Abstract
Noninvasive and practical techniques to longitudinally track viral infection are sought after in clinical practice. We report a proof-of-principle study to monitor the viral DNA copy number using a newly established mouse papillomavirus (MmuPV1) mucosal infection model. We hypothesized that viral presence could be identified and quantified by collecting lavage samples from cervicovaginal, anal and oral sites. Nude mice infected at these sites with infectious MmuPV1 were tracked for up to 23 weeks starting at 6 weeks post-infection. Viral DNA copy number was determined by SYBR Green Q-PCR analysis. In addition, we tracked viral DNA load through three complete oestrous cycles to pinpoint whether there was a correlation between the DNA load and the four stages of the oestrous cycle. Our results showed that high viral DNA copy number was reproducibly detected from both anal and cervicovaginal lavage samples. The infection and disease progression were further confirmed by histology, cytology, in situ hybridization, immunohistochemistry and transmission electron microscopy. Interestingly, the viral copy number fluctuated over the oestrous cycle, with the highest level at the oestrus stage, implying that multiple sampling might be necessary to provide a reliable diagnosis. Virus DNA was detected in oral lavage samples at a later time after infection. Lower viral DNA load was found in oral samples when compared with those in anal and vaginal tracts. To our knowledge, our study is the first in vivo study to sequentially monitor papillomavirus infection from mucosal anal, oral and vaginal tracts in a preclinical model.
Introduction
Human papillomavirus (HPV) is the most common sexually transmitted virus. It causes more than 5 % of human cancers and claims more than 270 000 lives from cervical cancer annually. HPV-associated anal and oral cancers are on the rise despite the advancement in the treatment of these cancers (Sathish et al., 2014; Stier et al., 2015). Concurrent anal and cervical high risk (HR) HPV infection was found in nearly half of women with CIN 2+ (Sehnal et al., 2014). According to NCI data, about 7270 new cases (4630 in women and 2640 in men) of anal cancer were reported in 2014 with 1010 deaths (610 in women and 400 in men). The prevalence of HR-HPV in the anus of HIV-positive women is higher than in the anus of HIV negative women (Brickman & Palefsky, 2015).
Two protective HPV vaccines hold promise to substantially reduce HPV related cancers in the coming years (Dochez et al., 2014). However, the uptake of these vaccines is still low worldwide due to social and economic barriers. In addition, these vaccines offer no therapeutic effect for pre-existing infections (Campo & Roden, 2010; Ting et al., 2014). Therefore, monitoring HPV infection and associated diseases and cancers will continue to be important, at least until an effective therapeutic treatment is available.
We are working to establish a proof-of-concept noninvasive protocol to track viral DNA copy numbers as well as abnormalities in cervicovaginal, anal and oral locations longitudinally. The Papinocolou (Pap) smear is a routine diagnostic tool for the detection of cervical dysplasia and has played a critical role in the decline of cervical cancer (Korn, 1996; Waxman, 2008). However, clinician-directed pelvic examinations require trained staff and are not readily acceptable to many women; this represents a challenge in both the research setting and in the clinic. To further strengthen cervical and anal cancer screening efforts, self-sampling techniques for the collection of cervical and anal cells have been proposed as an effective alternative to overcome these limitations. Several recent studies have demonstrated that self cervicovaginal lavage sampling, although less sensitive than clinician-acquired samples, is a reliable method for the detection of HPV DNA (Castell et al., 2014; Haguenoer et al., 2014; Igidbashian et al., 2011; Ortiz et al., 2013). Cervical self-sampling for HPV is acceptable to women and has been acknowledged as a way to increase the likelihood of participation in cervical cancer screening programmes, particularly among underserved populations (Castell et al., 2014; Deleré et al., 2011; Haguenoer et al., 2014; Stewart et al., 2007). This noninvasive and convenient method would facilitate the screening of large populations. Although data for anal samples are very limited, studies in women in Puerto Rico and in men who have sex with men in the US have also shown good agreement between self-sampling and clinician-sampling methods (Lampinen et al., 2006; Rosenberger et al., 2011). For oral infection, few studies on noninvasive sampling were found in the literature (Meyer et al., 2014).
Papillomavirus displays a strictly species-specific selectivity that constrains the opportunity to develop a preclinical animal model to study HPV infection. Several natural animal papillomavirus infection models including cow, horse, dog and rabbit have been widely used to study mechanisms of viral pathogenesis (Brandsma, 2005; Campo & Roden, 2010; Hu et al., 2007, 2009; Nicholls et al., 2001). These models have played a pivotal role in the development of the current vaccines (Bergot et al., 2011). However, these large animal models are not optimal to study mucosal infection because of limited resources and reagents (Campo, 2002; Doorbar, 2013). A small rodent model would be the most appropriate for this purpose. We have demonstrated that the newly established mouse model is susceptible to MmuPV1 infection at several mucosal sites, including oral, cervical, vaginal and anal tract in addition to the cutaneous sites (Cladel et al., 2013, 2015; Handisurya et al., 2014; Joh et al., 2012; Sundberg et al., 2014). In the current study, we aimed to establish a proof-of-concept protocol to track virus from these mucosal sites which are applicable to clinical settings. We hypothesized that the collection of lavage samples from anogenital and oral cavities was feasible for viral detection as well as pathological determination. HSD outbred Foxn1nu/+ nude mice were infected with MmuPV1 at vaginal, anal or oral cavity and tracked for viral infection by collecting non-invasive lavages and followed by Q-PCR analysis. The cytology analysis was used to determine the oestrous cycle stages and abnormalities of the infected cells exfoliated from the cervicovaginal tract. The infected tissues were examined by histology, in situ hybridization (ISH), immunohistochemistry (IHC) and transmission electron microscopy (TEM) for viral presence.
Results
Viral copy numbers can be monitored in cervicovaginal lavage samples by Q-PCR from both wounded and unwounded vaginally MmuPV1 infected animals
Wounding prior to inoculation was recommended from a previous study using HPV pseudovirus to deliver reporter genes to the vaginal tract of mice (Roberts et al., 2007). Our published and preliminary studies demonstrated that wounding is not essential for vaginal infection (Cladel et al., 2013). In the current study, seven nude mice were infected at the vaginal tract with MmuPV1. Among these animals, we tracked two with wounding (4-3L and 4-3R) and two without wounding (5-3L and 5-3R) before MmuPV1 infection by harvesting lavage samples as early as 6 weeks post-infection. Cervicovaginal lavage samples are an effective source for detection of HPV infection in humans (Deleré et al., 2011). Using SYBR Green Q-PCR analysis, we monitored viral copy numbers in 1 μl of 50 μl DNA extract from a lavage sample of the genital tract from infected animals up to week 23 (Table 1). The data were converted into equivalent DNA load using the formula 1 ng viral DNA = 1.2 × 108 copies of viral DNA (http://cels.uri.edu/gsc/cndna.html). To account for the variation from sample collection, a housekeeping gene that encoded 18sRNA was also used as the internal control. A fold change between viral copy number and 18sRNA gene was calculated using the difference between their cycle time (ΔC t) and subsequently fold change (2ΔCt) (Fig. 1). Wounding, although not essential, did promote infection at an earlier stage (Fig. 1, Table 1). However, once infections were launched in the non-wounded animals, extensive infection could be detected throughout the vaginal tract (Fig. 2a, b) to the cervix (Fig. 2c, d) of mouse 5-3L by ISH.
