Abstract
Throughout their lifetimes, messenger RNAs (mRNAs) associate with proteins to form ribonucleoproteins (mRNPs). Since the discovery of the first mRNP component more than 40 years ago, what is known as the mRNA interactome now comprises >1,000 proteins. These proteins bind mRNAs in myriad ways with varying affinities and stoichiometries, with many assembling onto nascent RNAs in a highly ordered process during transcription and precursor mRNA (pre-mRNA) processing. The nonrandom distribution of major mRNP proteins observed in transcriptome-wide studies leads us to propose that mRNPs are organized into three major domains loosely corresponding to 5′ untranslated regions (UTRs), open reading frames, and 3′ UTRs. Moving from the nucleus to the cytoplasm, mRNPs undergo extensive remodeling as they are first acted upon by the nuclear pore complex and then by the ribosome. When not being actively translated, cytoplasmic mRNPs can assemble into large multi-mRNP assemblies or be permanently disassembled and degraded. In this review, we aim to give the reader a thorough understanding of past and current eukaryotic mRNP research.
Keywords: ibonucleoproteins, RNA processing, coupling, mRNP packaging, RNP remodeling, RNA granules
INTRODUCTION
All expression of endogenous eukaryotic genes involves transcribing permanent genetic information stored in DNA into perishable RNA copies, many of which serve as messenger RNA (mRNA) templates for protein synthesis. As do all RNAs in living cells, mRNAs interact with proteins to form ribonucleoprotein (RNP) complexes. In higher eukaryotes, these mRNA-containing RNPs (mRNPs) comprise tens of thousands of different RNA sequences and hundreds of different RNA-binding proteins (RBPs), making them the most compositionally diverse of all RNPs. Whereas the sequence of an mRNA’s open reading frame (ORF) dictates the sequence of the encoded polypeptide, every other aspect of an mRNA’s metabolism—including its nucleocytoplasmic export, its subcellular localization in the cytoplasm, where and when it functionally engages with the translation machinery, and its mode and rate of degradation—is dictated by its complement of bound proteins. Some mRNP proteins recognize common mRNA structural elements [e.g., the 5′ 7-methyl-guanosine (m7G) cap and the 3′ polyadenosine (polyA) tail shared by most mRNAs]. Others recognize specific sequence motifs, sometimes with the help of short guide RNAs [e.g., Argonautes and microRNAs (miRNAs)]. Still others associate in a sequence-independent manner with either secondary or tertiary structural elements or as a consequence of some processing reaction. With regard to association dynamics, mRNP proteins run the gamut from stably bound architectural elements that help define overall mRNP organization to highly transient factors that modulate just one step of posttranscriptional gene expression.
The earliest observations of proteins on RNA polymerase II (Pol II) transcripts date back to the 1950s, when direct electron microscopic visualization of elongating transcripts on lampbrush chromosomes first suggested rapid packaging of emerging nascent RNAs with proteins to form RNPs (1; reviewed in Reference (2). In the 1970s, several laboratories began focusing on biochemical purification and compositional/structural analysis of nonribosomal RNPs. These studies led to early identification of such key mRNP components as the nuclear and cytoplasmic cap–binding proteins, polyA-binding protein (PABP), and heterogeneous nuclear ribonucleoproteins (hnRNPs). The following three decades saw a steady stream of new mRNP components appearing with ever-increasing regularity. These new proteins were usually found as a result of some specific property or activity such as an ability to modulate mRNA localization, translation, and/or decay via binding to a previously defined cis-regulatory element [e.g., zipcode-binding protein 1 (also known as IGF2BP1) and iron regulatory protein 1]; sequence-specific binding via small guide RNAs (e.g., Ago2 and GW182); or association with localized mRNP complexes. Some proteins were discovered via their ability to modulate expression of many mRNAs in a seemingly sequence-independent manner (e.g., FMRP, Staufen, and DDX5) or their recruitment as a consequence of some precursor mRNA (pre-mRNA) processing event [e.g., exon junction complex (EJC) proteins]. In the past few years, however, confluence of these more classical biochemical and molecular approaches with new high-throughput technologies has turned this steady stream into a virtual torrent.
