Summary
Information processing relies on precise patterns of synapses between neurons. The cellular recognition mechanisms regulating this specificity are poorly understood. In the medulla of the Drosophila visual system, different neurons form synaptic connections in different layers. Here, we sought to identify candidate cell recognition molecules underlying this specificity. Using RNA sequencing (RNA-seq), we show that neurons with different synaptic specificities express unique combinations of mRNAs encoding hundreds of cell surface and secreted proteins. Using RNA-seq and protein tagging, we demonstrate that 21 paralogs of the Dpr family, a subclass of immunoglobulin (Ig)-domain containing proteins, are expressed in unique combinations in homologous neurons with different layer-specific synaptic connections. Dpr interacting proteins (DIPs), comprising nine paralogs of another subclass of Ig-containing proteins, are expressed in a complementary layer-specific fashion in a subset of synaptic partners. We propose that pairs of Dpr/DIP paralogs contribute to layer-specific patterns of synaptic connectivity.
Keywords: DIP, Dpr, Drosophila, synaptic specificity, visual system
Graphical abstract
Introduction
Neural circuits typically comprise many different neurons linked in precise ways by synaptic connections. How neurites discriminate between one another during circuit assembly remains a central issue in neuroscience. Through regeneration studies in vertebrates, Langley (1895) and Sperry (1963) proposed that molecular differences between neurons determine the specificity of synaptic connections. In its simplest formulation, Sperry’s chemoaffinity hypothesis (Sperry, 1963) suggested that a lock and key mechanism mediates recognition between synaptic partners.
Over the past 30 years, biochemical and genetic approaches have led to the identification of the cell recognition molecules and the intercellular signaling pathways regulating the patterning of axons and dendrites. From these studies, three general molecular strategies underlying circuit assembly have emerged. First, combinatorial use of a limited set of conserved cell surface and secreted molecules regulates axon guidance in many different regions of the developing invertebrate and vertebrate nervous systems. These include netrins, slits, semaphorins, and various cell adhesion molecules, such as cadherins and immunoglobulin superfamily proteins (O’Donnell et al., 2009). Second, gradients of cell surface proteins, notably Ephs and Ephrins as well as Wnts, play a crucial role in the establishment of topographic maps (Cang and Feldheim, 2013, Schmitt et al., 2006 and Triplett and Feldheim, 2012) a widespread organizational principle in the vertebrate brain (Cang and Feldheim, 2013). Third, molecular diversity contributed by large families of related proteins with different recognition specificities impart unique identities to neurons and mediate self-avoidance (repulsion between neurites of the same cell) (Zipursky and Grueber, 2013 and Lefebvre et al., 2012). These include Dscam1 proteins in Drosophila (Schmucker et al., 2000) and clustered protocadherins in vertebrates (Kohmura et al., 1998 and Wu and Maniatis, 1999). The molecular diversity of both Dscam1 and protocadherins coupled with their exquisite isoform-specific homophilic binding specificities raised the possibility that they could directly specify patterns of synaptic specificity through a lock and key mechanism. As Dscam1 is largely, if not exclusively, expressed in a probabilistic manner (Miura et al., 2013), and protocadherins also appear to be expressed in this way, it is unlikely that these protein families mediate synaptic matching.
Important progress has been made in identifying cell surface molecules regulating synaptic specificity, including Syg1 and Syg2 in the worm (Shen and Bargmann, 2003 and Shen et al., 2004), Toll and Teneurin proteins in the fly olfactory system (Hong et al., 2012 and Ward et al., 2015) and Sidekick proteins in the mouse retina (Krishnaswamy et al., 2015). Studies by Yamagata and Sanes (Yamagata et al., 2002, Yamagata and Sanes, 2008 and Yamagata and Sanes, 2012) raised the possibility that related Ig superfamily proteins regulate layer-specific patterns of synaptic connections between different neurons in the chick retina (see Discussion). As a step toward identifying a common molecular logic underlying synaptic specificity, we sought to identify families of cell surface proteins expressed in a cell-type-enriched fashion in closely related neurons with different patterns of synaptic specificity. Here, we set out to do this using RNA sequencing (RNA-seq) and molecular genetic approaches in Drosophila.
The Drosophila visual system is well suited to uncovering the molecular recognition mechanisms regulating synaptic specificity. The cellular organization and circuitry has been described in detail (Fischbach and Dittrich, 1989 and Morante and Desplan, 2008) including serial electron microscopy (EM) reconstruction to reveal connections between neurons (Takemura et al., 2008, Takemura et al., 2013 and Takemura et al., 2015). In addition, molecular markers for many cell types are readily available (Jenett et al., 2012 and Kvon et al., 2014), genetic tools facilitate gain and loss of function studies at the level of single identified cells in developing and adult tissue (Lee and Luo, 1999 and Venken and Bellen, 2014), and an extensive protein interaction network of extracellular proteins has been assembled (Özkan et al., 2013).
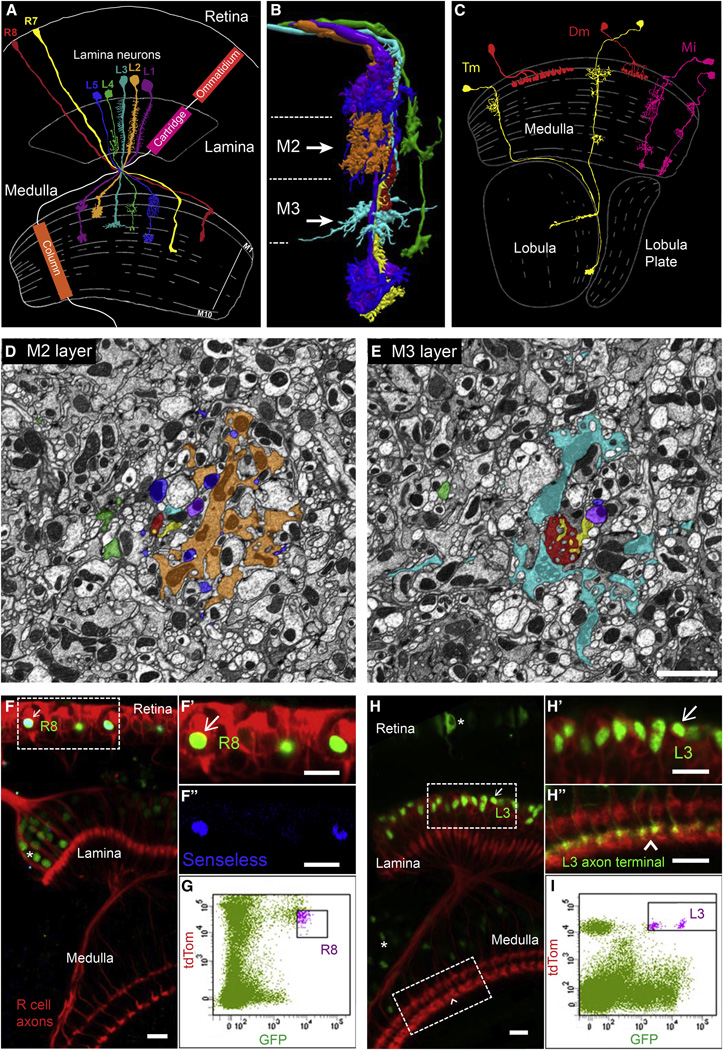
In this paper, we focus on the medulla region of the fly visual system. It comprises columns and layers (Figures 1A–1C). In a broad sense, columns process information from different points in space and layers process different types of visual information (e.g., ON versus OFF responses). The cell bodies of medulla neurons lie outside the neuropil and synaptic specificity is elaborated within a dense meshwork of axonal and dendritic processes. There are over 100 different types of neurons forming synapses in the medulla. These neurons fall into a few general categories based primarily on their morphology and location of their arbors (Fischbach and Dittrich, 1989, Morante and Desplan, 2008 and Takemura et al., 2013) (Figures 1A–1C). In a landmark study, the synaptic connectivity between neurons in the medulla was determined using serial section electron microscopic reconstruction (Takemura et al., 2013). The shaded electron micrographic sections through the adult column shown in Figures 1D and 1E are included to emphasize the complexity of the neuropil in one medulla column comprising the processes of on the order of 100 different neuronal cell types (A. Nern, personal communication) (Figures 1D and 1E). These patterns of synaptic connections are complex, specific, and reproducible (Takemura et al., 2015). In addition, these studies revealed that within a layer, neurons form synapses with multiple neuronal types (Takemura et al., 2013 and Takemura et al., 2015), but these represent only a subset of neurons with processes in the layer. Although some progress has been made in identifying genes regulating layer-specific targeting (Hadjieconomou et al., 2011), genes controlling synaptic specificity within layers have not been identified.
Figure 1. FACS Isolation of Developing Neurons with Different Synaptic Specificities.
(A) Schematic of the adult morphologies of R7 and R8 photoreceptors and lamina neurons L1–L5. The visual system comprises topographically matched modules (i.e., ommatidia, cartridges and columns). Adapted from Fischbach and Dittrich (1989).
(B) Axons of R7, R8, and L1–L5 are shown together within a single column as determined from serial EM reconstruction. The color of different neurons is the same as in (A). Dotted lines represent layer boundaries. Arrowheads indicate the plane of section shown in electron micrographs in (D) and (E). Courtesy of S. Takemura, I. Meinertzhagen, and L. Scheffer (JRC/HHMI).
(C) Examples of three general classes of medulla neurons that are synaptic targets for R7, R8, and L1–L5 neurons (two examples are shown for each class). Within each class, there are many cell types that display similar morphologies and branch in different layers. Adapted from Fischbach and Dittrich (1989).
(D and E) Cross-sections through medulla columns reconstructed by serial EM within the M2 and M3 layers (see arrows in B). Axons are colored as in (A) and (B). Each column contains processes from over 100 different neuronal cell types (A. Nern, personal communication). Scale bar, 2 µm. Courtesy of S. Takemura, I. Meinertzhagen, and L. Scheffer (JRC/HHMI).
(F and G) Isolation of R8 neurons at 40 hr APF using fluorescence-activated cell sorting (FACS). Only R8 neurons express both retinal-specific TdTom and R8-specific GFP. Senseless is an R8-specific transcription factor. Scale bars, 10 µm. In (F), arrows indicate double-labeled cells in developing tissue and the asterisks indicate single GFP-labeled cells of different cell types (i.e., contaminants).