Table 1. Tracking viral DNA in nude mice infected with MmuPV1 at vagina, anus and tongue. Viral DNA load (in 1 μl from 50 μl DNA extract of a lavage sample) in the vaginal, anal and oral cavity over time was determined by SYBR Green Q-PCR analysis.
| Mouse | Infection site | Wounding | Week 6 | Week 9 | Week 14 | Week 16 | Week 18 | Week 20 | Week 23 |
|---|---|---|---|---|---|---|---|---|---|
| 4-3L | Vagina | + | 9.5 × 104 | 1.3 × 104 | 1.2 × 107 | 1.6 × 107 | 1.3 × 107 | 3.3 × 107 | 2.9 × 107 |
| 4-3R | Vagina | + | 1.7 × 106 | 3.4 × 106 | 1.4 × 107 | 1.3 × 107 | 7 × 106 | 1.3 × 107 | 1.6 × 107 |
| 5-3L | Vagina | − | 9 × 104 | < 1 × 104 | 8.5 × 104 | 1 × 105 | 8.5 × 104 | 2.2 × 106 | 8.5 × 106 |
| 5-3R | Vagina | − | 7.5 × 104 | < 1 × 104 | 1.1 × 105 | 1.4 × 107 | 2 × 106 | 6 × 105 | 1.2 × 105 |
| 1-3L | Anus | + | 1.1 × 105 | 5 × 104 | 4.5 × 106 | 1.2 × 107 | 8.9 × 106 | 1.4 × 107 | 1 × 107 |
| 1-3R | Anus | + | 4 × 104 | < 1 × 104 | 8 × 104 | 2 × 106 | 1 × 105 | 6 × 106 | 1.7 × 107 |
| Week 18 | Week 23 | Week 30 | Week 33 | ||||||
| 2-3L | Tongue | + | 5.1 × 105 | 5.9 × 105 | 7.0 × 105 | 9.8 × 105 | 1.1 × 106 | ||
| 2-3R | Tongue | + | 6.7 × 105 | 6.7 × 105 | 7.7 × 105 | 6.6 × 105 | 1.0 × 106 | ||
| 3-3R | Tongue | + | 8.1 × 105 | 3.1 × 105 | 1.4 × 106 | 1.4 × 106 | 1.1 × 106 |
Fig. 1.
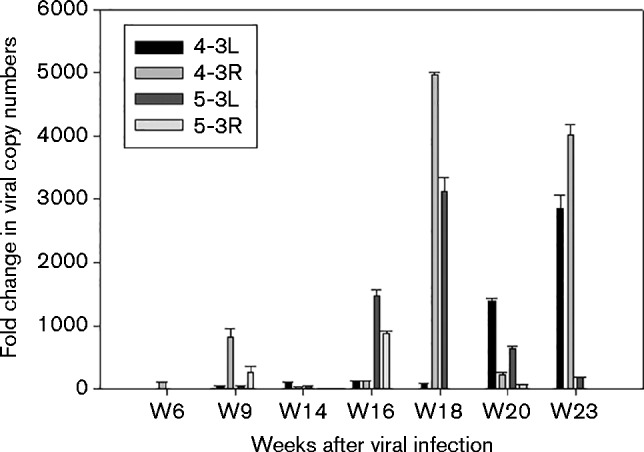
Fold changes in viral copy number versus 18sRNA gene from the vaginal lavage samples by tracking up to 23 weeks post-infection. All mice were treated with Depo-Provera at 3 days before MmuPV1 infection. The vaginal tracts of two mice (4-3L and 4-3R) were wounded with cytobrush and N-9 before MmuPV1 infection and the other two mice (5-3L and 5-3R) were not wounded before infection. SYBR Green Q-PCR was conducted with DNA samples extracted from the vaginal lavage. We calculated the difference between cycle time (C t) between the 18sRNA gene and viral DNA (ΔC t) and subsequently the fold change (2ΔCt). Error bars represent se.
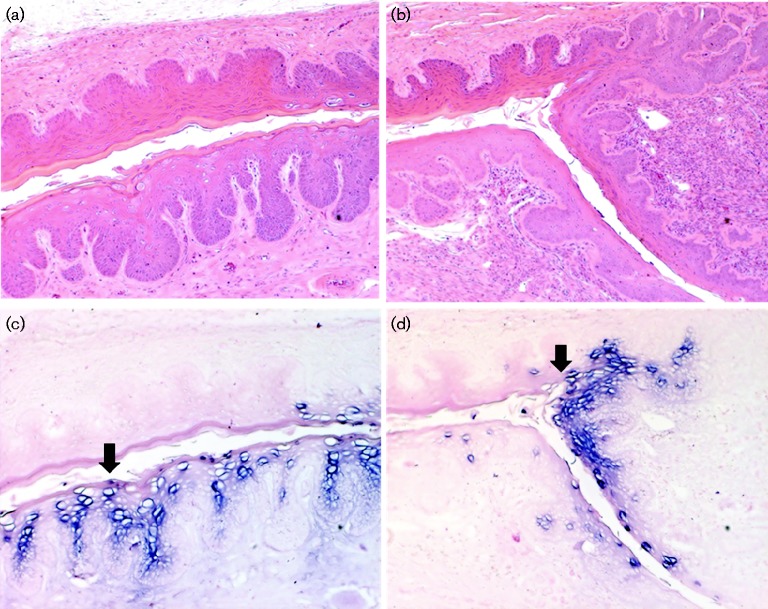
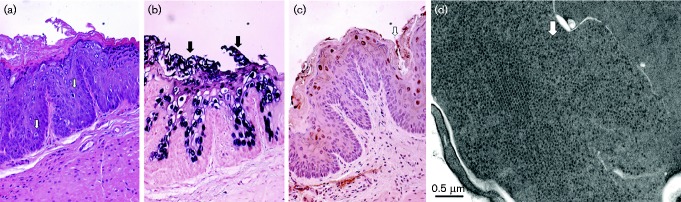
Fig. 2.
Histology and ISH of 5-3L at week 37 after MmuPV1 infection in the vaginal tract without wounding. (a) Vaginal tract by H&E at × 10; (b) cervix by H&E at × 10; viral DNA positive cells were present in (c) the vaginal tract, × 10, arrow, as well as (d) the cervix, × 10, arrow, by ISH.
Viral detection in the vaginal tract varies according to the oestrous cycle
In humans, the reproductive cycle, called the menstrual cycle, lasts approximately 28 days; in mice, this cycle, named the oestrous cycle, lasts approximately 4–5 days. The short cycle length makes mice an ideal animal model for investigating viral infection during the reproductive cycle (Teepe et al., 1990). Previous studies suggested that HPV detection might change during the menstrual cycle (Liu et al., 2013; Schmeink et al., 2010). We hypothesized that the mouse oestrous cycle might play a role in MmuPV1 detection because of the difference in viral shedding at different stages. Of seven vaginally infected animals, two wounded (4-3L and 4-3R) and two unwounded animals (5-3L and 5-3R) were followed for three oestrous cycles (5 continuous days to cover the complete cycle). Cervicovaginal lavage samples were collected and DNA was extracted for SYBR Green Q-PCR analysis. Viral copy numbers, as well as the relative fold change in viral DNA load of two 5-day cycles (weeks 20 and 27) are shown in Fig. 3. Fig. 3(a, c) shows the viral DNA copy numbers/lavage sample, while Fig. 3(b, d) shows the relative fold change between viral copy numbers and 18sRNA gene per lavage sample. Four naïve nude mice were also tested for viral presence. Viral DNA was not detectable from these naïve animals (data not shown). The relative fold change between copy numbers of viral DNA vs corresponding 18sRNA gene was below 5 (Fig. S1, available in the online Supplementary Material). Our infected animals showed a change with larger magnitude (Fig. 3b, d). Interestingly, viral detection fluctuated over the 5-day cycle. For the wounded mice (4-3L and 4-3R), the DNA load was above the detection threshold at all the time points while virus was undetectable at some of the time points for the unwounded animals (5-3L and 5-3R).
