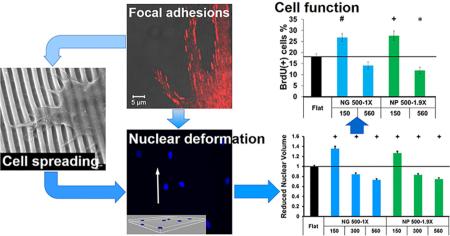
Abstract
Although nanotopography has been shown to be a potent modulator of cell behavior, it is unclear how the nanotopographical cue, through focal adhesions, affects the nucleus, eventually influencing cell phenotype and function. Thus, current methods to apply nanotopography to regulate cell behavior are basically empirical. We, herein, engineered nanotopographies of various shapes (gratings and pillars) and dimensions (feature size, spacing and height), and thoroughly investigated cell spreading, focal adhesion organization and nuclear deformation of human primary fibroblasts as the model cell grown on the nanotopographies. We examined the correlation between nuclear deformation and cell functions such as cell proliferation, transfection and extracellular matrix protein type I collagen production. It was found that the nanoscale gratings and pillars could facilitate focal adhesion elongation by providing anchoring sites, and the nanogratings could orient focal adhesions and nuclei along the nanograting direction, depending on not only the feature size but also the spacing of the nanogratings. Compared with continuous nanogratings, discrete nanopillars tended to disrupt the formation and growth of focal adhesions and thus had less profound effects on nuclear deformation. Notably, nuclear volume could be effectively modulated by the height of nanotopography. Further, we demonstrated that cell proliferation, transfection, and type I collagen production were strongly associated with the nuclear volume, indicating that the nucleus serves as a critical mechanosensor for cell regulation. Our study delineated the relationships between focal adhesions, nucleus and cell function and highlighted that the nanotopography could regulate cell phenotype and function by modulating nuclear deformation. This study provides insight into the rational design of nanotopography for new biomaterials and the cell–substrate interfaces of implants and medical devices.
Keywords: nanotopography, cell spreading, focal adhesions, nuclear deformation, proliferation, transfection, type I collagen
INTRODUCTION
The extracellular matrix (ECM), constructed from diverse biomacromolecules with sizes ranging from several to hundreds of nanometers,1 often manifests significant nanoscale topographies.2–5 Mammalian cells can sense and actively respond to the nanotopography on a synthetic surface, with sensitivity to variations of a few nanometers.6 In addition to inducing pronounced changes to cell morphology, migration, proliferation, nanotopography could potentially regulate cell function, for instance, facilitating the differentiation of stem cells into certain lineages.7–10 Nanotopography, thereby, can be a potent modulator of cell phenotype and function.
The use of nanotopography can dramatically enlarge the surface area; however, the apparent surface that cells can perceive is determined by the shape (e.g., pillars, wells and gratings), dimensions and arrangement of nanofeatures, which is cell type-specific.7,9–13 For instance, human fibroblasts responded differently to demixed isotropic nanoislands of 13, 35, and 95 nm in height. Fibroblasts displayed highly spread morphology containing well-defined cytoskeleton on the 13 nm islands, but had a reduced cell spreading morphology with diffuse actin and less stress fibers when the height of the islands increased.14,15 Anisotropic gratings usually enhance cell alignment, elongation and migration, while they may result in a smaller cell size and lower proliferation rate, even leading to apoptosis.8,11,16–19 On polydimethylsiloxane (PDMS) nanogratings (350 nm in width, 700 nm in pitch and 280 nm in depth), human mesenchymal stem cells (MSCs) exhibited smaller (3.2 ± 0.3 versus 5.3 ± 0.6 μm2 on flat controls) and more dynamic focal adhesions, and significantly downregulated expression of focal adhesion protein zyxin and nuclear proteins lamin C and retinoblastoma protein.20,21 The smaller zyxin-positive adhesions and a higher turnover rate of zyxin proteins led to faster and more directional migration on the gratings, 15.6 μm/h, compared with 8.3 μm/h observed on flat controls.20 The proliferation of MSCs on the nanogratings was significantly lower, 26.9 ± 3.1% of 5-bromo-2-deoxyuridine (BrdU) incorporation than 35.7 ± 7.6% on flat surfaces.8 Notably, on equally spaced nanogratings, neonatal rat ventricular myocytes extended downward toward but did not reach the bottom of 400 nm wide gratings, thus limiting the cell-substrate adhesion. By contrast, the cells could fill the 800 nm gratings completely, with focal adhesions present throughout the cell–groove interface.22 The different cell behavior may result from the alteration in cell-nanotopography adhesion.
Physical signals of ECM/substrates can be transmitted through the adhesion-cytoskeleton-nucleus mechanotransduction pathway to regulate cell phenotype and function.23,24 The molecular connections between focal adhesions, cytoskeletal filaments and nuclear scaffolds may produce integrated changes in cellular and nuclear structure.20,21,25 It has been shown that the differentiation of murine MSCs into a chondrocytic phenotype required a rounded cell shape. Direct comparison of cellular and nuclear shape of MSCs indicated that a more rounded nuclear shape was associated with the greatest expression of molecular markers associated with chondrogenesis.26 Nuclear deformation can result in conformational adaptation in chromatin structure and organization, which affects transcriptional regulation,27 gene expression and protein synthesis,24,28 eventually leading to the changes in proliferation, differentiation or cell death.29,30 Nanotopography can rearrange focal adhesion distribution and cytoskeleton assembly, which is speculated to regulate cellular and nuclear shape and polarity. Modulation of cellular and nuclear shape through the spatial confinement of focal adhesions is crucial in the context of mechanotransduction.31 Although efforts have been made to elucidate the mechanisms by which nanotopographical cues influence cell phenotype and function in the past decade, the relationships between focal adhesions, nuclear volume, phenotype and function of cells on nanotopography have not been fully explored. Thus, current methods to apply nanotopography to regulate cell behavior are basically empirical or even trial and error. The delineation of the relationships of these variables is critical to rational design of nanotopography to modulate cell behavior for specific applications.32,33
In the current study, we engineered PDMS nanotopographies of various shapes (anisotropic gratings and isotropic pillars) and dimensions (feature size, spacing and height), and measured the alignment, elongation and size of focal adhesions and nuclear volume of normal human lung fibroblasts (NHLFs) as an effect of nanotopography. We further examined the nanotopographical effects on cell proliferation, transfection and ECM protein type I collagen production, and correlated the cell functions with the nuclear deformation. The results indicate that nucleus serves as a critical mechanosensor and suggest that the nanotopography can be optimized to regulate cell phenotype and function by modulating nuclear deformation.
