Abstract
Human engineered heart tissues have potential to revolutionize cardiac development research, drug-testing, and treatment of heart disease; however, implementation is limited by the need to use pre-differentiated cardiomyocytes (CMs). Here we show that by providing a 3D poly(ethylene glycol)-fibrinogen hydrogel microenvironment, we can directly differentiate human pluripotent stem cells (hPSCs) into contracting heart tissues. Our straight-forward, ontomimetic approach, imitating the process of development, requires only a single cell-handling step, provides reproducible results for a range of tested geometries and size scales, and overcomes inherent limitations in cell maintenance and maturation, while achieving high yields of CMs with developmentally appropriate temporal changes in gene expression. Here we demonstrate that hPSCs encapsulated within this biomimetic 3D hydrogel microenvironment develop into functional cardiac tissues composed of self-aligned CMs with evidence of ultrastructural maturation, mimicking heart development, and enabling investigation of disease mechanisms and screening of compounds on developing human heart tissue.
Keywords: Engineered cardiac tissue, PEG-fibrinogen, embryonic, pluripotent, stem cell, developing heart tissue, maturation, microenvironment, hydrogel
Introduction
Limited access to samples of human cardiac tissue severely impedes cardiology research, drug discovery, and clinical cardiac regeneration efforts. Indeed, predicting cardiac toxicity and the triggering of arrhythmias represents a major hurdle for pharmaceutical compound development, resulting in 20-30% of drugs receiving a black box warning or being withdrawn from the market1, 2 and having devastating economic and societal consequences. Human pluripotent stem cells (hPSCs) provide the potential to produce large quantities of physiologically relevant, species-specific and even patient-specific cardiomyocytes (CMs) in vitro through directed differentiation. Engineered human heart tissues created from such cells can address the challenge of widespread cardiac tissue access, thereby providing the ability to study normal and abnormal human heart development, as well as revolutionizing high-throughput drug screening, modeling of human cardiac diseases, and the field of regenerative medicine. However, these tissues must develop structural and functional properties representative of the native human myocardium3, and their fabrication needs to be straightforward, automatable, scalable, and highly reproducible4. Achieving these goals has proven difficult.
Originally, hPSC differentiation protocols used cell aggregation to create 3D embryoid bodies (EBs)5, 6, which facilitated hPSC differentiation into spontaneously beating stem cell-derived cardiomyocytes (SC-CMs). To overcome the issues of inefficient CM production and irregular reproducibility using this EB cardiac differentiation protocol, researchers have recently focused on modulating the chemical environment of differentiating SC monolayers. Through the temporal introduction of soluble factors, this approach strives to replicate the cues directing native heart development6, 7. These highly efficient 2D differentiation protocols have revolutionized CM production from hPSCs8, 9; however, this monolayer-based approach does not replicate the fundamental 3D nature of myocardial development.
Tissue engineering offers a 3D solution to the 2D cell culture problem. The established paradigm for creation of engineered heart tissues requires a source of CMs, either isolated from rodent hearts or pre-differentiated from PSCs. Following dissociation, the CMs are combined with a biomaterial scaffold and re-assembled into cardiac tissues10-14. Although this approach has been successful in creating human cardiac tissue, the required pre-differentiation and subsequent dissociation of spontaneously contracting SC-CMs precludes direct production of mature cardiac tissues from hPSCs and hinders investigation of the role of cellular microenvironment during early human cardiac development. The multiple cell-handling steps involved create not only processing and fabrication challenges, and limit the ability for tissue biomanufacturing, but also disrupt important cell-cell junctions, and cause a high degree of cell loss. Establishing a simple workflow that reduces the number of cell-handling steps and provides a 3D microenvironment throughout differentiation would transform the fabrication of human cardiac tissues, which will be critical for their successful utilization in developmental biology research, high-throughput pharmaceutical screening, and generation of mature SC-CMs for basic science and clinical applications.
Natural biomaterials (e.g., fibrin, gelatin, Type-I collagen) have been used most frequently to support classic engineered cardiac tissue formation10, 13, 15 but present limitations due to their inherent batch-to-batch variability, lack of immediate structural support, and the degradation of contractility during long-term cardiac tissue maintenance in vitro. In comparison, synthetic materials are completely defined and rapidly crosslinkable with tunable properties, but typically lack biological components inherent to natural scaffolds. Hybrid biomaterials, having both natural and synthetic components, provide unique advantages for tissue engineering applications. Particularly relevant to forming cardiac tissues, hybrid biomaterials have tunable mechanical properties combined with natural sites to support cell survival, adhesion, proliferation, and differentiation, as well as degradation and remodeling of the scaffolds16, 17. Therefore, employing hybrid biomaterials, such as poly(ethylene glycol)-fibrinogen (PEG-fibrinogen), enables properties of both components to be exploited in guiding hPSC differentiation and the process of human engineered cardiac tissue formation (Table S1).
Here we asked whether it is possible to directly create a 3D tissue construct emulating key stages of cardiac embryologic development, growth, and maturation in an ontogeny-mimicking (i.e., ontomimetic) model of human heart muscle. We found that human induced pluripotent stem cells (hiPSCs) can successfully be differentiated into contracting CMs while encapsulated within PEG-fibrinogen hydrogels, thereby directly forming functional 3D developing human engineered cardiac tissues (3D-dhECTs) with a single cell-handling step during the cell-encapsulation process.
For the first time, we have successfully encapsulated and differentiated hiPSCs within a controlled biomimetic hydrogel microenvironment, achieving developmentally-appropriate temporal changes in gene expression, high CM yield, and calcium handling properties similar to age-matched CMs produced using high-efficiency 2D monolayer differentiation. Furthermore, CMs within our 3D-dhECTs became progressively anisotropic without external electromechanical stimuli and developed ultrastructural features characteristic of mature CMs. These results demonstrate that providing a 3D architecture during and after hiPSC differentiation without disturbing cell-cell junctions and providing continuous 3D cell-cell and cell-material interactions is advantageous for creating an ontomimetic model of native human myocardium.
Materials and methods
HiPSC expansion and culture
IMR-90 Clone 1 and 19-9-11 human induced pluripotent stem cells (hiPSCs) were purchased from WiCell and maintained at 37ºC, 5% CO2, and 85% relative humidity. HiPSCs were cultured as colonies on hESC qualified Matrigel (BD Biosciences) using mTeSR-1 medium (Stem Cell Technologies). HiPSCs were passaged using Versene (Life Technologies) and 5 μM ROCK inhibitor (Y-27632, R&D Systems) was added to the mTeSR-1 medium for 24 h post-seeding.
Glass acrylation and PDMS mold preparation
All chemicals were purchased from Sigma-Aldrich unless specified otherwise. In order to accurately track tissue development over time, microisland tissues were fabricated and immobilized on acrylated glass coverslips. Circular glass coverslips (21 mm, No. 1, Fisher Scientific) were cleaned using 30% hydrogen peroxide (H2O2) and 70% sulfuric acid (H2SO4), followed by thorough ethanol rinsing and air-drying prior to glass acrylation. Cleaned glass coverslips were incubated overnight using diluted acetic acid (9%), 3-(Trimethoxysilyl) propyl methacrylate, and 200-proof ethanol at 25°C. After incubation, the acrylated glass coverslips were rinsed in ethanol followed by air-drying.
Polydimethylsiloxane (PDMS) molds were prepared by combining SLYGARD 184 silicone elastomer curing agent and SLYGARD 184 elastomer base (Dow Corning Corporation). PDMS precursor solution was transferred onto a glass slide fixed with 200 μm thick spacers on all edges. A second slide was placed on top and the entire assembly was bound together by binder clips. The assembly was transferred into an oven at 70ºC for 2 h for curing of the PDMS. The cured PDMS was peeled off the assembly and three cylindrical holes with diameters of 6 mm were punched into the PDMS mold using a cork borer. The PDMS mold and acrylated glass coverslips were soaked in 70% ethanol for 24 h followed by complete air-drying under UV-light for at least 24 h to ensure complete sterilization. Before use in cell encapsulation, the PDMS mold with three cylindrical holes for tissue production was placed on the circular, acrylated glass coverslips and pressed down firmly to prevent leakage.
PEG-fibrinogen synthesis and precursor preparation
PEG-fibrinogen was prepared as previously described18. First, poly(ethylene glycol)-diacrylate (PEG-DA) was prepared by reacting linear PEG-OH (10 kDa) with acryloyl chloride and triethylamine. The product was precipitated in ice-cold diethyl ether, followed by vacuum drying for 48 h. Degree of acrylation was quantified by proton 1H NMR.
For PEG-fibrinogen synthesis, tris(2-carboxyethyl) phosphine hydrochloride (TCEP-HCl) was combined with 7 mg/ml fibrinogen in PBS with 8 M urea (1.5:1 TCEP to fibrinogen molar ratio). Next, for PEGylation of fibrinogen, PEG-DA was reacted with fibrinogen (4:1 molar ratio) for 3 h, precipitated in acetone and dissolved in PBS with 8 M urea. The reacted PEG-fibrinogen was dialyzed against PBS at 4ºC for 48 h followed by lyophilization. To characterize the PEGylated product, fibrinogen content was measured using Pierce BCA assay (Thermo Scientific).