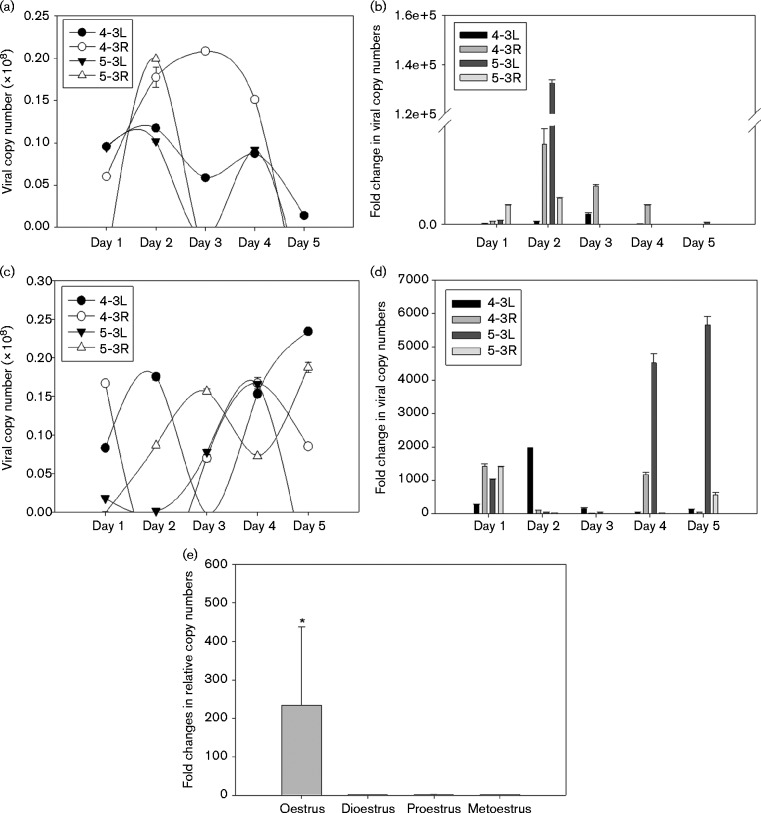
Fig. 3.
Viral copy numbers detected with SYBR Green Q-PCR at (a, b) week 20 and (c, d) week 27 post-infection. The equivalent copy number was calculated using the formula 1 ng viral DNA = 1.2 × 108 copy numbers and based on the detection of viral DNA in 1 μl of 50 μl DNA extracted from vaginal lavage samples. All mice were treated with Depo-Provera at 3 days before the viral infection. The vaginal tracts of two mice (4-3L and 4-3R) were wounded with cytobrush and N-9 before MmuPV1 infection and the other two mice (5-3L and 5-3R) were not wounded before MmuPV1 infection. DNA load from the vaginal lavage samples for weeks 20 and 27 post-infection is shown (a, c). The difference in cycle time between viral DNA and 18sRNA gene (ΔC t) was used to calculate the fold change (2ΔCt) shown in the y-axis (b, d, e). Significantly higher viral copy numbers were associated with oestrus than with the other three stages (one-way ANOVA, *P < 0.05 vs the other stages; error bars represent se).
We next examined whether viral copy numbers were correlated with specific oestrous stages. A typical oestrous cycle stage determination result is shown in Table S1. For the individual animals, we noticed that the oestrus stage was correlated with higher titre of viral copy numbers. Notably, significantly higher viral copy numbers were detected at the oestrus stage when compared with the other three stages (Fig. 3e, P < 0.05, one way ANOVA analysis).
Abnormal cell identification from the lavage samples at different oestrous cycle stages
The oestrous cycle as well as cytological changes due to viral infection can be monitored by collecting cervicovaginal lavages (Cladel et al., 2015). Frequent and very large numbers of atypical squamous cells with ribbon like central chromatin and abundant amphophilic cytoplasm (resembling inclusion) were found after 20 weeks post-infection (Fig. 4a–d). Subsequent examination conducted with IHC and ISH showed these abnormal cells to be virally infected cells (Cladel et al., 2015). MmuPV1 viral DNA and viral particles could be detected in the vaginal tract and cervix in these vaginally infected animals by ISH (Fig. 5a) as well as TEM (Fig. 5b, c).
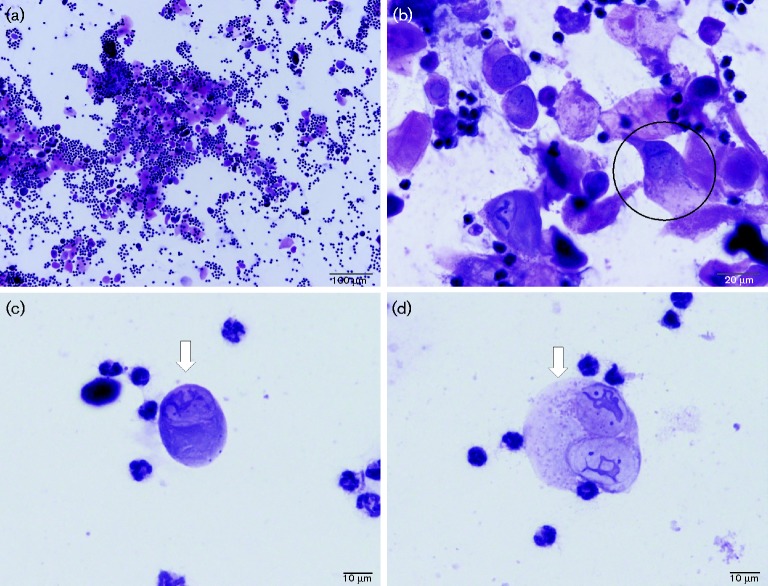
Fig. 4.
Cytology of vaginal lavage samples at week 20 post-infection. Oestrous cycle stages can be determined from lavage samples by H&E analysis. (a) This vaginal infected animal showed atypical metoestrus/dioestrus ( × 10). Frequently atypical squamous cells with ribbon like central chromatic and abundant amphophilic cytoplasm (arrows) resembling inclusion are seen. (b) A tri-nucleate cell (the circled cell, × 40). (c) Atypical cells with ribbon-like nuclei are shown in the lavage samples ( × 100, arrow) and (d) a bi-nucleate cell ( × 100, arrow) can be seen by H&E analysis.
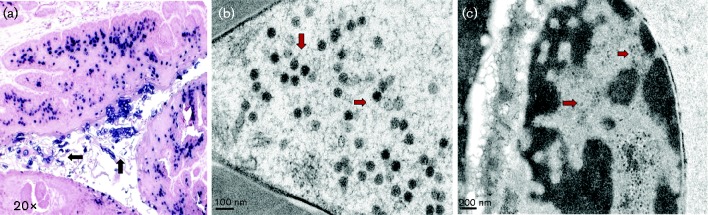
Fig. 5.
ISH at week 26 and TEM analyses at week 36 after MmuPV1 infection of the vaginal tract. (a) Strong viral DNA positivity is shown within vaginally infected cells as well as inside the vaginal tract ( × 20, indicating possible viral shedding, black arrows). (b, c) These viral particles can be visualized with TEM in the infected vaginal tissues and inside the tract (red arrows).