RESULTS AND DISCUSSION
Design and Fabrication of Nanotopographies
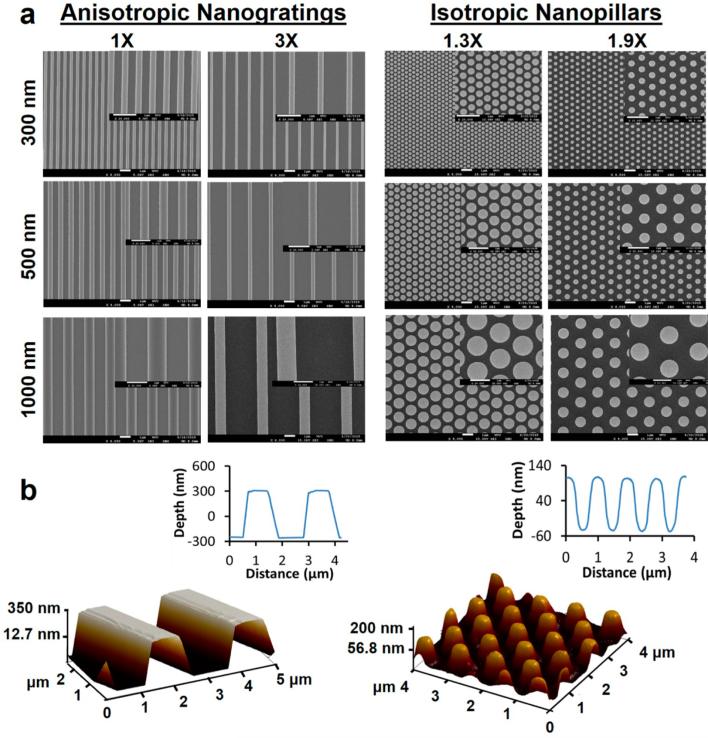
Nanoscale gratings and pillars were of interest because nanogratings (NGs) would provide anisotropic guidance to cells while hexagonally arranged nanopillars (NPs) were isotropic. The PDMS feature sizes were 300, 500, and 1000 nm for the line width of NGs and the diameter of NPs (Figure 1a), covering the interesting feature dimensions of 400–800 nm as previously reported.22 The edge-to-edge spacings of NGs were 1 and 3 times the line width for each feature size and the center-to-center spacings of NPs were 1.3 and 1.9 times the pillar diameter. The lateral dimensions were defined by electron beam lithography (EBL) and the measured values based on scanning electron microscopy (SEM) images (see Table S1 in the Supporting Information) agreed with the design. The height of nanotopographies was controlled mainly by the reaction ion etching (RIE) time, and the heights studied were 150, 300, and 560 nm as measured by using atomic force microscopy (AFM) (Figure 1b). To facilitate the discussion, the nanotopography was labeled by using its abbreviation (NG or NP) followed by a combination of the feature size (line width for NGs and diameter for NPs), the spacing and the height. For instance, “NG 500-3X-150” stood for the nanogratings of 500 nm in line width, 3× the line width (1500 nm) in spacing and 150 nm in height, and “NP 500-1.9X-560” for the nanopillars of 500 nm in diameter, 1.9× diameter in the center-to-center spacing (or 450 nm in the edge-to-edge distance) and 560 nm in height. When the height was not specified the nanotopography was 150 nm in height.
Figure 1.
Characterization of nanotopographies. (a) SEM micrographs of the nanotopographies. The nanotopographies in the same row have the same feature size (the line width of NGs and the diameter of NPs). The nanotopographies in the same column have the same ratio of spacing (the edge-to-edge spacing for NGs and the center-to-center spacing for NPs) to feature size. The inset provides the enlarged image. All scale bars are 1 μm. (b) AFM 3-D image and 2-D profile of NG 500-3X-560 (left) and NP 500-1.9X-150 (right).
By varying the combination of spacing and height, the surface area that the cell could perceive was altered from smaller to larger than the flat surface, which was expected to modulate cell spreading and nuclear deformation from less to more than those on the flat control. The current design allowed us to study systematically the effects of shape and dimensions of nanotopographies on cell spreading, focal adhesion organization, nuclear deformation and the downstream function.
Nanotopographical Modulation of Cell Spreading and Focal Adhesions
Prior to cell seeding, the fibroblasts were serum starved for 18 h to give rise to a 92% population of cells possessing a synchronized G1 phase (see Figure S1 in the Supporting Information), which reduced the variation in starting nuclear size.
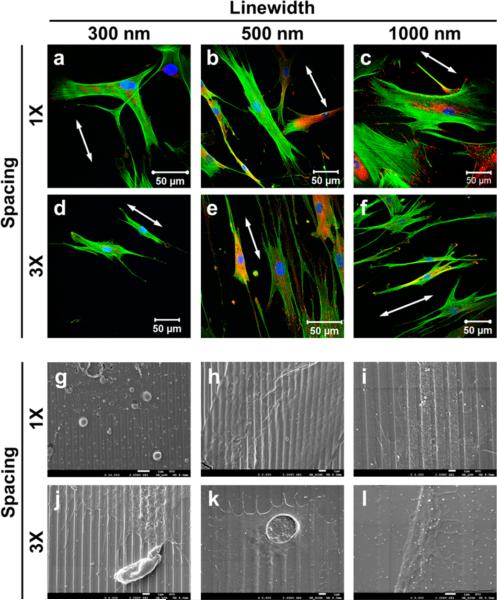
Although the fibroblasts displayed multipolar morphology on the nanopillars, similar to those on the flat control surfaces, they might orient along the nanograting direction (see Figure S2 in the Supporting Information). Surprisingly, the fibroblasts did not always align on the nanogratings. The cells did not display obvious alignment on the equally spaced nanogratings (Figure 2a–c), but preferentially oriented along the nanograting direction when the spacing was 3× the line width (Figure 2d–f). A close SEM examination showed that the cells stayed on the top surface of NG 300-1X without showing a preferential orientation (Figure 2g), but reached the trench of other five nanogratings (Figure 2h–l). On other equally spaced nanogratings (NG 500-1X and NG 1000-1X), the cells overcame the grating barrier and the filopodia reached out to the neighboring gratings. When the spacing was 3× the line width, the cells preferred to follow contact guidance and the filopodia extended along the grating direction. When the spacing was further increased, i.e., 3 μm for NG 1000-3X, the total surface area increased by 4%, the lamellipodia reached the groove bottom and the cell spreading was similar to the flat controls.
Figure 2.
Fibroblasts spreading on nanogratings of 150 nm in height. (a–f) Confocal images of the fibroblasts. The nuclei were stained with DAPI in blue, the actin filaments were stained with phalloidin in green, and focal adhesions were stained with paxillin in red. The white arrows point to the nanograting orientation. (g–l) SEM micrographs of the cell spread on nanogratings. The scale bars are 1 μm.
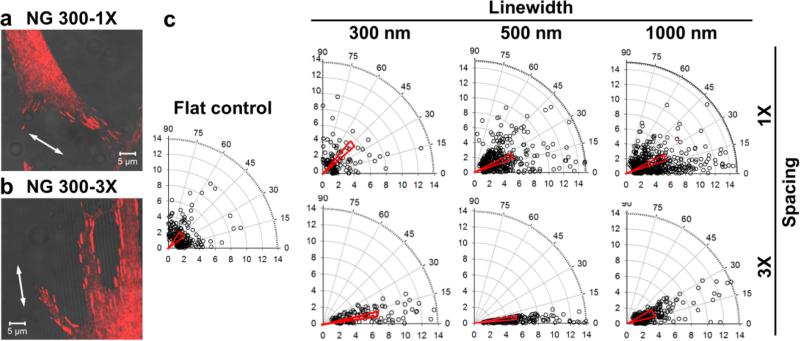
Next, we investigated the organization of focal adhesions, the first and essential step of cellular responses to the substrate, of the fibroblasts on the nanotopographies. The early cell–substrate interactions are critical to mechanosensing.34 The focal adhesions matured from early stage focal complexes could reach several micrometers in length. It was reported that focal adhesions of MC3T3-E1 preosteoblasts grew in length from 4 to 12 h culture.31 We thus quantified the alignment, elongation, and size of focal adhesions protein paxillin ranging between 2 and 10 μm2 (see Figure S3 in the Supporting Information), the typical size for mature focal adhesions after 12 h of cell culture.35 On the flat controls, the focal adhesions were randomly distributed with an average alignment angle of 42.6 ± 1.3 and an average elongation of 2.7 ± 0.1. On the nanogratings of 150 nm in height the focal adhesion alignment displayed similar patterns to the cell spreading except for NG 300-1X, where the focal adhesions did not align along the nanogratings (the alignment angle: 41.0 ± 1.9, Figure 3a). However, the focal adhesions displayed significant alignment on other nanogratings (p < 0.001, Figure 3 and Figure S4a–c in the Supporting Information), and the focal adhesion alignment was enhanced when the spacing increased from 1× line width to 3× line width (exemplified in Figure 3b). For instance, the alignment angle decreased from 25.6 ± 0.8 on NG 500-1X to 13.0 ± 0.8 on NG 500-3X. The enhancement in focal adhesion alignment for 1000 nm gratings was not significant, 23.0 ± 0.8 on NG 1000-1X versus 20.4 ± 0.7 on NG 1000-3X. In addition, the focal adhesions on all these nanogratings were largely elongated (aspect ratios: 4–5) except for NG 1000-3X (see Figure S4d in the Supporting Information).
