Highlights
-
•
Oxidation and reduction of aristolochic acid I (AAI) dictate its (geno)toxicity in vivo.
-
•
Cytochrome P450 (CYP) 1A1 and 1A2 are induced in rats treated with Sudan I and AAI.
-
•
Induced CYP1A enzyme activity resulted in decreased AAI-DNA adduct levels in vivo.
-
•
CYP1A1 and 1A2 mainly detoxify AAI and attenuate its genotoxicity in vivo.
Abbreviations: AA, aristolochic acid; AAI, aristolochic acid I; AAII, aristolochic acid II; AAIa, aristolochic acid Ia; AAN, aristolochic acid nephropathy; BEN, Balkan endemic nephropathy; bw, body weight; cT, cycle threshold; CYP, cytochrome P450; dA-AAI, 7-deoxyadenosine-N6-yl)aristolactam I; dA-AAII, 7-deoxyadenosine-N6-yl)aristolactam II; dG-AAI, 7-deoxyguanosin-N2-yl)aristolactam I; HPLC, high performance liquid chromatography; HUFs, Hupki (human TP53 knock-in) mouse fibroblasts; MROD, methoxyresorufin O-demethylation; NQO1, NAD(P)H:quinone oxidoreductase; POR, P450 oxidoreductase; PEI-cellulose, polyethylenimine-cellulose; RAL, relative adduct labeling; RT-PCR, real-time polymerase chain reaction; r.t., retention time; SD, standard deviation; TLC, thin-layer chromatography; UUC, upper urothelial tract carcinoma; UV–vis, ultraviolet–visible
Keywords: Aristolochic acid I, Cytochromes P450 1A1 and 1A2, Oxidative detoxification, Reductive activation, DNA adducts
Abstract
Aristolochic acid I (AAI) is a natural plant alkaloid causing aristolochic acid nephropathy, Balkan endemic nephropathy and their associated urothelial malignancies. One of the most efficient enzymes reductively activating AAI to species forming AAI-DNA adducts is cytosolic NAD(P)H:quinone oxidoreductase 1. AAI is also either reductively activated or oxidatively detoxified to 8-hydroxyaristolochic acid (AAIa) by microsomal cytochrome P450 (CYP) 1A1 and 1A2. Here, we investigated which of these two opposing CYP1A1/2-catalyzed reactions prevails in AAI metabolism in vivo. The formation of AAI-DNA adducts was analyzed in liver, kidney and lung of rats treated with AAI, Sudan I, a potent inducer of CYP1A1/2, or AAI after pretreatment with Sudan I. Compared to rats treated with AAI alone, levels of AAI-DNA adducts determined by the 32P-postlabeling method were lower in liver, kidney and lung of rats treated with AAI after Sudan I. The induction of CYP1A1/2 by Sudan I increased AAI detoxification to its O-demethylated metabolite AAIa, thereby reducing the actual amount of AAI available for reductive activation. This subsequently resulted in lower AAI-DNA adduct levels in the rat in vivo. Our results demonstrate that CYP1A1/2-mediated oxidative detoxification of AAI is the predominant role of these enzymes in rats in vivo, thereby suppressing levels of AAI-DNA adducts.
1. Introduction
Aristolochic acid (AA) is a herbal drug prepared from plants of the Aristolochia genus containing nitrophenanthrene carboxylic acids, of which 8-methoxy-6-nitro-phenanthro-(3,4-d)-1,3-dioxolo-5-carboxylic acid (aristolochic acid I, AAI) (Fig. 1) and 6-nitro-phenanthro-(3,4-d)-1,3-dioxolo-5-carboxylic acid (AAII) are the predominant components (Arlt et al., 2002b). Over twenty years ago, AA was shown to be the cause of a unique renal disease formerly called Chinese herbs nephropathy, now referred to as aristolochic acid nephropathy (AAN) (for a review, see Arlt et al., 2002b, Schmeiser et al., 2009, Gökmen et al., 2013). AAN is a rapidly progressive renal fibrosis with a high risk for upper urothelial tract carcinoma (UUC) and, subsequently, bladder urothelial carcinoma (Vanherweghem et al., 1993, Nortier et al., 2000, Arlt et al., 2002b, Yun et al., 2012, Gökmen et al., 2013). AA has been classified as a Group I carcinogen by IARC (IARC, 2012). Exposure to AA has also been found to be the cause of a similar type of renal disease, Balkan endemic nephropathy (BEN) and its associated occurrence of urothelial malignancy (Arlt et al., 2007, Grollman et al., 2007). This disease is endemic in certain rural areas of Balkan countries near the tributaries of the Danube river (Schmeiser et al., 2012).
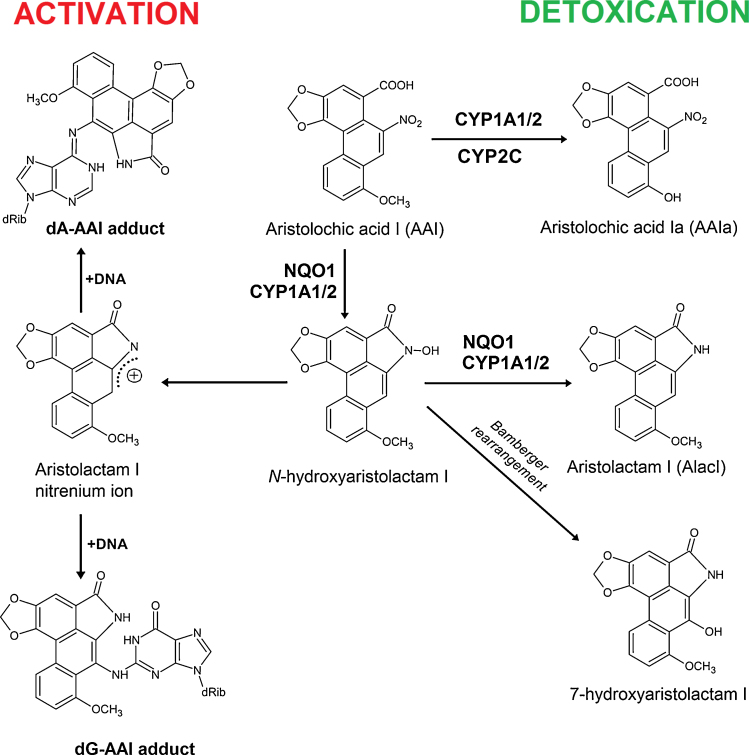
Fig. 1.
Activation and detoxification pathways of AAI. dA-AAI, 7-(deoxyadenosin-N6-yl) aristolactam I; dG-AAI, 7-(deoxyguanosin-N2-yl) aristolactam I; CYP1A1/2, cytochrome P450 1A1 and 1A2; CYP2C9, cytochrome P450 2C9; CYP3A4, cytochrome P450 3A4; NQO1, NAD(P)H:quinone oxidoreductase.
Characteristic AA-DNA adducts in renal tissue of AAN and BEN patients are biomarkers of exposure to AA even long after AA exposure, the 7-(deoxyadenosin-N6-yl) aristolactam I (dA-AAI) adduct being the most abundant adduct formed and the most persistent (Nortier et al., 2000, Arlt et al., 2002a, Arlt et al., 2002b, Schmeiser et al., 2012, Schmeiser et al., 2014). This deoxyadenosine adduct causes characteristic A–T transversion mutations and such mutations were found in the TP53 tumour suppressor gene in tumors from AAN and BEN patients (Lord et al., 2004, Grollman et al., 2007) and in immortalized Hupki (human TP53 knock-in) mouse fibroblasts (HUFs) exposed to AAI (Nedelko et al., 2009). This feature indicates a molecular mechanism of AA-mediated carcinogenesis (Arlt et al., 2007, Kucab et al., 2010). More recently, these A–T transversion mutations were also observed in loci of other genes by whole-genome and exome sequencing analyzing AA-associated UUC and AAI-treated HUFs (Poon et al., 2013, Hoang et al., 2013, Nik-Zainal et al., 2015).
Nitro-reduction of AAI, the compound that is considered as the major cause for AA-mediated development of AAN and BEN, is required to exert its carcinogenic properties (i.e. UUC development) (Schmeiser et al., 1996, Schmeiser et al., 2009, Arlt et al., 2002b, Stiborová et al., 2008a, Gökmen et al., 2013). Such nitro-reduction leads to the formation of N-hydroxylated aristolactam I which either converts to a reactive cyclic acylnitrenium ion generating DNA adducts or rearranges to 7-hydroxyaristolactam I (Schmeiser et al., 2009). The product of AAI oxidation, 8-hydroxyaristolochic acid I (aristolochic acid Ia, AAIa), is formed by O-demethylation of the methoxy group (Fig. 1), and is a detoxification product of this carcinogen. AAIa is excreted either in its free form or conjugated (Chan et al., 2006, Shibutani et al., 2010, Arlt et al., 2011, Stiborová et al., 2012).
Various enzymes are involved in the metabolism of AAI. A variety of studies by us and others have shown that the cytosolic nitroreductase, NAD(P)H:quinone oxidoreductase (NQO1), is the most efficient enzyme activating AAI to DNA adducts (Stiborová et al., 2002a, Stiborová et al., 2003, Stiborová et al., 2008a, Stiborová et al., 2008b, Stiborová et al., 2011a, Stiborová et al., 2013b, Stiborová et al., 2008a, Stiborová et al., 2008b, Martinek et al., 2011, Chen et al., 2011). In human and rodent hepatic microsomes AAI is also activated by cytochrome P450 (CYP) 1A2 and, to a lesser extent, by CYP1A1 and NADPH:cytochrome P450 oxidoreductase (POR) (Stiborová et al., 2001, Stiborová et al., 2005a, Stiborová et al., 2005b, Stiborová et al., 2005a, Stiborová et al., 2005b, Stiborová et al., 2011b, Stiborová et al., 2012, Stiborová et al., 2013b, Stiborová et al., 2005a, Stiborová et al., 2005b, Arlt et al., 2011, Arlt et al., 2015, Levová et al., 2011, Levová et al., 2012, Jerabek et al., 2012) (Fig. 1). However, human and rodent CYP1A1 and 1A2 play a dual role in the metabolism of AAI. Under anaerobic conditions they reductively activate AAI, while under oxidative conditions they are the predominant enzymes catalyzing O-demethylation of AAI to AAIa (i.e. detoxication) (Stiborová et al., 2001, Stiborová et al., 2005a, Stiborová et al., 2005b, Stiborová et al., 2005a, Stiborová et al., 2005b, Stiborová et al., 2011b, Stiborová et al., 2012, Stiborová et al., 2013b, Stiborová et al., 2005a, Stiborová et al., 2005b, Sistkova et al., 2008, Rosenquist et al., 2010, Arlt et al., 2011, Levová et al., 2011). Beside CYP1A/2, rat and human CYPs of the 2C and 3A subfamilies also oxidize AAI (Sistkova et al., 2008, Rosenquist et al., 2010, Levová et al., 2011, Stiborová et al., 2012, Stiborová et al., 2015a, Stiborová et al., 2015b) (Fig. 1). The CYP-mediated AAI oxidation leads to a decrease in AAI-induced renal injury (Xiao et al., 2008, Xue et al., 2008).