Viral detection in the anal tract
Our previous studies demonstrated active infection at the anal infection site, especially at the transformation zone area (Cladel et al., 2013, 2015). We hypothesized that it was possible to monitor viral DNA in the infected anal canal by collecting anal lavage. Two (1-3L and 1-3R) of five anally infected animals were followed up to 23 weeks post-infection. The viral copy number per lavage sample is reported in Table 1. The fold change in viral DNA copy number is shown in Fig. S2. We also tested anal lavage samples from four naïve nude mice for viral presence and all mice showed negative results. The relative fold change in copy numbers between viral DNA and corresponding 18sRNA gene was less than five (Fig. S1). The DNA load in these anally infected animals was comparable to that in the genital tract, indicating that possible viral shedding was also occurring in the anal tract. Proliferative cells were detected from anally infected animals at the transition zone, a site that has been identified as being susceptible to viral infection in both humans and mice (Baandrup et al., 2014; Cladel et al., 2015) (Fig. 6a). Productive infection was further confirmed by ISH (Fig. 6b) and IHC (Fig. 6c). Viral particles were found at the anally infected sites by TEM (Fig. 6d).
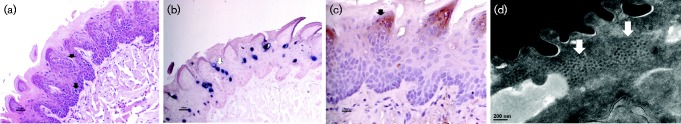
Fig. 6.
(a) H&E ( × 20), (b) ISH ( × 20), (c) IHC ( × 20) analyses of the anal tract at 26 weeks and (d) TEM at week 36 after viral infection. (a) Koilocytes can be seen in the infected anal tissues (white arrows). (b) These infected cells showed strong viral DNA positivity; some infected cells might be shedding from the anal tract (black arrows). Viral capsid protein and viral particles can be detected by both (c) IHC with a goat Group Specific Antibody (GSA) to a conserved region of L1 (ViroStat) (white arrows) and (d) TEM, respectively.
Viral detection in the oral cavity
Our previous study demonstrated that MmuPV1 infection appeared at some oral sites in animals initially infected at cutaneous sites (Cladel et al., 2013). This finding indicates that the oral cavity is susceptible to MmuPV1 infection. We hypothesized that viral presence could be detected from the oral rinse in mice infected with MmuPV1 in the tongue. Three (2-3L, 2-3R and 3-3R) out of the seven orally infected animals were monitored up to 33 weeks. We were not able to detect viral DNA until week 14 post-infection. Viral copy number per sample was much lower than that of cervicovaginal and anal lavage samples through all the time points we examined (Table 1). The DNA load did not change thereafter over time. The fold change in viral copy number versus 18sRNA gene is shown in Fig. S3. ISH demonstrated active infection in the infected tongue (Fig. 7b). Viral capsid and viral particles could be detected in the infected cells by either IHC (Fig. 7c) or TEM (Fig. 7d).
Fig. 7.
(a) Analysis of tongue by H&E ( × 20), (b) ISH ( × 20), (c) IHC ( × 40) with an in house monoclonal antibody MPV.B9 at week 14 after infection and (d) TEM at week 30, respectively, after viral infection. (a) Koilocytes can be seen in the infected tongue tissue ( × 20, black arrows); (b) some of these infected cells showed strong viral DNA positivity ( × 20, white arrows); (c) viral capsid positive cells are shown with an in-house monoclonal antibody (MPV.B9) targeting denatured viral capsid protein ( × 40, black arrows) and (d) viral particles are seen in infected cells (white arrows).
Discussion
We reported here a proof-of-concept protocol to track virus infection from mucosal sites including the oral cavity, the vaginal and anal canals, which are applicable to clinical settings. Viral copy numbers could be detected as early as week 6 after viral infection and possibly earlier from vaginal and anal tracts according to our most recent studies (unpublished observations). The viral DNA detected in our Q-PCR analysis may be arising from free viral particles as well as from the infected exfoliated cells of these mucosal tissues. Furthermore, ISH, IHC and TEM confirmed the presence of viral DNA, viral capsid protein as well as viral particles at these mucosal sites. Cytology of cervicovaginal lavage was used to examine abnormalities in exfoliated cells. In summary, this noninvasive sampling is feasible and effective for evaluating mucosal viral infection longitudinally.
For cutaneous infection, we found that pre-wounding did enhance the viral infectivity in our cottontail rabbit papillomavirus model (Cladel et al., 2008). Wounding has been reported to be necessary for an HPV pseudovirus to deliver reporter genes in the mouse vaginal tract (Roberts et al., 2007). However, our published studies showed that non-wounded mucosal sites including vagina, anus and oral cavity were also susceptible for MmuPV1 infection (Cladel et al., 2013). In the current study, we compared vaginal MmuPV1 infection with and without pre-wounding. Consistent with the previous findings, wounding promoted infection at an earlier time with a higher viral DNA load. Due to the small number of animals in our study, we could not determine the level of enhancement via wounding. However, we also confirmed that wounding was not necessary for MmuPV1 infection in the vaginal tract (Cladel et al., 2013). We hypothesize that the infectivity of natural virions is much higher than that of the pseudoviruses and thus significant wounding is not required for natural infection in vivo. We examined viral DNA encapsidated in the naturally produced virions and found the virions contained only viral DNA. In contrast, pseudoviruses contained mostly cellular DNA (our unpublished observations) (Holmgren et al., 2005). Therefore, the efficiency of natural viral infection is predicted to be much higher when compared with the pseudovirus delivery system. While we have shown that deliberate wounding is not necessary for MmuPV1 infection at mucosal sites, we cannot exclude the possibility that natural ‘wounding’ at the micro level during the continuous oestrous cycle facilitated viral infection in these animals. In the current study, we used Depo-Provera to synchronize all the animals at the dioestrus stage, which was found to be the most susceptible stage for other viral infections such as HSV2 (Gallichan & Rosenthal, 1996; Kaushic et al., 2003, 2011; Teepe et al., 1990). At the dioestrus stage, large numbers of leukocytes accumulate in the mouse cervicovaginal tract. The potential for direct viral entry into epithelial cells in this microenvironment needs further study.
Cytology of cervicovaginal lavage has allowed us to scrutinize the changes in the exfoliated cells (McLean et al., 2012). We not only detected abnormal cells from these samples but also confirmed that some of these abnormal cells were viral positive (Cladel et al., 2015). This finding suggests that viral shedding is possible in the cervicovaginal tract. In patient studies, a systematic quantitative review also demonstrated that infants born through vaginal delivery were at higher risk of exposure to HPV (Medeiros et al., 2005). Although viral infection was initiated in the lower vaginal tract in this study, we detected viral infection and abnormalities at the upper vaginal tract as well as the cervix. These findings suggest that the vaginal tract can serve as a viral reservoir for cervical infection (Medeiros et al., 2005). It's unclear whether this virus travelled to the cervix. Although Pap smear is a routine practice to examine cervical abnormality, we show that lavage samples can be used as a prescreening tool prior to the need for a Pap smear. This finding will be of special importance in areas such as developing countries with limited resources.
Normal female mice usually have a 4–5 day oestrous cycle and four distinguishable stages (proestrus, oestrus, metoestrus and dioestrus) can be identified by cytological determination (Byers et al., 2012; McLean et al., 2012). The oestrous cycle can be interrupted by pregnancy or hormonal treatment (such as the Depo-provera used in the experiment) (Caligioni, 2009). Other factors can play a role in an abnormal oestrous cycle in mice. For example, grouping females can trigger pseudo-pregnancy in which all the animals remained at dioestrus (Ryan & Schwartz, 1977). Repeated lavage was found to attenuate cocaine-stimulated behaviours in female rats (Walker et al., 2002). One of our vaginally infected animals stayed at the oestrus stage for the 5 consecutive days of sampling. Whether repeated lavage induced the abnormal oestrous cycle in this animal is unclear.