(H and I) Isolation of L3 neurons at 40 hr APF using FACS. Only L3 neurons express lamina-specific TdTom and L3-specific GFP. Scale bars, 10 µm. In (H), arrows indicate double-labeled cells in developing tissue and the asterisks indicate single GFP-labeled cells of different cell types (i.e., contaminants). See Experimental Procedures for purification protocols for other cells and additional details.
In the work described here, we set out to identify proteins regulating synaptic specificity using RNA-seq to determine the transcriptome of developing R7 and R8 photoreceptor neurons and five lamina monopolar neurons, L1–L5 (Figures 1A and 1B). The axon of each type of neuron elaborates a unique morphology, including layer-specific branches, and forms characteristic patterns of synaptic connections (Fischbach and Dittrich, 1989, Takemura et al., 2008 and Takemura et al., 2013). In most cases, synapses formed by each neuron type occur in the layer in which the axon terminates or in which interstitial branches arborize. We show that each cell type expresses mRNAs from hundreds of genes encoding cell surface proteins and unique combinations of them. Using a protein interaction database (Özkan et al., 2013), Minos-mediated integration cassette (MiMIC)-based protein traps to visualize protein expression (Nagarkar-Jaiswal et al., 2015 and Venken et al., 2011) and genetic markers for identified medulla cell types, we present evidence that two families of heterophilic recognition molecules of the Ig superfamily, the Dprs and DIPs, are expressed in discrete subsets of synaptic partners within layers. These families are promising candidates for regulating synaptic specificity within the developing Drosophila CNS.
Results
Purification of Developing Neurons Using Fluorescence-Activated Cell Sorting
As a first step toward identifying cell surface and secreted molecules as candidates involved in cellular recognition events regulating synaptic specificity through RNA-seq, we developed methods to purify seven neuronal cell types (R7, R8, and L1–L5 neurons) with different layer and synaptic specificities. These neurons were isolated at 40% after puparium formation (APF), just prior to (i.e., R7 and R8) or during early stages of synapse formation (i.e., L1–L5) (Chen et al., 2014; M.Y.P. and S.L.Z., unpublished data). To purify each cell type at this stage in development, we used a dual labeling approach. We generated transgenes expressing tandem tomato in all cells in the retina or in all cells in the lamina and combined these with a separate GFP marker expressed selectively in a specific cell type (e.g., R8 in the retina or L3 in the lamina) (Figures 1F–1I). Each cell type was isolated in a highly purified form as assessed using qPCR for several diagnostic markers (data not shown) and post hoc analysis of RNA-seq data (Figure 2C).
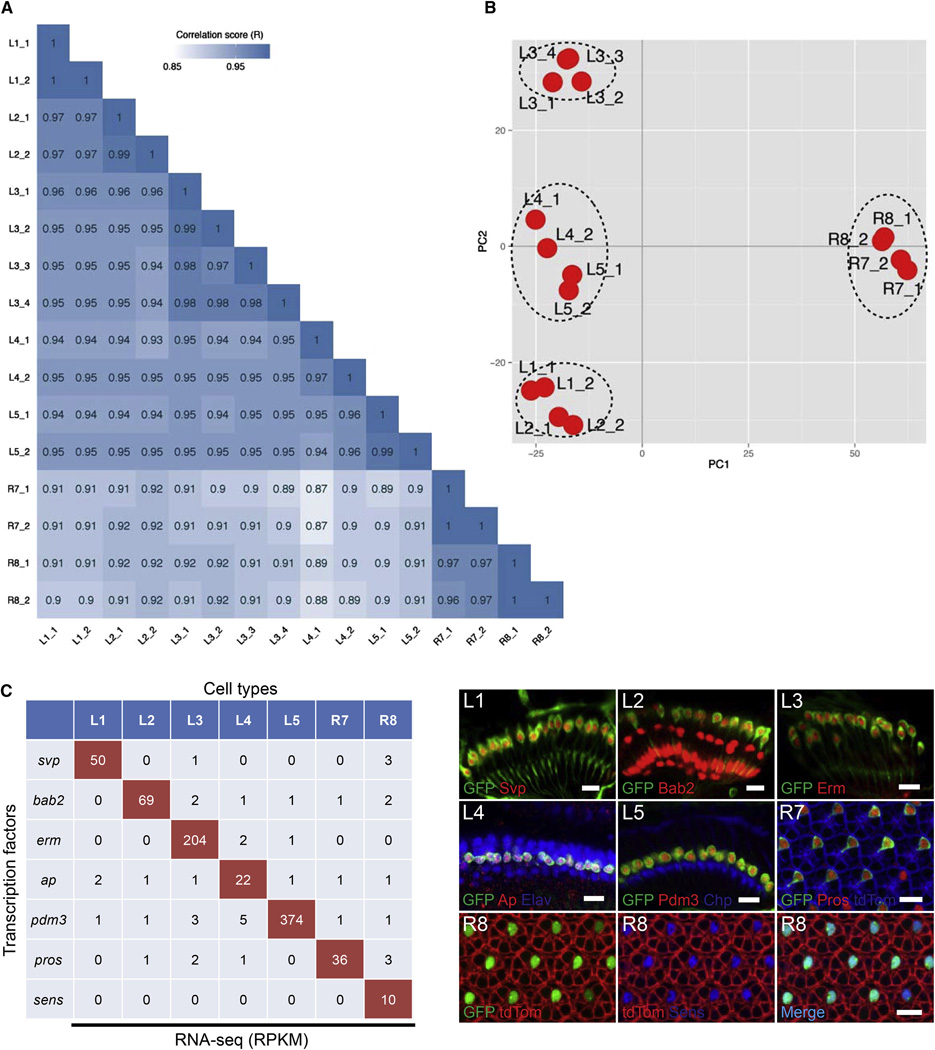
Figure 2. RNA-Seq of Visual System Neurons.
(A) Correlograms showing the correlation score matrix across all libraries of all seven cell types (R, Pearson correlation coefficient).
(B) Principal component analysis plot of the RNA-seq data for the indicated cell types. Each red dot represents an RNA-seq sample.
(C) RPKM values (left) and antibody staining (right) for transcription factors in the lamina (L1–L5) and retina (R7, R8) at 40 hr APF. Cell-type-specific markers are shown in green and antibodies for cell-type-specific transcription factors are shown in red (L1–L5 and R7) or blue (R8). Arrows in L2 panels indicate glial cells also stained with antibody to Bab2. A general retinal marker is shown in blue and red in the R7 and R8 panels, respectively. Scale bars, 10 µm.
See also Table S1.
Identification of Cell-Type-Specific Differences in Gene Expression
To obtain the transcriptomes of purified cell populations, we isolated total RNA, linearly amplified polyA-mRNA using T7 polymerase, and generated cDNA libraries that were analyzed in a single lane by 50 bp paired-end sequencing on an Illumina HiSeq 2000 platform. At least two independent biological replicate libraries were sequenced for each cell type. We obtained between 221 and 441 million reads from each library, with a percentage of uniquely mapped reads ranging between 33% and 74%. Of these, 19% to 32% were intergenic and 60% to 75% mapped to exons. A small fraction of reads mapped to intronic regions (Table S1).
The correlation in the distribution of normalized raw reads between biological replicates for each cell type was high (Figure 2A) and ranged from 0.97 to 1 for the L4 and R7 libraries, respectively. The correlation coefficients between libraries of different cell types ranged from 0.87 for R7 versus L4, to 0.97 for R7 versus R8. Pairs of neurons from either the retina or the lamina were more closely related to each other than any given retinal to lamina neuron pair. The L1 and L2 pair, both required for the optomotor response (Borst, 2014), are more closely related than other pairs of lamina neurons. These data are consistent with principle component analysis in which R7/R8 are distinct from L1–L5 and that the L1 and L2 pair, as well as L4 and L5 pair, are more closely related to each other than to L3 neurons (Figure 2B).
To assess whether differences revealed through RNA-seq reliably reflect differences in expression between cell types, we compared the reads per kilobase of a specific mRNA per million reads (RPKM) values for seven transcripts expressed specifically in each of the seven neuronal cell types as determined by immunohistology. There was an excellent correlation between antibody staining and RPKM values (Figure 2C). Thus, RNA-seq provides a reliable method to identify transcripts differentially expressed between these neurons.
Many Genes Are Differentially Expressed between Closely Related Neurons
We set out to gain a global perspective on gene expression differences between R7, R8, and L1–L5 using two different approaches. First, we performed pairwise comparisons of their transcriptomes and identified differentially expressed genes between different cell types. As we obtained independent verification of cell-type-enriched expression for transcripts with a maximum RPKM slightly below five using protein traps (Nagarkar-Jaiswal et al., 2015 and Venken et al., 2011) (see later in the text), we set conservative criteria for genes differentially expressed (DE) between cell-types.
We selected genes exhibiting a difference of greater than five times between one neuronal cell type and other neurons with expression in at least one cell type exhibiting an RPKM >5 with an adjusted p value of <0.05 (Table S2A). Even with these criteria, the number of DE genes between two cell types was substantial, ranging from 217 to 1,159. In summary, marked differences in gene expression between different neurons were observed at this stage in development.
In a second approach, we explored the relationship between patterns of gene expression and specific cell types using a weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008). This unsupervised and unbiased analysis identified distinct co-expression modules by clustering transcripts with similar expression patterns across all samples (see Supplemental Experimental Procedures). Cell-type-specific modules were preferentially enriched in cell surface membrane and secreted molecules (CSMs) (Figure S1; Table S3). This is consistent with an important role for intercellular interactions as important determinants of patterns of synaptic connectivity.