Lyophilized PEG-fibrinogen powder was re-dissolved in PBS to obtain a final fibrinogen concentration of 10 mg/ml. PEG-fibrinogen precursor solution was prepared by combining PEG-fibrinogen with 1.5% triethanolamine (TEOA), 3.96 μl/ml N-vinyl pyrrolidone (NVP), and 10 mM eosin Y (Fisher Scientific) photoinitiator (in PBS)19.
HiPSC dissociation and 3D-dhECT microisland formation
HiPSCs were dissociated using Versene (cluster hiPSC encapsulation for IMR90-1 and 19-9-11 hiPSC lines) or Accutase (single IMR90-1 hiPSC encapsulation, Innovative Cell Technologies) to form 3D developing human engineered cardiac tissue (3D-dhECT) microislands (day -3). Cluster or single hiPSCs were resuspended in mTeSR-1 medium and centrifuged for 5 min at 200 g. The supernatant was aspirated using a glass Pasteur pipette and complete aspiration was insured through inversion of the tube without disturbing the cell pellet. Cluster or single hiPSCs were resuspended uniformly at a density of 55 ± 8.5 × 106 hiPSCs/ml of PEG-fibrinogen precursor solution using a wide orifice pipette tip. 10 μl of this mixture was added to each of the three cylindrical holes in the PDMS mold on acrylated glass coverslips and crosslinked using visible light (intensity of 48 mW/cm2 at a distance 3 cm away) for 1 min. The PDMS mold surrounding the three crosslinked hydrogels (3D-dhECT microislands) was then carefully detached and the acrylated glass coverslip with covalently coupled microislands was transferred to a 12-well plate (three tissues per well) and cultured in 2 ml mTeSR-1 medium supplemented with 5 μM ROCK inhibitor for 24 h (day -3). On the two days following encapsulation (day -2, day -1), 3D-dhECT microislands were cultured in mTeSR-1 medium with daily medium exchange. On the third day post-encapsulation (day 0), cardiac differentiation was initiated. All encapsulations resulted into successfully contracting cardiac tissues.
2D monolayer differentiation of hiPSCs
For 2D monolayer differentiation of hiPSCs (IMR90-1 hiPSC line), the composition of the media and timeline of differentiation was based on a previously published method8; 2D differentiating monolayers were used as controls. Briefly, hiPSCs were dissociated using Accutase, resuspended in mTeSR-1 medium, counted, and centrifuged. HiPSCs were seeded at 1 × 106 hiPSCs/well in a Matrigel coated 6-well plate with 4 ml mTeSR-1 medium + 5 μM ROCK inhibitor for 24 h (day -4). From day -3 until day 0, mTeSR-1 medium was replaced daily. On day 0 of differentiation, medium was changed to 4 ml RPMI/B27 without insulin (Life Technologies) + 12 μM CHIR99021 (Selleckchem) for 24 h. Medium was changed to 4 ml RPMI/B27 without insulin for an additional 48 h. On day 3, 2 ml RPMI/B27 without insulin and 5 μM IWP2 (Tocris) were combined with 2 ml old RPMI/B27 without insulin (“combined medium”) and cells were cultured until day 5, when medium was changed back to RPMI/B27 without insulin. On day 7 and successively every three days, medium was replaced with RPMI/B27 (Life Technologies).
3D cardiac differentiation of cluster and single hiPSCs in PEG-fibrinogen hydrogels
Cluster and single encapsulated hiPSCs underwent differentiation following the same protocol used for the 2D monolayer differentiation of hiPSCs. On day 0 of differentiation, medium was changed from mTeSR-1 to 2 ml RPMI/B27 without insulin + 12 μM CHIR99021 per well. After 24 h (day 1), medium was changed to 2 ml RPMI/B27 without insulin. On day 3 of differentiation, medium was replaced with 1 ml fresh RPMI/B27 without insulin, 5 μM IWP2, and 1 ml old RPMI/B27 without insulin and cultured for an additional 48 h. On day 5, medium was switched back to 2 ml RPMI/B27 without insulin and followed by 2 ml RPMI/B27 medium on day 7, medium being replaced thereafter every three days.
HiPSC viability and immunofluorescence
HiPSC viability on day -2 (24 h post-encapsulation) was assessed using a LIVE/DEAD® viability kit (Molecular Probes) following manufacturer’s instructions. Confocal Z-stacks (step size = 5 μm) were taken through the entire tissue thickness at several randomly selected locations within each 3D-dhECT microisland (n = 3) using a Nikon A1R laser-scanning confocal microscope and NIS Elements software (Nikon). To assess 3D-dhECT protein expression of proliferation markers proliferating cell nuclear antigen (PCNA) and Ki67, cardiac markers cardiac troponin T (cTnT) and sarcomeric α-actinin (αSA), and gap junction protein connexin 43 (Cx43), tissue samples were prepared for immunofluorescence. To assess the area and circularity of single CMs, dissociated 3D-dhECT cells were immunostained using αSA and Caveolin 3 (T-tubules). First, samples were fixed using methanol for PCNA, 4% paraformaldehyde (Electron Microscopy Sciences) for Ki67, cTnT, αSA, and Caveolin 3, or 50/50 ice-cold acetone/ethanol for Cx43. Fixed tissues and dissociated cells were permeabilized with PBS-T (PBS with 1% bovine serum albumin (BSA) and 0.2% Triton X-100) and blocked (3% fetal bovine serum (FBS, Atlanta Biologicals) in PBS). Samples were consecutively incubated in primary and secondary antibody (Table S2). All primary and secondary antibodies were applied for at least 24 hours at 4ºC. Cell nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI, Molecular Probes). All fluorescently labeled samples were visualized using a Nikon A1si confocal microscope.
For subsequent characterization studies and comparisons to 2D monolayers, cluster encapsulated 3D-dhECTs were used.
Tissue area growth
Throughout the initial stages of hiPSC encapsulation, images of entire tissues from both hiPSC lines were acquired daily at low magnification using a phase contrast microscope (Ti Eclipse, Nikon) equipped with an Andor Luca S camera. Tissue edges were identified and the lateral surface area of 3D-dhECTs was analyzed in ImageJ with standard analysis plugins (n = 3-4 tissues per hiPSC line). Tissue growth was based on normalized day 0 tissue surface area.
Flow cytometry
On day 20, 2D monolayers were washed with PBS and incubated in 0.25% trypsin (EDTA, Mediatech) at 37ºC for 5 min. Age-matched 3D-dhECTs from both hiPSC lines were washed with PBS followed by 2 h incubation at 37ºC on a rotator (Boekel Orbitron Rotator, Model 260250, Boekel Scientific) with collagenase Type 2 (1 mg/ml, Worthington) in 120 mM NaCl, 5.4 mM KCl, 5 mM MgSO4, 5 mM Na-pyruvate, 20 mM glucose, 20 mM taurine, and 10 mM HEPES (pH 6.9) supplemented with 30 μM CaCl2, followed by incubation in 0.25% trypsin (EDTA) at 37ºC for 5 min. All cells (in 2D monolayers and 3D-dhECTs) were then singularized by pipetting (using a 1000 μl pipette), transferred to a centrifuge tube with RPMI20 medium (RPMI 1640 medium with 20% FBS) and centrifuged for 5 min at 200 g. The supernatant was removed and the cell pellet was resuspended in 4% paraformaldehyde and incubated for 20 min at 25°C. Cells were centrifuged for 5 min at 200 g, supernatant was aspirated and the cell pellet was resuspended in 90% cold methanol and incubated at 4ºC for 15 min. The fixed cells were blocked with 5% BSA in PBS for 5 min and centrifuged for 5 min at 200 g. After washing two more times with 5% BSA in PBS, cells were incubated in 100 μl primary antibody (Table S2) diluted in 0.5% BSA and 0.1% Triton X-100 in PBS. To analyze CMs, proliferating cells, and fibroblasts, the primary antibody combinations cTnT/Ki67 and cTnT/P4HB were chosen. After incubation at 4ºC overnight, cells were washed with 0.5% BSA and 0.1% Triton X-100 in PBS, which was repeated twice. Cells were further incubated in 100 μl secondary antibody (Table S2) diluted in 0.5% BSA and 0.1% Triton X-100 in PBS for 30 min at room temperature, protected from light. Finally, cells were washed with 0.5% BSA and 0.1% Triton X-100 in PBS three times and resuspended for analysis in 500 μl 5% BSA in PBS. Samples were run on a BD Accuri C6 (BD Biosciences) and were analyzed using FlowJo V10.
Reverse transcription quantitative PCR
Total RNA was extracted from hiPSCs, 2D monolayers, and 3D-dhECTs on days 0, 10, 20, and 30 of differentiation (n = 3 independent differentiations) using Nucleospin RNA kit (Macherey-Nagel). Reverse transcription quantitative PCR (RT-qPCR) was performed using SuperScript III Platinum One-Step RT-qPCR kit (Invitrogen) in conjunction with Taqman probes (Integrated DNA Technologies). Equal amounts of RNA (50 ng/19 μl) were used for each measurement. The RT-qPCR protocol consisted of 1 cycle at 50 ºC (15 min), 1 cycle of 95ºC (3 min), 45 cycles of 95ºC (15 s) and 55ºC (30 s). Gene expression levels were normalized to the housekeeping gene GAPDH using the 2-ΔCt method. Duplex RT-qPCR was used for Oct4, MYL2v, and Cx43 genes; αMHC, βMHC, and GAPDH were quantified separately (Table S3).