Different viruses have shown different patterns of shedding in reproductive-aged women. HIV shedding was reported greatest immediately following menses and lowest at ovulation (Curlin et al., 2013). HSV shedding did not vary with the menstrual cycle, but CMV shedding was significantly more frequent in the luteal phase (Mostad et al., 2000). The correlation between HPV detection and menstrual cycle has been inconsistent in the literature (Liu et al., 2013; Schmeink et al., 2010; Tota et al., 2013). In one study, HR HPV detection was significantly influenced by sample timing in the menstrual cycle (Schmeink et al., 2010). Another study found all HPV DNA likely peaks at the periovulatory phase (Liu et al., 2013). However, no correlation was found in Tota's study (Tota et al., 2013). In our current study, even with a small number of animals, we found that animals at the oestrous stage, the only stage at which the animals mate, had the highest viral DNA load in the vaginal lavage samples. This may indicate high MmuPV1 shedding occurs at the oestrus stage.
The dual tissue tropism of MmuPV1 that is evident in nude mice may be due to the deficiency of strong host defence in the nude mice (García-Sastre et al., 1998; McFadden et al., 2009; O'Brien, 1992). We did test mucosal MmuPV1 infection in immunocompetent animals such as B6 mice. Transient viral DNA was detected in lavage samples at week 3 after MmuPV1 infection in partially CD4/CD8 depleted B6 mice and subsequently became undetectable (unpublished observations). Antibodies were generated in these animals, indicating the possible clearance of viral infections (unpublished observations). These findings suggest that active viral infection was cleared by a strong host adaptive immune response in B6 mice, a response that is lacking in the nude mice (Taghian & Huang, 1995). Given the evidence of possible viral shedding from the anal and vaginal sites, we were not surprised to see that these mucosally infected animals also showed secondary visible lesions at cutaneous sites including tail, muzzle and other skin sites over a period of time (unpublished observations). Grooming may play a role in the spread of the virus shed from the mucosal sites to the cutaneous sites. We hypothesize that MmuPV1 could be a mucosa-tropic virus in nature but manifested dual tropism in the immunocompromised nude mice. There is also the possibility that MmuPV1 is a virus with a broader tissue tropism (such as the gamma papillomaviruses) than previously appreciated (Forslund et al., 2013; Ure & Forslund, 2014). Further studies will address these unknowns.
Viral DNA load in the anal lavage was comparable to that in the vaginal lavage, indicating possible viral shedding in the anal tract. Few studies have attempted to monitor viral infection in the anal tract (Bown et al., 2014; Liszewski et al., 2014; Wells et al., 2014). Our study demonstrated the feasibility of using anal lavage samples as a diagnostic tool. This observation is not surprising because the anal transition zone, located just distally to the rectum, supports viral replication and production and can be accessed easily (Assi et al., 2014). Our study suggests that HPV-associated anal infection can be tracked with a noninvasive method and that early detection of viral presence is achievable.
Viral DNA copy numbers were much lower in the oral cavity lavage samples when compared with those obtained from the vaginal and anal tracts in our study, although infection could be easily detected by ISH and IHC. Viral particles in the infected tongue tissues were much lower in numbers when compared with those in the vaginal and anal tracts, indicating the possibility of limited viral shedding in the oral cavity. One complication of the study was the limited number of cells harvested from the oral cavity. When we delivered the saline rinse solution to the inside of the mouth, some animals simply swallowed the liquid. Therefore, we might not have harvested sufficient numbers of exfoliated cells for adequate viral DNA extraction. This finding indicates that lavage might not be the ideal method for sampling viral infection in the oral cavity of mice. However, swallowing may not be a problem in adult patients and therefore could still be a valid technique for humans as shown in the literature (Boutaga et al., 2007; Meyer et al., 2014). An alternative method to increase the chance of detecting oral HPV infection might be a mouth swab. This technique has been demonstrated to be effective for PCR detection of HPV DNA (Sun et al., 2000).
In summary, our study demonstrated that persistent papillomavirus infection can be monitored longitudinally by collecting lavage samples from mucosal sites such as vaginal, anal and possibly the oral cavity. Given the fact that there is no general screening for HPV associated anal cancer, tracking papillomavirus infection in the anal tract would be very useful. These preclinical observations will pave the way for self-testing as a more acceptable and readily available tool for large scale screening for HPV infections.
Methods
Viral stock
Infectious virus was isolated from lesions on the tails of mice from our previous study (Cladel et al., 2013). In brief, the lesions were scraped from the tail with a scalpel blade and homogenized in PBS using a Polytron homogenizer (Brinkman PT10-35) at highest speed for 3 minutes while chilling in an ice bath. The homogenate was spun at 10 000 r.p.m. and the supernatant was decanted into Eppendorf tubes for storage at − 20 °C. For these experiments, the virus was diluted 1 : 5 in PBS and 200 μl was passed through a 0.2 μm cellulose acetate sterile syringe filter. This was chased by the addition of 200 μl PBS. The PBS filtrate was added to the filtered virus to give a total of 250 μl sterile virus solution when taking into account loss in the filter. Viral DNA was quantified by extraction of the DNA from 5 μl of this stock. 1 μl of the DNA extract contains 1.4 × 107 viral genome equivalents (Cladel et al., 2015).
Viral infections at tongue, vaginal and anal tracts
All mouse work was approved by the Institutional Animal Care and Use Committee of Pennsylvania State University's College of Medicine. HSD outbred Foxn1nu/+ nude mice (6–8 weeks) were obtained from Harlan Laboratories and were housed in sterile cages within sterile filter hoods and were fed sterilized food and water at the BL2 animal core facility. Mice were sedated i.p. with 0.1 ml/10 g body mass with ketamine/xylazine mixture (100 mg/10 mg in 10 ml ddH2O). For vaginal infection, mice were inoculated subcutaneously with 3 mg Depo-Provera (Pfizer) in 100 μl PBS 3 days before the viral infection as described previously (Cladel et al., 2015). Depo was not administered for anal and oral infections. Vaginal and anal tract were wounded with Doctors' Brush Picks coated with Conceptrol (ortho options, over the counter). Twenty-four hours after wounding, the mice were again anaesthetized and challenged with 25 μl (3.5 × 108) and 10 μl (1.4 × 108) of the sterilized viral suspension at vaginal and anal tract, respectively. For vaginal infection with no wounding, an aliquot of 25 μl of viral suspension was added inside the vaginal tract with a pipette and an additional 25 μl of viral suspension was added 3 days after (Cladel et al., 2015). For tongue infection, tongues were withdrawn using sterile forceps and microneedles were used to wound both the dorsal and ventral surfaces of the tongues (Gill & Prausnitz, 2008; Gill et al., 2008). Some bleeding occurred but care was taken to minimize the bleeding. The following day, each animal was again anaesthetized. Tongues were again gently abraded and 10 μl of sterile virus (1.4 × 108) was applied to the freshly abraded surfaces (10 μl each for the upper and under sides). Animals were placed on their backs during recovery to minimize loss of virus from the infection sites. Monitoring was conducted weekly and a photographic log was created for each animal.
Vaginal and anal lavage for DNA extraction
Vaginal and anal lavage was conducted using 30 μl sterile 0.9 % NaCl introduced into the vaginal or anal canal with a disposable filtered tip. The rinse was gently pipetted in and out of the vaginal canal and stored at − 20 °C before being processed. For DNA extraction, we used the DNeasy kit (Qiagen) and followed the instructions of the manufacturer. All DNA samples were eluted into 50 μl EB buffer.
Oestrous cycle determination and cytology analysis
About 30 μl sterile 0.9 % NaCl was used to rinse the vaginal tract gently with a filtered pipette tip (McLean et al., 2012). The wash was spread onto a glass slide for cytology with H&E staining. Dr Timothy Cooper, a veterinary pathologist in the Department of Comparative Medicine, and three independently trained researchers independently defined the oestrous stage. Abnormal cells can be determined in these samples (Cladel et al., 2015). The final results represented the agreements among all after the final examination with a teaching microscope system.
