Abstract
Decorin is generally recognized as a tumor suppressing molecule. Nevertheless, although decorin has been shown to be differentially expressed in malignant tissues, it has often remained unclear whether, in addition to non-malignant stromal cells, cancer cells also express it. Here, we first used two publicly available databases to analyze the current information about decorin expression and immunoreactivity in normal and malignant human colorectal tissue samples. The analyses demonstrated that decorin expression and immunoreactivity may vary in cancer cells of human colorectal tissues. Therefore, we next examined decorin expression in normal, premalignant and malignant human colorectal tissues in more detail using both in situ hybridization and immunohistochemistry for decorin. Our results invariably demonstrate that malignant cells within human colorectal cancer tissues are devoid of both decorin mRNA and immunoreactivity. Identical results were obtained for cells of neuroendocrine tumors of human colon. Using RT-qPCR, we showed that human colon cancer cell lines are also decorin negative, in accordance with the above in vivo results. Finally, we demonstrate that decorin transduction of human colon cancer cell lines causes a significant reduction in their colony forming capability. Thus, strategies to develop decorin-based adjuvant therapies for human colorectal malignancies are highly rational.
Keywords: adenoviral transduction, antitumorigenic, colon cancer, decorin, neuroendocrine tumor
Introduction
Decorin, the prototypic member of the small leucine-rich proteoglycans (SLRPs), is known to be not only a key regulator of fibrillogenesis and matrix assembly (Brown and Vogel 1989; Dugan et al. 2006; Järveläinen et al. 2004; Reed and Iozzo 2002) but also an important modulator of various cellular functions, particularly adhesion, migration, proliferation and apoptosis (Bi et al. 2012; De Luca et al. 1996; Kinsella et al. 2000; Seidler et al. 2006; Van Bockstahl et al. 2014; Winnemöller et al. 1991; Yamaguchi and Ruoslahti 1988). Furthermore, decorin has an established association with the regulation of cell differentiation (Gasimli et al. 2013; Gasimli et al. 2014; Ma et al. 2014). Decorin has also been shown to play a role in inflammation; for example, via its ability to interact with toll-like receptors 2 and 4 (Buraschi et al. 2012; Merline et al. 2011). In addition, decorin has recently been shown to contribute to autophagy of endothelial cells via paternally expressed gene 3 (Peg3) in response to vascular endothelial growth factor receptor 2 (VEGFR2)-mediated activation of AMP-activated protein kinase (AMPK) (Buraschi et al. 2013; Goyal et al. 2014). Consequently, it is not surprising that decorin is centrally involved in several physiological and pathological processes, including tumorigenesis (Bi et al. 2012; Horváth et al. 2014; Iozzo and Schaefer 2010; Shi et al. 2014; Sofeu Feugaing et al. 2013).
Initially, decorin was associated with cancer when it was discovered that decorin/p53 double knockout mice developed tumors faster than their wild-type counterparts (Iozzo et al. 1999). The study also indicated that, although the disruption of the decorin gene did not lead to the spontaneous development of tumors, the lack of decorin was permissive for tumorigenesis (Bi et al. 2008; Iozzo et al. 1999). Thereafter, a great number of studies have focused on the antitumorigenic role of decorin in cancers (Goldoni and Iozzo 2008; Horvath et al. 2014; Ma et al. 2014; Moscatello et al. 1998; Sainio et al. 2013; Sainio and Järveläinen 2014; Sofeu Feugaing et al. 2013).
The expression of decorin has previously been shown to be reduced in colorectal cancer, one of the leading causes of cancer mortality worldwide (Augoff et al. 2008; Bi et al. 2008; Ferlay et al. 2008; Suhovskih et al. 2015). Decreased expression of decorin has also been reported in several other cancers, such as prostate and ovarian cancers (Banerjee et al. 2003; Shridhar et al. 2001). In addition, it has been demonstrated that the de novo expression of decorin suppresses the malignant phenotype of human colon cancer cells (Santra et al. 1995). In agreement with these findings, reduced expression of decorin in villous adenomas is associated with high malignant potency (Augoff et al. 2008). On the other hand, increased expression of decorin, for example, in hepatocellular carcinoma tissues, has also been described (Jia et al. 2012).
Even though decorin has been shown to reside in various amounts in the stroma of cancers, including colon cancer, its exact cellular origin in the stroma of different malignancies has not been definitely demonstrated. This is due to the fact that, in previous studies, particularly in studies on colon cancer, immunohistochemistry has been applied to study decorin expression in malignant tissues (Adany et al. 1990, Augoff et al. 2008). However, immunohistochemistry only allows the localization of decorin in tissues and does not inevitably demonstrate the cellular origin of decorin; i.e., which cells express decorin mRNA. Recently, using in situ hybridization for decorin, we showed that malignant cells of human breast and bladder cancers completely lack decorin expression both in vivo and in vitro (Boström et al. 2013; Sainio et al. 2013). Furthermore, we have demonstrated that the malignant behavior of decorin-negative human breast cancer and bladder cancer cells can be markedly modulated via decorin transduction (Boström et al. 2013; Sainio et al. 2013).
In this study, we first used the publicly available IST Online and Human Protein Atlas databases (http://ist.medisapiens.com/ and http://www.proteinatlas.org/) to assess the currently available information about decorin expression in various human colorectal carcinomas (Kilpinen et al. 2008; Uhlen et al. 2010). Next, using both in situ hybridization (ISH) and immunohistochemistry (IHC) for decorin, we investigated whether cells of human colorectal cancers or those forming neuroendocrine tumors or carcinomas within human colons expressed decorin. Decorin expression of different human colon cancer cell lines was also examined. Furthermore, because methylation of the decorin gene has previously been shown to be involved in the control of decorin expression in human colon cancer, the methylation status of the decorin gene promoter of human colon cancer cell lines was analyzed (Adany and Iozzo 1991). Finally, using soft agar colony formation assays, we explored whether adenovirus-mediated decorin transduction of human colon cancer cell lines influences their malignant behavior.
Materials & Methods
Data Analysis Using IST Online and Human Protein Atlas Databases
Two publicly available databases were used for preliminary screening regarding the current information about decorin gene expression and decorin immunoreactivity in normal and various malignant human colorectal tissue samples (Kilpinen et al. 2008; Uhlen et al. 2010). IST Online (http://ist.medisapiens.com/) is a publicly available database of the human transcriptome, and represents the largest, integrated and annotated human gene expression data source in the world (Kilpinen et al. 2008). In this study, the database was used to evaluate the gene expression of decorin gene in tissue samples representing healthy human colon and different carcinomas of the human colon as well as different colorectal cell lines (n=22). The database statistics included tissue samples of healthy colons (n=47), colorectal adenocarcinomas (n=32), colon carcinomas (n=251), colon adenocarcinomas (n=497), and mucinous colon adenocarcinomas (n=28). The expression of decorin was compared between benign and different malignant colon tissues. Also, all 22 colorectal cancer cell lines included in the database were examined for decorin expression.