Figure 3.
Alignment and elongation of focal adhesions on nanogratings of 150 nm in height. (a, b) Overlay of the confocal image of paxillin (red) and the bright field image of nanogratings for the fibroblasts on (a) NG 300-1X and (b) NG 300-3X. (c) Polar plots of focal adhesion alignment (angular coordinate) and elongation (radial coordinate) of the fibroblasts on nanogratings as well as the flat control. Each data point represents a focal adhesion structure. In the plot, the narrow triangle on the top of the data points the mean θ, the length of the short side represents the standard error of mean (S.E.M.), and the length is defined as the elongation aspect ratio r of the focal adhesions.
On the nanopillars of 150 nm in height, the fibroblasts spread in all directions. Except for NP 300-1.3X, where the PDMS pillars were densely packed and collapsed into bigger aggregates because of the dimensional instability, the cells confined on all other pillar arrays and the filopodia extended on the top of both small (300 nm) and large (1000 nm) pillars (see Figure S5 in the Supporting Information). Focal adhesion protein paxillin displayed random orientation on the isotropic pillars (see Figure S6 in the Supporting Information). However, the focal adhesions were also significantly elongated on the nanopillars with the aspect ratio of 4–6 (p < 0.001, see Figure S7 in the Supporting Information).
In the current study, both nanogratings and nanopillars provided anchoring sites to facilitate focal adhesion elongation, but only nanogratings could provide contact guidance for focal adhesions to align along the nanograting direction. The organization of focal adhesions determined cell spreading. As shown in Figures 2 and 3, the alignment of focal adhesions along the nanograting direction was enhanced when the spacing increased from 1× to 3× line width; as a consequence, the cell alignment was promoted on the nanogratings with a spacing of 3× line width. When the spacing was large enough and the lamellipodia extend on the floor of nanotopography (e.g., NG 1000-3X-150), the nanotopographical effects diminished. Therefore, the nanograting-induced focal adhesion alignment and cell spreading were spacing dependent.
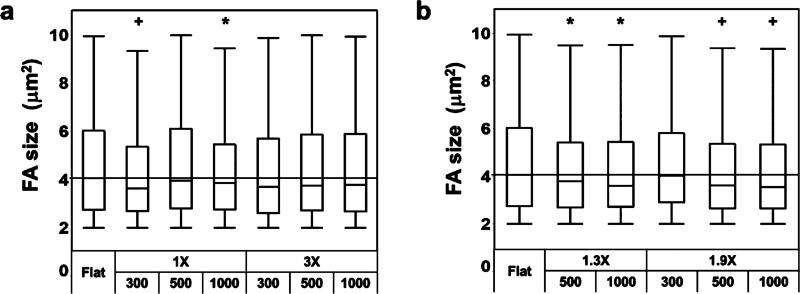
Moreover, we quantified the size of focal adhesions. As summarized in Figure 4, the average focal adhesion sizes on the nanotopographies were smaller than that on the flat controls. No clear trend was observed regarding the effects of feature size or spacing of nanotopographies on the average focal adhesion size. Although the elongation aspect ratios of focal adhesions on all the nanotopographies (except NG 1000-3X with a large spacing) were larger, 4–6 than 2.7 on the flat controls (see Figures S4d and S7 in the Supporting Information), their average focal adhesion sizes were smaller than the flat controls. Evidently, the elongation of focal adhesions did not warrant their size growth. Intriguingly, the decrease in the focal adhesion size was more pronounced on the nanopillars compared to the nanogratings. For example, NG 500-1X had a similar spacing to NP 500-1.9X (500 nm versus 450 nm). However, the average focal adhesion size on the nanogratings was comparable to the flat controls while the size on the nanopillars was significantly smaller than the flat controls (p < 0.001). Considering that the size of mature focal adhesions (2 to 10 μm2) is larger than a single nanofeature, it is understandable that the formation of a focal adhesion structure requests the involvement of multiple nanofeatures and the nanopillars are more effective in disrupting the growth and maturation of focal adhesions than nanogratings.
Figure 4.
Focal adhesion sizes of fibroblasts on the nanotopographies of 150 nm in height. Box plots of focal adhesion size on (a) nanogratings and (b) nanopillars. The straight horizontal lines indicate the median focal adhesion size on the flat control (4.1 ± 0.1 μm2). Significant difference from the flat controls is indicated by * where p < 0.05 and + where p < 0.001.
Nanotopography Modulated Nuclear Deformation
We next examined the nuclear alignment and elongation on the nanotopographies. Compared with the flat controls, the nuclear alignment angle significantly decreased on all nanogratings (p < 0.001, most alignment angles <25, see Figure S8 and Figure S9a–c in the Supporting Information). Different from focal adhesions, the nuclear elongation was less sensitive to the nanogratings (see Figure S9d in the Supporting Information). On the isotropic nanopillars, the nuclei did not show any alignment or significant elongation (data not shown).
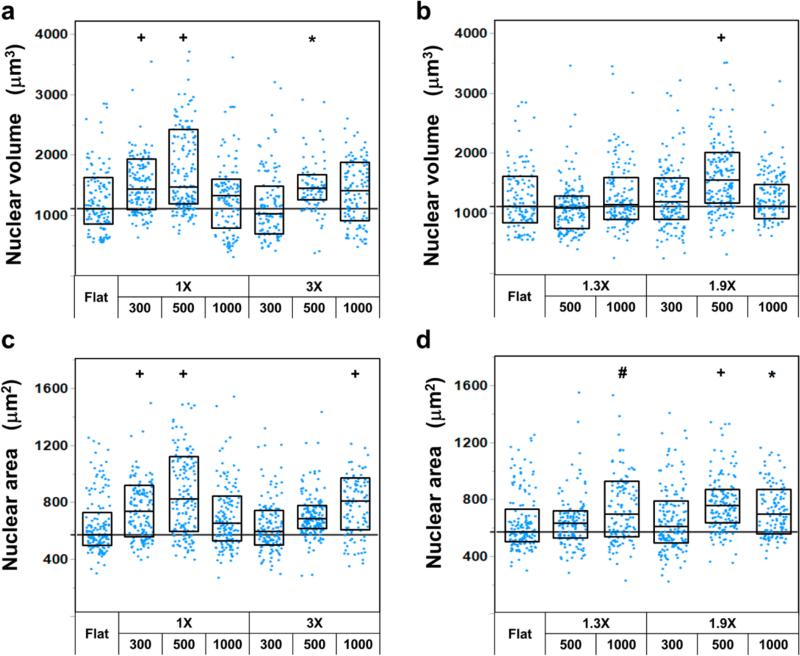
Of greater interest were the nanotopographical effects on nuclear volume. To validate the accuracy of the confocal z-stack measurement of nuclear volume, we measured the FITC-spheres of 15.4 ± 0.13 μm, which were embedded in type I collagen gel. The measured volume of the spheres was 1893.8 ± 71 μm3 (see Figure S10 in the Supporting Information), which agreed well with the calculated volume of 1912.3 ± 48 μm3 (measurement error <1%). On the nanotopographies of 150 nm in height, the fibroblasts showed larger nuclear volumes than those on the flat controls (Figure 5a,b). Generally, the nanogratings had a stronger influence on the nuclear volume than the nanopillars. It was worth noting that the nanotopographical effects on nuclear volume and area showed similar trends regardless of the level of significance (Figure 5a vs 5c and Figure 5b vs 5d). However, no general trends were observed regarding the influence of the feature size, spacing or the surface area on the nuclear deformation.