The crucial role of CYP1A1 and 1A2 enzymes in AAI metabolism in vitro was unambiguously proven using several systems containing these enzymes [i.e. microsomal systems, inhibitors of these enzymes and correlation analyses, recombinant human and rat CYP1A1/2 heterologously expressed in microsomes of insect cells (Supersomes™), purified enzymes reconstituted with POR and other components of the monooxygenase system] (Stiborová et al., 2001, Stiborová et al., 2005a, Stiborová et al., 2005b, Stiborová et al., 2011b, Stiborová et al., 2012, Stiborová et al., 2013b, Stiborová et al., 2005a, Stiborová et al., 2005b, Sistkova et al., 2008, Arlt et al., 2011, Levová et al., 2011). In addition, the importance of CYP1A1 and 1A2 in AAI metabolism has been demonstrated in vivo using Cyp1a1/2-knock-out (single and double knock-outs) and CYP1A-humanized mouse lines (Rosenquist et al., 2010, Arlt et al., 2011, Stiborová et al., 2012, Stiborová et al., 2014a, Stiborová et al., 2014b, Stiborová et al., 2014c). Based on current knowledge we proposed that AAI metabolism by CYP1A1/2 in vivo is determined by the binding affinity of AAI to these CYPs, and their enzymatic turnover as well as by the oxygen levels in the organs (Stiborová et al., 2012, Stiborová et al., 2013b, Stiborová et al., 2014a, Stiborová et al., 2014b). Even though several studies considered CYP1A1/2 to be enzymes that detoxify AAI in vivo (Xiao et al., 2008, Rosenquist et al., 2010, Arlt et al., 2011, Stiborová et al., 2012, Stiborová et al., 2014a, Stiborová et al., 2014b, Stiborová et al., 2014c), the question which of their two opposing roles in AAI metabolism (AAI detoxification to AAIa versus activation of AAI to form AAI-DNA adducts) prevails in vivo remains to be answered.
To elucidate the roles of CYP1A this study was performed. AAI was administered to Wistar rats pretreated with Sudan I (1-phenylazo-2-naphthol), a strong inducer of CYP1A1 and CYP1A2 (Refat et al., 2008, Stiborová et al., 2013a), and AAI-DNA adduct levels in target and non-target organs were determined by 32P-postlabeling and compared to those in organs of rats treated with AAI only. The amounts of CYP1A1/2 enzymes expressed in rats at transcriptional and translational levels were analyzed by real-time polymerase chain reaction (RT-PCR) and Western blotting, and their activities determined with their marker substrates. The formation of AAIa, the detoxification metabolite of AAI, was analyzed using high performance liquid chromatography (HPLC).
2. Materials and methods
2.1. Chemicals
NADPH, AAI (sodium salt), Sudan I [1-(phenylazo)-2-hydroxynaphthalene], menadione (2-methyl-1,4-naphthoquinone), cytochrome c and calf thymus DNA were from Sigma Chemical Co. (St. Louis, MO, USA). 7-Methoxyresorufin was purchased from Fluka Chemie AG (Buchs, Switzerland). All these and other chemicals were reagent grade or better. Enzymes and chemicals for the 32P-postlabeling assay were from sources already described (Stiborová et al., 2005a).
2.2. Animal experiments and sample preparation
The study was conducted in accordance with the Regulations for the Care and Use of Laboratory Animals (311/1997, Ministry of Agriculture, Czech Republic), which is in compliance with the Declaration of Helsinki. Animals were purchased from AnLab (Prague, Czech Republic), acclimatized for 5 days and maintained at 22 °C with a 12 h light/dark period. Standardized diet and water were provided ad libitum. One group of five weeks old male Wistar rats (∼125–150 g, n = 3/group) was treated i.p. with a single dose of AAI dissolved in 1% NaHCO3 (20 mg/kg body weight, bw), the second group with two doses of Sudan I dissolved in maize oil (i.p., always with 30 mg/kg bw) in two consecutive days, and the third group, where rats were treated i.p. with two doses of Sudan I (always with 30 mg/kg bw in two consecutive days) and with AAI (20 mg/kg bw) 24 h after the second dose of Sudan I-treatment. Three control rats received the same volume of both vehicles only. Animals were killed 1 day after the treatment by cervical dislocation. Livers, kidneys and lungs were removed, immediately after sacrifice, frozen in liquid nitrogen and stored at −80 °C. DNA from livers, kidneys and lungs was isolated by extraction with phenol/chloroform (Schmeiser et al., 1996). Total RNA was isolated from another aliquot of frozen organs using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) according to the procedure supplied by the manufacturer. The quality of isolated RNA was verified by horizontal agarose gel electrophoresis, RNA quantity was assessed by UV–vis spectrophotometry on a Carry 300 spectrophotometer (Varian, Palo Alto, CA, USA). Microsomes and cytosols were isolated from the rat tissues by a procedure described previously (Stiborová et al., 2003, Stiborová et al., 2005a). Protein concentration in the microsomal and cytosolic fractions was measured using bicinchoninic acid protein assay (Wiechelman et al., 1988) with bovine serum albumin as a standard. Pooled microsomal and cytosolic samples (n = 3 rats/group) were used for analyses. All microsomal and cytosolic samples were free of residual Sudan I, AAI or their metabolites as determined by HPLC (Stiborová et al., 1988, Stiborová et al., 2002b, Stiborová et al., 2005c, Levová et al., 2011).
2.3. DNA adduct analysis by 32P-postlabeling
The nuclease P1 enrichment version of 32P-postlabeling analysis, and thin-layer chromatography (TLC) on polyethylenimine-cellulose (PEI) plates were carried out and DNA adduct levels (RAL, relative adduct labeling) were calculated as described previously (Schmeiser et al., 1996, Schmeiser et al., 2013). AAI-DNA adducts were identified using reference standards as described (Schmeiser et al., 1996).
2.4. CYP1A and NQO1 mRNA content in rat livers, kidneys and lungs
RNA samples (1 μg) were reverse transcribed using 200 U of reverse transcriptase per sample with random hexamer primers utilizing RevertAid™ First Strand cDNA Synthesis Kit (MBI Fermentas, Vilnius, Lithuania) according to the manufacturer’s instructions. The prepared cDNA was used for real-time (RT) polymerase chain reaction (PCR) performed in RotorGene 2000 (Corbett Research, Sydney, Australia) under the following cycling conditions: incubation at 50 °C for 2 min and initial denaturation at 95 °C for 10 min, then 50 cycles of denaturation at 95 °C for 15 s and annealing at 60 °C for 1 min, and elongation for 30 s at 72 °C. Gain was set to 7 and fluorescence was acquired after elongation step. The PCR reaction mixtures (20 μl) contained 9 μl cDNA diluted 10-times in Milli-Q ultrapure water (Biocel A10, Millipore, Billerica, MA, USA), 10 μl TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA) and 1 μl TaqMan Gene Expression Assay Mix (commercially available unlabeled PCR primers and FAM™ dye-labelled probe for rat CYP1A1/2 or NQO1 as target genes and β-actin as reference internal standard gene). Each sample was analysed in two parallel aliquots. Negative controls had the same compositions as samples but cDNA was omitted from the mixture. Data were analyzed by the program RotorGene v6 (Corbett Research, Sydney, Australia) and evaluated by comparative cycle threshold (cT) method for relative quantitation of gene expression. Cycle thresholds, at which a significant increase in fluorescence signal was detected, were measured for each sample. Then ΔΔcT was evaluated according to following equations: ΔcT = cT (target) − cT (internal standard), ΔΔcT = ΔcTtreated − ΔcTcontrol, where ΔcTtreated is ΔcT for treated rats and ΔcTcontrol is ΔcT for untreated rats. ΔcT is positive if the target is expressed at a lower level than the internal standard (β-actin), and negative if expressed at a higher level. The induction of mRNA expression of studied target genes in treated animals was evaluated as 2−(ΔΔcT).
2.5. Preparation of antibodies and estimation of CYP1A1, 1A2, and NQO1 protein content in microsomal and cytosolic fractions isolated from rat liver and kidney
The chicken anti-rat CYP1A1, anti-rabbit CYP1A2 and anti-rat NQO1 antibodies were prepared as described previously (Stiborová et al., 2002b, Stiborová et al., 2006). Immunoquantification of microsomal CYP1A1 and 1A2 and cytosolic NQO1 was performed using Western blotting (Stiborová et al., 2006). Rat CYP1A1, rat CYP1A2 and human NQO1 (Sigma) were used to identify the CYP1A1, 1A2 and NQO1 bands, respectively. The antigen-antibody complex was visualized with an alkaline phosphatase-conjugated rabbit anti-chicken IgG antibody and 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium as dye and bands are expressed as arbitrary units (AU)/mg protein (Stiborová et al., 2002b, Stiborová et al., 2006). Glyceraldehyde phosphate dehydrogenase was used as loading control and detected by its antibody (1:750, Millipore; MA, USA).