The Human Protein Atlas portal (http://www.proteinatlas.org/) is a publicly available database with high-resolution images of immunohistochemically analyzed tissue specimens (Uhlen et al. 2010). The database provides information about the spatial distribution of different proteins in various tissues representing normal and malignant phenotypes. In this study, the database was used to evaluate decorin immunoreactivity in tissue samples representing healthy human colons (n=4), colon adenocarcinomas (n=10) and adenocarcinomas derived from the rectum (n=11).
Samples of Patients With Colonic Neoplasia
For decorin analyses, we used tumor tissue samples from eight patients with human colon adenocarcinoma, including five WHO grade II carcinomas and three WHO grade I carcinomas. In addition, we used tissue material from colonic adenomas from eight patients, including three tubulovillous adenomas with low grade dysplasia, one tubulovillous adenoma with high grade dysplasia, three tubular adenomas with low grade dysplasia, and one serrated adenoma with low grade dysplasia. Furthermore, we included tumor tissue samples from two patients with neuroendocrine tumor (WHO grade I) and one neuroendocrine carcinoma (WHO grade III). Three control samples were taken from normal colon area representing healthy colon. All samples were obtained from the archives of Turku University Hospital, Department of Pathology, and from Auria Biobank, Turku, Finland. The samples were fixed in 10% neutral-buffered formalin, embedded in paraffin and cut into 5-µm or 3.5-µm transverse sections. Sections were used for hematoxylin and eosin (HE) staining, in situ hybridization (ISH), and immunohistochemistry (IHC) analyses. The local ethics committee approved the use of the samples.
Human Colon Cancer Cell Lines
For in vitro studies, we used the following eight human colon cancer cell lines derived from the American Type Culture Collection (ATCC): CO115, HCT-116, DLD-1, HT-29, Vaco-5, LS180, SW620 and RKO. Cell lines CO115, DLD-1, HT-29, SW620 and LS180 represent human colorectal adenocarcinoma cells; RKO and Vaco-5 represent colonic carcinoma cells; and HCT-116 represents colorectal carcinoma cells. Of these eight cell lines, three have been graded as follows: SW620 and DLD-1 are graded Dukes type C and LS180 is graded Dukes type B. The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Paisley, Scotland) containing 10% fetal bovine serum (FBS; Biochrom AG, Berlin, Germany), penicillin (100 IU/ml) and streptomycin (100 µg/ml) (Sigma-Aldrich, St Louis, MO) at 37°C with 5% CO2.
Decorin In Situ Hybridization
Decorin ISH was performed on 5 µm tissue sections by probing them with human decorin antisense and sense single-stranded digoxigenin (DIG)-labeled RNA riboprobes, as previously described in detail (Salomäki et al. 2008). The dotslide (.slide) System (Soft imaging System, Olympus Company; Münster, Germany) was used to create images of representative tissue samples.
Immunohistochemistry
IHC for decorin was performed with a rabbit polyclonal antibody (H-80, Santa Cruz Inc., Dallas, TX; diluted 1:400) on all healthy, premalignant and malignant tissue sample sections, as previously described (Sainio et al. 2013). To localize neuroendocrine tumor cells, we used IHC for synaptophysin, a marker for neuroendocrine cells (Wiedenmann et al. 1986; Klimstra et al. 2010). IHC for synaptophysin was performed with rabbit monoclonal CONFIRM anti-synaptophysin antibody (SP11; Ventana Medical Systems, Inc. Tuscon, AZ). Immunohistochemistry was performed on 3.5-µm with BenchMark XT (Ventana/Roche) using ultra/VIEW Universal DAB Detection Kit (Ventana/Roche). As with ISH, the dotslide (.slide) System (Soft imaging System, An Olympus Company, Münster, Germany) was used to create images of representative tissue samples.
RT-qPCR for Decorin
Total RNA was extracted from the cultures of the aforementioned eight human colon cancer cell lines using a NucleoSpin RNA II Kit (Macherey-Nagel; Düren, Germany) according to the manufacturer’s protocol. The concentrations of the RNA samples were determined using a NanoDrop spectrophotometer (ThermoScientific; Waltham, MA). RT-qPCR analyses were performed as previously described (Boström et al. 2013).
Methylation Analyses of Decorin Gene Promoter
Total DNA was extracted from the cultures of randomly selected human colon cancer cell lines, namely DLD-1 and HCT-116, using QIAampH genomic DNA kit (QIAGEN GmbH; Hilden, Germany) following the manufacturer’s protocol. Total DNA extracted from human skin fibroblasts (HSFs) was used as a control. The methylation status of the decorin promoter region was determined using an automated MeDIP assay (Diagenode; Liège, Belgium) followed by RT-qPCR, as previously described in detail (Sainio et al. 2013).
Soft Agar Colony Formation Assay
Soft agar colony formation assays were performed in 6-well plates (Cellstar; Greiner Bio-One, GmbH, Frickenhausen, Germany) in three replicate cultures as follows. First, the wells were coated with a bottom layer (1 ml) of DMEM containing 0.5% agar (Bacto-agar, Difco; Detroit, MI), 10% FBS, penicillin (100 IU/ml), and streptomycin (100 μg/ml). After the bottom layer had solidified, 1 ml of a solution containing 60,000 cells in DMEM with 0.35% agar, 10% FBS, penicillin (100 IU/ml), and streptomycin (100 μg/ml) was added on top. The top layer was allowed to solidify and was then layered with DMEM (2 ml) containing 10% FBS, penicillin (100 IU/ml), and streptomycin (100 μg/ml). Cultures were maintained at 37°C with 5% CO2. The culture medium was replaced with fresh medium every four days. On day 8, the cultures were fixed with 4% paraformaldehyde and the number of developed colonies was counted using light microscopy. The number of colonies was determined by counting five fields of view in three layers in three replicate cultures.