Frequency of contraction
Video recordings of spontaneously contracting 3D-dhECTs were acquired on a phase contrast microscope (Ti Eclipse, Nikon) using a high speed camera (Andor Luca S) to determine frequency of contraction. Number of contractions per minute at early (day 8-11), intermediate (day 20-25), late (day 30-33, n = 3 independent differentiation batches), and long-term (days 50, 60, and 90, n = 2 tissues) time points were analyzed using NIS Elements and ImageJ software. Contractility differences between single and cluster encapsulated hiPSCs were determined by video analysis of the relative changes in transmitted light intensity of spontaneously contracting cardiac tissues on day 14 of differentiation.
Sarcomere alignment and spacing in 3D-dhECT CMs
Throughout the differentiation process, sarcomere alignment of days 20, 30, and 124 αSA-stained cardiac tissues was analyzed using ImageJ software version 1.48q (NIH) in affiliation with the fast Fourier transform (FFT) analysis tool. Images were imported into ImageJ, and sarcomere pattern formation over time was assessed by increasing periodicity, alignment, and pattern convergence in the FFT output spectra. Sarcomere alignment and spacing was also determined by manually drawn linear paths along visible sarcomeres. In addition to sarcomere alignment, day 124 αSA-stained CMs were used to quantify sarcomere spacing; for this, manually drawn linear paths along well-defined sarcomeres (n = 200 CMs) were used to collect an intensity profile for each chosen CM where distances between peaks were analyzed using Microscoft Excel. Sarcomeres chosen for alignment and spacing were selected based on continuity of sarcomeres in a single field of view.
3D-dhECT CM size, circularity, length-to-width ratio and Caveolin 3 immunofluorescence staining
For size and circularity measurements, day 52-60 tissues were dissociated into single cells (see tissue dissociation section in Flow Cytometry) and re-plated onto fibronectin coated PDMS coverslips. After three days, cells were immunofluorescently stained and imaged. Cells were analyzed for their cell area and circularity using ImageJ software with standard analysis plugins (n = 32 CMs). These samples were also used for αSA/Caveolin 3 staining. Four month old 3D-dhECTs were enzymatically dissociated and seeded onto fibronectin-coated well plates. 24 hrs after dissociation, phase contract images were taken of three separately dissociated tissues. Length and width of 60 cells were measured using ImageJ and the ratio was calculated. Cell clusters of two or more cells were excluded for this analysis.
Calcium handling
To immobilize cells for proper calcium transient acquisition, glass coverslips (21 mm, No. 1, Fisher Scientific) were coated with PDMS using a WS-400-6NPP spin coater (Laurell Technologies Corporation). Briefly, SLYGARD 184 silicone elastomer curing agent was mixed with SLYGARD 184 elastomer base (Dow Corning Corporation) at a ratio of 1:10, applied on the glass coverslips and evenly spin-coated to obtain PDMS coated coverslips. These were cured at 60ºC for several hours, sterilized using 70% ethanol, and then dried under sterile conditions for at least 24 h. In order for successful single CM attachment, 25μg/ml fibronectin in ice-cold ultrapure water was applied on PDMS coated coverslips and incubated for at least one hour at room temperature.
Day 14 2D monolayers and 3D-dhECTs were dissociated using 0.25% trypsin (EDTA) at 37ºC for 5 and 8 min, respectively. Cells were singularized by pipetting, added to RPMI20 medium20, and centrifuged for 5 min at 200 g. Singularized cells were resuspended in RPMI20 + 5 μM ROCK inhibitor and transferred onto fibronectin-coated PDMS-glass coverslips. Cells were incubated for 48 h to ensure uniform cell attachment. Medium was then switched to RPMI/B27 and cells maintained for 3 to 5 days. CM calcium transient recordings were obtained using an IonOptix Myocyte Calcium and Contractility Recording System. Samples were incubated in 5 μM Fura-2AM dye (Molecular Probes) in 37ºC warm Tyrode’s solution (1.8 mM CaCl2, 5 mM glucose, 5 mM HEPES, 1 mM MgCl2, 5.4 mM KCl, 135 mM NaCl, and 0.33 mM NaH2PO4, pH 7.4) for 30 min. CMs were paced at frequencies from 0.5 - 2.0 Hz (0.5 Hz increments) and stimulated at 30 V; maximum capture rate was defined as the highest pacing frequency for which there was a 1:1 correspondence between exogenous pacing and cell response. Calcium transients were recorded 100 frames per second and calcium transient duration at 50% and 80% time to baseline was measured. In addition to responses to outside electrical stimuli, spontaneously contracting 3D-dhECT CM response to 0.5 μM isoproterenol (Molecular Devices) was examined. Representative calcium transient traces were processed by temporal averaging of five time steps.
Multielectrode array (MEA)
Day 20 3D-dhECTs were dissociated for 2 hrs in collagenase solution (as previously described for flow cytometry), resuspended in RPMI20 medium supplemented with 5 μM ROCK inhibitor, and cultured on a S2 type MEA200/30-Ti-grfor (Multichannel Systems) for at least 24 hrs. Spontaneously contracting CMs were perfused with Tyrode’s solution at 37 ºC using a MEA system with Multichannel Systems 1060-Inv-BC amplifier. When stabilized, baseline field potential recordings were acquired at a sampling frequency of 10 kHz. CMs were exogenously paced at 0.5, 1, 2, and 3 Hz. To assess drug response, 1 μM isoproterenol was added with a subsequent addition of 1 μM propranolol (Molecular Devices). Drugs were washed out to recover the baseline field potential profile.
Transmission electron microscopy (TEM)
Day 24 and day 124 3D-dhECTs were fixed in 3% glutaraldehyde (Electron Microscopy Science) at 4ºC before shipping to the pathology core at Icahn School of Medicine at Mount Sinai, NY. Tissue samples were sectioned (50 - 60 nm) and stained with Uranyl Acetate Solution and Reynold’s Lead Citrate Solution. Processed tissue slices were examined using a transmission electron microscope (H-7650, Hitachi High Technologies).
Quantification of sarcomere structure
The prevalence of H-zones and I-bands were measured to quantify the sarcomere structural features. The numerical ratio of H-zones to sarcomeres, and the ratio of I-bands to Z-lines were analyzed from TEM images from day 124 3D-dhECTs using ImageJ software version 1.49v (NIH) with the Plot Profile analysis tool. Each image was imported into ImageJ and a linear region of interest was manually drawn across the length of a selected sarcomere; a 25-pixel line width was used to improve signal to noise, with the live plot option allowing real-time evaluation of the gray scale intensity pattern. The variation in gray scale values was used to identify Z-lines (pronounced decrease in the gray scale value), I-bands (pronounced increase in the gray scale value bordering the Z-lines) and H-zones (increase in gray value in the central region between each pair of Z-lines). A total of 101 sarcomeres and 142 z-lines were measured from 3 TEM images at 2-2.5K magnification.
Statistics
Results were analyzed from a minimum of three replicates for each experiment using Minitab 16. All values presented are mean ± SD. Data were compared using one-way analysis of variance (ANOVA) followed by Tukey’s test, P < 0.05 was considered statistically significant.
Results
PEG-fibrinogen hydrogels support stem cell survival and proliferation
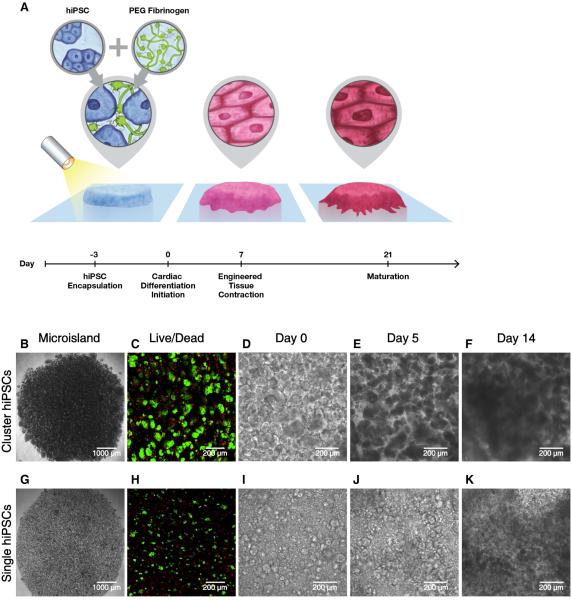
HiPSCs are frequently cultured and passaged in multi-cellular clusters, which has been reported to enhance cell viability and maintenance of pluripotency when compared to single cell dissociation21. On the other hand, the inherent variability in cluster size might present a challenge for creating highly reproducible 3D-dhECTs from hiPSCs. In the presented study, we established the ability to create 3D tissues using both cluster and single dissociated hiPSCs. In both cases, a cell suspension of 55 ± 8.5 × 104 hiPSCs per tissue was combined with aqueous PEG--fibrinogen precursor solution and photocrosslinked using visible light to create 200 μm thick 3D “microisland” tissues (Fig. 1A). Both cluster and single hiPSCs were uniformly distributed throughout the 3D-dhECT microislands (Fig. 1B, G); tissues formed using single hiPSCs appeared more homogeneous compared to cluster encapsulated hiPSCs (Fig. S1A). HiPSC viability 24 h post-encapsulation (day -2, Fig. 1C, H) was comparable to standard hiPSC passage and culture22.
Fig. 1.