Figure 5.
Nanotopographical effects on nuclear deformation of fibroblasts. (a) Nuclear volume and (c) projected nuclear area of the fibroblasts on nanogratings. (b) Nuclear volume and (d) projected nuclear area of the fibroblasts on nanopillars. Each data point represents a nucleus. The straight horizontal lines indicate the median nuclear volume (1109.9 ± 45.4 μm3) in (a, b) or area (571.5 ± 16.8 μm2) in (c, d) of the fibroblasts on the flat control. Significant difference from the flat controls is indicated by * where p < 0.05, # where p < 0.01 and + where p < 0.001.
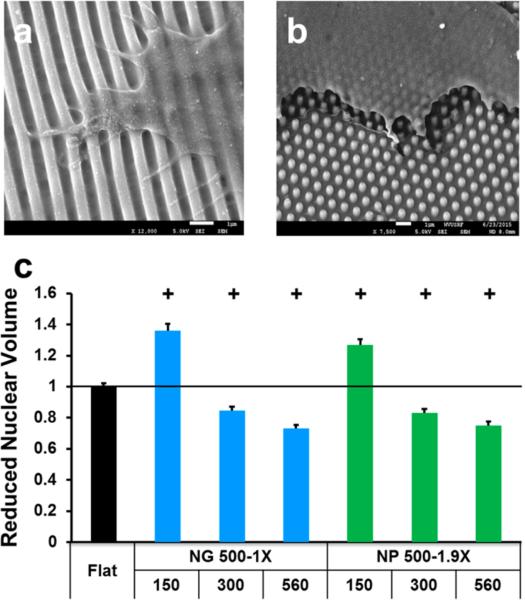
In addition to the shape and lateral dimensions of nanotopographies, we further investigated the influence of the nanotopography height on nuclear volume. On the basis of the results shown in Figure 5, we focused NG 500-1X and NP 500-1.9X, which had significantly higher nuclear volume at a height of 150 nm than the flat controls (p < 0.001). When the nanotopographies were shallow (150 nm), the cells confined on the nanotopographies (Figure 2 and Figure 5S in the Supporting Information) and well spread. With the increase in the height from 150 to 300 and 560 nm, the cells could not completely confine on the feature but bridged over the gaps (Figure 6a,b). The decrease in the cell-nanotopography adhesion reduced cell spreading, leading to a decrease in the nuclear volume. When the nanograting height increased from 150 to 560 nm, the cell spreading area decreased from 2773.4 ± 199.3 to 2081.9 ± 113.3 μm2 (p < 0.0001) and the nuclear volume varied from significantly larger (p < 0.001) to smaller (p < 0.001) than the flat controls (Figure 6c). The nanopillars also demonstrated similar effects to the nanogratings. To verify the nanotopography height effect, we further examined the difference in nuclear size on the representative nanotopographies using polystyrene (PS), the most commonly used cell culture substrate. We focused on nuclear area because of the similarity between nuclear volume and area demonstrated in Figure 5. The nuclear areas on PS nanotopographies of 560 nm height, i.e., NG 500-1X-560 and NP 500-1.9X-560, were significantly smaller than the corresponding nanotopography of 150 nm height (p < 0.0001) as well as the flat controls, similar to what was observed on PDMS nanotopographies (see Figure S11 in the Supporting Information). The cells could deflect pliant PDMS but not rigid PS nanotopographies, in particular the high features, as reported here and in a previous study.29 The deflection of nanofeatures might mediate intracellular tension and affect cell behavior, and as a consequence, the extent that nanotopography affected cell behavior such as nuclear deformation could differ. Nonetheless, the nanotopography could exert similar effects on cell behavior regardless of the substrate materials used.
Figure 6.
Effects of nanotopography height on cell spreading and nuclear volume. SEM micrographs of fibroblasts grown on (a) NG 500-1X-560 and (b) NP 500-1.9X-560. Scale bars are 1 μm. (c) Height-dependent nuclear volume. The first and second rows of x-axis labels provide the lateral dimensions and the height (both in nm) of nanotopographies. Significant difference from the flat controls is indicated by + where p < 0.001.
Our observations suggested that the nuclear size could be modulated more effectively by the height of nanotopography. Previously, we have shown that the average nuclear area of human MSCs decreased to 145.1 ± 4.1 μm2 on the PDMS nanogratings of 350 nm line width and 280 nm height from 194.8 ± 4.8 μm2 on the flat control.36 Recently, McKee et al. measured the nuclear volume of several human corneal epithelial cells grown on equally spaced nanogratings with the line width of 200, 400, 600, 800, and 1000 nm and a constant height of 300 nm. They found that the nuclear volume increased as the line width increased from 200 to 1000 nm, and the 200 nm nanogratings resulted in the smallest volume and other nanogratings had higher nuclear volume than the flat surface.37 Our observations agreed with the previous reports,36,37 and showed with solid data (sample size n > 150) that with an increase in height, the cell-nanotopography adhesion decreased, leading to a reduction in the nuclear volume. Moreover, McKee et al. showed that there was a significant decrease in the nuclear elastic modulus on 200 nm gratings compared to the moduli for flat surfaces and for larger gratings.37 It was expected that the nanotopography height could effectively modulate nuclear mechanics through nuclear deformation.
The external physical signals can be transmitted through the adhesion-cytoskeleton-nucleus mechanotransduction pathway to regulate cell phenotype and function. Therefore, in addition to investigating the nanotopographical effects on the initial and final components of this pathway, we examined the correlation between the two components. Note that the fibroblasts on the nanotopographies of 150 nm height had smaller average focal adhesion sizes (Figure 4) but larger nuclear volumes (Figure 5) compared with the flat controls. The nuclear volume became significantly smaller than the flat controls when the nanotopography height increased to 560 nm (Figure 6c). We postulated that the total area of focal adhesions instead of the average focal adhesion size determined cell spreading and nuclear deformation. This was supported by the observations reported by Jean and the co-workers.38 They measured nuclear deformation in response to the alteration of cell shape as cells detached from a flat surface, and found that the nuclear deformation appeared to be in direct and immediate response to the alteration of the cell adhesion area.
Nanotopography Height-Dependent Cell Function
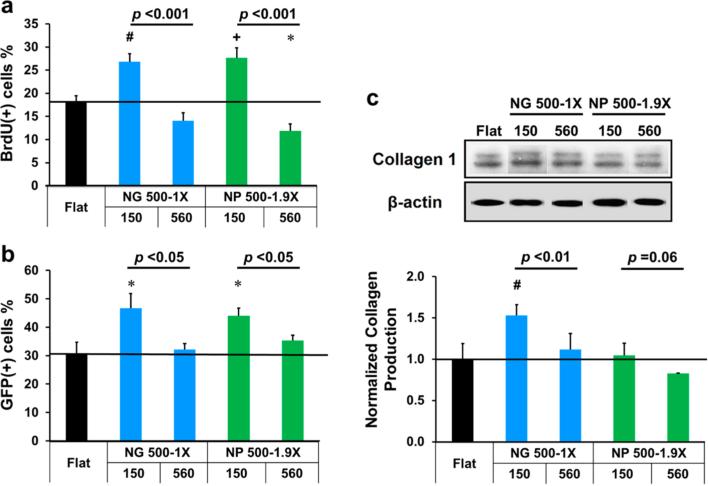
We further studied nanotopographical effects on cell functions such as cell proliferation, transfection and type I collagen production, with the focus on NG 500-1X and NP 500-1.9X of different heights, 150 and 560 nm. Interestingly, the nanotopographies showed similar influences on these functions to that on nuclear volume. As shown in Figure 7a, when the height of NG 500-1X increased from 150 to 560 nm, the percentage of BrdU positive cells decreased from higher (26.8 ± 1.8%) to lower (14.1 ± 1.7%) than the flat controls (18.1 ± 1.3%) while the nuclear volume changed from 37% larger to 27% smaller than the flat controls. The cell proliferation displayed a similar trend on NP 500-1.9X.