Effect of Adenovirus-mediated Decorin Transduction on Colony Formation Capability of Colon Cancer Cells
Before soft agar assays, colon cancer cells of all eight cancer cell lines were transduced with a decorin adenovirus vector (Ad-DCN) and control vector (Ad-LacZ), as previously described (Boström et al. 2013). Briefly, the cells were maintained in DMEM containing 10% FBS, penicillin (100 IU/mL), and streptomycin (100 μg/mL) and grown at 37°C with 5% CO2. The cells were plated into 6-well plates (Cellstar, Greiner bio-one, Germany) with 500,000 cells per well. The next day, the cells were transduced with 10 pfu/cell of Ad-DCN or Ad-lacZ in DMEM containing 10% FBS. Non-transduced cells were used as a negative control. After 24 hr, the cells were trypsinized, counted, and used for soft agar colony formation assays, as described above. The number of colonies formed by transduced and non-transduced cells was counted as described above. Furthermore, the sizes of the colonies were measured manually from photographs of these same cultures. Three photographs were taken of each of the three replicates and the sizes of all colonies were measured. The average size of the colonies was also calculated.
Statistical Analysis
An unpaired Student’s t-test was employed for statistical analyses. A p-value <0.05 was considered statistically significant.
Results
Decorin Expression in Colorectal Cancer According to IST Online and Human Protein Atlas Databases
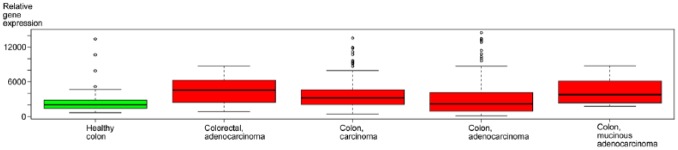
The analysis of the IST Online database showed that the overall decorin expression in previous studies has mostly been found to be upregulated in different human colon carcinoma tissue samples as compared with healthy colon samples (Fig. 1). However, of the 22 different colorectal cell lines included in the database, weak decorin expression was detected only in one cell line, namely, SW480 cells. The analysis of Human Protein Atlas database demonstrated that, in healthy colon samples, decorin immunoreactivity is localized to the cytoplasm of mesenchymal cells, whereas endothelial cells and glandular cells are decorin negative. Regarding decorin immunoreactivity in human colorectal carcinoma tissue samples, the results of Human Protein Atlas database are somewhat contradictory. Namely, in 8 of the 21 tissue samples, cancer cells were found to be variably positive for decorin immunoreactivity.
Figure 1.
Decorin expression in healthy and malignant human colorectal tissue samples, based on the IST Online database. Box plot analysis of relative decorin gene expression in healthy and malignant human colorectal cancer tissue samples based on IST Online in silico database (http://ist.medisapiens.com/). The continuous lines in the box plot images represent the median expression level of decorin in different colon tissues. Note that the relative decorin expression is increased in different carcinoma samples as compared with healthy samples. Capped bars in the box plot images indicate standard deviations of the results included in the databank.
Localization of Decorin mRNA and Immunoreactivity in Human Colorectal Cancer Tissues and in Neuroendocrine Tumors of Human Colon
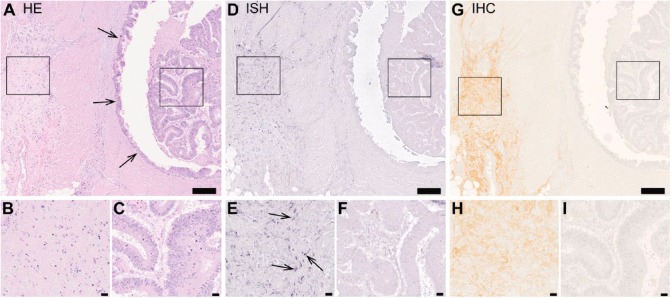
To localize decorin mRNA in human colorectal carcinoma tissue samples, we used ISH with DIG-labeled RNA probes for decorin. The ISH results indisputably demonstrated that cancer cells of human colorectal malignancies are devoid of decorin mRNA (Fig. 2D, 2F). This finding was also true for adenoma-forming cells of human colon (Fig. 3D, 3E), including cells of serrated adenoma (data not shown). Instead, decorin mRNA was localized merely to the original stromal areas in both adenocarcinoma (Fig. 2D, 2E) and adenoma (Fig. 3D, 3F) tissue samples of human colon. These results were verified by IHC analyses of the same tissue samples showing that decorin immunoreactivity and mRNA are overlapping (Figs. 2 and 3).
Figure 2.
Human colorectal adenocarcinoma cells do not express decorin. Images of a representative human colon adenocarcinoma tissue sample. Panel of serial sections of the same tissue sample representing hematoxylin and eosin staining (A–C), in situ hybridization (ISH) (D–F), and immunohistochemistry (G–I) for decorin. Left frames in A, D and G mark areas of non-malignant tissue and are magnified in B, E and H, respectively. Right frames in A, D and G mark areas of malignant adenocarcinoma tissue and are magnified in C, F and I, respectively. Arrows in A indicate the border between non-malignant and malignant tissues. In D and E, positive ISH signal for decorin can be seen in purple. Arrows in E indicate representative examples of positive ISH signals for decorin. In G and H, decorin immunoreactivity is visible in brown color and localizes to the non-malignant stromal areas of the colon tissue. Note that there is a complete deficiency of decorin mRNA expression and immunoreactivity in areas of adenocarcinoma tumor. Scale in A, D and G 200 µm; in B, C, E, F, H and I, 20 µm.
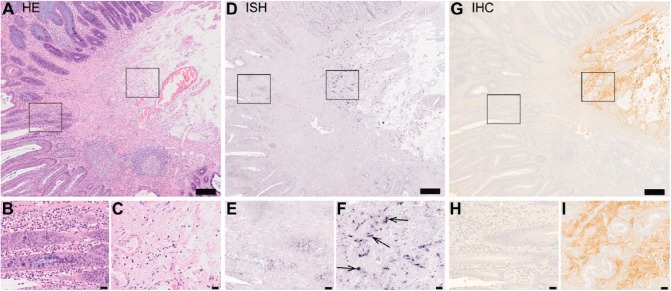
Figure 3.
Human colorectal adenoma cells are devoid of decorin expression. Images of a representative human colorectal adenoma tissue sample. Panel consists of images of serial sections of the same tissue sample representing hematoxylin and eosin staining (A-C), in situ hybridization (ISH) (D-F) and immunohistochemistry (G-I) for decorin. Left frames in A, D and G mark areas of basal adenoma tissue and are magnified in B, E and H, respectively. Right frames in A, D and G mark stroma tissue and are magnified in C, F and I, respectively. In D and F, positive ISH signal for decorin can be seen in purple. Arrows in F indicate representative examples of positive ISH signals for decorin. In G and H, decorin immunoreactivity can be seen in brown color and localizes to the stroma of the tissue. Note that decorin mRNA and decorin immunoreactivity can only be seen in the stroma and they are totally absent from adenoma cells. Scale in A, D and G, 200 µm; in B, C, E, F, H, and I, 20 µm.