Each Neuronal Cell Type Expresses a Unique Combination of mRNAs Encoding CSMs during Synapse Formation
We next sought to identify genes encoding CSMs that are expressed in a cell-type-enriched fashion, as these are candidates for regulating the cellular interactions underlying synaptic specificity. With a threshold of an RPKM greater than five and an adjusted p value <0.05, we observed that each cell type expresses between a quarter to a third (i.e., between 247 [for R7] and 322 [for L3]) of the 976 genes encoding CSMs predicted to be encoded in the fly genome (Figure S2) (Kurusu et al., 2008) (see Supplemental Experimental Procedures for the criteria used to establish the list of CSMs) and each cell type exhibits a unique pattern of expression (Figure 3A). To gain an appreciation of the differences in genes encoding CSMs expressed between cell types, we carried out a pairwise comparison of RPKM values for each pair of cell types. Here, we observed marked differences in expression, as each pair differentially expressed between 49 (between R7 and R8) and 168 (between R7 and L4) CSM genes (RPKM greater than five in at least one cell type and greater than five times difference between the two cell types) (Table S2B). Further analyses revealed that only a small fraction of the CSM transcripts are selectively enriched in only one cell type of the seven profiled. Thus, each cell type expresses many genes encoding CSMs, the majority of which are expressed in multiple cell types, and there are marked differences in expression between cell types.
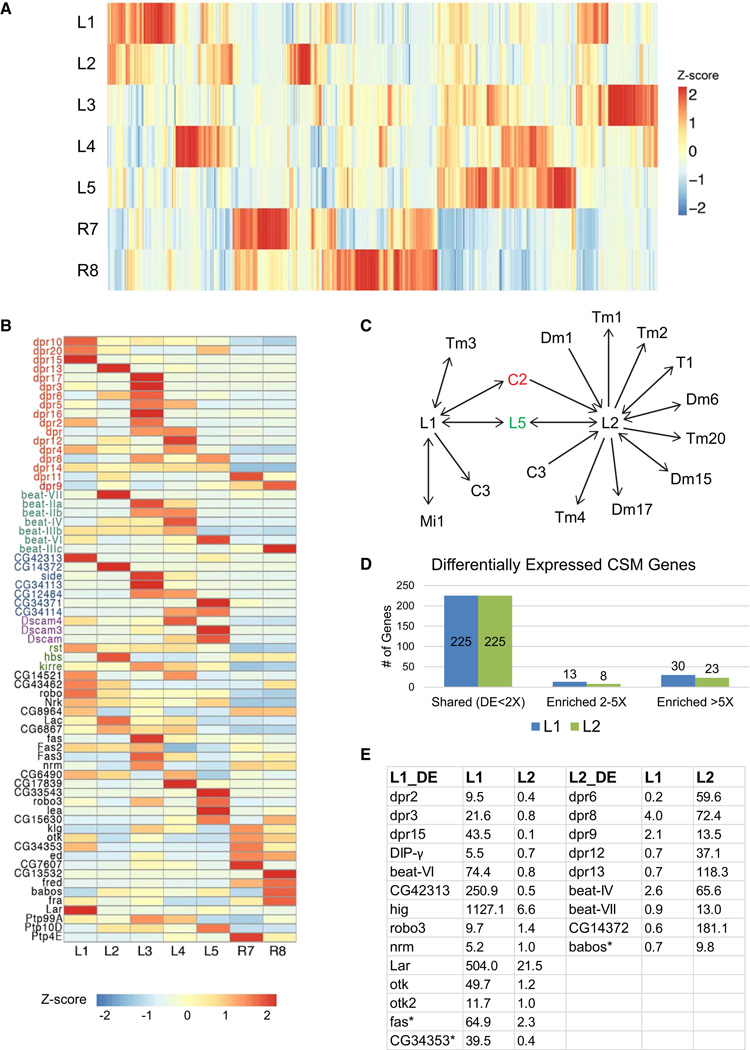
Figure 3. Gene Expression Patterns of CSMs in Each Cell Type.
(A) Heat map showing expression of all genes encoding CSMs expressed in at least one cell with an RPKM > 5 (n = 444). See also Figure S2.
(B) Heat map representing expression of genes encoding immunoglobulin (Ig) superfamily of cell surface proteins. Each gene in this list is expressed in at least one cell type with an RPKM greater than five and five times greater in one cell type than at least one of the six other cell types. Genes shown in color are members of gene sub-families. Note all Side family members (with the exception of Side) are shown as CG numbers. See also Figure S3 and Table S4.
(C) Synaptic connections of L1 and L2 in medulla neuropil. They are largely different. Arrows indicate directionality of connection from pre-synaptic neuron to post-synaptic neuron. For example, L1 is pre-synaptic to C3. And L1 is both pre- and post-synaptic to L5. C2 in red means that C2 is a shared synaptic partner with both L1 and L2. L5 in green means that L5 is also a shared synaptic partner for both L1 and L2.
(D) Numbers of genes exhibiting differences of less than two times (shared), two to five times, and greater than five times in expression between L1 and L2 with RPKM greater than five in at least one cell type, with an adjusted p value < 0.05. Enriched means the level of a gene in one cell type is higher than the other cell type. Numbers of genes in each category are shown. See also Table S2.
(E) Lists of genes encoding Ig superfamily cell surface proteins that are enriched in L1 and L2 by greater than five times. RPKM values in L1 and L2 are also listed. CG42313 and CG14372 are Side protein family members. Asterisk indicates that the interacting partner of the protein is not known yet. Interacting partners for all other proteins in this table have been identified (Johnson et al., 2006, Linnemannstöns et al., 2014, Özkan et al., 2013 and Winberg et al., 2001).
Several families of genes encoding CSMs known to regulate cellular interactions during circuit assembly were expressed in a cell-type-enriched fashion. These included genes encoding immunoglobulin (Ig) (Fischbach et al., 2009 and Zipursky et al., 2006), leucine-rich repeat (LRR) (de Wit et al., 2011), and epidermal growth factor (EGF) domain-containing proteins (Kenzelmann et al., 2007 and Serafini et al., 1994), as well as many members of the large tetraspanin protein family (Fradkin et al., 2002 and Kopczynski et al., 1996) (Figures 3B and S3).
Differential Expression of Ig Superfamily Proteins in Two Closely Related Lamina Neurons
As an additional step toward identifying candidates for regulating synaptic specificity in the medulla, we compared the pattern of expression of CSMs between two closely related neurons in more detail. To do this, we focused on L1 and L2. These neurons have similar patterns of gene expression and morphologies, particularly in the lamina, where their dendrites are postsynaptic to photoreceptor neurons. L1 and L2 are key components of the ON and OFF pathways, respectively, and play overlapping roles in motion detection (Borst, 2014). Their morphologies, layer specificity and patterns of synaptic connections in the medulla sharply diverge. For example, L1 and L2 are presynaptic to five and nine classes of neurons in the medulla, respectively, only one of which is in common (Figure 3C) (S. Takemura, I. Meinertzhagen, and L. Scheffer, personal communication). Thus, we reasoned that differences in the CSMs expressed between these neurons would be candidates for regulating their synaptic specificity in the medulla.
L1 and L2 express a similar number of CSM genes (i.e., ~260, with an RPKM greater than five, Figure S2). Of these, 225 were expressed at a similar level (i.e., less than two times difference) in both neurons. By contrast, 21 and 53 CSM genes were expressed two to five times and greater than five times between them, respectively (Figure 3D). Among the 53 CSM genes with greater than five times difference, 23 encode Ig superfamily cell surface proteins (Figure 3E). This enrichment is highly unlikely to arise by chance (p values = 6.681e-06). The Dpr sub-family of Ig proteins was also enriched within this category with 8 of the 21 family members differentially expressed (i.e., greater than five times) (p value = 2.853e-04). Furthermore, as seen in Figure 3B, each lamina neuron expresses a unique combination of dpr genes (Figure 3B; Table S4). Based on these findings, we speculated that Dprs were good candidates for regulating cell-type-specific patterns of synaptic connectivity in the medulla neuropil.
Many Dpr Proteins Are Expressed in a Cell-Type-Enriched Fashion
The 21 Dpr proteins are cell surface proteins comprising two Ig domains (Nakamura et al., 2002). They show a complex pattern of interactions in vitro, with another family, the Dpr interacting proteins or DIPs, comprising three Ig domains. These interactions were discovered in an ELISA-based assay and the interactions are presumed to occur in trans (Özkan et al., 2013). Their functional significance remains unclear, but they are expressed in the embryonic nervous system (Fisher et al., 2012 and Özkan et al., 2013). Each Dpr binds to between one and four DIPs and each DIP binds to between one and seven Dprs (Özkan et al., 2013) (Figure 6F).
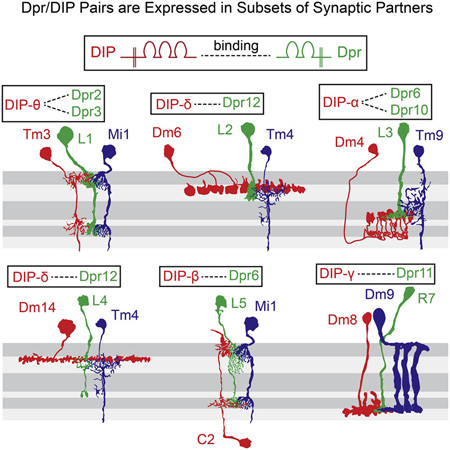
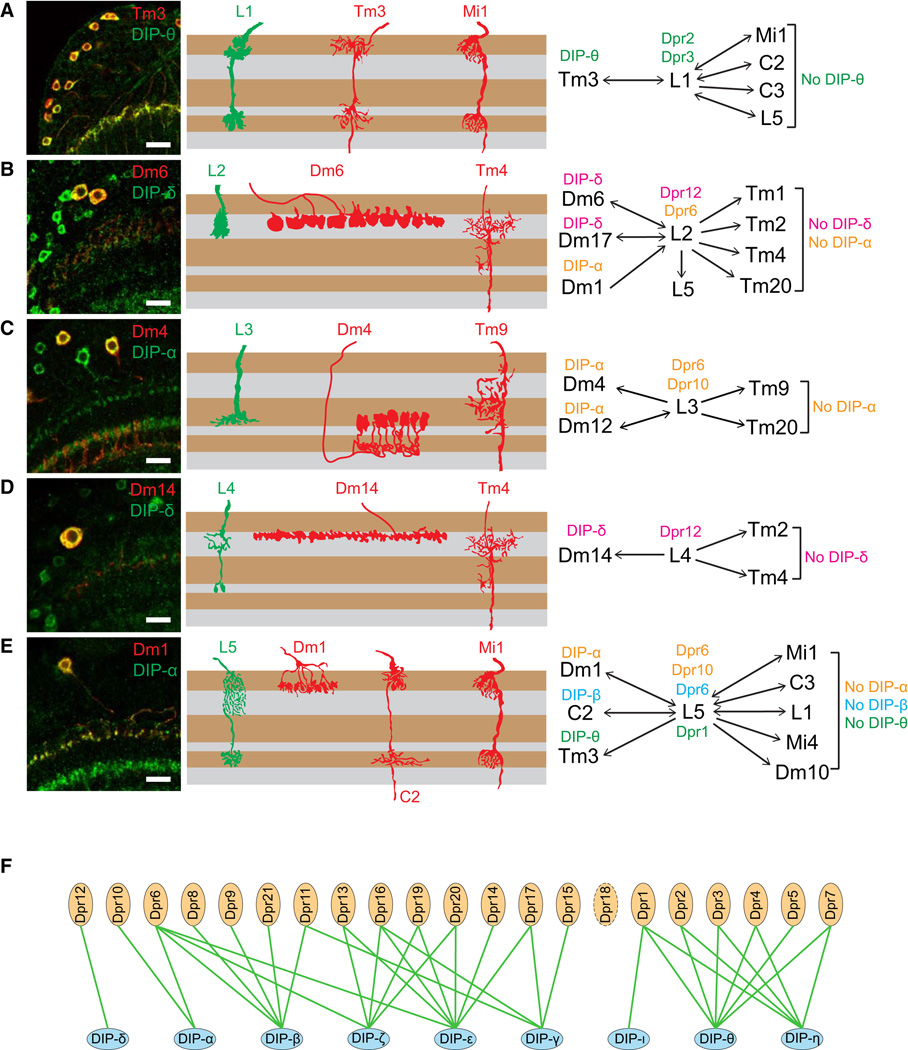
Figure 6. Matching of Cognate Dpr and DIP Expression in Synaptic Partners.