2.6. NQO1, CYP1A1/2 and 2C6/11 enzyme activity assays
In hepatic, renal and pulmonary cytosols NQO1 activity was measured using menadione (2-methyl-1,4-naphthoquinone) as a substrate; the assay was improved by the addition of cytochrome c and NQO1 activity expressed as nmol cytochrome c reduced (Levová et al., 2011, Levová et al., 2012). Microsomal samples were characterized for specific CYP1A1 and 1A2 activities: Sudan I hydroxylation to 4′-hydroxy-, 6-hydroxy-, and 4′,6-dihydroxy-Sudan I (CYP1A1) (Stiborová et al., 1988, Stiborová et al., 2002b, Stiborová et al., 2005c) and methoxyresorufin O-demethylation (MROD) (CYP1A2) (Burke et al., 1994). Hepatic microsomal samples were also characterized for specific CYP2C6 and 2C11 activities with their marker substrates determining diclofenac 4′-hydroxylation and testosterone 16α-hydroxylation, respectively (Kobayashi et al., 2002, Yamazaki et al., 2006).
2.7. Microsomal incubations to study AAI demethylation
Incubation mixtures contained 100 mM potassium phosphate buffer (pH 7.4), 1 mM NADPH, 1 mg rat hepatic, renal or pulmonary microsomal protein and 10 μM AAI in a final volume of 250 μl and were incubated at 37 °C for 20 min; AAI O-demethylation to AAIa was determined to be linear up to 25 min. Control incubations were carried out either (i) without microsomes, (ii) without NADPH or (iii) without AAI. AAI and its metabolite AAIa were separated by reverse-phase HPLC, identified by mass spectrometry and quantified as described previously (Levová et al., 2011). Briefly, HPLC was carried out with an Nucleosil 100-5C18, 250 × 4.0 mm, 5 mm (Macherey-Nagel) column, using a linear gradient of acetonitrile (20–60% acetonitrile in 55 min) in 100 mM triethylamonium acetate with a flow rate of 0.6 ml/min. A Dionex HPLC pump P580 with UV/VIS UVD 170S/340S spectrophotometer detector set at 254 nm was used. Peaks were integrated with CHROMELEON™ 6.01 integrator. A peak eluting at retention time (r.t.) 22.7 min was identified as AAIa using mass-spectroscopy analysis (Levová et al., 2011). A typical HPLC chromatogram is shown in Supplementary Fig. 1.
2.8. Microsomal and cytosolic formation of AAI-DNA adducts
The de-aerated and nitrogen-purged incubation mixtures, in which microsomes were used to activate AAI contained 50 mM potassium phosphate buffer (pH 7.4), 1 mM NADPH, 1 mg of hepatic or renal microsomal protein, 0.5 mg of calf thymus DNA (2 mM dNp) and 0.5 mM AAI in a final volume of 750 μl. Microsomal incubations were carried out at 37 °C for 60 min; AAI-DNA adduct formation was found to be linear up to 2 h in microsomes (Stiborová et al., 2005a). Control incubations were carried out either (i) without microsomes, (ii) without NADPH, (iii) without DNA or (iv) without AAI. After extraction with ethyl acetate, DNA was isolated from the residual water phase as described above (Stiborová et al., 2005a, Stiborová et al., 2011a, Stiborová et al., 2012, Arlt et al., 2011).
The de-aerated and nitrogen-purged incubation mixtures, in which cytosols were used to activate AAI contained 50 mM Tris–HCl buffer (pH 7.4), 0.2% Tween 20, 1 mM NADPH, 1 mg rat hepatic or renal cytosolic protein, 0.5 mg calf thymus DNA (2 mM dNp) and 0.5 mM AAI in a final volume of 750 μl. Incubations with cytosols were performed at 37 °C for 60 min; AAI-derived DNA adduct formation was found to be linear up to 2 h (Stiborová et al., 2003). Control incubations were performed either (i) without cytosol, (ii) without NADPH, (iii) without DNA or (iv) without AAI. After extraction with ethyl acetate DNA was isolated from the residual water phase by the phenol/chloroform extraction method as described above.
2.9. Statistical analyses
For statistical data analysis we used Student’s t-test. All P-values are two-tailed and considered significant at the 0.05 level.
3. Results
3.1. DNA adduct formation in rats treated with AAI and Sudan I compared to adduct formation in rats treated with AAI alone
AAI-DNA adduct formation was determined by 32P-postlabeling in liver, kidney and lung of male Wistar rats treated i.p. with AAI, Sudan I, or AAI after pretreatment with Sudan I. Using the nuclease P1 version of 32P-postlabeling assay, all liver, kidney and lung samples from rats treated with AAI showed an adduct pattern similar to that found in kidney tissue from AAN and BEN patients (Arlt et al., 2002b, Nortier et al., 2000, Schmeiser et al., 1996, Schmeiser et al., 1997, Schmeiser et al., 2012). As shown in Fig. 2, the adduct pattern consisted of three adduct spots. These spots have been identified as 7-(deoxyguanosin-N2-yl) aristolactam I (dG-AAI), 7-(deoxyadenosin-N6-yl) aristolactam I (dA-AAI) and 7-(deoxyadenosin-N6-yl) aristolactam II (dA-AAII). We have shown previously that the dA-AAII adduct can also be generated from AAI, probably via a demethoxylation reaction of AAI or dA-AAI (Stiborová et al., 1994, Schmeiser et al., 1997). No AAI-derived DNA adducts were found in DNA of control rats treated either with vehicle or Sudan I only (data not shown).
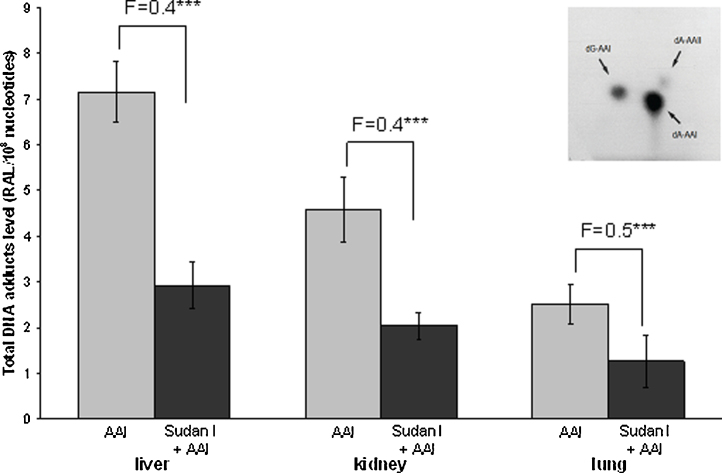
Fig. 2.
Quantitative TLC 32P-postlabeling analysis of AAI-DNA adduct levels in organs of rats treated with AAI, Sudan I or AAI after exposure to Sudan I. Numbers above columns (“F”) indicate fold changes in DNA adduct levels in animals treated with AAI combined with Sudan I compared to animals treated with AAI alone. Values are given as the means ± SD (n = 3); each DNA sample was determined by two postlabeled analyses. RAL, relative adduct labeling. Comparison was performed by t-test analysis; ***P < 0.001, different from animals treated with AAI alone. Insert: Autoradiographic profile of AAI-DNA adducts formed in liver of rats treated with AAI, determined by the nuclease P1 enrichment version of the 32P-postlabeling assay.
Generally, AAI-DNA adduct levels were higher in liver, the organ predominantly responsible for biotransformation of xenobiotics including AAI, as well as kidney, the target organ of AAI genotoxicity (Stiborová et al., 2008a, Stiborová et al., 2008b, Stiborová et al., 2008a, Stiborová et al., 2008b), than in lung (Fig. 2). In all organs of rats treated with AAI after pretreatment with Sudan I, the levels of AAI-DNA adducts were only half of those in rats exposed to AAI alone (Fig. 2 and Supplementary Table 1). Therefore, Sudan I, when administered to rats before their exposure to AAI, shifts the metabolic pathway of AAI that finally leads to a decrease in AAI-DNA adduct levels in all three organs.
Because CYP1A1/2 enzymes both oxidize (i.e. detoxify AAI) and reduce (i.e. activate AAI to form to AAI-DNA adducts) AAI, their expression might determine the balance between activation and detoxification pathways of AAI (Stiborová et al., 2008a, Stiborová et al., 2008b, Stiborová et al., 2013b, Stiborová et al., 2008a, Stiborová et al., 2008b). Therefore, we investigated whether expression levels of these enzymes influence AAI-DNA adduct formation found in vivo (Fig. 2 and Supplementary Table 1).
3.2. The effect of AAI treatment with or without Sudan I upon CYP1A1/2 and NQO1 mRNA and protein levels and their enzymatic activities in rat liver, kidney and lung
The effect of exposure to AAI, Sudan I and both compounds on expression of CYP1A1 and 1A2 at the mRNA and protein levels, was examined in liver, kidney and lung.
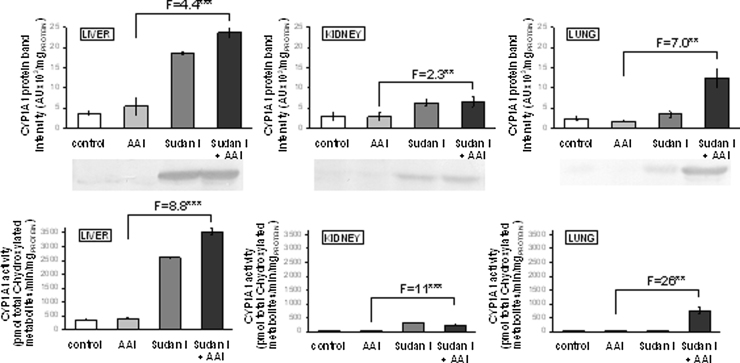
The mRNA and protein of CYP1A1 (Table 1 and Fig. 3) were expressed in all organs of control rats. Sudan I oxidation, a marker for CYP1A1 enzyme activity (Stiborová et al., 2002b, Stiborová et al., 2005c), was also detectable in all organs studied, but only very low Sudan I oxidation was measurable in kidney and lung, the organs expressing the lower protein levels of CYP1A1 than liver (Fig. 3).
Table 1.