Next, we localized decorin mRNA and immunoreactivity in human colon tissue samples representing low-grade (WHO grade I) neuroendocrine tumors and high-grade (WHO grade III) neuroendocrine carcinomas to examine whether decorin expression might also be lacking in cells of neuroendocrine gastrointestinal tumors (Fig. 4). Neuroendocrine tumors are a distinct subgroup of gastrointestinal tumors, and synaptophysin is one of the established immunohistochemical markers of these tumors (Klimstra et al. 2010; Wiedenmann et al. 1986). The results clearly showed that the cells of neuroendocrine tumors and carcinomas are negative for decorin mRNA and immunoreactivity (Fig. 4E–4H). As shown for colorectal adenocarcinoma and adenoma samples, decorin mRNA and immunoreactivity were detected solely in the original stromal areas surrounding the neuroendocrine tumor mass (Fig. 4E–4H).
Figure 4.
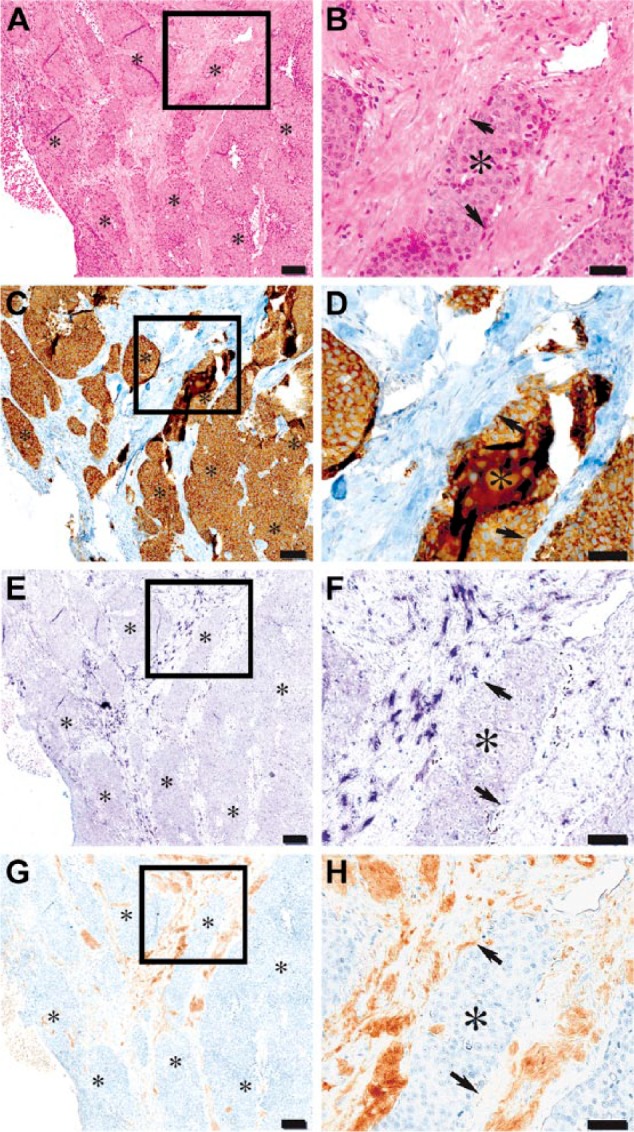
High-grade neuroendocrine carcinoma cells of human colon are decorin negative. Panel of serial sections of the same tissue sample representing hematoxylin and eosin staining (A, B), immunohistochemistry (IHC) for synaptophysin (C, D), and in situ hybridization (ISH) (E, F) and IHC (G, H) for decorin. Framed areas in A, C, E and G are shown magnified in B, D, F and H, respectively. Asterisks in all images indicate carcinoma areas and arrows in B, D, F and H point to the borders of the carcinoma. Synaptophysin staining for neuroendocrine cells can be seen in brown (C and D). Purple color indicates positive ISH signal for decorin (E and F) and decorin immunoreactivity can be seen in brown color (G and H). Note that neuroendocrine carcinoma cells of human colon completely lack decorin mRNA and immunoreactivity. Decorin expression is localized merely to the non-malignant stromal tissue areas surrounding neuroendocrine tumor masses. Scale in A, C, E and G, 100 µm; in B, D, F and H, 50 µm.
Expression of Decorin and Methylation of Decorin Gene Promoter in Human Colon Cancer Cell Lines
To confirm the above in vivo results that human colon cancer cells do not express decorin, eight different cell lines representing human colon cancer were examined for decorin expression using RT-qPCR analysis. The results showed that none of the human colon cancer cell lines used in this study expresses decorin.
We also determined the methylation status of the decorin gene promoter in representative human colon cancer cell lines DLD-1 and HCT-116. As shown in Fig. 5, the decorin gene promoter isoforms extracted from these human cancer cell lines were markedly methylated. As a control for decorin methylation, the decorin gene promoter of HSFs was not methylated (Fig. 5). These cells expressed significant amounts of decorin (data not shown).
Figure 5.
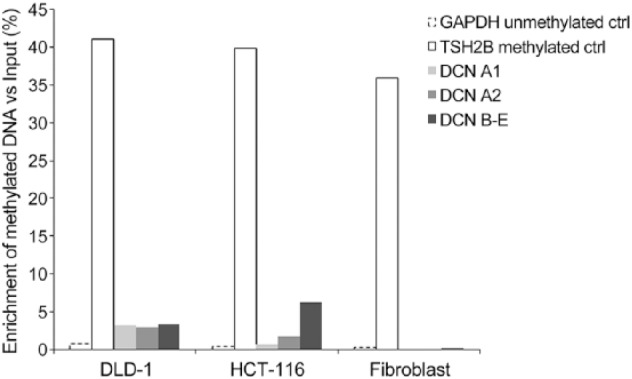
Decorin gene promoter is methylated in human colon cancer cell lines. Methylation status of decorin isoforms (DCN A1, A2, B–E) in human colon cancer cell lines (DLD-1 and HCT-116) was studied using the MeDIP assay. Total DNA isolated from human skin fibroblasts (HSFs) was used as a control. RT-qPCR results for MeDIP assay show the percentage of DNA methylation enrichment versus input DNA. In addition to decorin gene promoter isoforms, the results are shown for a positive control testis/sperm-specific histone 2B (TSH2B) and a negative control GAPDH promoter. Furthermore, the methylation status of the decorin gene promoter isoforms isolated from decorin expressing HSFs is included.