Figure 7.
Nanotopography height-dependent (a) proliferation, (b) transfection efficiency and (c) type I collagen production of fibroblasts. The first and second rows of x-axis labels provide the lateral dimensions and the height (nm) of nanotopographies, respectively. Significant difference from the flat controls is indicated by * where p < 0.05, # where p < 0.01 and + where p < 0.001. The p value between two groups (same lateral dimensions but different heights) is provided.
Similar nanotopographical effects were observed on the transfection efficiency of the fibroblasts (Figure 7b). The transfection efficiency on the nanotopographies of 150 nm hight was significantly higher than the flat control (p < 0.05), 46.8 ± 5.0% and 44.0 ± 2.6% versus 30.7 ± 4.1% for NG 500-1X and NP 500-1.9X, respectively. When the height increased from 150 to 560 nm, the transfection efficiency decreased and the difference between them and the flat controls was not significant.
We also examined the production of type I collagen, an important ECM protein as a critical cell function of fibroblasts. Western blot analysis (Figure 7c) showed that the type I collagen expression was higher on nanotopographies of 150 nm height than those on 560 nm nanotopographies and the type I collagen expression on NG 500-1X-150 was significantly higher than the flat controls, indicating that both height and shape of nanotopography had profound effects on type I collagen production.
Nuclear deformation can alter gene and protein expression,28,39 and is strongly associated with cell proliferation and differentiation.30,40 Roca-Cusachs et al. suggested that cell spreading promoted DNA synthesis by inducing nuclear swelling, thereby induced an increase in proliferation, actin polymerization (F-actin content), and stress fiber formation.40 The enhancement in cell proliferation, made by applying stiff substrates41 or micro/nanotopographies,42,43 can promote the efficiency of nonviral transfection. Our study demonstrated close correlations between nuclear deformation and cell proliferation, transfection and type I collagen production. By mediating cell spreading and cytoplasm membrane tension, nanotopography modulates nuclear deformation and thus alters cell proliferation and nuclear accessibility to nonviral vectors.44,45 Nanotopography-mediated cell spreading also affects cell contractility and then type I collagen production.46
In this study, we examined the correlations between focal adhesions, cell spreading, nuclear deformation, and cell function as an effect of nanotopography. Cells adhere and respond to the substrate through adhesive proteins/ligands. We postulate that nanotopography regulates cell behavior by restraining the adhesive ligands at the substrate surface that cells can perceive. At nanoscale, the cell membrane elasticity may prevent a radical bending,47 and hence favoring the plasma membrane to bridge over the top of nanofeatures rather than reaching the bottom when the spacing is small and/or the features are tall. The reduced cell-nanotopography adhesion restricts focal adhesion formation, leading to reduced cell spreading and decreased nuclear volume. With an increased spacing and/or a decreased height, the cell is able to reach partially, or even fully the bottom of the nanotopography and the cell would “see” a larger surface area, resulting in enhanced cell spreading and increased nuclear volume. Briefly, nanotopography modulates cell spreading by organizing focal adhesions through adhesive ligand rearrangement, which has implications in nuclear deformation. Our study suggests that the height of nanotopography is an effective regulator of nuclear deformation. Compared with nanogratings which provide continuous, anisotropic contact guidance to the cell, discrete, isotropic nanopillars tend to disrupt the formation and growth of focal adhesions and thus have less profound influence on nuclear volume. Alteration in nuclear volume and elastic modulus or plasticity is strongly associated with cell function.30,37 This study provides solid evidence that nucleus serves as a critical mechanosensor in nanotopographical modulation of cell phenotype and function.
More important, the understanding of the adhesion-cytoskeleton-nucleus mechanotransduction pathway provides insight into the rational design of nanotopography for regenerative medicine. The shape and dimensions, in particular the height of nanotopography can be optimally combined to largely enhance cell spreading through focal adhesions rearrangement and thus to increase the nuclear volume, which is favorable in bone regeneration (osteogenesis) and gene medicine.42 On the other hand, the shape and dimensions can be optimized to reduce cell spreading and nuclear volume, which may be desirable for maintaining the stemness of stem cells, as previously reported that the nanogratings which resulted in reduced nuclear volume and enhanced alignment facilitated neuronal differentiation of human MSCs.8
CONCLUSIONS
We engineered a variety of nanotopographies covering primary shapes and important dimensional parameters, and thoroughly investigated cell spreading, focal adhesion organization and nuclear deformation of human primary fibroblasts as the model cell grown on the nanotopographies, and examined the correlations between nuclear deformation and cell proliferation, transfection and type I collagen production. It was found that the nanoscale gratings and pillars could facilitate focal adhesion elongation by providing anchoring sites, and the nanogratings could orient focal adhesions and nuclei along the nanograting direction, depending on not only the feature size but also the spacing of the nanogratings. Compared with continuous nanogratings, discrete nanopillars tended to disrupt the formation and growth of focal adhesions and thus had less profound effects on nuclear deformation. Notably, the height of nanotopography was shown to be an effective regulator of nuclear deformation. Our observations suggest that the nucleus serves as a critical mechanosensor for cell regulation. By optimizing the shape and dimensions, in particular height of nanotopography, the nuclear volume can be modulated through focal adhesion rearrangement to regulate cell function for the end application. Delineation of the relationships between cell adhesion, nucleus and cell function provides insight into the rational design of nanotopography for new biomaterials and the cell–substrate interfaces of implants and medical devices.
EXPERIMENTAL SECTION
Fabrication of Nanotopographies
Nanotopographies were generated by EBL. Briefly, a poly(methyl methacrylate) (495 PMMA A4; MicroChem, Westborough, MA, USA) thin film was spin-coated onto a silicon substrate and a focused electron beam of 20 nm in diameter was used to directly write the designed pattern on the PMMA film. After developed, the nanopatterned PMMA surface was deposited with a thin nickel layer using a Kurt J. Lesker E-beam evaporator (Jefferson Hills, PA, USA). The PMMA part was then lifted off in Remover PG (MicroChem) at 80 °C, thus leaving a reversed nickel pattern on the silicon substrate, which functioned as a mask for the following RIE process performed in a Trion Minilock III ICP/RIE (Clearwater, FL, USA).
A mixture of PDMS resin and curing agent (Sylgard 184 kit, Dow Corning, Midland, MI, USA) in a 10:1.05 w/w ratio was poured onto the EBL-written mold. Different from the manufacturer recommended ratio of 10:1, a slightly higher concentration of curing agent was added to reduce the amount of uncured oligomers.48 After curing at 70 °C for 4 h, the inverse PDMS nanotopography was peeled from the EBL mold. To facilitate the separation of PDMS nanotopography from the EBL mold, the EBL mold was silanized with 1H,1H,2H,2H-perfluorooctylmethyldichlorosilane (Oakwood Products, West Columbia, SC, USA) for 5 h under vacuum prior to the PDMS casting.