Schematic and progression of hiPSC encapsulation to produce 3D-dhECTs. (A) HiPSCs were combined with aqueous PEG-fibrinogen precursor, added into a PDMS mold (not shown) on acrylated glass, and photocrosslinked using a non-toxic photoinitiator and visible light to form 200 μm thick “microisland” tissues. All encapsulated hiPSCs were maintained in their pluripotent state for three days, followed by initiation of cardiac differentiation to produce uniformly contracting 3D-dhECTs. Encapsulated (B) cluster and (G) single hiPSCs formed “microisland” tissues (images taken on day 0 of differentiation). (C, H) HiPSCs remained viable (green) 24 h post-encapsulation (n = 3 tissues) and (D-F, I-K) proliferated during early stages of cardiac differentiation. F, K represent Movies S3 and S4.
Encapsulated cluster and single dissociated hiPSCs grew and proliferated within the PEG--fibrinogen hydrogels both prior to (day -2 to day 0, Fig. S1A) and following initiation of differentiation (day 0), which resulted in an expanded microisland lateral surface area of 20.1 ± 8.0% (IMR90-1 hiPSC line) and 25.5 ± 8.0% (19-9-11 hiPSC line) by day 5 of differentiation (Fig. S1B). Cell proliferation on day 10 was observed by immunostaining for proliferating cell nuclear antigen (PCNA, Movie S1), with the highest number of proliferating cells located on the tissue edges (Fig. S1C). During early stages of cardiac differentiation, cluster dissociated hiPSCs continued to grow as aggregates. Cells occupied the entire hydrogel volume quickly (Fig. 1D, F) and tissues exhibited more growth at the edges than in the center. Tissue thickness increased from ~200 μm to 300 μm between days 3 and 5, with a dense tissue ring forming around the perimeter of the microisland (Fig. S1D). In comparison, encapsulated single hiPSCs first formed small colonies and then subsequently formed interconnected tissue structures (Fig. 1I, K).
Encapsulated hiPSCs form uniformly contracting, functional cardiac tissues
This direct encapsulation and cardiac differentiation process resulted in uniformly contracting cardiac tissues with a high degree of repeatability, while requiring only one cell-handling step. All 3D-dhECTs were observed to spontaneously contract, with contraction initiating consistently on day 6 for the cluster 19-9-11 hiPSC line encapsulations (n = 3 independent differentiations, Movie S2), day 7 for all cluster IMR90-1 hiPSC line encapsulations (n > 40 independent differentiations), and between days 9-11 for all single encapsulated hiPSCs (n = 3 independent differentiations), which compares to day 8-9 for control 2D monolayers (n > 20 independent differentiations). The number of spontaneously contracting areas and synchronicity of contraction increased over time for both cluster and single cell 3D-dhECTs, consistently resulting in essentially uniform contracting tissues by day 14 (Movies S3 and S4). Frequency of spontaneous contraction for cluster 3D-dhECTs was slowest during early differentiation (0.60 ± 0.21 Hz, days 8-11) and increased throughout the differentiation process to 1.37 ± 0.04 Hz by days 30-33 (n = 3 separate differentiations, Fig. S2A). During long-term culture, the spontaneous contraction rate reached its highest frequency (2.2 ± 0.1 Hz) on day 50 (n = 2 tissues) and then began to decrease, stabilizing at 1.35 ± 0.05 Hz at day 90 (Fig. S2A). Frequency of contraction for single cell 3D-dhECTs tended to be slower than for cluster 3D-dhECTs (0.48 ± 0.50 Hz, n = 2 vs. 0.60 ± 0.21 Hz) during early stages of differentiation, and contractility did not appear to be as strong as that of age-matched cluster 3D-dhECTs (Fig. S2B). Cluster hiPSC encapsulation was chosen for all subsequent experiments due to the higher batch-to-batch consistency of cluster 3D-dhECTs, particularly with respect to the time point for initiation of contraction, as compared to single cell 3D-dhECTs.
3D-dhECTs demonstrate differentiation efficiency and cardiac gene expression similar to 2D monolayers
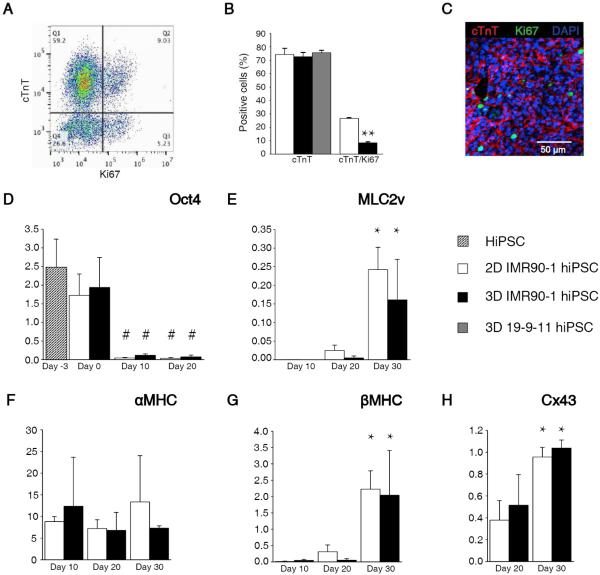
Cardiac differentiation within the 3D-dhECTs resulted in high CM yield and temporal changes in gene expression that were analogous to those in age-matched 2D monolayers. To determine the efficiency of cardiac differentiation within 3D-dhECTs, the percentage of cells expressing cardiac troponin T (cTnT, cardiac specific marker) was evaluated by flow cytometry and immunofluorescence on day 20. The 3D-dhECTs were composed of 72.5 ± 3.2% (IMR90-1 hiPSCs) and 75.65 ± 1.8% (19-9-11 hiPSCs) cTnT positive cells, of which 8.4 ± 0.7% also expressed the proliferation marker Ki67 (IMR90-1 hiPSCs, Fig. 2A); cardiac differentiation was similar to age-matched 2D monolayers which had 74.3 ± 4.4% cTnT positive cells, 26.6 ± 0.5% of which also expressed Ki67 (IMR90-1, Fig. 2B). Immunofluorescence of 3D-dhECTs also confirmed the expression of cTnT and Ki67 (Fig. 2C). Of the remaining 3D-dhECT cells, 5.24 ± 2.72% were fibroblasts (P4HB positive, Fig. S3) and 2.96 ± 1.98% expressed Ki67 (Fig. 2A).
Fig. 2.
3D-dhECTs enabled efficient cardiac differentiation and gene expression. (A) Representative flow cytometry results from day 20 3D-dhECTs. (B) 3D-dhECTs and aged-matched 2D monolayers (control) showed comparable differentiation efficiency on day 20 of differentiation. n = 3-5 biological replicates per group. Mean ± s.d. ANOVA P < 0.05, ** vs. 2D (C) Cardiac marker cTnT and proliferation marker Ki67 were also observed using immunofluorescence staining of day 20 cardiac tissues. (D) Pluripotency gene Oct4 decreased following initiation of differentiation, while cardiac genes (E) MLC2v, (F) αMHC, (G) βMHC, as well as (H) the functional gene Cx43 showed trends towards CM maturation over time. All mRNA levels were normalized to the housekeeping gene GAPDH. n = 3 biological replicates per group. Mean ± s.d. ANOVA P < 0.05, # vs hiPSCs and day 0; * vs. earlier time point.
Temporal changes in pluripotency and cardiac gene expression in 3D-dhECTs paralleled that of age-matched control 2D monolayers. Three days post-encapsulation (day 0), expression of the pluripotency gene Oct4 continued to be similar to hiPSCs (day -3) in both culture systems, as assessed by RT-qPCR. By day 10 of differentiation, Oct4 expression had decreased significantly compared to hiPSCs and remained low (Fig. 2D). The decrease in pluripotency was paired with a simultaneous upregulation in cardiac and functional gene expression. MLC2v, an early cardiac marker, increased in both 2D monolayers and 3D-dhECTs from day 10 to day 30 of differentiation (Fig. 2E). Cardiac development was also tracked through assessment of αMHC and βMHC expression, where βMHC is the predominant isoform in the healthy adult human ventricle23. 2D monolayers and 3D-dhECTs expressed αMHC from day 10 through day 30. Expression of αMHC in 3D-dhECTs did not change significantly between days 10-30 whereas βMHC expression increased by day 30 (Fig. 2F, G). This increase in βMHC gene expression by day 30 of differentiation suggests that cardiac maturation occurs over time within the 3D-dhECTs, similar to 2D differentiation. Expression of the Cx43 gene, which encodes for a gap junction protein important for cardiac function, increased significantly from day 20 to day 30 in 3D-dhECTs and control 2D monolayers and could contribute to maturation of cell-cell junctions between adjacent CMs (Fig. 2H).