Relative expression of mRNA of hepatic, renal and pulmonary CYP1A1, CYP1A2 and NQO1 in liver, kidney and lung from untreated (control) animals and animals treated with AAI, Sudan I or AAI combined with Sudan I.
| Liver |
Kidney |
Lung |
|||||
|---|---|---|---|---|---|---|---|
| ΔcTa | Fold change over control | ΔcTa | Fold change over control | ΔcTa | Fold change over control | ||
| CYP1A1 | Control | 12.84 ± 0.44 | 1.00 | 7.22 ± 0.22 | 1.00 | 15.20 ± 0.15 | 1.00 |
| AAI | 9.93 ± 0.44 | 7.54*** | 7.56 ± 0.27 | 0.791 | 13.53 ± 0.29 | 3.19** | |
| Sudan I | 0.36 ± 0.06 | 5710*** | 4.53 ± 0.35 | 6.45*** | 2.08 ± 0.04 | 8930*** | |
| Sudan I + AAI | 1.56 ± 0.31 | 2490*** | 4.53 ± 0.17 | 6.63*** | 2.05 ± 0.22 | 9090*** | |
| CYP1A2 | Control | 0.75 ± 0.34 | 1.00 | 16.38 ± 0.42 | 1.00 | 19.89 ± 0.18 | 1.00 |
| AAI | −2.22 ± 0.08 | 7.86*** | 14.60 ± 0.32 | 3.43** | 12.26 ± 0.26 | 198*** | |
| Sudan I | −5.23 ± 0.44 | 63.2*** | 7.85 ± 0.25 | 370*** | 7.30 ± 0.26 | 6170*** | |
| Sudan I + AAI | −5.76 ± 0.16 | 91.5*** | 8.72 ± 0.82 | 202*** | 10.67 ± 0.43 | 595*** | |
| NQO1 | Control | 6.03 ± 0.24 | 1.00 | 7.51 ± 0.16 | 1.00 | 5.98 ± 0.46 | 1.00 |
| AAI | 2.10 ± 0.29 | 15.2*** | 7.27 ± 0.18 | 1.18 | 5.66 ± 0.27 | 1.25 | |
| Sudan I | 1.06 ± 0.22 | 31.2*** | 5.88 ± 0.28 | 3.10** | 2.97 ± 0.08 | 8.06*** | |
| Sudan I + AAI | 1.47 ± 0.28 | 23.5*** | 6.05 ± 0.31 | 2.76** | 3.42 ± 0.44 | 5.92*** | |
Values relative to β-actin are means ± S.D. from data found for three male rats (n = 3) (control and treated with AAI, Sudan I and AAI with Sudan I). The induction of mRNA expression of studied target genes in treated animals was evaluated as 2−(ΔΔcT) (see Section 2). Comparison was performed by Student’s t-test analysis.
P > 0.01.
P > 0.001 significantly different from controls.
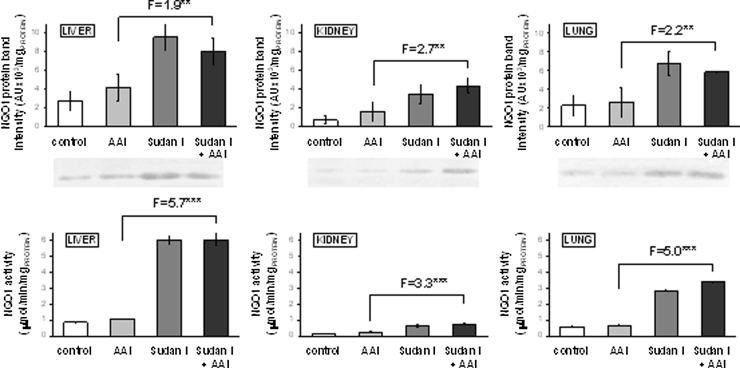
Fig. 3.
CYP1A1 protein levels (upper panels) in rat microsomes isolated from untreated (control) animals and animals treated with AAI, Sudan I or AAI after exposure to Sudan I. Microsomes isolated from liver, kidney and lung were analyzed by Western blotting in the same blot (insert) and, therefore, can be compared directly. Values are given as the means of arbitrary units (AU per mg protein) ± SD (n = 3). CYP1A1 enzyme activity as measured by Sudan I oxidation (nmol total C-hydroxylated Sudan I metabolites/min × mg protein) (lower panels). All values are given as the means ± SD (n = 3). Numbers above columns (“F”) indicate fold changes in protein level or enzyme activity in microsomes of rats treated with AAI with Sudan I compared to those with AAI alone. Comparison was performed by t-test analysis; **P < 0.01, ***P < 0.001, different from data found in microsomes form rats treated with AAI alone.
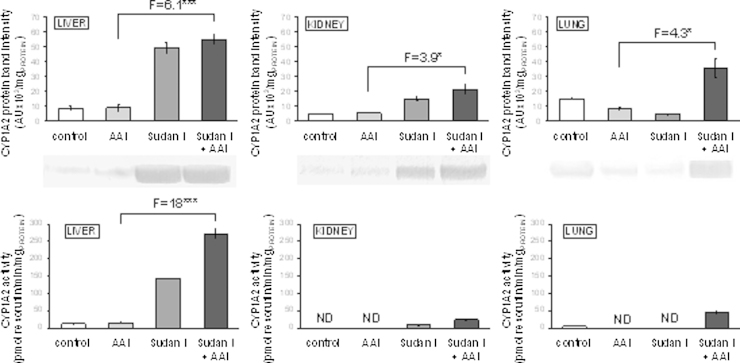
The CYP1A2 mRNA was expressed mainly in liver (Table 1), whereas the CYP1A2 protein expression levels were higher in liver and lung than in kidney (Fig. 4). In concordance, MROD activity, a marker reaction of CYP1A2, was found in liver and lung, with no activity in kidney (see Fig. 4).
Fig. 4.
CYP1A2 protein levels (upper panels) in rat microsomes isolated from untreated (control) animals and animals treated with AAI, Sudan I or AAI after exposure to Sudan I. Microsomes isolated from liver, kidney and lung were analyzed by Western blotting in the same blot (insert) and, therefore, can be compared directly. Values are given as the means of arbitrary units (AU per mg protein) ± SD (n = 3). CYP1A2 enzyme activity as measured by MROD (pmol resorufin/min × mg protein) (lower panels). All values are given as the means ± SD (n = 3). Numbers above columns (“F”) indicate fold changes in protein level or enzyme activity in microsomes of rats treated with AAI with Sudan I compared to those with AAI alone. Comparison was performed by t-test analysis; *P < 0.05, ***P < 0.001, different from data found in microsomes form rats treated with AAI alone.
As shown in Table 1, treatment of rats with Sudan I alone or with this compound before exposure to AAI induced expression of CYP1A1 mRNA in all tested organs. Treatment of rats with AAI alone induced mRNA levels of this CYP only in the liver and lung. The effect of both compounds combined was either the same as of Sudan I alone (lung and kidney) or led to lower mRNA levels in the liver. The most drastic effect was seen in the lung where Sudan I alone or in combination with AAI increased levels of CYP1A1 mRNA 2900-times as compared to AAI alone (Table 1). Expression of CYP1A1 protein and oxidation of Sudan I, a marker for CYP1A1, were always higher in organs of rats treated with AAI after pretreatment with Sudan I than with AAI alone (Fig. 3).
Expression of mRNA and protein of CYP1A2 was also induced by treatment of rats with AAI, Sudan I or their combined administration (Table 1 and Fig. 4). In liver the mRNA, protein and CYP1A2 enzyme activities ran parallel, in kidney activities were detectable only in microsomes of rats treated with Sudan I or Sudan I combined with AAI. In lung the very high mRNA induction was not reflected in the phenotype; a decrease in amounts of CYP1A2 protein found in lung of rats treated with AAI or Sudan I did not correspond to a 198- or 6170-fold increase in the CYP1A2 mRNA expression levels (Fig. 4).
The results found confirmed that Sudan I is a strong inducer of CYP1A1/2 in rats and indicate that a combined treatment of rats with Sudan I and AAI leads to even higher enzyme levels than with Sudan I alone.
Treatment of rats with Sudan I and Sudan I combined with AAI also led to an increased expression of cytosolic NQO1, again at the mRNA, protein and enzyme activity levels in liver, kidney and lung (Table 1 and Fig. 5). Similarly to CYP1A, at the doses used, Sudan I resulted in greater increases at the protein level. Expression of mRNA, protein and enzyme activity of NQO1 measured with menadione as a substrate ran parallel in all three organs and were always higher in organs of rats treated with AAI and Sudan I than in those treated with AAI alone (Fig. 5). However, the efficacy of NQO1 induction by AAI with Sudan I compared to AAI alone was lower than that for CYP1A expression (compare Fig. 3, Fig. 4, Fig. 5). These findings indicate that both compounds administered to rats act as moderate inducers of NQO1.
Fig. 5.
NQO1 protein levels (upper panels) and NQO1 enzyme activity (lower panels) in rat cytosols isolated from untreated (control) animals and animals treated with AAI, Sudan I or AAI after pretreatment with Sudan I. Cytosol isolated from liver, kidney or lung was analyzed by Western blotting in the same blot (insert) and, therefore, can be compared directly. Human recombinant NQO1 was used to identify the rat NQO1 band in rat cytosol (data not shown). Values are given as the means of arbitrary units (AU per mg protein) ± SD (n = 3). NQO1 activity in hepatic, renal and pulmonary cytosols was determined using menadione and cytochrome c as substrate (expressed as nmol cytochrome c reduced/min × mg protein). Numbers above columns (“F”) indicate fold changes in protein level or enzyme activity in cytosols of rats treated with AAI with Sudan I compared to those with AAI alone. Values are given as the means ± SD (n = 3). Comparison was performed by t-test analysis; **P < 0.01, ***P < 0.001, different from data found in cytosols of rats treated with AAI alone.