To seed enough cells for subsequent biological analyses, the PDMS nanotopography was expanded to a large area of nanopatterned substrate by applying a stitch technique developed previously.36 Briefly, multiple PDMS nanotopographies, replicated from an EBL mold were aligned with the nanopatterned surface face-down on a silicon wafer, thus ensuring that all the nanopatterned surfaces were at the same level. The backside of these PDMS nanotopographies was glued with a PDMS prepolymer layer spin-coated on a glass plate. After curing at 80 °C for 2 h, a single large nanopatterned surface was created. The stitched mold was then imprinted into polystyrene (PS) substrates, which were directly used for cell culture and also served as a master mold to replicate working PDMS substrates.
The PDMS substrates including nanopatterned and the flat control surfaces were sterilized by using 70% ethanol and then UV exposure, each for 30 min, and incubated with fibronectin (human, BD Biosciences, San Jose, CA, USA) of 20 μg/mL in phosphate buffered saline (PBS, Life Technologies, Carlsbad, CA, USA) for 15 min prior to cell seeding.
Cell Culture
Normal human lung fibroblasts (NHLFs, CRL1490 cells from ATCC, Manassas, VA, USA) were cultured in fibroblast basal medium supplemented with 2% fetal bovine serum (FBS), 0.1% recombinant human fibroblast growth factor basic (rhFGF-B), 0.1% insulin, and 0.1% gentamicin/amphotecin-B (Lonza, Walkersville, MD, USA), and used in passages 3–6.
NHFLs were starved in the serum-free medium for 18 h to induce cell cycle synchronization,49 and then refreshed with complete culture medium. The cells were seeded on the substrates at a density of 4000 cells/cm2 and cultured at 37 °C and 5% CO2 for a predefined period of time before the analyses.
SEM Observations
The substrates with cells grown were washed with PBS and fixed in a mixture of 4% paraformaldehyde (PFA, Electron Microscopy Sciences, Hatfield, PA, USA) and 2% glutaraldehyde (Fisher Scientific, Pittsburgh, PA, USA) in PBS at ambient for 4 h. The samples were then rinsed with deionized water and progressively dehydrated using a Tousimis Autosamdri-815 CO2 Critical Point Dryer (Tousimis Co., Rockville, MD, USA).
The substrates with and without cells grown were sputter-coated with a gold layer of ~10 nm thick by using a Denton Vacuum Desk V sputter unit (Denton Vacuum, LLC, Moorestown, NJ, USA), and imaged in a JEOL JSM-7600F SEM.
AFM Measurement
AFM measurement was performed using a Veeco Nanoscope MultiMode scanning probe microscope in the tapping mode under ambient conditions. Topography images were recorded at the fundamental resonance frequency of the cantilever, with a typical scan rate of 1 Hz and a resolution of 512 samples per line.
Flow Cytometry
The cells were harvested as a single cell suspension in PBS and fixed in cold 70% ethanol overnight at 4 °C. Fixed cells were washed with PBS containing 2% FBS and resuspended in PBS containing 0.2% Tween 20 (Fisher Scientific) for 15 min at 37 °C. Next, the cells were washed and resuspended in 1.8% RNase-A (Sigma-Aldrich, St. Louis, MO, USA) in PBS for 15 min at room temperature. Propidium Iodide (50 mg/mL, Life Technologies) in PBS was added for 15 min at room temperature. After incubation, the volume of each sample was brought up to 500 mL. Samples were assayed using a LSRFortessa (BD Biosciences) and Diva 8 software immediately after staining. A minimum of 20 000 events were collected for each samples. Final cell cycle analysis (data modeling) was done using FCS Express 4 software (De Novo Software Los Angeles CA).
Immunofluorescence Staining
NHLFs were fixed in 4% PFA in PBS for 15 min at room temperature, and permeabilized in a blocking solution, which consisted of 0.03 g/mL bovine serum albumin (BSA, Sigma-Aldrich) and 0.1% goat serum (Sigma-Aldrich) in 0.2% Triton X-100 (Sigma-Aldrich) in PBS for 1 h. Primary antibody conjugation was performed in 0.2% Triton X-100 in PBS for 2 h, followed by secondary antibody labeling for 1 h, both at room temperature. Focal adhesions were stained with antipaxillin rabbit monoclonal antibody (1:200; Abcam, Cambridge, MA, USA), followed by the Alexa Fluor 555 goat antirabbit secondary antibody (1:200; Life Technologies). BrdU was stained with anti-BrdU mouse monoclonal antibody (1:200; Life technologies), followed by the Alexa Fluor 488 antimouse IgG secondary antibody (1:200; Life Technologies). F-actin was stained with Alexa Fluor 488 phalloidin (Life Technologies), and the nuclei were counterstained and mounted using ProLong Gold Antifade Reagent with 4,6-diamidino-2-phenylindole (DAPI, Life Technologies).
Imaging was performed with a Zeiss LSM 510 inverted confocal microscope. For the nuclear volume measurement, confocal z-stack imaging with 0.1 μm intervals between planar images was performed. To validate the accuracy of z-stack imaging of nuclei, FITC-labeled beads (diameter: 15.4 ± 0.13 μm, BD Bioscience) were used as the standard.
Proliferation Assay
22 h after NHLFs were cultured in complete culture medium on the nanotopographies and the flat controls, BrdU labeling reagent (1:100; Life technologies) was added and the cells were cultured for additional 2 h. The immunofluorescence staining for BrdU detection was described in the Immunofluorescence Staining Section. Prior to permeabilization, the cells were treated with 2 N hydrogen chloride for 30 min to separate DNA into single strands.
Cell Transfection
pEGFP-N1 (Clontech Laboratories, Mountain View, CA, USA) plasmid expressing green fluorescent protein (GFP) was employed for the cell transfection assay. NHFLs were first cultured with the complete medium on the nanotopographies and the flat controls for 24 h. Next, the cells were transfected with pEGFP-N1 plasmid using Lipofectamine 2000 (Life Technologies) in the absence of serum for 5 h. The medium was then replaced with the complete medium, and the cells were cultured for another 19 h, followed by 4% PFA fixation and DAPI staining.
Western Blot
The whole cell protein was extracted by lysing cells with radioimmune precipitation assay (RIPA) buffer (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) containing protease and phosphatase inhibitors for 30 min on ice, separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene difluoride (PVDF) membranes (EMD Millipore, Darmstadt, Germany). The PVDF membranes were then blocked with 5% nonfat milk in TBST (1× Tris-buffered saline with 0.1% Tween-20), followed by blotting with anticollagen I rabbit monoclonal antibody (Fitzgerald, Concord, MA, USA, 1:3000) at 4 °C overnight with shaking, and incubating with a horseradish peroxidase-conjugated secondary antibody (Sigma-Aldrich) for 1 h at room temperature. Protein bands were visualized using enhanced chemiluminescence detection reagents (EMD Millipore) and acquired by C DiGit Blot Scanner (LI-COR Biosciences, Lincoln, NE, USA).
Densitometric analysis was performed using ImageJ (http://rsb.info.nih.gov/ij/index.html). The collagen bands were corrected against β-actin, compared with those in cells cultured on flat PDMS surface.
Image Analysis
Focal adhesion size and nuclear area and volume were analyzed using Imaris (Bitplane, South Windsor, CT, USA). For focal adhesion analysis, superficial surfaces were first created to overlay the image. The threshold was set to the TRITC channel and was adjusted to best match the confocal image. Focal adhesions (surfaces) of sizes between 2 and 10 μm2 were extracted using a filter and analyzed. The focal adhesions of different size were further labeled using different colors (see Figure S3 in the Supporting Information). Nuclei were analyzed by using the same method except swapping the channel to DAPI (see Figure S10 in the Supporting Information).