3D-dhECT CMs exhibit similar calcium handling to CMs from 2D cardiac monolayers
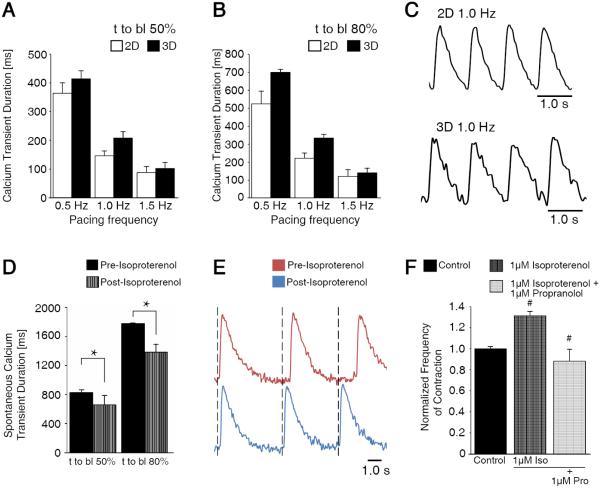
In addition to 3D-dhECTs demonstrating high CM yields and appropriate cardiac gene expression profiles similar to age-matched 2D monolayers, CMs from both systems had similar contractile function and calcium handling properties, which are critical features of differentiating CMs24. To study these properties, CMs were enzymatically dissociated from 3D-dhECTs and 2D monolayers at day 14. Isolated CMs continued to contract spontaneously and also responded to exogenous pacing, exhibiting a maximum 1:1 capture rate of 1.5 Hz for both 3D- and 2D-differentiated CMs Calcium transient durations for 3D-dhECT CMs were similar to those of control 2D monolayer CMs (Fig. 3A-C). Calcium transient duration decreased in response to increased pacing frequency for both groups of CMs, mimicking the frequency-dependent physiological response of healthy myocardium25. CMs also responded appropriately to treatment with isoproterenol (0.5 μM for 3 min), a β-adrenergic agonist; the frequency of spontaneous contraction increased by 1.2-fold and calcium transient duration shortened significantly (Fig. 3D, E). The frequency of 3D-dhECT spontaneous contraction slowed significantly after the addition of the β-adrenergic antagonist propranolol. First, 1 μM isoproterenol was administered to whole tissues, which caused a significant increase in contraction frequency. As previously observed from isolated cells26, 27, the addition of propranolol reversed this initial increase in beats per minute, demonstrating that our 3D-dhECTs respond to beta agonists and antagonists (Fig. 3F). These responses to exogenous pacing and β-adrenergic agonist and antagonist were further verified using MEA analysis (Fig. S4). Dissociated day 20 3D-dhECTs responded to outside pacing frequencies up to 3 Hz (Fig. S4A) and spontaneously contracting CMs increased in response to isoproterenol and was reversed after the addition of propranolol (Fig. S4B).
Fig. 3.
3D-dhECT CMs responded to exogenous pacing and beta adrenergic agonist and antagonist drug treatment. During exogenous pacing, time to baseline (t to bl) (A) 50% and (B) 80% were similar for 2D and 3D cultured CMs (day 14). (C) Representative calcium transient traces of 2D and 3D cultured cells at 1 Hz pacing frequency. (D) Addition of 0.5 μM isoproterenol shortened spontaneously contracting 3D-dhECT CM calcium transient t to bl 50% and 80%, suggesting that the chronotopic response to β-adrenergic signaling is operational. (E) In these early stage 3D-dhECT CMs, isoproterenol increased the rate of spontaneous contraction. n = 3. Mean ± s.d. ANOVA P < 0.05, * vs. Pre-Isoproterenol. (F) Rate of 3D-dhECT spontaneous contraction increased after the addition of isoproterenol (1 μM) and decreased after subsequent addition of the β-adrenergic antagonist propranolol (1 μM). # vs. earlier condition.
3D-dhECT CMs develop mature structural features over time
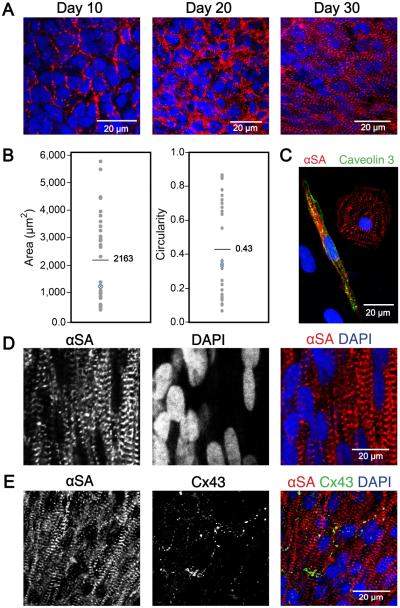
3D-dhECTs were able to be maintained long-term, with CMs developing aligned and more defined sarcomeres over time and establishing mature ultrastructural features. Cells within the 3D-dhECTs self-aligned, particularly on tissue edges, by day 30 of differentiation; immunofluorescence staining for the cardiac marker sarcomeric α-actinin (αSA) on days 10, 20, and 30 revealed progressive sarcomere development with better defined and aligned sarcomeres by day 30 of differentiation (Fig. 4A). This result was unexpected; extrinsic stimuli, such as mechanical stretching10, 28-30 or electrical pacing10, 31, 32, have often been used to enhance the ability of CMs to mature, align, and uniformly contract within engineered cardiac tissues assembled using pre-differentiated SC-CMs. However, the temporal upregulation of cardiac gene expression and increased organization of cardiac proteins suggested CM maturation within 3D-dhECTs was progressing spontaneously, which led us to further investigate CM organization and ultrastructural development during long-term tissue culture. Based on 2D fast Fourier Transform (FFT) analysis of immunofluorescence images from day 20, 30, and 124 3D-dhECTs, sarcomeres became more defined over time (Fig. S5A-C). Some discrete sarcomere structure was observed during early stages of differentiation (day 20); by day 124 3D-dhECT CMs had aligned, organized, well-defined sarcomeres with an average relaxed length of 1.8 ± 0.1 μm (Fig. S5D, n = 200 cells), similar to mature human CM sarcomere structure12, 33. Following extended in vitro culture (day 52-60), tissue dissociation into single CMs revealed a wide range of cell sizes (400-5,800 μm2, mean = 2,163 μm2) and circularities (0.10-0.86, mean = 0.43) (Fig. 4B, C). When maintained long-term, 3D-dhECTs CMs showed features of mature CMs (Fig. S6A) and 3D-dhECTs continued to have higher cell density on the tissue edges with CMs growing outward from the tissue onto the supporting substrate (Fig. S6B, C). Cellular organization within the long-term cultured 3D-dhECTs (day 124) exhibited large cell nuclei (Fig. 4D) and developed characteristics of mature electromechanically anisotropic cardiac tissues, including aligned CMs with the gap junction protein connexin 43 (Cx43) located on the transverse ends (Fig. 4E) of adjoining cells. When dissociated, 3D-dhECT CMs exhibited an elongated, anisotropic morphology with a length-to-width ratio of 6.2±2.2 (n = 60 CMs), similar to morphologies expected in mature CMs3, 32, 34.
Fig. 4.
Ontogenetic tissue development generating well-defined and aligned sarcomeres over time. (A) Sarcomere definition and alignment became more pronounced with culture time. Immunofluorescence staining with cardiac marker αSA on days 10, 20, and 30 of differentiation showed increased sarcomere definition and alignment in 3D-dhECT CMs. (B) Day 52-60 dissociated 3D-dhECT CM area and circularity ranged from 400-5,800 μm2 (mean = 2,163 μm2) and 0.10 – 0.86 (mean = 0.43), respectively (n = 32). Blue highlighted data points indicate medians. (C) Representative day 52 dissociated CMs stained with αSA (red) and Caveolin 3 (green) for T-tubule development. Long-term cultured 3D-dhECT (day 124) CMs (D) developed highly aligned sarcomeres and contained large and elongated cell nuclei. (E) Additionally, these CMs expressed Cx43 on their transverse ends between adjoining cells.
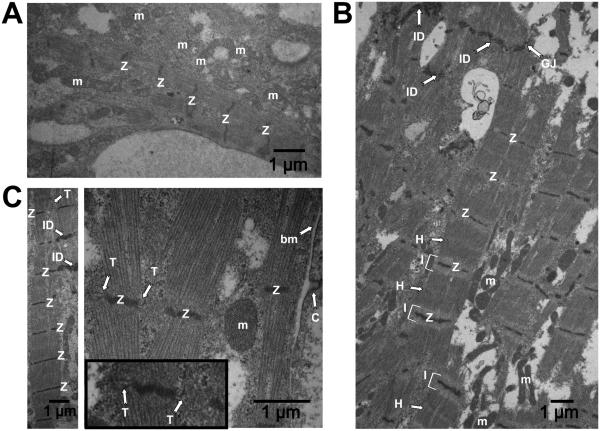
To further investigate 3D-dhECT CM maturation, cardiac ultrastructural features were visualized using TEM. Day 24 cardiac tissues showed the presence of Z-lines and mitochondria (Fig. 5A), reflecting early developmental stages of functional CMs. 3D-dhECTs maintained long-term (day 124) were composed of CMs with ultrastructural features approximating those of mature CMs. TEM images revealed numerous mitochondria, and myofibrils with well-defined sarcomeric Z-lines, I-bands, and H-zones, with a H-zone to sarcomere ratio of 0.257 and an I-band to Z-line ratio of 0.669 (Fig. S6D). Other features of 3D-dhECT CMs included intercalated discs on the transverse edges of CMs with adjacent gap junctions (Fig. 5B), caveolae in the plasma membrane, and presence of basement membrane. Most notably, 3D-dhECT CMs exhibited transverse tubules (T-tubules) which were structurally aligned with Z-lines (Fig. 5C); up until now, T-tubules have only been observed in primary CMs isolated from postnatal and adult mammalian hearts35, but have not been reported in any other in vitro human SC-CM study36-38.
Fig. 5.