Effect of Decorin Transduction on Colony Formation in Human Colon Cancer Cell Lines in Soft Agar Assay
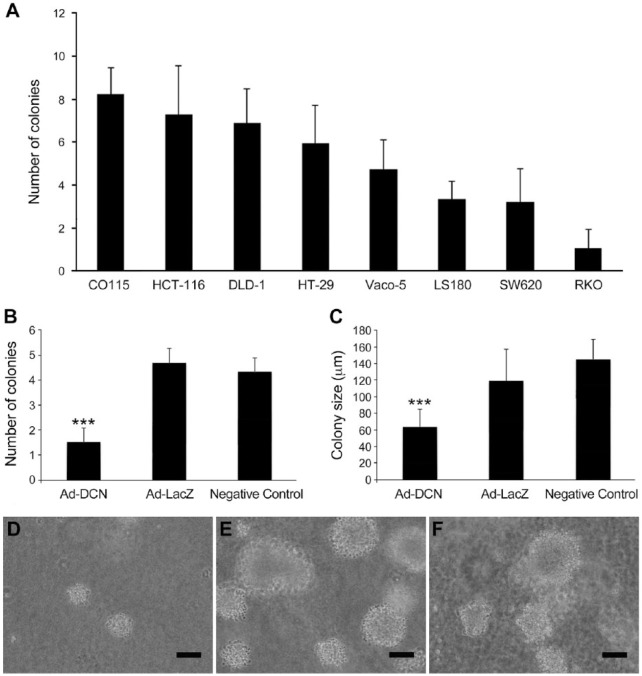
We used soft agar colony formation assays to examine whether decorin transduction might have an effect on the malignant behavior of human colon cancer cells. The assay was performed on eight human colon cancer cell lines, namely CO115, HCT-116, DLD-1, HT-29, Vaco-5, LS180, SW620 and RKO. The results showed a marked variation in the degree of colony formation among the cell lines, indicating that the capability of the cells to form colonies is cell line specific (Fig. 6A). Next, we chose five of the above cell lines, three of which showed high colony forming capability (HCT-116, DLD-1 and HT-29) and two with lower capability (Vaco-5 and LS180), and tested how adenovirus-mediated decorin transduction influences their capacity to form colonies. The above cell lines were chosen to examine whether the effect of decorin transduction on the colon cancer cells is similar independent of their colony forming capability. The results showed that decorin transduction drastically reduces the ability of all tested cell lines to form colonies. Both the number and the size of the colonies were significantly decreased in response to decorin transduction as compared with colonies formed by cells transduced with a control vector or non-transduced cells. The results for the cell line HCT-116 are shown in Fig. 6B–6F. In contrast, LacZ-transduced and negative control cells did not show any difference in their colony forming capability (Fig. 6B, 6C).
Figure 6.
Decorin transduction decreases colony formation of human colon cancer cell lines in soft agar assay. (A) Histogram showing colony formation of eight human colon carcinoma cell lines grown in soft agar. The columns represent the mean number of colonies in three replicate cultures. Note that the ability of cells to form colonies seems to be cell line specific. (B) Number of colonies formed by the HCT-116 cell line after transduction with decorin adenoviral vector (Ad-DCN) or control vector (Ad-LacZ). The negative control represents the number of colonies formed by non-transduced cells. Each column represents the mean of three replicate cultures. (C) Size of colonies formed by HCT-116 cell line after transduction with the decorin Ad-DCN or Ad-LacZ. The negative control represents the number of colonies formed by non-transduced HCT-116 cells. Each column represents the mean of three replicate cultures. Representative images of colonies formed by HCT-116 cells transduced with Ad-DCN (D) or Ad-LacZ (E) and by non-transduced cells (F). Note that adenovirus-mediated decorin transduction significantly inhibits the capability of HCT-116 cells to form colonies; both the number and size of the colonies formed by the cells are decreased in response to decorin transduction (B–F). Identical results were obtained with all five cell lines tested (see the text in Results). Capped bars on the top of all columns (A–C) mark standard deviations. ***p<0.001, Student’s t-test. Scale in D–F, 100 µm.
Discussion
The role of decorin in carcinogenesis, including colon carcinoma, has been the focus of numerous studies (Augoff et al. 2008; Bi et al. 2012; Bozoky et al. 2014; Goldoni and Iozzo 2008; Horváth et al. 2014; Santra et al. 1995; Suhovskih et al. 2015). Regarding colon carcinoma, the expression of decorin has previously been shown to be variously either increased or decreased (Adany et al. 1990; Augoff et al. 2008; Bi et al. 2008; Santra et al. 1995; Theocharis 2002). However, so far, the exact cellular origin of decorin in premalignant and malignant colorectal tissue stroma has remained partially unresolved due to the fact that immunohistochemistry and microarray analyses have been used to examine decorin expression in colorectal tissue samples.
In the present study, one of our aims was to clarify whether decorin residing in the stroma of malignant colorectal tissues is solely derived from stromal cells or whether it might also be derived from cancer cells. Using two publicly available databases, namely IST Online and Human Protein Atlas databases, we first screened the data with regard to the current knowledge about the overall decorin expression in normal, premalignant, and malignant colorectal tissues (Kilpinen et al. 2008; Uhlen et al. 2010). The IST Online database was also used to ascertain decorin expression in different colorectal cancer cell lines. Our analyses demonstrated that a marked variation in decorin expression has been observed to take place in human colorectal tissue samples. Furthermore, the databases revealed that decorin expression, particularly its immunoreactivity, can occasionally be detected also in colon cancer cells. These previously published results are somewhat contradictory with our present results that demonstrate that malignant cells within human colorectal cancer tissues are devoid of both decorin mRNA and immunoreactivity. This was true for all colon tissue samples used in our study ranging from adenoma with mild dysplasia to high-grade adenocarcinomas. The contradiction can be a consequence of various issues, including differences in tissue samples used in the studies as well as differences in the techniques and antibodies employed. The IST Online database, on the other hand, is based on gene expression array data from tissue samples consisting of a heterogeneous group of cells and, as such, it does not specify which cell types express the decorin gene. The Human Protein Atlas database is, in turn, based on IHC that is restricted; e.g., in its specificity. In our study, besides using IHC, we have also used ISH, which is a precise method for detecting cells that express mRNA for decorin. Notably, in our study, the results obtained with ISH and IHC analyses were parallel. Recently, we have utilized this same approach in human breast cancer and bladder cancer tissue samples and shown that different types of human breast cancer cells and urinary bladder cancer cells also completely lack decorin mRNA and immunoreactivity (Boström et al. 2013; Sainio et al. 2013). In the present study, we have also demonstrated that cells forming neuroendocrine tumors (WHO grade I) and high-grade (WHO grade III) neuroendocrine carcinomas in human colon are negative for decorin mRNA and immunoreactivity. Neuroendocrine tumors of the colon are extremely uncommon comprising less than 1% of all colorectal cancers (Bernick et al. 2004). To the best of our knowledge, our study is the first to examine decorin gene expression in neuroendocrine tumors and carcinomas of the human colon.