The alignment and elongation of focal adhesions and nuclei were analyzed using ImageJ. The nuclei (based on the projected area) and focal adhesions were highlighted by adjusting threshold of the brightness, outlined, and best-fitted with ellipses. From the ellipses, the major and minor axes were determined and the elongation was defined by the aspect ratio of major axis/minor axis. The alignment angle was defined by the angle between the major axis and the nanograting direction (see Figure S3 in the Supporting Information).
Fluorescence imaging (for proliferation and transfection assays) of stained cells was performed using a 10× objective on a Nikon Eclipse Ti-E inverted microscope. The proliferation and transfection efficiency were calculated by dividing BrdU positive cells or GFP positive cells by the total cell number indicated by DAPI staining.
Statistical Analysis
Dimensions of nanotopographies were expressed as mean ± standard error from 6 measurements. Focal adhesions (n > 1000, from 15 to 20 cells), nuclear area and volume (n > 150), proliferation (n > 200), and transfection (n > 200) of NHLFs were measured from three replicates for each condition and presented as mean ± standard error of mean (S.E.M.). The difference in cell behaviors is analyzed by one-way ANOVA and compared using Dunnet's method.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by NSF CBET 1227766, NSF CBET 1511759 and NSF EPS 1003907. We gratefully acknowledge use of the West Virginia University (WVU) Shared Research Facilities which are supported, in part, by NSF EPS-1003907, the WVU Microscope Imaging Facility, which has been supported by the Mary Babb Randolph Cancer Center (MBRCC) and NIH grants P30 GM103488 and P20 RR016477, and the WVU Flow Cytometry Core supported by MBRCC CoBRE grant GM103488/RR032138, Fortessa S10 grant OD016165 and WV InBRE grant GM103434. We thank X Yu for her help with nanotopography fabrication and cell imaging. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.5b10531.
Flow cytometry analysis of cell cycle synchronization of the fibroblasts, immunofluorescence and SEM images of fibroblasts spreading on nanotopographies and the flat control surface, image analysis of focal adhesions, summary of alignment and elongation of focal adhesions on nanogratings of 150 nm in height, SEM images of fibroblasts interacted with nanopillars of 150 nm in height, details and summary of focal adhesion alignment and elongation on nanopillars of 150 nm in height, details and summary of nuclear alignment and elongation on nanogratings of 150 nm in height, validation of confocal z-stack measurement of nuclear volume, effect of nanotopography height on nuclear area of fibroblasts grown on PDMS and PS nanotopographies, and comparison between the designed and measured dimensions of nanotopographies (PDF).
The authors declare no competing financial interest.
REFERENCES
- 1.Mwenifumbo S, Stevens MM. ECM Interactions with Cells from the Macro- to Nanoscale. In: Gonsalves K, Halberstadt C, Laurencin CT, Nair L, editors. Biomedical Nanostructures. 1 ed. Wiley-Interscience; Hoboken, NJ: 2008. pp. 225–260. [Google Scholar]
- 2.Abrams GA, Goodman SL, Nealey PF, Franco M, Murphy CJ. Nanoscale Topography of the Basement Membrane Underlying the Corneal Epithelium of the Rhesus Macaque. Cell Tissue Res. 2000;299:39–46. doi: 10.1007/s004419900074. [DOI] [PubMed] [Google Scholar]
- 3.Shirato I, Tomino Y, Koide H, Sakai T. Fine Structure of the Glomerular Basement Membrane of the Rat Kidney Visualized by High-resolution Scanning Electron Microscopy. Cell Tissue Res. 1991;266:1–10. doi: 10.1007/BF00678705. [DOI] [PubMed] [Google Scholar]
- 4.Hironaka K, Makino H, Yamasaki Y, Ota Z. Renal Basement Membranes by Ultrahigh Resolution Scanning Electron Microscopy. Kidney Int. 1993;43:334–345. doi: 10.1038/ki.1993.51. [DOI] [PubMed] [Google Scholar]
- 5.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF. Effects of Synthetic Micro- and Nano-structured Surfaces on Cell Behavior. Biomaterials. 1999;20:573–588. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 6.Brunetti V, Maiorano G, Rizzello L, Sorce B, Sabella S, Cingolani R, Pompa P. Neurons Sense Nanoscale Roughness with Nanometer Sensitivity. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6264–6269. doi: 10.1073/pnas.0914456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. The Control of Human Mesenchymal Cell Differentiation Using Nanoscale Symmetry and Disorder. Nat. Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 8.Yim EKF, Pang SW, Leong KW. Synthetic Nanostructures Inducing Differentiation of Human Mesenchymal Stem Cells into Neuronal Lineage. Exp. Cell Res. 2007;313:1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh S, Brammer KS, Li YSJ, Teng D, Engler AJ, Chien S, Jin S. Stem Cell Fate Dictated Solely by Altered Nanotube Dimension. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMurray R, Gadegaard N, Tsimbouri P, Burgess K, McNamara L, Tare R, Murawski K, Kingham E, Oreffo R, Dalby M. Nanoscale Surfaces for the Long-term Maintenance of Mesenchymal Stem Cell Phenotype and Multipotency. Nat. Mater. 2011;10:637–644. doi: 10.1038/nmat3058. [DOI] [PubMed] [Google Scholar]
- 11.Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. Epithelial Contact Guidance on Well-defined Micro- and Nanostructured Substrates. J. Cell Sci. 2003;116:1881–1892. doi: 10.1242/jcs.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W, Villa-Diaz LG, Sun Y, Weng S, Kim JK, Lam RH, Han L, Fan R, Krebsbach PH, Fu J. Nanotopography Influences Adhesion, Spreading, and Self-renewal of Human Embryonic Stem Cells. ACS Nano. 2012;6:4094–4103. doi: 10.1021/nn3004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeon H, Koo S, Reese WM, Loskill P, Grigoropoulos CP, Healy KE. Directing Cell Migration and Organization via Nanocrater-Patterned Cell-repellent Interfaces. Nat. Mater. 2015;14:918–923. doi: 10.1038/nmat4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalby MJ, Riehle MO, Johnstone H, Affrossman S, Curtis ASG. In vitro Reaction of Endothelial Cells to Polymer Demixed Nanotopography. Biomaterials. 2002;23:2945–2954. doi: 10.1016/s0142-9612(01)00424-0. [DOI] [PubMed] [Google Scholar]
- 15.Dalby MJ, Marshall GE, Johnstone HJH, Affrossman S, Riehle MO. Interactions of Human Blood and Tissue Cell Types with 95-nm-high Nanotopography. IEEE Trans. Nanobioscience. 2002;1:18–23. doi: 10.1109/tnb.2002.806933. [DOI] [PubMed] [Google Scholar]
- 16.Dunn GA, Brown AF. Alignment of Fibroblasts on Grooved Surfaces Described by a Simple Geometric Transformation. J. Cell Sci. 1986;83:313–340. doi: 10.1242/jcs.83.1.313. [DOI] [PubMed] [Google Scholar]