(A–E) Co-localization of DIPs in synaptic partners of L1–L5. Left panels: co-localization of indicated DIP (green) and cell-type-specific marker (red) in the adult. Middle panels: schematic of morphology of lamina neurons (green) and a subset of their synaptic partners (red) within the medulla neuropil. Right panels: summary of Dpr expression in L1–L5 and DIPs in synaptic partners. Layer patterns for DIPs in the medulla are the same as at 72 hr. Synaptic partner assignments from Takemura et al. (2015) and S. Takemura, I. Meinertzhagen, and L. Scheffer, personal communication.
(F) Summary of the Dpr/DIP interactome (Özkan et al., 2013 and Carrillo et al., 2015).
See also Table S5.
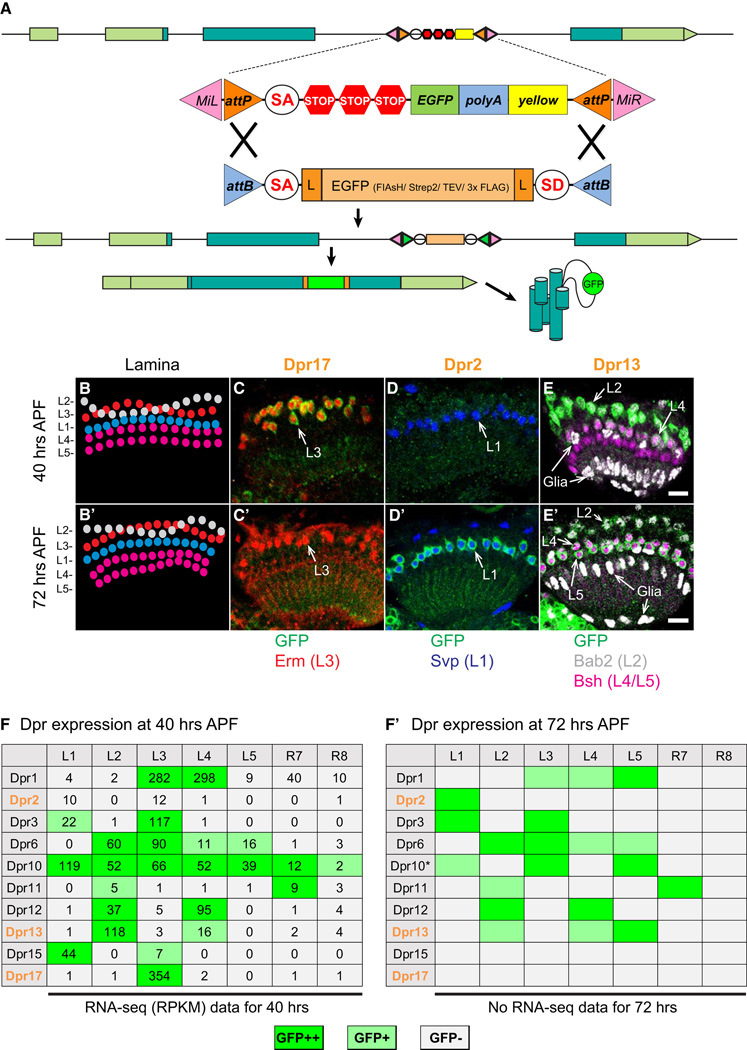
To independently assess the pattern of expression of Dprs in R7, R8, and L1–L5, we tagged the proteins produced from the endogenous locus with GFP using recombination-mediated cassette exchange of specific MiMIC insertions into Dpr genes (Venken et al., 2011) (Figure 4A). MiMIC insertions into introns separating coding exons for 10 of the 21 Dpr genes were identified and converted into protein traps (Nagarkar-Jaiswal et al., 2015) (see Supplemental Experimental Procedures). These contain an open reading frame encoding GFP flanked by splice acceptor and donor sites. Pupal brains were stained at 40 hr APF just prior to the onset of synapse formation and some 32 hr later (at 72 hr APF), a stage at which these neurons continue to add synaptic connections (Chen et al., 2014). In all cases, sufficient protein was detected in the cell body to identify the specific cells expressing the modified Dpr by co-staining the retina and lamina with antibodies to cell-type-specific nuclear proteins (Figures 4B–4E′). Cell-type-enriched expression in lamina neurons was also observed in Beat (Pipes et al., 2001) and Side (Sink et al., 2001) protein families using this method (Figures S4F–S4H). For each Dpr tested, the protein trap expression pattern correlated well with the RPKM values observed (Figures 4F–4F′). For some cell types, the pattern of expression was stable between 40 hr and 72 hr APF and for others marked changes were observed. For instance, Dpr15 and Dpr17 are expressed only at 40 hr APF. By contrast, Dpr2 is selectively expressed at 72 hr. Thus, from both RNA-seq studies and MiMIC expression analysis, all cells express more than one Dpr and they express different combinations of them.
Figure 4. Dpr Proteins Are Expressed in a Neuronal Cell-Type-Enriched Fashion in the Lamina.
(A) Schematic of a MiMIC-based protein trap. MiMIC protein traps contain GFP in frame flanked by splice acceptor and donor sites. They are generated by cassette exchange using ϕC31 recombinase to catalyze recombination with the Minos insertion between the AttP and AttB sites. The green inverted arrows after recombination represent recombined recombination sites (i.e., attR sites). Genes modified in this way encode chimeric proteins containing GFP.
(B and B′) Arrangement of lamina neuron cell bodies at 40 hr and 72 hr APF. L2 and L3 are intermingled at the top of lamina cell clusters. L4 and L5 make up the bottom two rows with L5 beneath L4.
(C–E′) Dpr17, Dpr2, and Dpr13 expression in lamina neurons visualized using MiMIC protein traps. See Figure 2 for lamina neuron markers. Scale bars, 10 µm.
(F and F′) Summary of Dpr expression using protein trap lines (10 of 21 dpr genes). RPKM values from the RNA-seq results indicating level of gene expression are included in (F). Dpr2, Dpr13, and Dpr17 are orange colored in bold to indicate changes in staining with the preceding panels. *Indicates Dpr10 expression level in L5 is variable at 72 hr APF.
See also Figure S4.
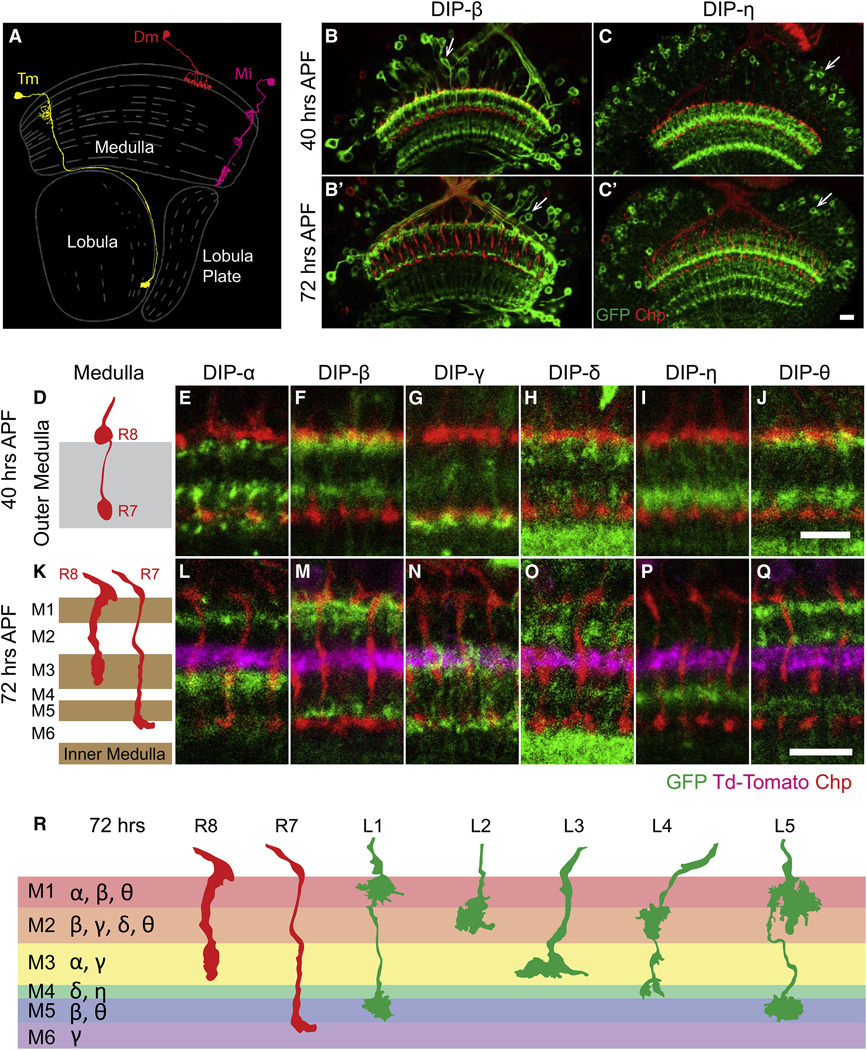
DIPs Are Expressed in a Layer-Specific Fashion
If Dpr proteins regulate interactions with specific neurons, we would anticipate that DIPs would be expressed in neurons with which R7, R8, and L1–L5 interact. To explore the in vivo expression of DIPs, we generated and analyzed GFP protein trap derivatives for six of the nine DIPs (asterisk in Figure S4A; Supplemental Experimental Procedures) and assessed their expression at 24, 40, and 72 hr APF, and in the adult. Consistent with our RNA-seq data, DIP-β and DIP-γ are expressed at low levels in a subset of lamina neurons (Figures S4C–S4D′) and the remaining DIPs are not expressed in these cells (Figures S4B, S4B′, S4E, and S4E′). Indeed, as specified earlier in the text, we used this independent verification of cell-type-enriched expression of transcripts slightly below five RPKM (e.g., DIP-β) to set conservative criteria for genes differentially expressed (DE) between cell-types.