After testing for the localization of decorin expression in vivo, we then examined decorin expression in vitro in eight human colon cancer cell lines using RT-qPCR analysis. Our results revealed that none of the examined cell lines expresses decorin; thus confirming our in vivo results. Previously, epigenetic mechanisms and, particularly, hypomethylation of the decorin gene have been shown to be associated with increased decorin gene activation in human colon carcinoma (Adany et al. 1990; Adany and Iozzo 1991). This led us to explore the methylation status of the decorin gene promoter in human colon cancer cell lines. We investigated whether the total absence of decorin expression in colon cancer cells could in fact be due to methylation of the decorin gene promoter. Indeed, our results showed that the promoter of the decorin gene is markedly methylated in the human colon cancer cell lines analyzed in this study. In contrast, in HSFs, which express abundant amounts of decorin, no methylation of the decorin gene promoter was detected. These results are in full agreement with the study by Adany and collaborators, who demonstrated that the decorin gene in colon carcinoma cells is not hypomethylated (Adany et al. 1990). Moreover, also in agreement with our study, the same study showed that decorin gene is hypomethylated in fibroblasts that are known to express abundant decorin. Furthermore, increased decorin mRNA levels in colon carcinoma tissues correlated with hypomethylation (Adany et al. 1990). Interestingly, we have previously shown that the decorin gene promoter is not methylated in human urinary bladder cancer cells, which also lack decorin expression, suggesting that different epigenetic mechanisms contribute to the regulation of decorin gene expression in various types of human malignancies (Sainio et al. 2013).
To examine whether decorin expression could influence the behavior of human colon cancer cells, we transduced the cells with a decorin adenoviral vector. Results from our previous studies have shown that decorin transduction into human breast cancer and bladder cancer cell lines leads to decreased proliferation of the cells (Boström et al. 2013; Sainio et al. 2013). Moreover, decorin transduction increases apoptosis in breast cancer cells (Boström et al. 2013). In line with our previous results, in the present study, we demonstrated decreased colony forming capability of the cells in response to decorin transduction. Decorin transduction reduced both the number and the size of colonies. Thus, decorin transduction alters the behavior of human colon cancer cells in vitro, further suggesting that decorin transduction could decrease cancer progression (Boström et al. 2013; Sainio et al. 2013; Santra et al. 1995; Sofeu Feugaing et al. 2013).
Several studies have shown the antitumoral potential of decorin in the intestinal tract (Bi et al. 2012; Reed et al. 2002; Santra et al. 1995). Indeed, decorin appears to act as a tumor suppressor molecule in mice, because inactivation of its gene can lead to intestinal tumor formation via disruption of intestinal maturation; this results in, for example, decreased differentiation of the cells (Bi et al. 2008). In addition, adenovirus-mediated decorin gene transfer has been shown to significantly inhibit the growth of colon carcinoma xenografts in mice via attenuation of epidermal growth factor receptor (EGFR) phosphorylation (Reed et al. 2002). Increased decorin expression has also been shown to attenuate the migration of colorectal cancer cells and to promote their apoptosis via the regulation of E-cadherin (Bi et al. 2012). Besides, in colon cancer, increased apoptosis after decorin treatment has been observed in other cancer types, such as in cholangiocarcinoma (Yu et al. 2014), hepatocellular carcinoma (Hamid et al. 2013), and squamous carcinoma (Seidler et al. 2006). The apoptosis promoting effect of decorin in human hepatocellular and squamous carcinoma cells has been shown to be mediated via activation of caspase-3 (Hamid et al. 2013; Seidler et al. 2006).
The effect of decorin in colon cancer cells can also be discussed from the perspective of its glycosaminoglycan (GAG) chain composition. In human colon adenocarcinoma, the GAG chains of decorin molecules are significantly modified; i.e., the GAG chains are of a smaller size and their sulfation pattern is altered so that they are mainly composed of 6-sulfated disaccharides (Theocharis 2002). These structural changes can markedly influence the biology of cancer cells via mechanisms including alterations in growth factor binding and cell signaling (Deepa et al. 2002; Weyers et al. 2012). On the other hand, regarding viral transduction of cancer cells, it also has to be acknowledged that cells that do not normally express a certain molecule do not necessarily have the capacity to set the required synthetic machinery in motion. Finally, other mechanisms, such as the capability of decorin to modulate the three-dimensional structure of the extracellular matrix, may also be behind decorin’s action as a tumor suppressing molecule (Bonnans et al. 2014; Järveläinen et al. 2004; Van Bockstal et al. 2014).
In sum, in this study, we have shown that human colon cancer cells are devoid of decorin expression in vivo and in vitro and that adenovirus-mediated decorin transduction of human colon cancer cell lines attenuates their malignant behavior towards a more benign demeanor. Thus, in the future, development of decorin-based adjuvant therapies in the treatment of colorectal malignancies is highly rational.
Acknowledgments
We would like to express our gratitude to Jaakko Liippo for his help with the images. We also thank the Finnish Microarray and Sequencing Centre at Turku Centre for Biotechnology, Turku, Finland for technical support with epigenetic analysis.
Footnotes
Declaration of Competing Interests: The authors declared no potential competing interests with respect to the research, authorship, and/or publication of this article.
Author Contributions: HJ conceived and designed the experiments. MCN, AOS, MMP, RJL and SV carried out the experiments. MCN, AOS, RJL, SV, JTTS and HTJ analyzed the data. MCN, AOS and HTJ wrote the manuscript.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Biocenter Finland, Academy of Finland, Medical Research fund (EVO) of Turku University Hospital, The Swedish Cultural Foundation in Finland, K. Albin Johansson Foundation, Cancer Foundations of South Western Finland and University of Turku.
