F, Bosse M, Crooijmans RPM a, Groenen M a M. Copy number variation in the speciation of pigs: a possible prominent role for olfactory receptors. BMC Genomics. 2015;16:1–14. [DOI] [PMC free article] [PubMed]
- 72.Sparks JT, Bohbot JD, Dickens JC. The genetics of chemoreception in the labella and tarsi of Aedes aegypti. Insect Biochem Mol Biol. 2014;48:8–16. doi: 10.1016/j.ibmb.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 73.Leal WS, Choo Y, Xu P, da Silva CSB, Ueira-Vieira C. Differential expression of olfactory genes in the southern house mosquito and insights into unique odorant receptor gene isoforms. Proc Natl Acad Sci U S A. 2013;110:18704–18709. doi: 10.1073/pnas.1316059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Getahun MN, Wicher D, Hansson BS, Olsson SB. Temporal response dynamics of Drosophila olfactory sensory neurons depends on receptor type and response polarity. Front Cell Neurosci. 2012;6. [DOI] [PMC free article] [PubMed]
- 76.Ai M, Blais S, Park J-Y, Min S, Neubert T a, Suh GSB. Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila. J Neurosci. 2013;33:10741–9. doi: 10.1523/JNEUROSCI.5419-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kronforst MR, Papa R. The functional basis of wing patterning in Heliconius butterflies: the molecules behind mimicry. Genetics. 2015;200:1–19. doi: 10.1534/genetics.114.172387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Counterman B, Araujo-Perez F, Hines HM, Baxter SW, Morrison CM, Lindstrom DP, Papa R, Ferguson L, Joron M, French-Constant RH, Smith CP, Nielsen DM, Chen R, Jiggins CD, Reed RD, Halder G, Mallet J, McMillan WO. Genomic hotspots for adaptation: the population genetics of Müllerian mimicry in Heliconius erato. PLoS Genet. 2010;6:e1000796. [DOI] [PMC free article] [PubMed]