Each DIP analyzed was expressed in neurons exhibiting unique layer-specific patterns of processes within the medulla neuropil (Figures 5A–5Q and S5). Prior to synapse formation (i.e., 24 hr APF) layered patterns are diffuse and overlap (Figures S5A–S5G). By 40% APF, the patterns are less diffuse with less overlap between different DIPs (Figures 5D–5J). At this stage of development, layers are still forming and most neurons have yet to form synapses. By contrast, some 32 hr later (at 72 hr APF) the medulla neuropil has expanded, many neurons have formed synapses (Chen et al., 2014), and the processes expressing DIPs are, in general, more clearly separated (Figures 5K–5Q). At this stage, the six DIPs are expressed in one to three layers and all layers are defined by a unique combination of them (Figure 5R). With one exception (DIP-θ) (Figure S5H), layer-specific expression patterns remained the same in the adult as they were at 72 hr APF. In summary, DIPs are differentially expressed in layers innervated by R7, R8, and L1–L5 neurons.
Figure 5. DIP Proteins Are Expressed in a Layer-Specific Fashion in the Medulla.
(A) Schematic of three classes of medulla neurons. A transmedullary neuron (Tm, in yellow), an amacrine-like distal medulla neuron (Dm, in red), and a medulla intrinsic neuron (Mi, in magenta) are shown. Each class of medulla neurons can be further divided into specific cell types based on different patterns of layer-specific branching. Adapted from Fischbach and Dittrich (1989).
(B–C′) DIP protein traps are expressed in scattered cells in the medulla cortex (arrows) and in layer-specific patterns in the medulla neuropil. DIP-β and DIP-η are shown as examples (green). Photoreceptor axons are visualized by staining for the cell surface protein Chp (red).
(D–J) All six DIP genes for which protein trap lines are available were expressed in neurons exhibiting unique layer-specific patterns of processes within the outer medulla neuropil at 40 hr APF. (D) Schematic of R8 and R7 axon morphology and layer distribution in the outer medulla. (E–J) Protein expression of six DIPs (green) in the outer medulla. The six DIPs are expressed in one to three layers; each layer is defined by a unique combination of DIPs.
(K–R) The DIP expression pattern at 72 hr is shown. (K) Schematic of R7 and R8 axons at 72 hr. The medulla expands and R7 and R8 layers change between 40 and 72 hr. (L–Q) The layered expression of DIPs is largely the same as at 40 hr. Expression in an additional layer, however, appears in DIP-θ. Dm3 axons are labeled with td-tomato (magenta). They run parallel to layers and mark the M2 and M3 border. (R) Summary of expression of DIPs in the medulla and the projection of R8, R7, and L1–L5 terminals at 72 hr APF. Scale bars, 10 µm.
See also Figure S5.
Dprs and DIPs Are Expressed by Synaptic Partners
We next sought to assess whether Dpr-expressing lamina neurons and DIP-expressing medulla neurons with processes in the same layers were synaptic partners. To assess the identity of medulla neurons expressing specific DIPs, we crossed flies carrying a DIP-GFP (DIP-α, DIP-δ, and DIP-θ) to a panel of GAL4 marker lines for specific medulla neurons and assessed co-localization of the markers (see Supplemental Experimental Procedures). For some DIPs (i.e., DIP-β), we used DIP-GAL4 derivatives of MiMICs in combination with FLP-mediated excision to express target UAS reporter constructs active in scattered DIP-expressing cells (Nern et al., 2015). This allowed us to visualize individual DIP-expressing neurons and to identify them by their morphologies. We then correlated the identification of cells expressing specific DIPs and Dprs with the pattern of synaptic connections within layers determined by serial EM reconstruction (Takemura et al., 2013 and Takemura et al., 2015).
Dense synaptic connectomes for a single column (Takemura et al., 2013) and, more recently, seven adjacent columns, comprising a central one surrounded by six additional ones (Takemura et al., 2015; S. Takemura, I. Meinertzhagen, and L. Scheffer, personal communication), have been determined. In general, these studies revealed that lamina neurons make synapses with multiple partners and show marked specificity. An example of the synaptic connections made by L5 neurons is shown in Table 1. L1–L5 neurons each express one or more Dpr proteins, which bind to DIPs expressed in a subset of their synaptic partners (Figures 6A–6E). For instance, L1 expresses Dpr2 and Dpr3 and these proteins bind to DIP-θ expressed on one of their synaptic partners, Tm3. Other synaptic partners of L1 do not express DIP-θ (Figure 6A; Table S5). The Dpr/DIP patterns of expression in L5 and its partners provide an example of a more complex relationship between these paralogs than L1 (Figure 6E). L5 expresses Dprs that bind to three different DIPs, DIP-α, DIP-β, and DIP-θ, in three synaptic partners: (1) like L1, L5 makes synapses with Tm3 and these neurons express a matching pair of Dpr1 and DIP-θ, respectively; (2) L5 expresses Dpr6, which binds to DIP-β, which is expressed in post-synaptic C2 neurons; and (3) Dpr6 and Dpr10 bind to DIP-α on Dm1 neurons. By contrast, five other synaptic partners of L5 do not express these DIPs. In addition, we also demonstrated that R7 neurons express Dpr11 and its cognate DIP is expressed in its synaptic partner Dm8 (Carrillo et al., 2015). Thus, Dprs in R7 and L1–L5 neurons match DIPs expressed in subsets of their synaptic partners.
Table 1.
Synaptic Partners of L5
| Synaptic Partners | Number of Synapses | |
|---|---|---|
| L5 Pre | L5 Post | |
| Dm1 | 39 | 13 |
| Dm10 | 48 | 7 |
| Dm18 | 39 | 13 |
| C2 | 35 | 37 |
| C3 | 13 | 24 |
| Mi1 | 56 | 11 |
| Mi4 | 58 | 0 |
| Tm3 | 83 | 0 |
| L1 | 29 | 127 |
A dense connectome of 7 medulla columns has been completed by serial EM reconstruction (Takemura et al., 2015; S. Takemura, I. Meinertzhagen, and L. Scheffer, personal communication.). This includes a central column and six surrounding ones. Here, the synaptic partners of L5 with the number of inputs and outputs are listed. Most synapses are made with partners in the same column but processes can also extend into neighboring columns and form synapses. Here, we are showing the sum of the synapses made by an L5 neuron in the central column to partners within the same and neighboring columns.
Discussion
Here, we used RNA-seq of mRNAs from different, but highly related neuronal cell types, to identify families of cell surface proteins as potential regulators of synaptic specificity. Using a conservative RPKM threshold for expression, we estimate that at the onset of synapse formation neurons express between a quarter and a third of the potential CSMs encoded by the fly genome. Many of these are expressed in a cell-type-enriched fashion; they are expressed at least five times greater in one cell type, than in one or more of the other cell types profiled. Thus, neurons express many different CSMs, and each neuronal cell type expresses a unique combination of them. Many of the differentially expressed proteins have known interacting partners. For instance, of the 23 Ig superfamily proteins differing in expression by more than five times between L1 and L2, all but three have known interacting partners. Thus, expression studies coupled with the cell surface interactome and genetic analysis provide a multipronged approach to dissecting the cellular interactions leading to neural circuit formation.
Several families of proteins were differentially expressed in different neuronal cell types through RNA-seq analysis and some of these were confirmed using MiMIC protein traps. The most dramatic and complex pattern of expression observed, however, was seen for the Dpr proteins with 17 of the 21 paralogs expressed in a cell-type-enriched fashion. This pattern and the extensive protein interaction network of these proteins defined by Özkan et al. (2013) prompted us to explore the expression patterns of these proteins and their ligands in further detail.
The cell-type-enriched pattern of Dpr expression observed in lamina or photoreceptor neurons was striking. By contrast, most DIPs were not expressed, or were expressed at only very low levels in these cells. Localization of DIP expression using protein traps, however, revealed that each of the outer six layers of the medulla neuropil was defined by the expression of one or more DIPs and that DIPs were expressed in only a subset of processes within a layer. And these, in turn, are specific subsets of synaptic targets of cells expressing an interacting Dpr. These observations raise the possibility that Dpr/DIP interactions specify patterns of synaptic connections between neurons within each layer.
These findings, in combination with previous genetic studies, suggest a model for mechanisms regulating the formation of layer-specific connections within the medulla. Previous work demonstrated that at early stages of medulla development, lamina neuron growth cones target to overlapping regions that are established by broadly expressed adhesive (i.e., N-cadherin) and repulsive cell surface molecules (i.e., plexin/semaphorin signaling). Growth cones then segregate into discrete domains as the medulla matures through interactions between signals localized to specific layers (Nern et al., 2008, Pecot et al., 2013 and Timofeev et al., 2012). We speculate that different combinations of Dpr and DIP proteins specify synaptic connections within a layer.