Ontomimetic environment resulted in 3D-dhECT CMs exhibiting mature ultrastructural features. (A) Day 24 3D-dhECT CMs developed first characteristics of a muscle cell including mitochondria (m) and Z-lines (Z). (B) Ultrastructural images of 124 day old 3D-dhECT CMs showed sarcomere structures, including Z-lines (Z), I-bands (I), H-zones (H), intercalated discs (ID), gap junctions (GJ), and mitochondria (m). (C) Additionally, CMs contained caveolae (C) adjacent to basement membrane (bm), mitochondria with well-defined cristae, and T-tubules (T) adjacent to Z-lines, a key component of functional mature CMs. Insert: magnified view processed to highlight T-tubules.
Multiple geometries support cardiac tissue formation through direct hydrogel encapsulation
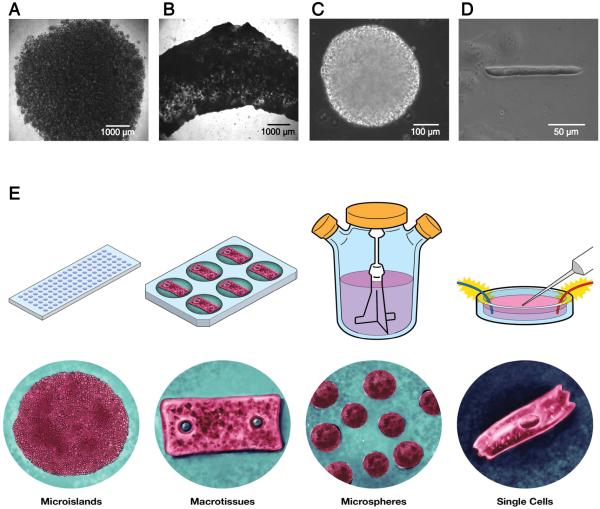
This straightforward 3D-dhECT formation process was extended to fabricate cardiac tissues across multiple size scales and geometries, demonstrating our ability to create tissues exploitable for a wide range of possible applications (Fig. 6). Furthermore, immobilization of the 3D-dhECTs through covalent attachment to the glass coverslips was found not to be necessary for cardiac tissue formation; 3D-dhECTs in suspension culture also formed contracting tissues that were consistently maintained for multiple months. In addition to the immobilized microislands (Fig. 6A) described in-depth above, tested geometries included macrotissues (Fig. 6B) and microspheres (Fig. 6C). Macrotissues (Fig. 6B) of varying lengths, thicknesses of 800-900 μm and widths greater than 2 mm were formed and cultured in suspension. Analogous to the microislands, contracting areas were first observed in the macrotissues on day 7 of differentiation, resulting in robust, uniform contraction early in the differentiation process (day 12) and appearing strong over the entire culture period (Movie S5). Similarly, suspension culture of 3D-dhECT microspheres of diameter 280 ± 70 μm (n = 25, Fig. 6C) resulted in uniformly contracting spheres that again could be maintained over extended time periods (Movie S6). Finally, all cardiac tissue geometries were successfully dissociated into individual CMs (Fig. 6D), enabling subsequent single-cell experiments or collection and re-suspension as might be required for delivery as a cell therapy.
Fig. 6.
Using this single cell-handling approach, different tissue sizes and geometries of 3D-dhECTs can be formed. Uniformly contracting tissues, in addition to (A) immobilized microislands, included: (B) macrotissues and (C) microspheres, all of which were able to be used to obtain (D) 3D-dhECT-dissociated single CMs. (E) Tissue sizes and geometries can be varied based on desired future applications, e.g., microislands for high-throughput drug-screening, macrotissues for large-scale mechanical testing and cardiac patches, injectable cardiac tissue spheroids, or single CMs post-tissue digestion for automated patch clamping.
Discussion
Differentiation of encapsulated hiPSCs to directly produce cardiac tissues in vitro opens a wide range of possibilities in the field of cardiac tissue engineering. Here we show that hiPSCs can be readily and efficiently encapsulated within PEG-fibrinogen hydrogels and directly differentiated to produce uniformly contracting 3D-dhECTs. Gene expression, CM yield, and calcium handling were similar to hiPSC differentiation using the current gold-standard, high efficiency 2D monolayer differentiation. Moreover, using 3D-dhECTs to produce cardiac tissues has distinct advantages over 2D monolayer differentiation39: it enables long-term CM culture and maturation without having to disrupt pre-differentiated SC-CMs, and it does not require the re-formation of functionally critical cell-cell junctions, which is notoriously problematic for CMs, particularly as they mature. The minimal cell-handling requirements and applicability of this cardiac tissue production technique to a wide range of tissue sizes and geometries facilitates high-throughput and cost-effective formation of human cardiac tissue for an array of future applications. PEG-fibrinogen can be rapidly photocrosslinked using a non-toxic photoinitiator and visible light, creating a controlled 3D microenvironment for encapsulated cells with potential for automation of the tissue production process using bioprinting of structurally organized microislands and other printed 3D tissue geometries40, 41 or microfluidics for microsphere production42, 43. Previous studies have optimized the initial mechanical properties of the PEG-fibrinogen hydrogels for neonatal CM encapsulation16, pre-differentiated SC-CMs encapsulation16, 44, or SC-CMs seeded onto PEG-fibrinogen hydrogels45 (Table S1). However, encapsulation and maintenance of hiPSCs within PEG-fibrinogen, and their direct differentiation in the presence of PEG-fibrinogen, had not previously been investigated. The encapsulation procedure established in this study allows for hiPSC survival with maintenance of pluripotency for at least three days within this optimized PEG-fibrinogen hydrogel formulation. Maintenance of encapsulated hiPSC pluripotency during extended culture periods has not been investigated; however, other biomaterials have previously been used for large-scale hPSC culture and expansion46-48.
Even when varying the initial hiPSC seeding density, cell proliferation and contracting cardiac tissue formation was attainable in PEG-fibrinogen hydrogels (Fig. S7). This robustness is not applicable to the production of contracting 2D monolayers, where having a tightly defined initial cell-seeding density is critical to success8. We believe the biomimetic microenvironment provided by the hydrogel plays a central role in successful hiPSC 3D culture, proliferation, and cardiac differentiation, while initial cell-seeding density may influence the uniformity of contraction within the cardiac tissue over time. Both of these factors directly influence the rate of degradation and temporal remodeling of PEG-fibrinogen hydrogels, allowing cells to secrete ECM proteins and form a continuous tissue. Having a platform that replicates key features of ontogenic cardiac tissue development and CM maturation will enable new opportunities for investigating the process of human heart tissue development.
During initial stages of SC differentiation, we have seen a steady increase in frequency of spontaneous contraction over time (day 8-50), which could be due to the temporal development of cell-cell junctions and calcium handling capabilities of maturing SC-CMs. Based on published results using the small molecule protocol employed in this study8, our 3D-dhECTs are likely composed of a distribution of ventricular, atrial, and nodal CM subtypes, which is appropriate for mimicking heart development. To enrich for ventricular myocytes, recently published small molecule differentiation protocols9, 49 could potentially be adapted for use with our 3D-dhECT system. 3D-dhECT contraction frequency decreased from its maximum of 2.2 ± 0.1 Hz on day 50 to 1.35 ± 0.05 Hz on day 90. A reduction in the rate of spontaneous contraction is anticipated when modeling the developing human heart; human heart rate in vivo slows down significantly during maturation (~3.0 Hz during fetal stage to 1.0-1.5 Hz in adults)50.
CM proliferation and hyperplasia is a phenomenon seen during fetal heart growth and early post-natal development51, after which it diminishes over time52, 53. On day 20 of cardiac differentiation, both 2D monolayers and 3D-dhECTs were composed of a similar percentage of CMs. However, the percentage of proliferating CMs in 2D was higher than that in the 3D-dhECTs. This difference in CM proliferation was also seen previously between engineered “biowires” and embryoid bodies10. Alternatively, considering the low rates of CM proliferation during in vivo development53, 54 and that cell proliferation rates generally differ in 2D versus 3D culture55, the observed lower CM proliferation might be a consequence of a more physiologic microenvironment in the 3D-dhECTs.
Having a platform that replicates key features of ontogenic CM maturation will enable new opportunities for investigating this critical process, and thereby could provide important insights about how developing human heart tissue remodels after birth. Furthermore, achieving CM maturation within an in vitro 3D environment will provide more physiologically relevant tools for toxicology screening and disease modeling using patient-specific hiPSCs56-58 to engineer mature cardiac tissues3 that current differentiation systems cannot provide. In addition, there is a high demand for cardiac tissues that can be used in patients for myocardial repair59; to avoid induction of deadly arrhythmias, clinical applications require CMs that can achieve appropriate mechanical and electrical integration with native tissues. Balancing the needs for both CM plasticity and suitable maturation to achieve this goal is an area of intense research60; we have demonstrated that 3D-dhECTs can be used to produce CMs from early time points in stem cell differentiation through ultrastructural maturation, providing the flexibility to meet future specifications for optimizing cardiac cell therapy.
T-tubule formation and other mature ultrastructural features that are important in excitation-contraction coupling61, 62 have been detected in our long-term cultured 3D-dhECT CMs; day 52-60 dissociated CMs showed Caveolin 3 positive staining, indicating the development of T-tubule formation. When incorporated into engineered heart tissue, neonatal rat ventricular myocytes previously have been shown to support formation of T-tubules44, however, T-tubule formation in SC-CMs has not been previously reported10, 38, 39, 61, 63. Initiation of differentiation and long-term culture within an in vitro 3D environment may better support CM maturation, which is important for establishment of more physiologically relevant tools for toxicology screening and disease modeling of adult human myocardium3.
