Fig. 4.
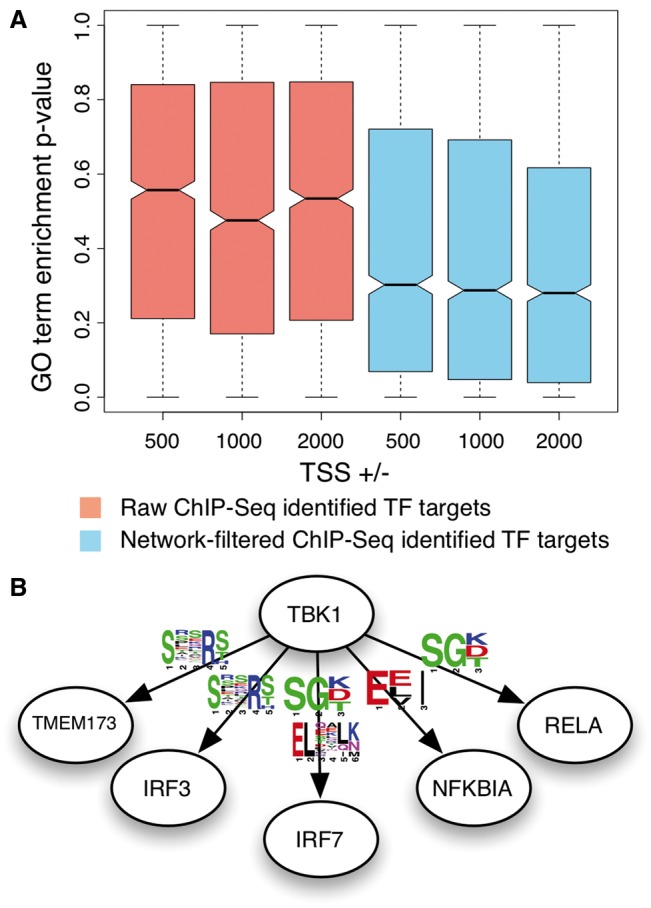
Improving the interpretability of primary experimental data. Our interaction networks can be used to help investigators increase the interpretability of condition-specific primary experimental data. In panel A, ChIP-Seq datasets generated for the ENCODE project (109 TFs) were processed to identify TF and potential regulatory target genes. We observe a significant increase in enrichment of TF associated GO terms among its putative regulator targets, when removing TF-targets pairs that have low predicted probability in our TF regulatory network (skyblue). In panel B, represents our predicted phospho-binding motifs that are present in known TBK1 substrates not identified in the phospho-proteomics study. Such results support the potential biological relevance of these motifs and the accuracy of our phosphorylation network that enabled the analysis