In support of this notion, Carrillo et al. (2015) have shown that loss of Dpr11 and its binding partner DIP-γ show abnormalities in the M6 target layer. Dpr11 is expressed in a discrete subset of R7 neurons and these form synapses with DIP-γ-expressing Dm8 neurons. These include abnormalities in the R7 terminal morphology consistent with a role in synapse formation and a reduction in Dm8 neurons. Interestingly, we have also recently observed a reduction in the number of DIP-α expressing neurons in DIP-α null mutants suggesting a commonality in the function of Dprs and DIPs in the medulla. The simplest interpretation of the matching of Dprs and DIPs in synaptic partners is that these proteins regulate synaptic specificity. It would not be surprising, however, if these proteins play different roles such as contributing to layer-specific targeting, as with N-cadherin (Lee et al., 2001) or netrin (Timofeev et al., 2012), or cell-type-specific trophic support as we described previously for Jeb/Alk signaling (Pecot et al., 2014). Detailed phenotypic analyses of null mutants lacking Dprs and DIPs, and given the redundancy within these families, perhaps genetic analysis of animals lacking combinations of them, will be required to ascertain the precise functions of this family of ligand/receptor pairs in circuit assembly.
The two-step model for synaptic connectivity in the medulla shares intriguing similarities to, and indeed was significantly influenced by, models for layer specificity in the analogous structure in the mouse retina, the inner plexiform layers. Here, cadherin and semaphorin/plexin proteins direct processes to layers (Duan et al., 2014 and Matsuoka et al., 2011). In a subsequent step, Ig superfamily proteins then promote synaptic matching within them. Important support for this second step comes from recent genetic studies from Krishnaswamy et al. (2015) demonstrating that homophilic interaction between Sdk2 proteins (an Ig superfamily protein) is required for synapses between a specific pair of amacrine and retinal ganglion cell neurons. That this may represent a general strategy for synaptic-specificity in the vertebrate retina is suggested by the layer-specific expression of Sdk2 and related homophilic Ig superfamily proteins Sdk1, Dscam1, Dscam2, and Contactins 1–5 (Yamagata and Sanes, 2008, Yamagata and Sanes, 2012 and Yamagata et al., 2002). Thus, the studies in the mouse IPL and the medulla region of the fly allude to a common strategy for achieving synaptic specificity.
Conclusion
Dprs and DIPs are likely to be only a part of the story of synaptic specificity in the medulla. Indeed, a striking feature of the synaptic connectome in the medulla column is its complexity (Takemura et al., 2013 and Takemura et al., 2015), with synapses between the processes of >100 neuronal cell types (A. Nern, personal communication). This complexity is mirrored by the unique combination of hundreds of cell surface and secreted molecules expressed by each of the photoreceptor and lamina neurons profiled in this study. How this complexity contributes to specificity remains elusive, but the convergence of improved histological, genetic, physiological, and molecular tools promises to provide important insights into the molecular recognition strategies controlling synaptic specificity.
Experimental Procedures
Fly Husbandry and Stocks
Flies were reared at 25°C on standard medium. For developmental analysis and sorting experiments white pre-pupae were collected and incubated for the indicated number of hours. See Supplemental Experimental Procedures for the list of stocks used in different experiments.
Sorting Cell Types and Library Construction
For tissue dissociation, pupal brain tissue dissected at 40 hr after pupal formation was incubated with a Papain (Worthington) and Liberase TM protease cocktail (Roche) at 25°C for 15 min in a microfuge shaker at 1,000 rpm. At 5 and 10 min into this incubation, the tissue was pipetted up and down with a P200. At 15 min, the sample was passed through a 25G 5/8-gauge needle until the tissue was completely dissociated. Digestion was inactivated by the addition of rich media with serum, and the cell suspension was passed through a 70 µm filter. To concentrate the cells, the suspension was spun down at 1,600 rpm for 8 min at 4°C. After decanting the supernatant, cells were re-suspended in ~500 µl of rich media and sorted in a BD FACSAria II.
RNA was then isolated from sorted cells using the RNA-min elute kit from QIAGEN. mRNA was amplified in a linear fashion using Arcuturus RiboAmp HS kit (Life Technologies). cDNA was then generated for quality assessment and paired-end Illumina sequencing libraries were prepared.
Detailed protocols are available upon request.
Microscopy and Image Analysis
Confocal images were acquired on a Zeiss LSM780 confocal microscope. The staining patterns were reproducible between samples. However, some variation on the overall fluorescence signal and noise levels existed between sections and samples. Thus, proper adjustments of laser power, detector gain, and black level settings were made to obtain similar overall fluorescence signals. Single plane or maximum intensity projection confocal images were exported into TIFF files using ImageJ software.
See Supplemental Experimental Procedures for bioinformatics analysis and immunohistochemistry.
Supplementary Material
(A) Heatmap of the relationship of modules to each cell type (see text). WGCNA was carried out to identify genes expressed in a common pattern. Twenty-five expression modules were identified (see Supplemental Experimental Procedures and Table S3). These were then assessed for their correlation with an idealized cell-type specific pattern (see text). Cells are shown in columns and modules in rows. Here, cell-type specific modules are defined as r > 0.8 and p value < 0.001. Class refers to the comparison of retinal cells (i.e., R7 and R8 together) to lamina neurons (i.e., L1–L5 together).
(B) Heatmap showing the significance of gene overlaps between hand-curated biological categories and cell-type specific modules. Each column represents a cell-type specific module and each row represents a different biological category. A p value of the intersection is based on the permutation test. Number of overlapping genes between the indicated modules and categories are only labeled in cells with p value < 0.001.
Note Dpr18 RPKM values are considerable below five RPKM in all cell types and thus the only Dpr we consider not expressed in our dataset.
DIP protein traps were tested for co-localization with a set of GAL4 lines driving marker expression in the indicated medulla neurons (see text). Medulla neuron classes tested for each DIP-GFP are shown. Neuron in red indicates that the cell expresses the DIP and black indicates that it does not. Note question marks for DIP-η to indicate that staining was very weak.
The number of reads in each range is shown in color. Note, for instance, that for each cell type there are approximately 250 CSMs with levels of expression >5.
The tetraspanin family of proteins has been implicated in synapse formation. Mutations in late bloomer (lbm) (Kopczynski et al., 1996) lead to delayed innervation of muscle in the Drosophila embryo and mutations in a human tetraspanins are associated with intellectual disability and synapse formation (Bassani et al., 2012). Proteins containing EGF and LRR domains play roles in axon guidance (e.g., netrin) (Timofeev et al., 2012) and synapse formation (e.g., Toll-6,) (Ward et al., 2015) respectively.
(A) RPKM values of DIP protein families in L1–L5, R7 and R8 at 40 hr APF. Asterisks indicate DIP genes for which protein trap or Gal4 trap lines are available.
(B–E′) DIP-α, DIP-β, DIP-γ and DIP-η expression in retina and lamina neurons at 40 hr and 72 hr APF using MiMIC derivatives. For DIP-α and DIP-η, MiMIC protein traps were used. For DIP-β and DIP-γ, Gal4 trap derivatives were used in combination with UAS-myr-GFP to visualize DIP expression in lamina neurons. (B and B′) DIP-α is not expressed in retina or lamina neurons at 40 and 72 hr APF. (C and C′) DIP-β (green) is specifically expressed in L4 (red) at 40 hr and 72 hr APF. (D and D′) DIP-γ (green) is expressed strongly in L1 (red) and weakly in L2 (gray) at 40 hr APF. The expression in L1 and L2 at 72 hr APF is close to threshold of detection. Although DIP-γ is expressed with an RPKM in L4 similar to the RPKM in L1, we did not detect expression using the MiMIC derivative. DIP-γ is also expressed in glia. (E and E′) DIP-η is not expressed in retina or lamina neurons at 40 and 72 hr APF. Scale bars, 10 microns.
(F–H) Beat-IIb expression in lamina neurons and Beat-IIIc expression in retina at 40 hr APF using MiMIC protein traps. (F) Beat-IIb (green) is specifically expressed in L3 (red) and L4 (upper magenta) in lamina neurons. It is also expressed in glia beneath L5 (lower magenta). The massive green staining is likely to be due to expression in the processes of glial cells within the lamina neuropil. (G and G′) Beat-IIIc (green) is specifically expressed in R1 (ˆ), R4 (*), R6 (+) and R8 (blue), but not in R7 (red, arrowhead) in the retina. Bristles (arrow) are also labeled by Pros staining (red). (H) CG34114 (green), a member of the Side protein family is specifically expressed in L4 and L5 (magenta), with higher level in L5 than in L4. The strong staining just beneath these cells is likely to be the proximal region of the L5 axon. Scale bars, 10 microns.
(A–G) Expression in the medulla at 24 hr APF of the six DIP genes for which protein trap lines are available. They were expressed in neurons exhibiting layer-specific patterns of processes within the outer medulla neuropil. However, prior to synapse formation (i.e., 24 hr APF), layered patterns are diffuse and overlap. (A) Schematic of R8 and R7 axon morphology in the outer medulla at 24 hr APF to visualize the boundaries of the outer medulla. (B–G) Protein expression of six DIPs (green) in the outer medulla. The six DIPs are expressed in layer-specific patterns. (H) Expression of DIP-θ in the medulla neuropil in the adult. The axons of Dm3 neurons are labeled with td-tomato (magenta) to mark the M2 and M3 border. Note that the pattern is different from the one at 72 hr APF (see Figure 5Q); expression at the M2 layer is missing. Layer-specific expression patterns for all other 5 DIPs remained the same in the adult as they were at 72 hr APF. Scale bars, 5 microns.
(A) Differentially expressed genes with RPKM value greater than five (for at least one neuron in each pairwise comparison) and a five times difference between pairs of neurons (see text for results with RPKM values greater than five) and an adjusted p value of <0.05.
(B) Differentially expressed genes encoding CSM genes. The criteria for a differentially expressed gene are an RPKM value greater than five for a least one of the pair and at least a five times difference in RPKM values with an adjusted p value <0.05.
Highlights.