3.3. The effect of treatment of rats with AAI, Sudan I and both agents in combination on oxidation of AAI to AAIa by rat hepatic, renal and pulmonary microsomes
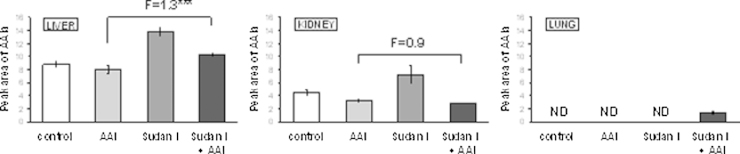
Since microsomal CYP1A1 and 1A2 detoxify AAI to its oxidative O-demethylated metabolite AAIa (Sistkova et al., 2008, Rosenquist et al., 2010, Arlt et al., 2011, Levová et al., 2011, Stiborová et al., 2012, Stiborová et al., 2013b, Stiborová et al., 2014a, Stiborová et al., 2014b, Stiborová et al., 2015b), AAIa formation from AAI was investigated ex vivo in hepatic, renal and pulmonary microsomes of all treatment groups. AAIa was formed by liver microsomes from the AAI plus Sudan I group at moderately higher levels as compared to microsomes of rats treated with AAI alone. But in kidney only Sudan I treatment alone increased AAIa formation 1.6-fold (P < 0.01), AAI had no effect or even inhibited oxidation of AAI (Fig. 6). In lung the low activity of CYP1A enzymes detectable essentially only in microsomes of rats exposed to both Sudan I and AAI (see the CYP1A1/2 activities determined with their marker substrates shown in Fig. 3, Fig. 4) was confirmed also by formation of AAIa, as AAIa was only detectable at low levels in pulmonary microsomes of this group (Fig. 6). These results indicate that CYP1A1/2 enzymes catalyze AAI demethylation to AAIa in test rat organs, but this activity does not seem to be very effectively induced by Sudan I either alone or in combination with AAI.
Fig. 6.
Formation of AAIa (peak area per minute per miligram protein) in rat microsomes isolated from untreated (control) animals and animals treated with AAI, Sudan I or AAI after exposure to Sudan I with AAI as a substrate. All values are given as the means ± SD (n = 3). Numbers above columns (“F”) indicate fold changes in AAIa levels in microsomes of rats treated with AAI with Sudan I compared to those with AAI alone. ND, not detected. Comparison was performed by t-test analysis; ***P < 0.001, different from data found in microsomes of rats treated with AAI alone.
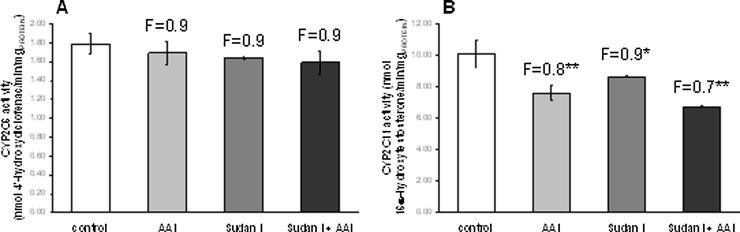
A probable reason for this observation is that not only CYP1A1/2, but also enzymes of the 2C subfamily, which are highly expressed in the livers of male rats, accounting for approximately 55% of the rat liver CYP complement (Nedelcheva and Gut, 1994), can oxidize AAI. CYP2C11 with ∼50% and CYP2C6 at ∼20% are the main members of the hepatic CYP2C family in rats (Večeřa et al., 2011, Zachařová et al., 2012). Both have been shown to be capable of efficiently oxidizing AAI to AAIa (Levová et al., 2011, Stiborová et al., 2014c, Stiborová et al., 2015a, Stiborová et al., 2015b), and the contribution of the CYP2C enzymes to AAIa formation in rat liver microsomes is more than 4-times higher than that of CYP1A (Stiborová et al., 2015b). Upon induction of CYP1A with Sudan I the relative amount of the CYP2C enzymes in the microsomes will decrease leading to lower CYP2C activity if analyzed based on mg protein, as was the case in our study. To test this, CYP2C activity was also analyzed in hepatic microsomes using diclofenac 4′-hydroxylation for CYP2C6 and testosterone 16α-hydroxylation as a marker for CYP2C11 (Kobayashi et al., 2002, Yamazaki et al., 2006). As shown in Fig. 7 exposure of rats to Sudan I, either with or without AAI, decreased testosterone 16α-hydroxylation activities based on mg protein up to 33% relative to control while diclofenac 4′-hydroxylation was marginally lower. Therefore, decreased relative CYP2C activity could explain why AAIa formation in liver microsomes of rats treated with AAI, Sudan I or with a combination of both compounds did not run parallel to CYP1A induction tested with their marker activities, namely, Sudan I oxidation and MROD.
Fig. 7.
CYP2C6 (A) and CYP2C11 enzyme activities (B) in rat hepatic microsomes. CYP2C6 was measured as diclofenac 4′-hydroxylation (nmol 4′-hydroxydiclofenac/min × mg protein) and CYP2C11 as testosterone 16α-hydroxylation (nmol 16α-hydroxytestosterone/min × mg protein). All values are given as the means ± SD (n = 3). Numbers above columns (“F”) indicate fold changes in enzyme activities compared to control. Comparison was performed by t-test analysis; ***P < 0.001, different from control.
3.4. Microsomal versus cytosolic activation of AAI
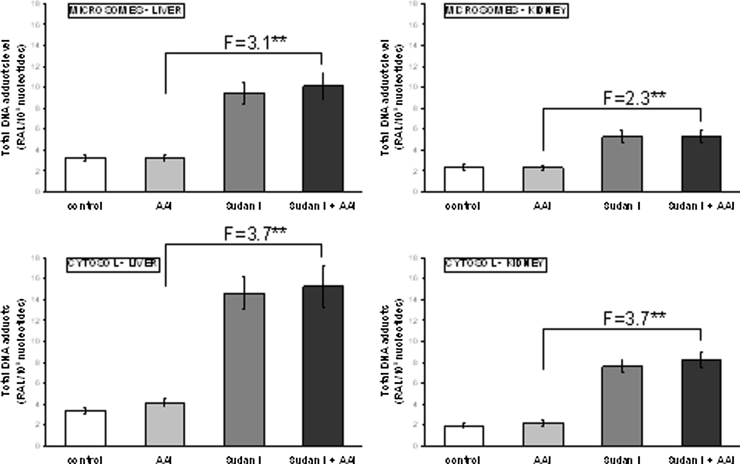
In further experiments we investigated whether induction of microsomal CYP1A1/2 and cytosolic NQO1 also influences the reductive activation of AAI to AAI-DNA adducts catalyzed by rat microsomal and cytosolic fractions ex vivo. For the investigations we focused on the liver and kidney (target organ for AAI genotoxicity).
AAI-DNA adduct formation was analyzed in ex-vivo incubations under hypoxic conditions. Incubation mixtures were purged with a stream of nitrogen for 2 minutes before the addition of AAI. AAI was reductively activated by both hepatic and renal microsomes from all treatment groups (Fig. 8). The adduct pattern generated was the same as that found in vivo (see Fig. 2). No adducts were observed in control incubations carried out in parallel (data not shown). A significant two to three-fold increase in AAI-DNA adduct formation was seen in incubations of DNA with AAI and hepatic or renal microsomes of rats exposed to Sudan I alone or in combination with AAI (Fig. 8). Overall, the increases in AAI-DNA adduct formation ex vivo corresponded to the induction of CYP1A1/2 at protein levels in rats and confirmed the participation of these CYPs in the reductive activation of AAI found previously (Stiborová et al., 2001, Stiborová et al., 2005a, Stiborová et al., 2005b, Stiborová et al., 2012, Stiborová et al., 2014b). The AAI-DNA adduct formation by microsomes under the oxidative (i.e. aerobic) conditions was not analyzed in this study. Namely, under these conditions the oxidation of AAI in microsomes (see Fig. 6) should compete with its reduction, which finally result in decreased levels of AAI-DNA adducts. Indeed, as shown in our previous study, an inhibition of AAI-DNA adduct formation occurred in the microsomal system under the aerobic conditions (Schmeiser et al., 1997).
Fig. 8.
DNA adduct formation ex vivo by AAI in rat microsomes (upper panels) and cytosols (lower panels) isolated from liver and kidney of untreated (control) animals and animals treated with AAI, Sudan I or AAI after exposure to Sudan I and incubated with DNA, AAI and NADPH. AAI-DNA adduct formation was determined by 32P-postlabeling. Values are given as the means ± SD (n = 3); each DNA sample was determined by two postlabeling analyses. RAL, relative adduct labeling. Numbers above columns (“F”) indicate fold changes in AAI-DNA adduct levels in microsomes and cytosols of rats treated with AAI with Sudan I compared to those with AAI alone. Comparison was performed by t-test analysis; ***P < 0.001, different from data found with microsomes or cytosols of rats treated with AAI alone.
Cytosols, where NQO1 is expressed, were also incubated with AAI, calf thymus DNA and NADPH, the cofactor of NQO1, and analyzed for DNA adduct formation by 32P-postlabeling. AAI was activated by hepatic cytosols as evidenced by specific AAI-DNA adduct formation (Fig. 8). No DNA adducts were observed in control incubations carried out in parallel (data not shown). Liver cytosols from rats treated with AAI, Sudan I and AAI after pretreatment with Sudan I produced AAI-DNA adduct levels which were 1.2-, 4.3- and 4.5-fold higher, respectively, relative to cytosols isolated from untreated animals (Fig. 8). The increase in AAI-DNA adduct formation ran parallel to higher NQO1 activity in these cytosols (compare Fig. 5). Renal cytosols isolated from AAI-treated rats, rats treated with Sudan I and rats treated with Sudan I plus AAI led to 1.1-, 3.9- and 4.2-fold higher AAI-DNA adduct levels relative to cytosols from control animals, respectively. Again, the observed adduct levels was consistent with the observed NQO1 enzyme activity (compare Fig. 5, Fig. 8).
4. Discussion
CYP1A1 and 1A2 have the dual function to catalyze AAI detoxification to AAIa and the activation of AAI to form AAI-DNA adducts. The aim of this study was to evaluate which of the two opposing functions prevails in an experimental rat model in vivo. Here we modulated the expression of CYP1A1/2 by Sudan I treatment which is a strong inducer of these enzymes (Stiborová et al., 2013a, Refat et al., 2008). As a measure of genotoxicity the formation of AAI-DNA adducts was determined. The formation of AAIa was used as a measure for AAI detoxification.