Although immobilized microislands are advantageous for high-throughput applications, this tissue size and geometry also had some drawbacks. At the tested diameter of 6 mm, cell growth and proliferation resulted in non-uniform tissue thickness and the generation of CMs with a range of levels of maturity. Since these regional differences in cell growth were observed prior to the onset of contraction, space limitations at the microisland center could potentially be a contributing factor; in comparison, hiPSCs on tissue edges grew both upwards and radially outwards from the original hydrogel boundaries. In a previous study, a similar tissue morphology (scaffold-free tissue patch) showed higher levels of cardiac maturation and CM purity on tissue edges compared to tissue centers64. Additional testing will be required to determine whether observed regional differences in microisland 3D-dhECTs are also observed at smaller diameters or when using alternative fabrication techniques, such as microprinting, versus molds.
Our study reveals the first 3D ontomimetic model of human heart development, enabling hiPSC growth and cardiac differentiation in a controlled 3D microenvironment. 3D-dhECTs have been used to produce CMs from early time points in SC differentiation through ultrastructural maturation, providing the flexibility to meet future specifications for optimizing cardiac cell therapy. Translatable to a variety of tissue sizes and geometries, this robust approach will ease and improve the cardiac tissue formation process, thereby impacting the ability to produce human heart tissue for a range of applications from investigating the role of 3D microenvironmental cues in human heart development to advancing clinical treatments for heart disease.
Supplementary Material
Acknowledgments
This work was funded by the National Science Foundation (NSF) CBET-1150854 (E.A.L.), and CBET-1403495 (C.J.E.), as well as the US National Institutes of Health (NIH)/NHLBI through grant 2T32HL007824-16A1 (I.C.T.) and NIH/NHLBI Program of Excellence in Nanotechnology (PEN) Award, Contract HHSN268201000045C (K.D.C.), and grant R01DK093810 (C.J.E.). P.K. held an American Heart Association pre-doctoral fellowship AHA -13PRE1470078 and A.J.H. an AL-EPSCoR GRSP fellowship. We would like to thank Dr. Ronald Gordon, Department of Pathology at Mount Sinai, for performing transmission electron microscopy; Dr. Juming Zhong, Auburn University, for technical support in calcium transient imaging; Dr. Elizabeth Schwartz and Allison Church Bird, Auburn University, for technical expertise and support in flow cytometry; Dr. Vishnu Suppiramaniam for providing assistance with multielectrode array (MEA) experiments. Scientific illustrations were graciously created by Brennen Reece.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lasser KE, Allen PD, Woolhandler SJ, Himmelstein DU, Wolfe SM, Bor DH. Timing of new black box warnings and withdrawals for prescription medications. Jama. 2002;287:2215–20. doi: 10.1001/jama.287.17.2215. [DOI] [PubMed] [Google Scholar]
- 2.Frank C, Himmelstein DU, Woolhandler S, Bor DH, Wolfe SM, Heymann O, Zallman L, Lasser KE. Era of faster FDA drug approval has also seen increased black-box warnings and market withdrawals. Health affairs. 2014;33:1453–9. doi: 10.1377/hlthaff.2014.0122. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Pabon L, Murry CE. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res. 2014;114:511–23. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertassoni LE, Cardoso JC, Manoharan V, Cristino AL, Bhise NS, Araujo WA, Zorlutuna P, Vrana NE, Ghaemmaghami AM, Dokmeci MR, Khademhosseini A. Direct-write bioprinting of cell-laden methacrylated gelatin hydrogels. Biofabrication. 2014;6:024105. doi: 10.1088/1758-5082/6/2/024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zwi L, Caspi O, Arbel G, Huber I, Gepstein A, Park IH, Gepstein L. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation. 2009;120:1513–23. doi: 10.1161/CIRCULATIONAHA.109.868885. [DOI] [PubMed] [Google Scholar]
- 6.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell stem cell. 2011;8:228–40. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Lev S, Kehat I, Gepstein L. Differentiation pathways in human embryonic stem cell-derived cardiomyocytes. Annals of the New York Academy of Sciences. 2005;1047:50–65. doi: 10.1196/annals.1341.005. [DOI] [PubMed] [Google Scholar]
- 8.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–75. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–60. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Masse S, Gagliardi M, Hsieh A, Thavandiran N, Laflamme MA, Nanthakumar K, Gross GJ, Backx PH, Keller G, Radisic M. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods. 2013;10:781–7. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbull IC, Karakikes I, Serrao GW, Backeris P, Lee JJ, Xie C, Senyei G, Gordon RE, Li RA, Akar FG, Hajjar RJ, Hulot JS, Costa KD. Advancing functional engineered cardiac tissues toward a preclinical model of human myocardium. FASEB J. 2014;28:644–54. doi: 10.1096/fj.13-228007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang D, Shadrin IY, Lam J, Xian HQ, Snodgrass HR, Bursac N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials. 2013;34:5813–20. doi: 10.1016/j.biomaterials.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaaf S, Shibamiya A, Mewe M, Eder A, Stohr A, Hirt MN, Rau T, Zimmermann WH, Conradi L, Eschenhagen T, Hansen A. Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PLoS One. 2011;6:e26397. doi: 10.1371/journal.pone.0026397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circulation research. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mihic A, Li J, Miyagi Y, Gagliardi M, Li SH, Zu J, Weisel RD, Keller G, Li RK. The effect of cyclic stretch on maturation and 3D tissue formation of human embryonic stem cell-derived cardiomyocytes. Biomaterials. 2014;35:2798–808. doi: 10.1016/j.biomaterials.2013.12.052. [DOI] [PubMed] [Google Scholar]
- 16.Shapira-Schweitzer K, Seliktar D. Matrix stiffness affects spontaneous contraction of cardiomyocytes cultured within a PEGylated fibrinogen biomaterial. Acta biomaterialia. 2007;3:33–41. doi: 10.1016/j.actbio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Mironi-Harpaz I, Wang DY, Venkatraman S, Seliktar D. Photopolymerization of cell-encapsulating hydrogels: crosslinking efficiency versus cytotoxicity. Acta Biomater. 2012;8:1838–48. doi: 10.1016/j.actbio.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 18.Dikovsky D, Bianco-Peled H, Seliktar D. The effect of structural alterations of PEG-fibrinogen hydrogel scaffolds on 3-D cellular morphology and cellular migration. Biomaterials. 2006;27:1496–506. doi: 10.1016/j.biomaterials.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 19.Franco CL, Price J, West JL. Development and optimization of a dual-photoinitiator, emulsion-based technique for rapid generation of cell-laden hydrogel microspheres. Acta Biomater. 2011;7:3267–76. doi: 10.1016/j.actbio.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 20.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109:E1848–57. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nature protocols. 2012;7:2029–40. doi: 10.1038/nprot.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nie Y, Walsh P, Clarke DL, Rowley JA, Fellner T. Scalable passaging of adherent human pluripotent stem cells. PloS one. 2014;9:e88012. doi: 10.1371/journal.pone.0088012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowes BD, Minobe W, Abraham WT, Rizeq MN, Bohlmeyer TJ, Quaife RA, Roden RL, Dutcher DL, Robertson AD, Voelkel NF, Badesch DB, Groves BM, Gilbert EM, Bristow MR. Changes in gene expression in the intact human heart. Downregulation of alpha-myosin heavy chain in hypertrophied, failing ventricular myocardium. The Journal of clinical investigation. 1997;100:2315–24. doi: 10.1172/JCI119770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itzhaki I, Rapoport S, Huber I, Mizrahi I, Zwi-Dantsis L, Arbel G, Schiller J, Gepstein L. Calcium handling in human induced pluripotent stem cell derived cardiomyocytes. PloS one. 2011;6:e18037. doi: 10.1371/journal.pone.0018037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lou Q, Fedorov VV, Glukhov AV, Moazami N, Fast VG, Efimov IR. Transmural heterogeneity and remodeling of ventricular excitation-contraction coupling in human heart failure. Circulation. 2011;123:1881–90. doi: 10.1161/CIRCULATIONAHA.110.989707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Planat-Benard V, Menard C, Andre M, Puceat M, Perez A, Garcia-Verdugo JM, Penicaud L, Casteilla L. Spontaneous cardiomyocyte differentiation from adipose tissue stroma cells. Circ Res. 2004;94:223–9. doi: 10.1161/01.RES.0000109792.43271.47. [DOI] [PubMed] [Google Scholar]
- 27.Matsuura K, Nagai T, Nishigaki N, Oyama T, Nishi J, Wada H, Sano M, Toko H, Akazawa H, Sato T, Nakaya H, Kasanuki H, Komuro I. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. J Biol Chem. 2004;279:11384–91. doi: 10.1074/jbc.M310822200. [DOI] [PubMed] [Google Scholar]