Viral DNA copy number analysis
Linearized MmuPV1 genome DNA was used for standard curve determination by SYBR Green Q-PCR analysis [FastStart Universal SYBR Green Master (Rox), Roche]. The primer pairs (5′GCCCGA AGACA ACACCG CCACG3′ and 5′CCTCCGCCTC GTCCCCA AATGG 3′) that amplify E2 were used. Viral copy numbers in 1 μl of 50 μl DNA extract from a lavage sample were converted into equivalent DNA load using a formula (1 ng viral DNA = 1.2 × 108 copy number, http://cels.uri.edu/gsc/cndna.html). Viral copy number per sample initially provides a simple positive or negative answer and is well adapted to a clinical setting (Massad et al., 2014; Meyer et al., 2014; Micalessi et al., 2015). However, false-negative results may occur due to sampling error (Liu et al., 2014). We acknowledge the variations in sampling and that some samples may have produced a negative result because too little DNA was collected at select sampling points. We thus determined that a housekeeping gene would improve quantification of viral DNA by comparing host DNA yield. Of several housekeeping genes that we pretested (GAPDH, TBP, β-actin and 18sRNA), the 18sRNA gene was shown to be the most consistent for this analysis and we selected it as the host target control for all the studies presented in the paper. Primer pairs to amplify 18sRNA were 5′ CGC CGC TAG AGG TGA AAT TC 3′ and 5′TTG GCA AAT GCT TTC GCT C 3′. DNA-free water was used as no template control (NTC). The Q-PCRs were run in a Stratagene Mx Pro-Mx3005P (Stratagene). Each reaction consisted of a final volume of 15 μl containing 7.5 μl of ultrapure water, 5 pmol of each primer, 7.5 μl of SYBR Green-PCR Master Mix (Roche) and 1 μl of DNA template. PCR conditions were: initial denaturation at 95 °C for 10 min, then 40 cycles at 95 °C for 15 s and at 60 °C for 1 min. All samples were tested in at minimum duplicates. We calculated the difference in cycle time (C t) between 18sRNA gene and viral DNA (ΔC t) and subsequently the fold change (2ΔCt). Fold change can determine the relative viral DNA load in each sample (Cui et al., 2015; Larionov et al., 2005; Provenzano & Mocellin, 2007).
IHC, ISH and TEM analyses of infected tissues
After termination of the experiment, the animals were euthanized and tissues of interest were fixed in 10 % buffered formalin as described previously (Cladel et al., 2013). H&E analysis, ISH and IHC were conducted as described in previous work (Cladel et al., 2013, 2015). For IHC, a goat Group Specific Antibody (GSA) to a conserved region of L1 (ViroStat) and an in-house made monoclonal antibody (MPV.B9) against denatured L1 were used on FFPE sections.
For TEM, the tissue was immersion fixed for 24 h in Karnovsky's fixative (5 % glutaraldehyde/4 % paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.3). Following fixation, the tissue was washed in 0.1 M sodium cacodylate buffer, post-fixed in buffered 1 % osmium tetroxide/1.5 % potassium ferrocyanide, washed again with buffer, dehydrated in a graded series of ethanol, transferred to propylene oxide and embedded in Spurr low viscosity resin. Sections (70–90 nm) were cut with a diamond knife, mounted on copper grids and stained with 2 % aqueous uranyl acetate and lead citrate. The sections were examined in a JEOL JEM 1400 electron microscope. The pictures were recorded.
Statistical analysis
The data were statistically analysed with one-way anova analysis in Sigmaplot 12 software. Standard errors were calculated and presented as error bars in all figures. Differences were considered to be significant at P < 0.05.
Acknowledgements
This work was supported by the Public Health Service, National Cancer Institute, National Institutes of Health (Grant R01 CA47622) and by the Jake Gittlen Memorial Golf Tournament.
Supplementary Data
Supplementary Data
References
- Assi R., Reddy V., Einarsdottir H., Longo W. E. (2014). Anorectal human papillomavirus: current concepts Yale J Biol Med 87 537–547 . [PMC free article] [PubMed] [Google Scholar]
- Baandrup L., Thomsen L. T., Olesen T. B., Andersen K. K., Norrild B., Kjaer S. K. (2014). The prevalence of human papillomavirus in colorectal adenomas and adenocarcinomas: a systematic review and meta-analysis Eur J Cancer 50 1446–1461 10.1016/j.ejca.2014.01.019 . [DOI] [PubMed] [Google Scholar]
- Bergot A. S., Kassianos A., Frazer I. H., Mittal D. (2011). New approaches to immunotherapy for HPV associated cancers Cancers (Basel) 3 3461–3495 10.3390/cancers3033461 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutaga K., Savelkoul P. H., Winkel E. G., van Winkelhoff A. J. (2007). Comparison of subgingival bacterial sampling with oral lavage for detection and quantification of periodontal pathogens by real-time polymerase chain reaction J Periodontol 78 79–86 10.1902/jop.2007.060078 . [DOI] [PubMed] [Google Scholar]
- Bown E., Shah V., Sridhar T., Boyle K., Hemingway D., Yeung J. M. (2014). Cancers of the anal canal: diagnosis, treatment and future strategies Future Oncol 10 1427–1441 10.2217/fon.14.23 . [DOI] [PubMed] [Google Scholar]
- Brandsma J. L. (2005). The cottontail rabbit papillomavirus model of high-risk HPV-induced disease Methods Mol Med 119 217–235 . [DOI] [PubMed] [Google Scholar]
- Brickman C., Palefsky J. M. (2015). Human papillomavirus in the HIV-infected host: epidemiology and pathogenesis in the antiretroviral era Curr HIV/AIDS Rep 12 6–15 10.1007/s11904-014-0254-4 . [DOI] [PubMed] [Google Scholar]
- Byers S. L., Wiles M. V., Dunn S. L., Taft R. A. (2012). Mouse estrous cycle identification tool and images PLoS One 7 e35538 10.1371/journal.pone.0035538 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligioni C. S. (2009). Assessing reproductive status/stages in mice Curr Protoc Neurosci 48 A.4I.1–A.4I.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo M. S. (2002). Animal models of papillomavirus pathogenesis Virus Res 89 249–261 10.1016/S0168-1702(02)00193-4 . [DOI] [PubMed] [Google Scholar]
- Campo M. S., Roden R. B. (2010). Papillomavirus prophylactic vaccines: established successes, new approaches J Virol 84 1214–1220 10.1128/JVI.01927-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castell S., Krause G., Schmitt M., Pawlita M., Deleré Y., Obi N., Flesch-Janys D., Kemmling Y., Kaufmann A. M. (2014). Feasibility and acceptance of cervicovaginal self-sampling within the German National Cohort (Pretest 2) Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 57 1270–1276 10.1007/s00103-014-2054-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cladel N. M., Hu J., Balogh K., Mejia A., Christensen N. D. (2008). Wounding prior to challenge substantially improves infectivity of cottontail rabbit papillomavirus and allows for standardization of infection J Virol Methods 148 34–39 10.1016/j.jviromet.2007.10.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cladel N. M., Budgeon L. R., Cooper T. K., Balogh K. K., Hu J., Christensen N. D. (2013). Secondary infections, expanded tissue tropism, and evidence for malignant potential in immunocompromised mice infected with Mus musculus papillomavirus 1 DNA and virus J Virol 87 9391–9395 10.1128/JVI.00777-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cladel N. M., Budgeon L. R., Balogh K. K., Cooper T. K., Hu J., Christensen N. D. (2015). A novel pre-clinical murine model to study the life cycle and progression of cervical and anal papillomavirus infections PLoS One 10 e0120128 10.1371/journal.pone.0120128 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Yu S., Tamhane A., Causey Z. L., Steg A., Danila M. I., Reynolds R. J., Wang J., Wanzeck K. C., other authors (2015). Simple regression for correcting DeltaCt bias in RT-qPCR low-density array data normalization BMC Genomics 16 82 10.1186/s12864-015-1274-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curlin M. E., Leelawiwat W., Dunne E. F., Chonwattana W., Mock P. A., Mueanpai F., Thep-Amnuay S., Whitehead S. J., McNicholl J. M. (2013). Cyclic changes in HIV shedding from the female genital tract during the menstrual cycle J Infect Dis 207 1616–1620 10.1093/infdis/jit063 . [DOI] [PubMed] [Google Scholar]