The results of this study demonstrate that AAI-DNA adducts are formed in vivo in all organs tested (liver, kidney and lung), both in rats treated with AAI alone or in combination with the inducer Sudan I. These findings suggest that AAI is distributed via the blood stream and that these tissues have the metabolic capacity to reductively activate this carcinogen. The levels of AAI-DNA adducts in individual organs therefore depend both on a distribution of AAI to individual organs and on the activities of enzymes catalyzing either its oxidative detoxification or its reductive activation to species forming AAI-DNA adducts. Indeed, our results demonstrate that expression levels of CYP1A enzymes modulate the metabolism of AAI in the rat organs, thereby dictating AAI-DNA adduct formation in vivo. Furthermore, it is probable that enhanced clearance of AAI in the liver of induced animals is also altering the levels of AAI-DNA adducts in the kidney.
In our study rats were exposed to AAI for 24 h only to resolve the role of CYP1A1/2 in AAI oxidative or reductive metabolism in vivo. We had previously shown the formation of AAI-DNA adducts in liver and kidney 24 h after administration (Pfau et al., 1990, Stiborová et al., 1994, Stiborová et al., 2014c, Arlt et al., 2002b). Therefore, for these experimental purposes and to study the acute effects we used this short exposure, in order to resolve the role of CYP1A1/2 in AAI oxidative or reductive metabolism in vivo. Our results indicate that under these conditions AAI genotoxicity (i.e. AAI-DNA adduct formation) is reduced after administration of the CYP1A1/2 inducer Sudan I. However, it is important to note that the doses to which humans are exposed to are orders of magnitude lower than the AAI dose administered to rats in this study and its effect at lower but chronic and life-long doses may be different. We found that only half of the AAI-DNA adduct levels were formed in liver, kidney and lung of rats treated with AAI after exposure to Sudan I, than in rats treated with AAI alone (see Fig. 2). These findings demonstrate that induction of CYP1A1 and 1A2 by Sudan I might increase AAI detoxification, leading to lower amounts of AAI available for activation. However, only 1.3-fold higher AAI detoxification (O-demethylation activity) was found ex vivo in microsomes of treated rats. Previous studies have shown that CYP2C enzymes are also capable in O-demethylating AAI (i.e. AAI detoxification), and are even more efficient than the CYP1A enzymes to catalyze this reaction in rat liver microsomes (Stiborová et al., 2014c, Stiborová et al., 2015b). CYP2C enzymes constitute about 55% of hepatic CYPs in male rats, Sudan I alone or in combination with AAI induces CYP1A about 4-fold, thereby reducing the relative amount of the other CYP enzymes. In microsomes from CYP1A induced rats, the contribution of CYP2C is therefore lower by a factor of approximately 4 explaining the relatively weak induction of AAIa formation we observed in such microsomes.
The results of the present study fit with the proposed scheme of AAI metabolism (see Fig. 1). If AAI is oxidized to AAIa, lower amounts of AAI are available to be activated by enzymes with nitroreductase activity like NQO1 (for a review, see Stiborová et al., 2008b, Stiborová et al., 2014a, Stiborová et al., 2014b, Stiborová et al., 2014c) which generate cyclic acylnitrenium ions that bind to DNA (i.e. DNA adduct formation) (Fig. 1). Our results are in accordance with two previous studies showing that AAI detoxification is lower in Cyp1a knockout mice (i.e. Cyp1a1(-/-), Cyp1a2(-/-) and Cyp1a1/2(-/-) mouse lines) leading to an increase in AAI (geno) toxicity (Rosenquist et al., 2010, Arlt et al., 2011).
Our results of the ex-vivo experiments also confirm previous findings (Stiborová et al., 2001, Stiborová et al., 2012, Arlt et al., 2011, Levová et al., 2011) that under hypoxic (anaerobic) conditions, rat hepatic and renal CYP1A enzymes are capable of reducing AAI to species forming DNA adducts. Induction of CYP1A proteins and their enzyme activities correlated with increased AAI-DNA adduct formation ex vivo (Fig. 8). Therefore, induction of CYP1A1 and 1A2 leads to both oxidation and reduction of AAI which indicates that in case of hypoxia AAI must act as a ligand of CYP1A heme iron under low pO2. Indeed, reduction of AAI as a ligand of heme iron of CYP1A1 and 1A2 could be confirmed by molecular modeling (Jerabek et al., 2012, Stiborová et al., 2014b). On the other hand, under aerobic conditions AAI acts as a classical substrate of CYP1A1 or 1A2, and takes one atom of atmospheric oxygen to O-demethylate the methoxy group of AAI to generate AAIa. In line with this suggestion is the finding that binding of AAI to the active site of the Compounds I of CYP1A1 and 1A2 indeed favors O-demethylation of AAI to AAIa (see Fig. 5 in Stiborová et al., 2015b). However, as shown in Fig. 2, the increased reductive activation of AAI ex vivo had no apparent impact on the reductive metabolism of AAI in vivo; AAI-DNA adduct formation was attenuated by induction of CYP1A enzymes. Likewise, induction of cytosolic NQO1, which led to an increase in AAI-DNA adduct formation ex vivo, had no significant effect in vivo, as a decrease in AAI-DNA adduct levels was observed. These findings demonstrate that in vivo the oxygen concentrations in rat tissues are sufficient to facilitate the process of the oxidative O-demethylation of AAI, which is thereafter the predominant reaction of CYP1A1/2 in AAI metabolism in vivo. Therefore, in addition to the influence of CYP1A expression, the in vivo pO2 in tissues is an important factor that affects the balance between nitroreduction and O-demethylation of AAI, thereby influencing its (geno) toxicity and carcinogenicity. Indeed, the presence of oxygen in the in-vitro incubations of AAI with DNA and microsomal or cytosolic enzymes strongly inhibits the levels of AAI-DNA adducts formed in these systems (Schmeiser et al., 1997).
Based on the present study and taking into account previous results obtained in Cyp1a-knock-out and CYP1A-humanized mouse lines (Rosenquist et al., 2010, Arlt et al., 2011, Stiborová et al., 2012, Stiborová et al., 2014a, Stiborová et al., 2014b, Stiborová et al., 2014c), we conclude that the efficiency of the CYP1A family to protectively oxidize AAI to AAIa prevails over its reducing activation in vivo. The evaluation of inter-individual variations in the human CYP1A enzymes, including their genetic polymorphisms, remains a major challenge to explain human individual susceptibility to AAI, and to predict the risk of cancer among patients suffering from AAN and BEN.
Conflict of interest
The authors declare that there are no conflicts of interest.
Funding
Financial support from Grant Agency of the Czech Republic (grant 14-18344S) and Charles University in Prague (grants UNCE 204025/2012 and 570513) is highly acknowledged. Work at King’s College London is supported by Cancer Research UK (grant C313/A14329).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.tox.2016.01.011.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Arlt V.M., Ferluga D., Stiborova M., Pfohl-Leszkowicz A., Vukelic M., Ceovic S., Schmeiser H.H., Cosyns J.P. Is aristolochic acid a risk factor for Balkan endemic nephropathy-associated urothelial cancer? Int. J. Cancer. 2002;101:500–502. doi: 10.1002/ijc.10602. [DOI] [PubMed] [Google Scholar]
- Arlt V.M., Stiborova M., Schmeiser H.H. Aristolochic acid as a probable human cancer hazard in herbal remedies: a review. Mutagenesis. 2002;17:265–277. doi: 10.1093/mutage/17.4.265. [DOI] [PubMed] [Google Scholar]
- Arlt V.M., Stiborova M., vom Brocke J., Simoes M.L., Lord G.M., Nortier J.L., Hollstein M., Phillips D.H., Schmeiser H.H. Aristolochic acid mutagenesis: molecular clues to the aetiology of Balkan endemic nephropathy-associated urothelial cancer. Carcinogenesis. 2007;28:2253–2261. doi: 10.1093/carcin/bgm082. [DOI] [PubMed] [Google Scholar]
- Arlt V.M., Levova K., Barta F., Shi Z., Evans J.D., Frei E., Schmeiser H.H., Nebert D.W., Phillips D.H., Stiborova M. Role of P450 1A1 and P450 1A2 in bioactivation versus detoxication of the renal carcinogen aristolochic acid I: studies in Cyp1a1-/- Cyp1a2-/-, and Cyp1a1/1a2-/- mice. Chem. Res. Toxicol. 2011;24:1710–1719. doi: 10.1021/tx200259y. [DOI] [PubMed] [Google Scholar]
- Arlt V.M., Henderson C.J., Wolf C.R., Stiborova M., Phillips D.H. The Hepatic Reductase Null (HRN™) and Reductase Conditional Null (RCN) mouse models as suitable tools to study metabolism, toxicity and carcinogenicity of environmental pollutants. Toxicol. Res. 2015;4:548–562. [Google Scholar]
- Burke M.D., Thompson S., Weaver R.J., Wolf C.R., Mayer R.T. Cytochrome P450 specificities of alkoxyresorufin O-dealkylation in human and rat liver. Biochem. Pharmacol. 1994;48:923–936. doi: 10.1016/0006-2952(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Chan W., Cu L., Xu G., Cai Z. Study of the phase I and phase II metabolism of nephrotoxin aristolochic acid by liquid chromatography/tandem mass spectrometry. Rapid. Commun. Mass Spectrom. 2006;20:1755–1760. doi: 10.1002/rcm.2513. [DOI] [PubMed] [Google Scholar]