RNA-seq was performed on seven different neuron classes during synapse formation
Each class expresses a unique set of hundreds of genes encoding cell surface proteins
The 21 Ig-containing Dpr mRNAs are differentially expressed
Different synaptic pairs express matching Dprs and Dpr interacting proteins (DIPs)
Acknowledgments
We thank Orkun Akin, John Carlson, Claude Desplan, Yasushi Hiromi, Frank Laski, Cheng-yu Lee for sharing reagents; members of our laboratory for discussion; Louis K. Scheffer and Ian Meinertzhagen for discussions, sharing results from the EM studies at Janelia Research Campus (HHMI) and comments on the manuscript; Kaushiki Menon and Robert Carrillo (Zinn group) for communicating results prior to publication; Donghui Cheng, Tanya Stoyanova and Owen Witte for assistance in FACS purification of cells; Dorian Gunning for generating Bsh antibody; and Barret Pfeiffer and Gerald Rubin (JRC/HHMI) for reagents to construct the markers for R7 neurons. The work was supported by a Graduate Student Fellowship from the China Scholarship Council (CSC) and a Graduate Student Fellowship from the University of California, Los Angeles (UCLA) Philip Whitcome Training Program (L.T.); the Canadian Institute of Health Research Fellowship (K.X.Z.); Jane Coffin Childs Memorial Fund for Medical Research (M.P.); the Robert A. and Renee E. Belfer Family Foundation (S.N.-J.); Target A.L.S. (P.-T.L.); a Long-Term Fellowship from the Human Frontiers Science Program (W.T.); grants from National Institute of Health (R01GM067858 (H.J.B.), R01NS62821 (K.Z.), and R01NS28182 (K.Z.); a Ramon y Cajal contract (RYC-2011-09479) and a Ministerio de Economia y Competitividad grant (BFU2012-32282) (M.M.). H.J.B. and S.L.Z. are investigators of Howard Hughes Medical Institute.
Footnotes
Author Contributions
S.L.Z., M.M., L.T., and K.X.Z. conceived the project and wrote the paper. S.L.Z., L.T., and M.M. designed the experiments. L.T. and M.M. conducted the experiments and analyzed data. K.X.Z. conducted bioinformatic analysis.
Accession Numbers
The accession number for the raw data reported in this paper is GEO: GSE68235.
References
- 1.Borst A. Fly visual course control: behaviour, algorithms and circuits. Nat Rev Neurosci. 2014 Sep;15(9):590–599. doi: 10.1038/nrn3799. [DOI] [PubMed] [Google Scholar]
- 2.Cang J, Feldheim DA. Developmental mechanisms of topographic map formation and alignment. Annu Rev Neurosci. 2013 Jul 8;36:51–77. doi: 10.1146/annurev-neuro-062012-170341. [DOI] [PubMed] [Google Scholar]
- 3.Carrillo RA, Özkan E, Menon KP, Nagarkar-Jaiswal S, Lee PT, Jeon M, Birnbaum ME, Bellen HJ, Garcia KC, Zinn K. Control of Synaptic Connectivity by a Network of Drosophila IgSF Cell Surface Proteins. Cell. 2015 Dec 17;163(7):1770–1782. doi: 10.1016/j.cell.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Akin O, Nern A, Tsui CY, Pecot MY, Zipursky SL. Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron. 2014 Jan 22;81(2):280–293. doi: 10.1016/j.neuron.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Wit J, Hong W, Luo L, Ghosh A. Role of leucine-rich repeat proteins in the development and function of neural circuits. Annu Rev Cell Dev Biol. 2011;27:697–729. doi: 10.1146/annurev-cellbio-092910-154111. [DOI] [PubMed] [Google Scholar]
- 6.Duan X, Krishnaswamy A, De la Huerta I, Sanes JR. Type II cadherins guide assembly of a direction-selective retinal circuit. Cell. 2014 Aug 14;158(4):793–807. doi: 10.1016/j.cell.2014.06.047. [DOI] [PubMed] [Google Scholar]
- 7.Fischbach KF, Dittrich AP. The optic lobe of Drosophila melanogaster. I: A. Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258:441–475. [Google Scholar]
- 8.Fischbach KF, Linneweber GA, Andlauer TF, Hertenstein A, Bonengel B, Chaudhary K. The irre cell recognition module (IRM) proteins. J Neurogenet. 2009;23(1–2):48–67. doi: 10.1080/01677060802471668. [DOI] [PubMed] [Google Scholar]
- 9.Fisher B, Weiszmann R, Frise E, Hammonds A, Tomancak P, Beaton A, Berman B, Quan E, Shu S, Lewis S, et al. BDGP in situ homepage. 2012 http://insitu.fruitfly.org/cgi-bin/ex/insitu.pl. [Google Scholar]
- 10.Fradkin LG, Kamphorst JT, DiAntonio A, Goodman CS, Noordermeer JN. Genomewide analysis of the Drosophila tetraspanins reveals a subset with similar function in the formation of the embryonic synapse. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13663–13668. doi: 10.1073/pnas.212511099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadjieconomou D, Timofeev K, Salecker I. A step-by-step guide to visual circuit assembly in Drosophila. Curr Opin Neurobiol. 2011 Feb;21(1):76–84. doi: 10.1016/j.conb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Hong W, Mosca TJ, Luo L. Teneurins instruct synaptic partner matching in an olfactory map. Nature. 2012 Mar 18;484(7393):201–207. doi: 10.1038/nature10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012 Oct 25;2(4):991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson KG, Tenney AP, Ghose A, Duckworth AM, Higashi ME, Parfitt K, Marcu O, Heslip TR, Marsh JL, Schwarz TL, Flanagan JG, Van Vactor D. The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron. 2006 Feb 16;49(4):517–531. doi: 10.1016/j.neuron.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 15.Kenzelmann D, Chiquet-Ehrismann R, Tucker RP. Teneurins, a transmembrane protein family involved in cell communication during neuronal development. Cell Mol Life Sci. 2007 Jun;64(12):1452–1456. doi: 10.1007/s00018-007-7108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998 Jun;20(6):1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 17.Kopczynski CC, Davis GW, Goodman CS. A neural tetraspanin, encoded by late bloomer, that facilitates synapse formation. Science. 1996 Mar 29;271(5257):1867–1870. doi: 10.1126/science.271.5257.1867. [DOI] [PubMed] [Google Scholar]
- 18.Krishnaswamy A, Yamagata M, Duan X, Hong YK, Sanes JR. Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature. 2015 Aug 27;524(7566):466–470. doi: 10.1038/nature14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurusu M, Cording A, Taniguchi M, Menon K, Suzuki E, Zinn K. A screen of cell-surface molecules identifies leucine-rich repeat proteins as key mediators of synaptic target selection. Neuron. 2008 Sep 25;59(6):972–985. doi: 10.1016/j.neuron.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kvon EZ, Kazmar T, Stampfel G, Yáñez-Cuna JO, Pagani M, Schernhuber K, Dickson BJ, Stark A. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014 Aug 7;512(7512):91–95. doi: 10.1038/nature13395. [DOI] [PubMed] [Google Scholar]
- 21.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008 Dec 29;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langley JN. Note on Regeneration of Prae-Ganglionic Fibres of the Sympathetic. J Physiol. 1895 Jul 18;18(3):280–284. doi: 10.1113/jphysiol.1895.sp000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999 Mar;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 24.Lee CH, Herman T, Clandinin TR, Lee R, Zipursky SL. N-cadherin regulates target specificity in the Drosophila visual system. Neuron. 2001 May;30(2):437–450. doi: 10.1016/s0896-6273(01)00291-4. [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012 Aug 23;488(7412):517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linnemannstöns K, Ripp C, Honemann-Capito M, Brechtel-Curth K, Hedderich M, Wodarz A. The PTK7-related transmembrane proteins off-track and off-track 2 are co-receptors for Drosophila Wnt2 required for male fertility. PLoS Genet. 2014 Jul 10;10(7):e1004443. doi: 10.1371/journal.pgen.1004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuoka RL, Nguyen-Ba-Charvet KT, Parray A, Badea TC, Chédotal A, Kolodkin AL. Transmembrane semaphorin signalling controls laminar stratification in the mammalian retina. Nature. 2011 Feb 10;470(7333):259–263. doi: 10.1038/nature09675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miura SK, Martins A, Zhang KX, Graveley BR, Zipursky SL. Probabilistic splicing of Dscam1 establishes identity at the level of single neurons. Cell. 2013 Nov 21;155(5):1166–1177. doi: 10.1016/j.cell.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morante J, Desplan C. The color-vision circuit in the medulla of Drosophila. Curr Biol. 2008 Apr 22;18(8):553–565. doi: 10.1016/j.cub.2008.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagarkar-Jaiswal S, Lee PT, Campbell ME, Chen K, Anguiano-Zarate S, Gutierrez MC, Busby T, Lin WW, He Y, Schulze KL, et al. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife. 2015 Mar 31;4 doi: 10.7554/eLife.05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura M, Baldwin D, Hannaford S, Palka J, Montell C. Defective proboscis extension response (DPR), a member of the Ig superfamily required for the gustatory response to salt. J Neurosci. 2002 May 1;22(9):3463–3472. doi: 10.1523/JNEUROSCI.22-09-03463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nern A, Zhu Y, Zipursky SL. Local N-cadherin interactions mediate distinct steps in the targeting of lamina neurons. Neuron. 2008 Apr 10;58(1):34–41. doi: 10.1016/j.neuron.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nern A, Pfeiffer BD, Rubin GM. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):E2967–E2976. doi: 10.1073/pnas.1506763112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Donnell M, Chance RK, Bashaw GJ. Axon growth and guidance: receptor regulation and signal transduction. Annu Rev Neurosci. 2009;32:383–412. doi: 10.1146/annurev.neuro.051508.135614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Özkan E, Carrillo RA, Eastman CL, Weiszmann R, Waghray D, Johnson KG, Zinn K, Celniker SE, Garcia KC. An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell. 2013 Jul 3;154(1):228–239. doi: 10.1016/j.cell.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pecot MY, Tadros W, Nern A, Bader M, Chen Y, Zipursky SL. Multiple interactions control synaptic layer specificity in the Drosophila visual system. Neuron. 2013 Jan 23;77(2):299–310. doi: 10.1016/j.neuron.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pecot MY, Chen Y, Akin O, Chen Z, Tsui CY, Zipursky SL. Sequential axon-derived signals couple target survival and layer specificity in the Drosophila visual system. Neuron. 2014 Apr 16;82(2):320–333. doi: 10.1016/j.neuron.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pipes GC, Lin Q, Riley SE, Goodman CS. The Beat generation: a multigene family encoding IgSF proteins related to the Beat axon guidance molecule in Drosophila. Development. 2001 Nov;128(22):4545–4552. doi: 10.1242/dev.128.22.4545. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt AM, Shi J, Wolf AM, Lu CC, King LA, Zou Y. Wnt-Ryk signalling mediates medial-lateral retinotectal topographic mapping. Nature. 2006 Jan 5;439(7072):31–37. doi: 10.1038/nature04334. [DOI] [PubMed] [Google Scholar]