- Deleré Y., Schuster M., Vartazarowa E., Hänsel T., Hagemann I., Borchardt S., Perlitz H., Schneider A., Reiter S., Kaufmann A. M. (2011). Cervicovaginal self-sampling is a reliable method for determination of prevalence of human papillomavirus genotypes in women aged 20 to 30 years J Clin Microbiol 49 3519–3522 10.1128/JCM.01026-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dochez C., Bogers J. J., Verhelst R., Rees H. (2014). HPV vaccines to prevent cervical cancer and genital warts: an update Vaccine 32 1595–1601 10.1016/j.vaccine.2013.10.081 . [DOI] [PubMed] [Google Scholar]
- Doorbar J. (2013). Latent papillomavirus infections and their regulation Curr Opin Virol 3 416–421 10.1016/j.coviro.2013.06.003 . [DOI] [PubMed] [Google Scholar]
- Forslund O., Johansson H., Madsen K. G., Kofoed K. (2013). The nasal mucosa contains a large spectrum of human papillomavirus types from the Betapapillomavirus and Gammapapillomavirus genera J Infect Dis 208 1335–1341 10.1093/infdis/jit326 . [DOI] [PubMed] [Google Scholar]
- Gallichan W. S., Rosenthal K. L. (1996). Effects of the estrous cycle on local humoral immune responses and protection of intranasally immunized female mice against herpes simplex virus type 2 infection in the genital tract Virology 224 487–497 10.1006/viro.1996.0555 . [DOI] [PubMed] [Google Scholar]
- García-Sastre A., Durbin R. K., Zheng H., Palese P., Gertner R., Levy D. E., Durbin J. E. (1998). The role of interferon in influenza virus tissue tropism J Virol 72 8550–8558 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill H. S., Prausnitz M. R. (2008). Pocketed microneedles for drug delivery to the skin J Phys Chem Solids 69 1537–1541 10.1016/j.jpcs.2007.10.059 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill H. S., Denson D. D., Burris B. A., Prausnitz M. R. (2008). Effect of microneedle design on pain in human volunteers Clin J Pain 24 585–594 10.1097/AJP.0b013e31816778f9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haguenoer K., Sengchanh S., Gaudy-Graffin C., Boyard J., Fontenay R., Marret H., Goudeau A., Pigneaux de Laroche N., Rusch E., Giraudeau B. (2014). Vaginal self-sampling is a cost-effective way to increase participation in a cervical cancer screening programme: a randomised trial Br J Cancer 111 2187–2196 10.1038/bjc.2014.510 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handisurya A., Day P. M., Thompson C. D., Bonelli M., Lowy D. R., Schiller J. T. (2014). Strain-specific properties and T cells regulate the susceptibility to papilloma induction by Mus musculus papillomavirus 1 PLoS Pathog 10 e1004314 10.1371/journal.ppat.1004314 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren S. C., Patterson N. A., Ozbun M. A., Lambert P. F. (2005). The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle J Virol 79 3938–3948 10.1128/JVI.79.7.3938-3948.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Cladel N. M., Balogh K., Budgeon L., Christensen N. D. (2007). Impact of genetic changes to the CRPV genome and their application to the study of pathogenesis in vivo Virology 358 384–390 10.1016/j.virol.2006.08.045 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Cladel N. M., Budgeon L., Balogh K. K., Christensen N. D., Papillomavirus D. N. A. (2009). Papillomavirus DNA complementation in vivo Virus Res 144 117–122 10.1016/j.virusres.2009.04.006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igidbashian S., Boveri S., Spolti N., Radice D., Sandri M. T., Sideri M. (2011). Self-collected human papillomavirus testing acceptability: comparison of two self-sampling modalities J Womens Health (Larchmt) 20 397–402 10.1089/jwh.2010.2189 . [DOI] [PubMed] [Google Scholar]
- Joh J., Jenson A. B., Proctor M., Ingle A., Silva K. A., Potter C. S., Sundberg J. P., Ghim S. J. (2012). Molecular diagnosis of a laboratory mouse papillomavirus (MusPV) Exp Mol Pathol 93 416–421 10.1016/j.yexmp.2012.07.001 . [DOI] [PubMed] [Google Scholar]
- Kaushic C., Ashkar A. A., Reid L. A., Rosenthal K. L. (2003). Progesterone increases susceptibility and decreases immune responses to genital herpes infection J Virol 77 4558–4565 10.1128/JVI.77.8.4558-4565.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushic C., Roth K. L., Anipindi V., Xiu F. (2011). Increased prevalence of sexually transmitted viral infections in women: the role of female sex hormones in regulating susceptibility and immune responses J Reprod Immunol 88 204–209 10.1016/j.jri.2010.12.004 . [DOI] [PubMed] [Google Scholar]
- Korn A. P. (1996). Interpretation of abnormal Pap smears Infect Med 13 405–406. [Google Scholar]
- Lampinen T. M., Latulippe L., van Niekerk D., Schilder A. J., Miller M. L., Anema A., Hogg R. S. (2006). Illustrated instructions for self-collection of anorectal swab specimens and their adequacy for cytological examination Sex Transm Dis 33 386–388 10.1097/01.olq.0000204747.66265.2c . [DOI] [PubMed] [Google Scholar]
- Larionov A., Krause A., Miller W. (2005). A standard curve based method for relative real time PCR data processing BMC Bioinformatics 6 62 10.1186/1471-2105-6-62 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski W., Ananth A. T., Ploch L. E., Rogers N. E. (2014). Anal Pap smears and anal cancer: what dermatologists should know J Am Acad Dermatol 71 985–992 10.1016/j.jaad.2014.06.045 . [DOI] [PubMed] [Google Scholar]
- Liu S. H., Brotman R. M., Zenilman J. M., Gravitt P. E., Cummings D. A. (2013). Menstrual cycle and detectable human papillomavirus in reproductive-age women: a time series study J Infect Dis 208 1404–1415 10.1093/infdis/jit337 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. H., Cummings D. A., Zenilman J. M., Gravitt P. E., Brotman R. M. (2014). Characterizing the temporal dynamics of human papillomavirus DNA detectability using short-interval sampling Cancer Epidemiol Biomarkers Prev 23 200–208 10.1158/1055-9965.EPI-13-0666 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massad L. S., Xie X., Burk R., Keller M. J., Minkoff H., D'Souza G., Watts D. H., Palefsky J., Young M., other authors (2014). Long-term cumulative detection of human papillomavirus among HIV seropositive women AIDS 28 2601–2608 10.1097/QAD.0000000000000455 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G., Mohamed M. R., Rahman M. M., Bartee E. (2009). Cytokine determinants of viral tropism Nat Rev Immunol 9 645–655 10.1038/nri2623 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean A. C., Valenzuela N., Fai S., Bennett S. A. (2012). Performing vaginal lavage, crystal violet staining, and vaginal cytological evaluation for mouse estrous cycle staging identification J Vis Exp 15 e4389 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros L. R., Ethur A. B., Hilgert J. B., Zanini R. R., Berwanger O., Bozzetti M. C., Mylius L. C. (2005). Vertical transmission of the human papillomavirus: a systematic quantitative review Cad Saude Publica 21 1006–1015 10.1590/S0102-311X2005000400003 . [DOI] [PubMed] [Google Scholar]