References
- Adany R, Heimer R, Caterson B, Sorrel JM, Iozzo RV. (1990). Altered expression of chondroitin sulfate proteoglycan in the stroma of human colon carcinoma. J Biol Chem 265:11389-11396. [PubMed] [Google Scholar]
- Adany R, Iozzo RV. (1991). Hypomethylation of the decorin proteoglycan gene in human colon cancer. Biochem J 276:301-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augoff K, Rabczynski J, Tabola R, Czapla L, Ratajczak K, Grabowski K. (2008). Immunohistochemical study of decorin expression in polyps and carcinomas of the colon. Med Sci Monit 14:CR530-535. [PubMed] [Google Scholar]
- Banerjee AG, Liu J, Yuan Y, Gopalakrishnan VK, Johansson SL, Dinda AK, Gupta NP, Trevino L, Vishwanatha JK. (2003). Expression of biomarkers modulating prostate cancer angiogenesis: differential expression of annexin II in prostata carcinomas from India and USA. Mol Cancer 2:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernick PE, Klimstra DS, Shia J, Minsky B, Salz L, Shi W, Thaler H, Guillem J, Paty P, Cohen AM, Wong WD. (2004). Neuroendocrine carcinomas of the colon and rectum. Dis Colon Rectum 47:163-169. [DOI] [PubMed] [Google Scholar]
- Bi X, Pohl NM, Qian Z, Yang GR, Gou Y, Guzman G, Kajdacsy-Balla A, Iozzo RV, Yang W. (2012). Decorin-mediated inhibition of colorectal cancer growth and migration is associated with E-cadherin in vitro and in mice. Carcinogenesis 33:326-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Tong C, Dockendorff A, Bancroft L, Gallagher L, Guzman G, Iozzo RV, Augenlicht LH, Yang W. (2008). Genetic deficiency of decorin causes intestinal tumor formation through disruption of intestinal cell maturation. Carcinogenesis 29:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnans C, Chou J, Werb Z. (2014). Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol 15:786-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boström P, Sainio A, Kakko T, Savontaus M, Söderström M, Järveläinen H. (2013). Localization of decorin gene expression in normal human breast tissue and in benign and malignant tumors of the human breast. Histochem. Cell Biol 139:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozoky B, Savchenko A, Guven H, Ponten F, Klein G, Szekely L. (2014). Decreased decorin expression in the tumor microenvironment. Cancer Med 3:485-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DC, Vogel KG. (1989). Characteristics of the in vitro interaction of a small proteoglycan (PG II) of bovine tendon with type I collagen. Matrix 9:468-478. [DOI] [PubMed] [Google Scholar]
- Buraschi S, Neill T, Owens RT, Iniguez LA, Purkins G, Vadigepalli R, Evans B, Schaefer L, Peiper SC, Wang ZX, Iozzo RV. (2012). Decorin protein core affects the global gene expression profile of the tumor microenvironment in a triple-negative orthotopic breast carcinoma xenograft model. PLoS One 7:e45559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buraschi S, Neill T, Goyal A, Poluzzi C, Smythies J, Owens RT, Schaefer L, Torres A, Iozzo RV. (2013). Decorin causes autophagy in endothelial cells via Peg3. Proc Natl Acad Sci U S A 110:E2582-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepa SS, Umehara Y, Higashiyama S, Itoh N, Sugahara K. (1992). Specific molecular interactions of oversulfated chondroitin sulfate E with various heparin-binding growth factors. Implications as a physiological binding partner in the brain and other tissues. J Biol Chem 277:43707-43716. [DOI] [PubMed] [Google Scholar]
- De Luca A, Santra M, Baldi A, Giordano A, Iozzo RV. (1996). Decorin-induced Growth Suppression Is Associated with Up-regulation of p21, an Inhibitor of Cyclin-dependent Kinases. J Biol Chem 271:18961-18965. [DOI] [PubMed] [Google Scholar]
- Dugan TA, Yang VW, McQuillan DJ, Höök M. (2006). Decorin modulated fibrin assembly and structure. J Biol Chem 281:38208-38216. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. (2008). Estimates of worldwide burden of cancer in 2008. Int J Cancer 127:2893-2917. [DOI] [PubMed] [Google Scholar]
- Gasimli L, Hickey AM, Yang B, Li G, dela Rosa M, Nairn AV, Kulik MJ, Dordick JS, Moremen KW, Dalton S, Linhardt RJ. (2014). Changes in glycosaminoglycan structure on differentiation of human embryonic stem cells towards mesoderm and endoderm lineages. Biochim Biophys Acta 840:1993-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasimli L, Stansfield HE, Nairn AV, Liu H, Paluh JL, Yang B, Dordick JS, Moremen KW, Linhardt RJ. (2013). Structural remodeling of proteoglycans upon retinoic acid-induced differentiation of NCCIT cells. Glycoconj J 30:497-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldoni S, Iozzo RV. (2008). Tumor microenvironment: Modulation by decorin and related molecules harboring leucine-rich tandem motifs. Int J Cancer 123:2473-2479. [DOI] [PubMed] [Google Scholar]
- Goyal A, Neill T, Owens RT, Schaefer L, Iozzo RV. (2014). Reprint of: Decorin activates AMPK, an energy sensor kinase, to induce autophagy in endothelial cells. Matrix Biol 35:42-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid AS, Li J, Wang Y, Wu X, Ali HA, Du Z, Bo L, Zhang Y, Zhang G. (2013). Recombinant human decorin upregulates p57KIP2 expression in HepG2 hepatoma cell lines. Mol Med Rep 8:511-516. [DOI] [PubMed] [Google Scholar]
- Horváth Z, Kovalszky I, Fullár A, Kiss K, Schaff Z, Iozzo RV, Baghy K. (2014). Decorin deficiency promotes hepatic carcinogenesis. Matrix Biology 35:194-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo RV, Chakrani F, Perrotti D, McQuillan DJ, Skorski T, Calabretta B, Eichstetter I. (1999). Cooperative action of germ-line mutations in decorin and p53 accelerates lymphoma tumorigenesis. Proc Natl Acad Sci U S A 96:3092-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo RV, Schaefer L. (2010). Proteoglycans in health and disease: novel regulatory signaling mechanisms evoked by the small leucine-rich proteoglycans. FEBS J 277:3864-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia XL, Li SY, Dang SS, Cheng YA, Zhang X, Wang WJ, Hughes CE, Caterson B. (2012). Increased expression of chondroitin sulphate proteoglycans in rat hepatocellular carcinoma tissues. World J Gastroenterol 18:3962-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järveläinen H, Vernon RB, Gooden MD, Francki A, Lara S, Johnson PY, Kinsella MG, Sage EH, Wight TN. (2004). Overexpression of decorin by rat arterial smooth muscle cells enhances contraction of type I collagen in vitro. Arterioscler Tromb Vasc Biol 24:76-72. [DOI] [PubMed] [Google Scholar]