- 40.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000 Jun 9;101(6):671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 41.Serafini T, Kennedy TE, Galko MJ, Mirzayan C, Jessell TM, Tessier-Lavigne M. The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell. 1994 Aug 12;78(3):409–424. doi: 10.1016/0092-8674(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 42.Shen K, Bargmann CI. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell. 2003 Mar 7;112(5):619–630. doi: 10.1016/s0092-8674(03)00113-2. [DOI] [PubMed] [Google Scholar]
- 43.Shen K, Fetter RD, Bargmann CI. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG–1. Cell. 2004 Mar 19;116(6):869–881. doi: 10.1016/s0092-8674(04)00251-x. [DOI] [PubMed] [Google Scholar]
- 44.Sink H, Rehm EJ, Richstone L, Bulls YM, Goodman CS. sidestep encodes a target-derived attractant essential for motor axon guidance in Drosophila. Cell. 2001 Apr 6;105(1):57–67. doi: 10.1016/s0092-8674(01)00296-3. [DOI] [PubMed] [Google Scholar]
- 45.Sperry RW. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc Natl Acad Sci U S A. 1963 Oct;50:703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takemura SY, Lu Z, Meinertzhagen IA. Synaptic circuits of the Drosophila optic lobe: the input terminals to the medulla. J Comp Neurol. 2008 Aug 10;509(5):493–513. doi: 10.1002/cne.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013 Aug 8;500(7461):175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takemura SY, Xu CS, Lu Z, Rivlin PK, Parag T, Olbris DJ, Plaza S, Zhao T, Katz WT, Umayam L, et al. Synaptic circuits and their variations within different columns in the visual system of Drosophila. Proc Natl Acad Sci U S A. 2015 Nov 3;112(44):13711–13716. doi: 10.1073/pnas.1509820112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timofeev K, Joly W, Hadjieconomou D, Salecker I. Localized netrins act as positional cues to control layer-specific targeting of photoreceptor axons in Drosophila. Neuron. 2012 Jul 12;75(1):80–93. doi: 10.1016/j.neuron.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Triplett JW, Feldheim DA. Eph and ephrin signaling in the formation of topographic maps. Semin Cell Dev Biol. 2012 Feb;23(1):7–15. doi: 10.1016/j.semcdb.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venken KJ, Bellen HJ. Chemical mutagens, transposons, and transgenes to interrogate gene function in Drosophila melanogaster. Methods. 2014 Jun 15;68(1):15–28. doi: 10.1016/j.ymeth.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, Bellen HJ. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011 Sep;8(9):737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward A, Hong W, Favaloro V, Luo L. Toll receptors instruct axon and dendrite targeting and participate in synaptic partner matching in a Drosophila olfactory circuit. Neuron. 2015 Mar 4;85(5):1013–1028. doi: 10.1016/j.neuron.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winberg ML, Tamagnone L, Bai J, Comoglio PM, Montell D, Goodman CS. The transmembrane protein Off-track associates with Plexins and functions downstream of Semaphorin signaling during axon guidance. Neuron. 2001 Oct 11;32(1):53–62. doi: 10.1016/s0896-6273(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 55.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999 Jun 11;97(6):779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 56.Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008 Jan 24;451(7177):465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- 57.Yamagata M, Sanes JR. Expanding the Ig superfamily code for laminar specificity in retina: expression and role of contactins. J Neurosci. 2012 Oct 10;32(41):14402–14414. doi: 10.1523/JNEUROSCI.3193-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamagata M, Weiner JA, Sanes JR. Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell. 2002 Sep 6;110(5):649–660. doi: 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
- 59.Zipursky SL, Grueber WB. The molecular basis of self-avoidance. Annu Rev Neurosci. 2013 Jul 8;36:547–568. doi: 10.1146/annurev-neuro-062111-150414. [DOI] [PubMed] [Google Scholar]
- 60.Zipursky SL, Wojtowicz WM, Hattori D. Got diversity? Wiring the fly brain with Dscam. Trends Biochem Sci. 2006 Oct;31(10):581–588. doi: 10.1016/j.tibs.2006.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Heatmap of the relationship of modules to each cell type (see text). WGCNA was carried out to identify genes expressed in a common pattern. Twenty-five expression modules were identified (see Supplemental Experimental Procedures and Table S3). These were then assessed for their correlation with an idealized cell-type specific pattern (see text). Cells are shown in columns and modules in rows. Here, cell-type specific modules are defined as r > 0.8 and p value < 0.001. Class refers to the comparison of retinal cells (i.e., R7 and R8 together) to lamina neurons (i.e., L1–L5 together).
(B) Heatmap showing the significance of gene overlaps between hand-curated biological categories and cell-type specific modules. Each column represents a cell-type specific module and each row represents a different biological category. A p value of the intersection is based on the permutation test. Number of overlapping genes between the indicated modules and categories are only labeled in cells with p value < 0.001.
Note Dpr18 RPKM values are considerable below five RPKM in all cell types and thus the only Dpr we consider not expressed in our dataset.
DIP protein traps were tested for co-localization with a set of GAL4 lines driving marker expression in the indicated medulla neurons (see text). Medulla neuron classes tested for each DIP-GFP are shown. Neuron in red indicates that the cell expresses the DIP and black indicates that it does not. Note question marks for DIP-η to indicate that staining was very weak.
The number of reads in each range is shown in color. Note, for instance, that for each cell type there are approximately 250 CSMs with levels of expression >5.
The tetraspanin family of proteins has been implicated in synapse formation. Mutations in late bloomer (lbm) (Kopczynski et al., 1996) lead to delayed innervation of muscle in the Drosophila embryo and mutations in a human tetraspanins are associated with intellectual disability and synapse formation (Bassani et al., 2012). Proteins containing EGF and LRR domains play roles in axon guidance (e.g., netrin) (Timofeev et al., 2012) and synapse formation (e.g., Toll-6,) (Ward et al., 2015) respectively.
(A) RPKM values of DIP protein families in L1–L5, R7 and R8 at 40 hr APF. Asterisks indicate DIP genes for which protein trap or Gal4 trap lines are available.
(B–E′) DIP-α, DIP-β, DIP-γ and DIP-η expression in retina and lamina neurons at 40 hr and 72 hr APF using MiMIC derivatives. For DIP-α and DIP-η, MiMIC protein traps were used. For DIP-β and DIP-γ, Gal4 trap derivatives were used in combination with UAS-myr-GFP to visualize DIP expression in lamina neurons. (B and B′) DIP-α is not expressed in retina or lamina neurons at 40 and 72 hr APF. (C and C′) DIP-β (green) is specifically expressed in L4 (red) at 40 hr and 72 hr APF. (D and D′) DIP-γ (green) is expressed strongly in L1 (red) and weakly in L2 (gray) at 40 hr APF. The expression in L1 and L2 at 72 hr APF is close to threshold of detection. Although DIP-γ is expressed with an RPKM in L4 similar to the RPKM in L1, we did not detect expression using the MiMIC derivative. DIP-γ is also expressed in glia. (E and E′) DIP-η is not expressed in retina or lamina neurons at 40 and 72 hr APF. Scale bars, 10 microns.
(F–H) Beat-IIb expression in lamina neurons and Beat-IIIc expression in retina at 40 hr APF using MiMIC protein traps. (F) Beat-IIb (green) is specifically expressed in L3 (red) and L4 (upper magenta) in lamina neurons. It is also expressed in glia beneath L5 (lower magenta). The massive green staining is likely to be due to expression in the processes of glial cells within the lamina neuropil. (G and G′) Beat-IIIc (green) is specifically expressed in R1 (ˆ), R4 (*), R6 (+) and R8 (blue), but not in R7 (red, arrowhead) in the retina. Bristles (arrow) are also labeled by Pros staining (red). (H) CG34114 (green), a member of the Side protein family is specifically expressed in L4 and L5 (magenta), with higher level in L5 than in L4. The strong staining just beneath these cells is likely to be the proximal region of the L5 axon. Scale bars, 10 microns.
(A–G) Expression in the medulla at 24 hr APF of the six DIP genes for which protein trap lines are available. They were expressed in neurons exhibiting layer-specific patterns of processes within the outer medulla neuropil. However, prior to synapse formation (i.e., 24 hr APF), layered patterns are diffuse and overlap. (A) Schematic of R8 and R7 axon morphology in the outer medulla at 24 hr APF to visualize the boundaries of the outer medulla. (B–G) Protein expression of six DIPs (green) in the outer medulla. The six DIPs are expressed in layer-specific patterns. (H) Expression of DIP-θ in the medulla neuropil in the adult. The axons of Dm3 neurons are labeled with td-tomato (magenta) to mark the M2 and M3 border. Note that the pattern is different from the one at 72 hr APF (see Figure 5Q); expression at the M2 layer is missing. Layer-specific expression patterns for all other 5 DIPs remained the same in the adult as they were at 72 hr APF. Scale bars, 5 microns.
(A) Differentially expressed genes with RPKM value greater than five (for at least one neuron in each pairwise comparison) and a five times difference between pairs of neurons (see text for results with RPKM values greater than five) and an adjusted p value of <0.05.
(B) Differentially expressed genes encoding CSM genes. The criteria for a differentially expressed gene are an RPKM value greater than five for a least one of the pair and at least a five times difference in RPKM values with an adjusted p value <0.05.