- Meyer M. F., Huebbers C. U., Siefer O. G., Vent J., Engbert I., Eslick G. D., Valter M., Klussmann J. P., Preuss S. F. (2014). Prevalence and risk factors for oral human papillomavirus infection in 129 women screened for cervical HPV infection Oral Oncol 50 27–31 10.1016/j.oraloncology.2013.10.009 . [DOI] [PubMed] [Google Scholar]
- Micalessi M. I., Boulet G. A., Bogers J. (2015). A real-time PCR approach based on SPF10 primers and the INNO-LiPA HPV genotyping extra assay for the detection and typing of human papillomavirus Methods Mol Biol 1249 27–35 10.1007/978-1-4939-2013-6_2 . [DOI] [PubMed] [Google Scholar]
- Mostad S. B., Kreiss J. K., Ryncarz A., Chohan B., Mandaliya K., Ndinya-Achola J., Bwayo J. J., Corey L. (2000). Cervical shedding of herpes simplex virus and cytomegalovirus throughout the menstrual cycle in women infected with human immunodeficiency virus type 1 Am J Obstet Gynecol 183 948–955 10.1067/mob.2000.106589 . [DOI] [PubMed] [Google Scholar]
- Nicholls P. K., Moore P. F., Anderson D. M., Moore R. A., Parry N. R., Gough G. W., Stanley M. A. (2001). Regression of canine oral papillomas is associated with infiltration of CD4+ and CD8+ lymphocytes Virology 283 31–39 10.1006/viro.2000.0789 . [DOI] [PubMed] [Google Scholar]
- O'Brien W. A. (1992). Viral determinants of cellular tropism Pathobiology 60 225–233 10.1159/000163727 . [DOI] [PubMed] [Google Scholar]
- Ortiz A. P., Romaguera J., Pérez C. M., Otero Y., Soto-Salgado M., Méndez K., Valle Y., Da Costa M., Suarez E., other authors (2013). Human papillomavirus infection in women in Puerto Rico: agreement between physician-collected and self-collected anogenital specimens J Low Genit Tract Dis 17 210–217 10.1097/LGT.0b013e318260e312 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano M., Mocellin S. (2007). Complementary techniques: validation of gene expression data by quantitative real time PCR Adv Exp Med Biol 593 66–73 10.1007/978-0-387-39978-2_7 . [DOI] [PubMed] [Google Scholar]
- Roberts J. N., Buck C. B., Thompson C. D., Kines R., Bernardo M., Choyke P. L., Lowy D. R., Schiller J. T. (2007). Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan Nat Med 13 857–861 10.1038/nm1598 . [DOI] [PubMed] [Google Scholar]
- Rosenberger J. G., Dodge B., Van Der Pol B., Reece M., Herbenick D., Fortenberry J. D. (2011). Reactions to self-sampling for ano-rectal sexually transmitted infections among men who have sex with men: a qualitative study Arch Sex Behav 40 281–288 10.1007/s10508-009-9569-4 . [DOI] [PubMed] [Google Scholar]
- Ryan K. D., Schwartz N. B. (1977). Grouped female mice: demonstration of pseudopregnancy Biol Reprod 17 578–583. [DOI] [PubMed] [Google Scholar]
- Sathish N., Wang X., Yuan Y. (2014). Human papillomavirus (HPV)-associated oral cancers and treatment strategies J Dent Res 93 (Suppl), 29S–36S 10.1177/0022034514527969 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeink C. E., Massuger L. F., Lenselink C. H., Quint W. G., Melchers W. J., Bekkers R. L. (2010). Effect of the menstrual cycle and hormonal contraceptives on human papillomavirus detection in young, unscreened women Obstet Gynecol 116 67–75 10.1097/AOG.0b013e3181e238f0 . [DOI] [PubMed] [Google Scholar]
- Sehnal B., Dusek L., Cibula D., Zima T., Halaska M., Driak D., Slama J. (2014). The relationship between the cervical and anal HPV infection in women with cervical intraepithelial neoplasia J Clin Virol 59 18–23 10.1016/j.jcv.2013.11.004 . [DOI] [PubMed] [Google Scholar]
- Stewart D. E., Gagliardi A., Johnston M., Howlett R., Barata P., Lewis N., Oliver T., Mai V., HPV Self-collection Guidelines Panel (2007). Self-collected samples for testing of oncogenic human papillomavirus: a systematic review J Obstet Gynaecol Can 29 817–828 . [DOI] [PubMed] [Google Scholar]
- Stier E. A., Sebring M. C., Mendez A. E., Ba F. S., Trimble D. D., Chiao E. Y. (2015). Prevalence of anal human papillomavirus infection and anal HPV-related disorders in women: a systematic review Am J Obstet Gynecol 213 278–309 10.1016/j.ajog.2015.03.034 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. D., Weatherly R. A., Koopmann C.F.,, Jr, Carey T. E. (2000). Mucosal swabs detect HPV in laryngeal papillomatosis patients but not family members Int J Pediatr Otorhinolaryngol 53 95–103 10.1016/S0165-5876(00)00304-9 . [DOI] [PubMed] [Google Scholar]
- Sundberg J. P., Stearns T. M., Joh J., Proctor M., Ingle A., Silva K. A., Dadras S. S., Jenson A. B., Ghim S. J. (2014). Immune status, strain background, and anatomic site of inoculation affect mouse papillomavirus (MmuPV1) induction of exophytic papillomas or endophytic trichoblastomas PLoS One 9 e113582 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghian A., Huang P. (1995). The nude and SCID mice as a tumor model in experimental cancer biology Cancer J 8 52–58. [Google Scholar]
- Teepe A. G., Allen L. B., Wordinger R. J., Harris E. F. (1990). Effect of the estrous cycle on susceptibility of female mice to intravaginal inoculation of herpes simplex virus type 2 (HSV-2) Antiviral Res 14 227–235 10.1016/0166-3542(90)90004-Q . [DOI] [PubMed] [Google Scholar]
- Ting J., Rositch A. F., Taylor S. M., Rahangdale L., Soeters H. M., Sun X., Smith J. S. (2014). Worldwide incidence of cervical lesions: a systematic review Epidemiol Infect 143 225–241 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tota J. E., Ramanakumar A. V., Mahmud S. M., Trevisan A., Villa L. L., Franco E. L., Ludwig-McGill Cohort Study Group (2013). Cervical human papillomavirus detection is not affected by menstrual phase Sex Transm Infect 89 202–206 10.1136/sextrans-2012-050610 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ure A. E., Forslund O. (2014). Characterization of human papillomavirus type 154 and tissue tropism of gammapapillomaviruses PLoS One 9 e89342 10.1371/journal.pone.0089342 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker Q. D., Nelson C. J., Smith D., Kuhn C. M. (2002). Vaginal lavage attenuates cocaine-stimulated activity and establishes place preference in rats Pharmacol Biochem Behav 73 743–752 10.1016/S0091-3057(02)00883-3 . [DOI] [PubMed] [Google Scholar]
- Waxman A. G. (2008). Cervical cancer screening in the early post vaccine era Obstet Gynecol Clin North Am 35 537–548. [DOI] [PubMed] [Google Scholar]
- Wells J. S., Holstad M. M., Thomas T., Bruner D. W. (2014). An integrative review of guidelines for anal cancer screening in HIV-infected persons AIDS Patient Care STDS 28 350–357 10.1089/apc.2013.0358 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data