- Kilpinen S, Autio R, Ojala K, Iljin K, Bucher E, Sara H, Pisto T, Saarela M, Skotheim RI, Björkman M, Mpindi JP, Haapa-Paananen S, Vainio P, Edgren H, Wolf M, Astola J, Nees M, Hautaniemi S, Kallioniemi O. (2008). Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol 9:R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella MG, Fischer JW, Mason DP, Wight TN. (2000). Retrovirally mediated expression of decorin by macrovascular endothelial cells. Effects on cellular migration and fibronectin fibrillogenesis in vitro. J Biol Chem 275:13924-13932. [DOI] [PubMed] [Google Scholar]
- Klimstra DS, Modlin IR, Adsay NV, Chetty R, Deshpande V, Gönen M, Jensen RT, Kidd M, Kulke MH, Lloyd RV, Moran C, Moss SF, Oberg K, O’Toole D, Rindi G, Robert ME, Suster S, Tang LH, Tzen CY, Washington MK, Wiedenmann B, Yao J. (2010). Pathology reporting of neuroendocrine tumors: application of the Delphic consensus process to the development of a minimum pathology data set. Am J Surg Pathol 34:300-313. [DOI] [PubMed] [Google Scholar]
- Ma HI, Hueng DY, Shui HA, Han JM, Wang CH, Lai YH, Cheng SY, Xiao X, Chen MT, Yang YP. (2014). Intratumoral decorin gene delivery by AAV vector inhibits brain glioblastomas and prolongs survival of animals by inducing cell differentiation. Int J Mol Sci 15:4393-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merline R, Moreth K, Beckmann J, Nastase MV, Zeng-Brouwers J, Tralhão JG, Lemarchand P, Pfeilschifter J, Schaefer RM, Iozzo RV, Schaefer L. (2011). Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and microRNA-21. Sci Signal 4:ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscatello DK, Santra M, Mann DM, McQuillan DJ, Wong AJ, Iozzo RV. (1998). Decorin suppresses tumor cell growth by activating the epidermal growth factor receptor. J Clin Invest 101:406-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed CC, Gauldie J, Iozzo RV. (2002). Suppression of tumorigenecity by adenovirus-mediated gene transfer of decorin. Oncogene 21:3688-3695. [DOI] [PubMed] [Google Scholar]
- Reed CC, Iozzo RV. (2002). The role of decorin in collagen fibrillogenesis and skin homeostasis. Glycoconj J 19:249-255. [DOI] [PubMed] [Google Scholar]
- Sainio A, Järveläinen H. (2014). Extracellular matrix macromolecules: potential tools and targets in cancer gene therapy. Mol Cell Ther 2:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainio A, Nyman M, Lund R, Vuorikoski S, Boström P, Laato M, Boström PJ, Järveläinen H. (2013). Lack of decorin expression by human bladder cancer cells offers new tools in the therapy of urothelial malignancies. Plos One 8:e 76190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomäki HH, Sainio AO, Söderström M, Pakkanen S, Laine J, Järveläinen HT. (2008). Differential expression of decorin by human malignant and benign vascular tumors. J Histochem Cytochem 56:639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra M, Skorski T, Calabretta B, Lattime EC, Iozzo RV. (1995). De novo decorin gene expression suppresses the malignant phenotype in human colon cancer cells. Proc Natl Acad Sci U S A 92:7016-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler DG, Goldoni S, Agnew C, Cardi C, Thakur ML, Owens RT, McQuillan DJ, Iozzo RV. (2006). Decorin protein core inhibits in vivo cancer growth and metabolism by hindering epidermal growth factor receptor function and triggering apoptosis via caspase-3 activation. J Biol Chem 281:26408-26418. [DOI] [PubMed] [Google Scholar]
- Shi X, Liang W, Yang W, Xia R, Song Y. (2014). Decorin is responsible for progression of non-small-cell lung cancer by promoting cell proliferation and metastasis. Tumour Biol 36:3345-3354. [DOI] [PubMed] [Google Scholar]
- Shridhar V, Lee J, Pandita A, Iturria S, Avula R, Staub J, Morrissey M, Calhoun E, Sen A, Kalli K, Keeney G, Roche P, Cliby W, Lu K, Schmandt R, Mills GB, Bast RC, Jr, James CD, Couch FJ, Hartmann LC, Lillie J, Smith DI. (2001). Genetic analysis of early-versus late stage ovarian tumors. Cancer Res 61:5895-5904. [PubMed] [Google Scholar]
- Sofeu Feugaing DD, Götte M, Viola M. (2013). More than matrix: the multifaceted role of decorin in cancer. Eur J Cell Biol 92:1-11. [DOI] [PubMed] [Google Scholar]
- Suhovskih AV, Aidagulova SV, Kashuba VI, Grigorieva EV. (2015). Proteoglycans as potential microenvironmental biomarkers for colon cancer. Cell Tissue Res doi: 10.1007/s00441-015-2141-8. [DOI] [PubMed] [Google Scholar]
- Theocharis AD. (2002). Human colon adenocarcinoma is associated with specific post-translational modifications of versican and decorin. Biochim Biophys Acta 1588:165-172. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, Ponten F. (2010). Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28:1248-1250. [DOI] [PubMed] [Google Scholar]
- Van Bockstal M, Lambein K, Van Gele M, De Vlieghere E, Limame R, Braems G, Van den Broecke R, Cocquyt V, Denys H, Bracke M, Libbrecht L, De Wever O. (2014). Differential regulation of extracellular matrix protein expression in carcinoma-associated fibroblasts by TGF-β1 regulates cancer cell spreading but not adhesion. Oncoscience 15:634-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyers A, Yang B, Yoon DS, Park J-H, Zhang F, Lee KB, Linhardt RJ. (2012). A structural analysis of glycosaminoglycans from lethal and nonlethal breast cancer tissues: Toward a novel class of theragnosctics for personalized medicine in oncology? OMICS 16:79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenmann B, Franke W, Kuhn C, Moll R, Gould VE. (1986). Synaptophysin: A marker protein for neuroendocrine cells and neoplasms. Proc Natl Acad Sci U S A 83:3500-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnemöller M, Schmidt G, Kresse H. (1991). Influence of decorin on fibroblast adhesion to fibronectin. Eur J Cell Biol 54:10-17. [PubMed] [Google Scholar]
- Yamaguchi Y, Ruoslahti E. (1988). Expression of human proteoglycan in Chinese hamster ovary cells inhibits cell proliferation. Nature 336:244-246. [DOI] [PubMed] [Google Scholar]
- Yu X, Zou Y, Li Q, Mao Y, Zhu H, Huang G, Ji G, Luo X, Yu C, Zhang X. (2014). Decorin-mediated inhibition of cholangiocarcinoma cell growth and migration and promotion of apoptosis are associated with E-cadherin in vitro. Tumour Biol 35:3103-3112. [DOI] [PubMed] [Google Scholar]