- 28.Zimmermann WH, Schneiderbanger K, Schubert P, Didie M, Munzel F, Heubach JF, Kostin S, Neuhuber WL, Eschenhagen T. Tissue engineering of a differentiated cardiac muscle construct. Circulation research. 2002;90:223–30. doi: 10.1161/hh0202.103644. [DOI] [PubMed] [Google Scholar]
- 29.Fink C, Ergun S, Kralisch D, Remmers U, Weil J, Eschenhagen T. Chronic stretch of engineered heart tissue induces hypertrophy and functional improvement. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2000;14:669–79. doi: 10.1096/fasebj.14.5.669. [DOI] [PubMed] [Google Scholar]
- 30.Tiburcy M, Didie M, Boy O, Christalla P, Doker S, Naito H, Karikkineth BC, El-Armouche A, Grimm M, Nose M, Eschenhagen T, Zieseniss A, Katschinksi DM, Hamdani N, Linke WA, Yin X, Mayr M, Zimmermann WH. Terminal differentiation, advanced organotypic maturation, and modeling of hypertrophic growth in engineered heart tissue. Circulation research. 2011;109:1105–14. doi: 10.1161/CIRCRESAHA.111.251843. [DOI] [PubMed] [Google Scholar]
- 31.Radisic M, Park H, Shing H, Consi T, Schoen FJ, Langer R, Freed LE, Vunjak-Novakovic G. Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:18129–34. doi: 10.1073/pnas.0407817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirt MN, Boeddinghaus J, Mitchell A, Schaaf S, Bornchen C, Muller C, Schulz H, Hubner N, Stenzig J, Stoehr A, Neuber C, Eder A, Luther PK, Hansen A, Eschenhagen T. Functional improvement and maturation of rat and human engineered heart tissue by chronic electrical stimulation. J Mol Cell Cardiol. 2014;74:151–61. doi: 10.1016/j.yjmcc.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Bird SD, Doevendans PA, van Rooijen MA, Brutel de la Riviere A, Hassink RJ, Passier R, Mummery CL. The human adult cardiomyocyte phenotype. Cardiovascular research. 2003;58:423–34. doi: 10.1016/s0008-6363(03)00253-0. [DOI] [PubMed] [Google Scholar]
- 34.Gerdes AM, Kellerman SE, Moore JA, Muffly KE, Clark LC, Reaves PY, Malec KB, McKeown PP, Schocken DD. Structural remodeling of cardiac myocytes in patients with ischemic cardiomyopathy. Circulation. 1992;86:426–30. doi: 10.1161/01.cir.86.2.426. [DOI] [PubMed] [Google Scholar]
- 35.Ziman AP, Ward CW, Rodney GG, Lederer WJ, Bloch RJ. Quantitative measurement of Ca(2)(+) in the sarcoplasmic reticulum lumen of mammalian skeletal muscle. Biophysical journal. 2010;99:2705–14. doi: 10.1016/j.bpj.2010.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knollmann BC. Induced pluripotent stem cell-derived cardiomyocytes: boutique science or valuable arrhythmia model? Circ Res. 2013;112:969–76. doi: 10.1161/CIRCRESAHA.112.300567. discussion 976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson C, Tran DD, George SC. Concise review: maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem Cells. 2013;31:829–37. doi: 10.1002/stem.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Priori SG, Napolitano C, Di Pasquale E, Condorelli G. Induced pluripotent stem cell-derived cardiomyocytes in studies of inherited arrhythmias. J Clin Invest. 2013;123:84–91. doi: 10.1172/JCI62838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lundy SD, Zhu WZ, Regnier M, Laflamme MA. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem cells and development. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol. 2014;32:773–85. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 41.Stanton MM, Samitier J, Sanchez S. Bioprinting of 3D hydrogels. Lab Chip. 2015;15:3111–5. doi: 10.1039/c5lc90069g. [DOI] [PubMed] [Google Scholar]
- 42.Workman VL, Dunnett SB, Kille P, Palmer DD. Microfluidic chip-based synthesis of alginate microspheres for encapsulation of immortalized human cells. Biomicrofluidics. 2007;1:14105. doi: 10.1063/1.2431860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung BG, Lee KH, Khademhosseini A, Lee SH. Microfluidic fabrication of microengineered hydrogels and their application in tissue engineering. Lab Chip. 2012;12:45–59. doi: 10.1039/c1lc20859d. [DOI] [PubMed] [Google Scholar]
- 44.Shapira-Schweitzer K, Habib M, Gepstein L, Seliktar D. A photopolymerizable hydrogel for 3-D culture of human embryonic stem cell-derived cardiomyocytes and rat neonatal cardiac cells. Journal of molecular and cellular cardiology. 2009;46:213–24. doi: 10.1016/j.yjmcc.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 45.Bearzi C, Gargioli C, Baci D, Fortunato O, Shapira-Schweitzer K, Kossover O, Latronico MV, Seliktar D, Condorelli G, Rizzi R. PlGF-MMP9-engineered iPS cells supported on a PEG-fibrinogen hydrogel scaffold possess an enhanced capacity to repair damaged myocardium. Cell Death Dis. 2014;5:e1053. doi: 10.1038/cddis.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei Y, Schaffer DV. A fully defined and scalable 3D culture system for human pluripotent stem cell expansion and differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E5039–48. doi: 10.1073/pnas.1309408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amit M, Laevsky I, Miropolsky Y, Shariki K, Peri M, Itskovitz-Eldor J. Dynamic suspension culture for scalable expansion of undifferentiated human pluripotent stem cells. Nature protocols. 2011;6:572–9. doi: 10.1038/nprot.2011.325. [DOI] [PubMed] [Google Scholar]
- 48.Zweigerdt R, Olmer R, Singh H, Haverich A, Martin U. Scalable expansion of human pluripotent stem cells in suspension culture. Nature protocols. 2011;6:689–700. doi: 10.1038/nprot.2011.318. [DOI] [PubMed] [Google Scholar]
- 49.Weng Z, Kong CW, Ren L, Karakikes I, Geng L, He J, Chow MZ, Mok CF, Keung W, Chow H, Leung AY, Hajjar RJ, Li RA, Chan CW. A simple, cost-effective but highly efficient system for deriving ventricular cardiomyocytes from human pluripotent stem cells. Stem Cells Dev. 2014;23:1704–16. doi: 10.1089/scd.2013.0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arduini D. Fetal Cardiac Function. 1995:43–49. [Google Scholar]
- 51.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. Journal of molecular and cellular cardiology. 1996;28:1737–46. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 52.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–35. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mollova M, Bersell K, Walsh S, Savla J, Das LT, Park SY, Silberstein LE, Dos Remedios CG, Graham D, Colan S, Kuhn B. Cardiomyocyte proliferation contributes to heart growth in young humans. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1446–51. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bensley JG, Stacy VK, De Matteo R, Harding R, Black MJ. Cardiac remodelling as a result of pre-term birth: implications for future cardiovascular disease. European heart journal. 2010;31:2058–66. doi: 10.1093/eurheartj/ehq104. [DOI] [PubMed] [Google Scholar]
- 55.Mann BK, West JL. Cell adhesion peptides alter smooth muscle cell adhesion, proliferation, migration, and matrix protein synthesis on modified surfaces and in polymer scaffolds. Journal of biomedical materials research. 2002;60:86–93. doi: 10.1002/jbm.10042. [DOI] [PubMed] [Google Scholar]
- 56.Liang P, Lan F, Lee AS, Gong T, Sanchez-Freire V, Wang Y, Diecke S, Sallam K, Knowles JW, Wang PJ, Nguyen PK, Bers DM, Robbins RC, Wu JC. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation. 2013;127:1677–91. doi: 10.1161/CIRCULATIONAHA.113.001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doyle MJ, Lohr JL, Chapman CS, Koyano-Nakagawa N, Garry MG, Garry DJ. Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes as a Model for Heart Development and Congenital Heart Disease. Stem Cell Rev. 2015;11:710–27. doi: 10.1007/s12015-015-9596-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–4. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA, Fugate JA, Muskheli V, Gough GM, Vogel KW, Astley CA, Hotchkiss CE, Baldessari A, Pabon L, Reinecke H, Gill EA, Nelson V, Kiem HP, Laflamme MA, Murry CE. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–7. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunn DA, Hodge AJ, Lipke EA. Biomimetic materials design for cardiac tissue regeneration. Wiley interdisciplinary reviews Nanomedicine and nanobiotechnology. 2014;6:15–39. doi: 10.1002/wnan.1241. [DOI] [PubMed] [Google Scholar]
- 61.Lieu DK, Liu J, Siu CW, McNerney GP, Tse HF, Abu-Khalil A, Huser T, Li RA. Absence of transverse tubules contributes to non-uniform Ca(2+) wavefronts in mouse and human embryonic stem cell-derived cardiomyocytes. Stem Cells Dev. 2009;18:1493–500. doi: 10.1089/scd.2009.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei S, Guo A, Chen B, Kutschke W, Xie YP, Zimmerman K, Weiss RM, Anderson ME, Cheng H, Song LS. T-tubule remodeling during transition from hypertrophy to heart failure. Circulation research. 2010;107:520–31. doi: 10.1161/CIRCRESAHA.109.212324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baharvand H, Azarnia M, Parivar K, Ashtiani SK. The effect of extracellular matrix on embryonic stem cell-derived cardiomyocytes. Journal of molecular and cellular cardiology. 2005;38:495–503. doi: 10.1016/j.yjmcc.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 64.Stevens KR, Pabon L, Muskheli V, Murry CE. Scaffold-free human cardiac tissue patch created from embryonic stem cells. Tissue engineering Part A. 2009;15:1211–22. doi: 10.1089/ten.tea.2008.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.