- Chen M., Gong L., Qi X., Xing G., Luan Y., Wu Y., Xiao Y., Yao J., Li Y., Xue X., Pan G., Ren J. Inhibition of renal NQO1 activity by dicoumarol suppresses nitroreduction of aristolochic acid I and attenuates its nephrotoxicity. Toxicol. Sci. 2011;122:288–296. doi: 10.1093/toxsci/kfr138. [DOI] [PubMed] [Google Scholar]
- Gökmen M.R., Cosyns J.P., Arlt V.M., Stiborová M., Phillips D.H., Schmeiser H.H., Simmonds M.S.J., Look H.T., Vanherweghem J.L., Nortier J.L., Lord G.M. The epidemiology: diagnosis and management of aristolochic acid nephropathy: a narrative review. Ann. Intern. Med. 2013;158:469–477. doi: 10.7326/0003-4819-158-6-201303190-00006. [DOI] [PubMed] [Google Scholar]
- Grollman A.P., Shibutani S., Moriya M., Miller F., Wu L., Moll U., Suzuki N., Fernandes A., Rosenquist T., Medverec Z., Jakovina K., Brdar B., Slade N., Turesky R.J., Goodenough A.K., Rieger R., Vukelic M., Jelakovic B. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang M.L., Chen C.H., Sidorenko V.S., He J., Dickman K.G., Yun B.H., Moriya M., Niknafs N., Douville C., Karchin R., Turesky R.J., Pu Y.S., Vogelstein B., Papadopoulos N., Grollman A.P., Kinzler K.W., Rosenquist T.A. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci. Transl. Med. 2013;5:197ra102. doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC IARC Monogr. Eval. Carcinog. Risk Hum. 2012;100A [Google Scholar]
- Jerabek P., Martinek V., Stiborova M. Theoretical investigation of differences in nitroreduction of aristolochic acid I by cytochromes P450 1A1, 1A2 and 1B1. Neuro Endocrinol. Lett. 2012;33(3):25–32. [PubMed] [Google Scholar]
- Kobayashi K., Urashima K., Shimada N., Chiba K. Substrate specificity for rat cytochrome P450 (CYP) isoforms: screening with cDNA-expressed systems of the rat. Biochem. Pharmacol. 2002;63:889–896. doi: 10.1016/s0006-2952(01)00843-7. [DOI] [PubMed] [Google Scholar]
- Kucab J.E., Phillips D.H., Arlt V.M. Linking environmental carcinogen exposure to TP53 mutations in human tumours using the human TP53 knock-in (Hupki) mouse model. FEBS J. 2010;277:2567–2583. doi: 10.1111/j.1742-464X.2010.07676.x. [DOI] [PubMed] [Google Scholar]
- Levová K., Mizerová M., Kotrbová V., Šulc M., Henderson C.J., Wolf C.R., Philips D.H., Frei E., Schmeiser H.H., Mareš J., Arlt V.M., Stiborová M. Role of cytochromes P450 1A1/2 in detoxication and activation of carcinogenic aristolochic acid I: studies with the hepatic NADPH:cytochrome P450 reductase null (HRN) mouse model. Toxicol. Sci. 2011;121:43–56. doi: 10.1093/toxsci/kfr050. [DOI] [PubMed] [Google Scholar]
- Levová K., Moserova M., Nebert D.W., Phillips D.H., Frei E., Schmeiser H.H., Arlt V.M., Stiborova M. NAD(P)H:quinone oxidoreductase expression in Cyp1a-knockout and CYP1A-humanized mouse lines and its effect on bioactivation of the carcinogen aristolochic acid I. Toxicol. Appl. Pharmacol. 2012;265:360–367. doi: 10.1016/j.taap.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Lord G.M., Hollstein M., Arlt V.M., Roufosse C., Pusey C.D., Cook T., Schmeiser H.H. DNA adducts and p53 mutations in a patient with aristolochic acid-associated nephropathy. Am. J. Kidney Dis. 2004;43:e11–e17. doi: 10.1053/j.ajkd.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Martinek V., Kubickova B., Arlt V.M., Frei E., Schmeiser H.H., Hudeček J., Stiborova M. Comparison of activation of aristolochic acid I and II with NADPH:quinone oxidoreductase, sulphotransferases and N-acetyltranferases. Neuro Endocrinol. Lett. 2011;32(1):57–70. [PubMed] [Google Scholar]
- Nedelcheva V., Gut I. P450 in the rat and man: methods of investigation, substrate specificities and relevance to cancer. Xenobiotica. 1994;24:1151–1175. doi: 10.3109/00498259409038673. [DOI] [PubMed] [Google Scholar]
- Nedelko T., Arlt V.M., Phillips D.H., Hollstein M. TP53 mutation signature supports involvement of aristolochic acid in the aetiology of endemic nephropathy-associated tumours. Int. J. Cancer. 2009;124:987–990. doi: 10.1002/ijc.24006. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S., Kucab J.E., Morganella S., Glodzik D., Alexandrov L.B., Arlt V.M., Weninger A., Hollstein M., Stratton M.R., Phillips D.H. The genome as a record of environmental exposure. Mutagenesis. 2015;30:763–770. doi: 10.1093/mutage/gev073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nortier J.L., Martinez M.C., Schmeiser H.H., Arlt V.M., Bieler C.A., Petein M., Depierreux M.F., De Pauw L., Abramowicz D., Vereerstraeten P., Vanherweghem J.L. Urothelial carcinoma associated with the use of a Chinese herb (Aristolochia fangchi) N. Engl. J. Med. 2000;342:1686–1692. doi: 10.1056/NEJM200006083422301. [DOI] [PubMed] [Google Scholar]
- Pfau W., Schmeiser H.H., Wiessler M. 32P-postlabelling analysis of the DNA adducts formed by aristolochic acid I and II. Carcinogenesis. 1990;11:1627–1633. doi: 10.1093/carcin/11.9.1627. [DOI] [PubMed] [Google Scholar]
- Poon S.L., Pang S.T., McPherson J.R., Yu W., Huang K.K., Guan P., Weng W.H., Siew E.Y., Liu Y., Heng H.L., Chong S.C., Gan A., Tay S.T., Lim W.K., Cutcutache I., Huang D., Ler L.D., Nairismägi M.L., Lee M.H., Chang Y.H., Yu K.J., Chan-On W., Li B.K., Yuan Y.F., Qian C.N., Ng K.F., Wu C.F., Hsu C.L., Bunte R.M., Stratton M.R., Futreal P.A., Sung W.K., Chuang C.K., Ong C.K., Rozen S.G., Tan P., Teh B.T. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci. Transl. Med. 2013;5:197ra101. doi: 10.1126/scitranslmed.3006086. [DOI] [PubMed] [Google Scholar]
- Refat N.A., Ibrahim Z.S., Moustafa G.G., Sakamoto K.Q., Ishizuka M., Fujita S. The induction of cytochrome P450 1A1 by Sudan dyes. J. Biochem. Mol. Toxicol. 2008;22:77–84. doi: 10.1002/jbt.20220. [DOI] [PubMed] [Google Scholar]
- Rosenquist T.A., Einolf H.J., Dickman K.G., Wang L., Smith A., Grollman A.P. Cytochrome P450 1A2 detoxicates aristolochic acid in the mouse. Drug Metab. Disp. 2010;38:761–768. doi: 10.1124/dmd.110.032201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeiser H.H., Bieler C.A., Wiessler M., van Ypersele de Strihou C., Cosyns J.P. Detection of DNA adducts formed by aristolochic acid in renal tissue from patients with Chinese herbs nephropathy. Cancer Res. 1996;56:2025–2028. [PubMed] [Google Scholar]
- Schmeiser H.H., Frei E., Wiessler M., Stiborová M. Comparison of DNA adduct formation by aristolochic acids in various in vitro activation systems by 32P-post-labelling: evidence for reductive activation by peroxidases. Carcinogenesis. 1997;18:1055–1062. doi: 10.1093/carcin/18.5.1055. [DOI] [PubMed] [Google Scholar]
- Schmeiser H.H., Stiborová M., Arlt V.M. Chemical and molecular basis of the carcinogenicity of Aristolochia plants. Curr. Opin. Drug Discov. Devel. 2009;12:141–148. [PubMed] [Google Scholar]
- Schmeiser H.H., Kucab J.E., Arlt V.M., Phillips D.H., Hollstein M., Gluhovschi G., Gluhovschi C., Modilca M., Daminescu L., Petrica L., Velciov S. Evidence of exposure to aristolochic acid in patients with urothelial cancer from a Balkan endemic nephropathy region of Romania. Environ. Mol. Mutagen. 2012;53:636–641. doi: 10.1002/em.21732. [DOI] [PubMed] [Google Scholar]
- Schmeiser H.H., Stiborova M., Arlt V.M. 32P-postlabeling analysis of DNA adducts. Methods Mol. Biol. 2013;1044:389–401. doi: 10.1007/978-1-62703-529-3_21. [DOI] [PubMed] [Google Scholar]
- Schmeiser H.H., Nortier J., L, Singh R., Gamboa da Costa G., Sennesael J., Cassuto-Viguier E., Ambrosetti D., Rorive S., Pozdzik A., Phillips D.H., Stiborova M., Arlt V.M. Exceptionally long-term persistence of DNA adducts formed by carcinogenic aristolochic acid I in renal tissue from patients with aristolochic acid nephropathy. Int. J. Cancer. 2014;135:562–567. doi: 10.1002/ijc.28681. [DOI] [PubMed] [Google Scholar]
- Shibutani S., Bonala R.R., Rosenquist T., Rieger R., Suzuki N., Johnson F., Miller F., Grollman A.P. Detoxification of aristolochic acid I by O-demethylation: less nephrotoxicity and genotoxicity of aristolochic acid Ia in rodents. Int. J. Cancer. 2010;127:1021–1027. doi: 10.1002/ijc.25141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistkova J., Hudecek J., Hodek P., Frei E., Schmeiser H.H., Stiborova M. Human cytochromes P450 1A1 and 1A2 participate in detoxication of carcinogenic aristolochic acid. Neuro Endocrinol. Lett. 2008;29:733–737. [PubMed] [Google Scholar]