- 17.Lenhert S, Meier M-B, Meyer U, Chi L, Wiesmann HP. Osteoblast Alignment, Elongation and Migration on Grooved Polystyrene Surfaces Patterned by Langmuir-Blodgett Lithography. Biomaterials. 2005;26:563–570. doi: 10.1016/j.biomaterials.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 18.Gerecht S, Bettinger CJ, Zhang Z, Borenstein JT, Vunjak-Novakovic G, Langer R. The Effect of Actin Disrupting Agents on Contact Guidance of Human Embryonic Stem Cells. Biomaterials. 2007;28:4068–4077. doi: 10.1016/j.biomaterials.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bettinger CJ, Zhang Z, Gerecht S, Borenstein JT, Langer R. Enhancement of in vitro Capillary Tube Formation by Substrate Nanotopography. Adv. Mater. 2008;20:99–103. doi: 10.1002/adma.200702487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulangara K, Yang Y, Yang J, Leong KW. Nanotopography as Modulator of Human Mesenchymal Stem Cell Function. Biomaterials. 2012;33:4998–5003. doi: 10.1016/j.biomaterials.2012.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulangara K, Yang J, Chellappan M, Yang Y, Leong KW. Nanotopography Alters Nuclear Protein Expression, Proliferation and Differentiation of Human Mesenchymal Stem/Stromal Cells. PLoS One. 2014;9:e114698. doi: 10.1371/journal.pone.0114698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh KY, Tung L, Levchenko A. Nanoscale Cues Regulate the Structure and Function of Macroscopic Cardiac Tissue Constructs. Proc. Natl. Acad. Sci. U. S. A. 2010;107:565–570. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wormer DB, Davis KA, Henderson JH, Turner CE. The Focal Adhesion-localized CdGAP Regulates Matrix Rigidity Sensing and Durotaxis. PLoS One. 2014;9:e91815. doi: 10.1371/journal.pone.0091815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering Gene Expression and Protein Synthesis by Modulation of Nuclear Shape. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1972–1977. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maniotis AJ, Chen CS, Ingber DE. Demonstration of Mechanical Connections between Integrins, Cytoskeletal Filaments, and Nucleoplasm that Stabilize Nuclear Structure. Proc. Natl. Acad. Sci. U. S. A. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McBride SH, Knothe Tate ML. Modulation of Stem Cell Shape and Fate A: The Role of Density and Seeding Protocol on Nucleus Shape and Gene Expression. Tissue Eng., Part A. 2008;14:1561–1572. doi: 10.1089/ten.tea.2008.0112. [DOI] [PubMed] [Google Scholar]
- 27.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear Shape, Mechanics, and Mechanotransduction. Circ. Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CDW, Curtis ASG. Nucleus Alignment and Cell Signaling in Fibroblasts: Response to a Micro-grooved Topography. Exp. Cell Res. 2003;284:272–280. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 29.Chalut KJ, Kulangara K, Giacomelli MG, Wax A, Leong KW. Deformation of Stem Cell Nuclei by Nanotopographical Cues. Soft Matter. 2010;6:1675–1681. doi: 10.1039/B921206J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical Plasticity of the Nucleus in Stem Cell Differentiation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natale CF, Ventre M, Netti PA. Tuning the Material-cytoskeleton Crosstalk via Nanoconfinement of Focal Adhesions. Biomaterials. 2014;35:2743–2751. doi: 10.1016/j.biomaterials.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 32.Frey MT, Tsai IY, Russell TP, Hanks SK, Wang YL. Cellular Responses to Substrate Topography: Role of Myosin II and Focal Adhesion Kinase. Biophys. J. 2006;90:3774–3782. doi: 10.1529/biophysj.105.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ankam S, Lim CK, Yim EK. Actomyosin Contractility Plays a Role in MAP2 Expression during Nanotopography-directed Neuronal Differentiation of Human Embryonic Stem Cells. Bio-materials. 2015;47:20–28. doi: 10.1016/j.biomaterials.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Gautrot JE, Malmstrom J, Sundh M, Margadant C, Sonnenberg A, Sutherland DS. The Nanoscale Geometrical Maturation of Focal Adhesions Controls Stem Cell Differentiation and Mechanotransduction. Nano Lett. 2014;14:3945–3952. doi: 10.1021/nl501248y. [DOI] [PubMed] [Google Scholar]
- 35.Zaidel-Bar R, Cohen M, Addadi L, Geiger B. Hierarchical Assembly of Cell-matrix Adhesion Complexes. Biochem. Soc. Trans. 2004;32:416–420. doi: 10.1042/BST0320416. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Kulangara K, Sia J, Wang L, Leong KW. Engineering of a Microfluidic Cell Culture Platform Embedded with Nanoscale Features. Lab Chip. 2011;11:1638–1646. doi: 10.1039/c0lc00736f. [DOI] [PubMed] [Google Scholar]
- 37.McKee CT, Raghunathan VK, Nealey PF, Russell P, Murphy CJ. Topographic Modulation of the Orientation and Shape of Cell Nuclei and their Influence on the Measured Elastic Modulus of Epithelial Cells. Biophys. J. 2011;101:2139–2146. doi: 10.1016/j.bpj.2011.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jean RP, Gray DS, Spector AA, Chen CS. Characterization of the Nuclear Deformation Caused by Changes in Endothelial Cell Shape. J. Biomech. Eng. 2004;126:552–558. doi: 10.1115/1.1800559. [DOI] [PubMed] [Google Scholar]
- 39.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering Gene Expression and Protein Synthesis by Modulation of Nuclear Shape. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1972–1977. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roca-Cusachs P, Alcaraz J, Sunyer R, Samitier J, Farre R, Navajas D. Micropatterning of Single Endothelial Cell Shape Reveals a Tight Coupling between Nuclear Volume in G1 and Proliferation. Biophys. J. 2008;94:4984–4995. doi: 10.1529/biophysj.107.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong HJ, Liu J, Riddle K, Matsumoto T, Leach K, Mooney DJ. Non-viral Gene Delivery Regulated by Stiffness of Cell Adhesion Substrates. Nat. Mater. 2005;4:460–464. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- 42.Adler AF, Speidel AT, Christoforou N, Kolind K, Foss M, Leong KW. High-throughput Screening of Microscale Pitted Substrate Topographies for Enhanced Nonviral Transfection Efficiency in Primary Human Fibroblasts. Biomaterials. 2011;32:3611–3619. doi: 10.1016/j.biomaterials.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang NC, Ji QM, Ariga K, Hsu SH. Nanosheet Transfection: Effective Transfer of Naked DNA on Silica Glass. NPG Asia Mater. 2015;7:e184. [Google Scholar]
- 44.Huang C, Ozdemir T, Xu LC, Butler PJ, Siedlecki CA, Brown JL, Zhang S. The Role of Substrate Topography on the Cellular Uptake of Nanoparticles. J. Biomed. Mater. Res., Part B. 2015 doi: 10.1002/jbm.b.33397. DOI: 10.1002/jbm.b.33397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medina-Kauwe LK, Xie J, Hamm-Alvarez S. Intracellular Trafficking of Nonviral Vectors. Gene Ther. 2005;12:1734–1751. doi: 10.1038/sj.gt.3302592. [DOI] [PubMed] [Google Scholar]
- 46.Balestrini JL, Chaudhry S, Sarrazy V, Koehler A, Hinz B. The Mechanical Memory of Lung Myofibroblasts. Integr. Biol. 2012;4:410–421. doi: 10.1039/c2ib00149g. [DOI] [PubMed] [Google Scholar]
- 47.Ohara PT, Buck RC. Contact Guidance in vitro: A Light, Transmission, and Scanning Electron Microscopic Study. Exp. Cell Res. 1979;121:235–249. doi: 10.1016/0014-4827(79)90002-8. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, Kulangara K, Lam RTS, Dharmawan R, Leong KW. Effects of Topographical and Mechanical Property Alterations Induced by Oxygen Plasma Modification on Stem Cell Behavior. ACS Nano. 2012;6:8591–8598. doi: 10.1021/nn301713d. [DOI] [PubMed] [Google Scholar]
- 49.Chen M, Huang J, Yang X, Liu B, Zhang W, Huang L, Deng F, Ma J, Bai Y, Lu R, Huang B, Gao Q, Zhuo Y, Ge J. Serum Starvation Induced Cell Cycle Synchronization Facilitates Human Somatic Cells Reprogramming. PLoS One. 2012;7:e28203. doi: 10.1371/journal.pone.0028203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.