- Stiborová M., Asfaw B., Anzenbacher P., Hodek P. A new way to carcinogenicity of azo dyes The benzenediazonium ion formed from a non-aminoazo dye, 1-phenylazo-2- hydroxynaphthalene (Sudan I) by microsomal enzymes binds to deoxyguanosine residues of DNA. Cancer Lett. 1988;40:327–333. doi: 10.1016/0304-3835(88)90092-4. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Fernando R.C., Schmeiser H.H., Frei E., Pfau W., Wiessler M. Characterization of DNA adducts formed by aristolochic acids in the target organ (forestomach) of rats by 32P-postlabelling analysis using different chromatographic procedures. Carcinogenesis. 1994;15:1187–1192. doi: 10.1093/carcin/15.6.1187. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Frei E., Wiessler M., Schmeiser H.H. Human enzymes involved in the metabolic activation of carcinogenic aristolochic acids: evidence for reductive activation by cytochromes P450 1A1 and 1A2. Chem. Res. Toxicol. 2001;14:1128–1137. doi: 10.1021/tx010059z. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Frei E., Sopko B., Wiessler M., Schmeiser H.H. Carcinogenic aristolochic acids upon activation by DT-diaphorase form adducts found in DNA of patients with Chinese herbs nephropathy. Carcinogenesis. 2002;23:617–625. doi: 10.1093/carcin/23.4.617. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Martínek V., Rýdlová H., Hodek P., Frei E. Sudan I is a potential carcinogen for humans: evidence for its metabolic activation and detoxication by human recombinant cytochrome P450 1A1 and liver microsomes. Cancer Res. 2002;62:5678–5684. [PubMed] [Google Scholar]
- Stiborová M., Frei E., Sopko B., Sopková K., Marková V., Laňková M., Kumstýřová T., Wiessler M., Schmeiser H.H. Human cytosolic enzymes involved in the metabolic activation of carcinogenic aristolochic acid: evidence for reductive activation by human NAD(P)H:quinone oxidoreductase. Carcinogenesis. 2003;24:1695–1703. doi: 10.1093/carcin/bgg119. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Frei E., Hodek P., Wiessler M., Schmeiser H.H. Human hepatic and renal microsomes, cytochromes P450 1A1/2, NADPH:CYP reductase and prostaglandin H synthase mediate the formation of aristolochic acid DNA-adducts found in patients with urothelial cancer. Int. J. Cancer. 2005;113:189–197. doi: 10.1002/ijc.20564. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Sopko B., Hodek P., Frei E., Schmeiser H.H., Hudeček J. The binding of aristolochic acid I to the active site of human cytochromes P450 1A1 and 1A2 explains their potential to reductively activate this human carcinogen. Cancer Lett. 2005;229:193–204. doi: 10.1016/j.canlet.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Martínek V., Rýdlová H., Koblas T., Hodek P. Expression of cytochrome P450 1A1 and its contribution to oxidation of a potential human carcinogen 1-phenylazo-2-naphthol (Sudan I) in human livers. Cancer Lett. 2005;220:145–154. doi: 10.1016/j.canlet.2004.07.036. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Dračínská H., Hájková J., Kadeřábková P., Frei E., Schmeiser H.H., Souček P., Phillips D.H., Arlt V.M. The environmental pollutant and carcinogen 3-nitrobenzanthrone and its human metabolite 3-aminobenzanthrone are potent inducers of rat hepatic cytochromes P450 1A1 and −1A2 and NAD(P)H:quinone oxidoreductase. Drug Metab. Disp. 2006;34:1398–1405. doi: 10.1124/dmd.106.009373. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Frei E., Arlt V.M., Schmeiser H.H. Metabolic activation of carcinogenic aristolochic acid: a risk factor for Balkan endemic nephropathy Mutat. Res. Rev. Mutat. Res. 2008;658:55–67. doi: 10.1016/j.mrrev.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Frei E., Schmeiser H.H. Biotransformation enzymes in development of renal injury and urothelial cancer caused by aristolochic acid. Kidney Int. 2008;73:1209–1211. doi: 10.1038/ki.2008.125. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Mareš J., Frei E., Arlt V.M., Martínek V., Schmeiser H.H. The human carcinogen aristolochic acid I is activated to form DNA adducts by human NAD(P)H:quinone oxidoreductase without the contribution of acetyltransferases or sulfotransferases. Environ. Mol. Mutagen. 2011;52:448–459. doi: 10.1002/em.20642. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Mareš J., Levová K., Pavlíčková J., Bárta F., Hodek P., Frei E., Schmeiser H.H. Role of cytochromes P450 in metabolism of carcinogenic aristolochic acid I: evidence of their contribution to aristolochic acid I detoxication and activation in rat liver. Neuro Endocrinol. Lett. 2011;32(1):121–130. [PubMed] [Google Scholar]
- Stiborová M., Levová K., Bárta F., Shi Z., Frei E., Schmeiser H.H., Nebert D.W., Phillips D.H., Arlt V.M. Bioactivation versus detoxication of the urothelial carcinogen aristolochic acid I by human cytochrome P450 1A1 and 1A2. Toxicol. Sci. 2012;125:345–358. doi: 10.1093/toxsci/kfr306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiborová M., Dračínská H., Martínek V., Svášková D., Hodek P., Milichovský J., Hejduková Ž, Brotánek J., Schmeiser H.H., Frei E. Induced expression of cytochrome P450 1A and NAD(P)H:quinone oxidoreductase determined at mRNA, protein and enzyme activity levels in rats exposed to the carcinogenic azo dye 1-phenylazo-2-naphthol (Sudan I) Chem. Res. Toxicol. 2013;26:290–299. doi: 10.1021/tx3004533. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Martínek V., Frei E., Arlt V.M., Schmeiser H.H. Enzymes metabolizing aristolochic acid and their contribution to the development of Aristolochic acid nephropathy and urothelial cancer. Curr. Drug Metab. 2013;14:695–705. doi: 10.2174/1389200211314060006. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Frei E., Arlt V.M., Schmeiser H.H. Knock-out and humanized mice as suitable tools to identify enzymes metabolizing the human carcinogen aristolochic acid. Xenobiotica. 2014;44:135–145. doi: 10.3109/00498254.2013.848310. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Frei E., Schmeiser H.H., Arlt V.M., Martínek V. 2014. Mechanisms of enzyme-catalyzed reduction of two carcinogenic nitro-aromatics: 3-nitrobenzanthrone and aristolochic acid I: experimental and theoretical approaches. Int. J. Mol. Sci. 2014;15:10271–10295. doi: 10.3390/ijms150610271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiborová M., Levová K., Bárta F., Šulc M., Frei E., Arlt V.M., Schmeiser H.H. The influence of dicoumarol on the bioactivation of the carcinogen aristolochic acid I in rats. Mutagenesis. 2014;29:189–200. doi: 10.1093/mutage/geu004. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Bárta F., Levová K., Hodek P., Frei E., Arlt V.M., Schmeiser H.H. The influence of ochratoxin A on DNA adduct formation by the carcinogen aristolochic acid in rats. Arch. Toxicol. 2015;89:2141–2158. doi: 10.1007/s00204-014-1360-1. [DOI] [PubMed] [Google Scholar]
- Stiborová M., Bárta F., Levová K., Hodek P., Schmeiser H.H., Arlt V.M., Martínek V. A mechanism of O-demethylation of aristolochic acid I by cytochromes P450 and their contributions to this reaction in human and rat livers: experimental and theoretical approaches. Int. J. Mol. Sci. 2015;16:27561–27562. doi: 10.3390/ijms161126047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanherweghem J.L., Depierreux M., Tielemans C., Abramowicz D., Dratwa M., Jadoul M., Richard C., Vandervelde D., Verbeelen D., Vanhaelen-Fastre R., Vanhaelen M. Rapidly progressive interstitial renal fibrosis in young women: association with slimming regimen including Chinese herbs. Lancet. 1993;341:387–391. doi: 10.1016/0140-6736(93)92984-2. [DOI] [PubMed] [Google Scholar]
- Večeřa R., Zachařová A., Orolin J., Strojil J., Skottová N., Anzenbacher P. Fenofibrate-induced decrease of expression of CYP2C11 and CYP2C6 in rat. Biopharm. Drug Dispos. 2011;32:482–487. doi: 10.1002/bdd.774. [DOI] [PubMed] [Google Scholar]
- Wiechelman K.J., Braun R.D., Fitzpatrick J.D. Investigation of the bicinchoninic acid protein assay: identified cation of the groups responsible for color formation. Anal. Biochem. 1988;175:231–237. doi: 10.1016/0003-2697(88)90383-1. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Ge M., Xue X., Wang C., Wang H., Wu X., Li L., Liu L., Qi X., Zhang Y., Li Y., Luo H., Xie T., Gu J., Ren J. Hepatic cytochrome P450s metabolize aristolochic acid and reduce its kidney toxicity. Kidney Int. 2008;73:1231–1239. doi: 10.1038/ki.2008.103. [DOI] [PubMed] [Google Scholar]
- Xue X., Xiao Y., Zhu H., Wang H., Liu Y., Xie T., Ren J. Induction of P450 1A by 3-methylcholanthrene protects mice from aristolochic acid-I-induced acute renal injury. Nephrol. Dial. Transplant. 2008;23:3074–3081. doi: 10.1093/ndt/gfn262. [DOI] [PubMed] [Google Scholar]
- Yamazaki H., Shimizu M., Nagashima T., Minoshima M., Murayama N. Rat cytochrome P450 2C11 in liver microsomes involved in oxidation of anesthetic agent propofol and deactivated by prior treatment with propofol. Drug Metab. Dispos. 2006;34:1803–1805. doi: 10.1124/dmd.106.011627. [DOI] [PubMed] [Google Scholar]
- Yun B.H., Rosenquist T.A., Sidorenko V., Iden C.R., Chen C.H., Pu Y.S., Bonala R., Johnson F., Dickman K.G., Grollman A.P., Turesky R.J. Biomonitoring of aristolactam-DNA adducts in human tissues using ultra-performance liquid chromatography/ion-trap mass spectrometry. Chem. Res. Toxicol. 2012;25:1119–1131. doi: 10.1021/tx3000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachařová A., Siller M., Spičáková A., Anzenbacherová E., Skottová N., Anzenbacher P., Večeřa R. Rosuvastatin suppresses the liver microsomal CYP2C11 and CYP2C6 expression in male Wistar rats. Xenobiotica. 2012;42:731–736. doi: 10.3109/00498254.2012.661099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.