Major advances in mass spectrometry and next-generation sequencing over the past decade now allow for precise quantification of both protein and mRNA copy numbers in samples as complex as whole-cell lysates. Thus, mRNP organization and architecture can now be analyzed on a whole new scale encompassing the entire transcriptome. For example, the combination of ultraviolet (UV) in vivo RNA–protein cross-linking with immunoprecipitation and high-throughput sequencing (CLIP-seq) has elucidated the transcriptome-wide RNA-binding sites of more than 100 RBPs, many at single-nucleotide (nt) resolution. Similarly, the combination of ribonuclease protection (RNP footprinting), immunoprecipitation, and high-throughput sequencing has revealed the transcriptome-wide landscapes of structural mRNP components such as the EJC and Staufen. Offshoots of these approaches have identified vast protein-occupied or otherwise inaccessible stretches of mRNAs, suggesting that mRNPs are much more highly structured and compact than previously appreciated. Finally, mass spectrometry analyses of proteins that UV-cross-link to and copurify with polyadenylated RNAs have yielded huge catalogs of proteins directly in contact with mRNA. These new mRNA interactomes are revealing both new and unexpected RBPs and providing novel insights into the properties of previously known RBPs. For example, the mRNA-bound proteome is disproportionately enriched in low-complexity, inherently disordered regions. These domains presumably confer upon mRNPs the ability to coalesce into useful and dynamic assemblages such as transport and stress granules. A downside, however, is the accompanying tendency of these unstructured domains to form pathogenic aggregates such as those associated with neurodegenerative disease.
While the explosion of high-throughput information is rapidly transforming our understanding of mRNP architecture, other equally exciting advances are transforming our understanding of mRNP biology. For example, it was recently revealed that different transcriptional promoters can shunt mRNAs into particular export and decay pathways, presumably by altering mRNP composition akin to EJC deposition by pre-mRNA splicing. Also notable are the recent discoveries of two new nuclear export pathways: one specifically targeting mRNAs to the endoplasmic reticulum or mitochondria and another wherein exceedingly large megaRNPs bud through the nuclear envelope instead of exiting via the nuclear pore complex (NPC). With regard to more classical nuclear export, researchers have also gained many new insights into the dynamics of mRNP interaction with and transit through the NPC, as well as compositional remodeling on the cytoplasmic side to ensure unidirectional transport. Using their nucleus-acquired proteins, mRNPs newly arrived in cytoplasm can engage the translation machinery preferentially over older mRNAs that have lost these proteins as a result of previous ribosome transit. Finally, we now know that active mRNP disassembly is an important prerequisite for efficient mRNA degradation.
These recent discoveries attest that these are exciting times in the ongoing quest to unravel the intricacies of eukaryotic mRNPs. In this review, we aim to present many recent advances in detail while placing them in context with several decades of preceding work. Because the functions of mRNP proteins in mediating the myriad steps of posttranscriptional gene expression have been well reviewed elsewhere, these aspects are not discussed here. Instead, we aim to provide a comprehensive and up-to-date view of mRNP assembly, composition, structure, and remodeling. In so doing, we hope to give the reader a greater understanding of the general themes that have emerged from more than 40 years of mRNP research, as well as the major unanswered questions that remain. Because the bulk of this research was carried out in mammalian and yeast cells, these systems are our primary focus.
THE mRNP PARTS LIST
Full biochemical and structural understanding of any complex macromolecular assemblage requires a complete parts list, knowledge about the relative stoichiometries of these parts, and a comprehensive structural understanding of how these parts fit together to form a functional whole. We thus begin with an up-to-date description of the major mRNP components, their organizing principles, and how these individual parts assemble into functional mRNPs. We focus primarily on RNPs containing polyadenylated mRNAs, although nonpolyadenylated mRNPs (e.g., those containing polyA-tail-less histone mRNAs) and polyadenylated long noncoding RNPs also exist and play crucial roles in eukaryotic biology.
First mRNP Proteins: PABP, YBX1, and hnRNPs
The first four decades of mRNP research (from approximately 1970 to 2010) saw the identification of many ubiquitous proteins common to all polyadenylated RNAs. In the early 1970s, several reports began describing proteins that purified with Pol II transcripts, originally distinguished as heterogeneous nuclear RNAs (hnRNAs) in the nuclear compartment and mRNAs in the cytoplasm. hnRNA-containing RNPs assembled into and purified as stable hnRNP particles that contain a complex mixture of proteins with apparent molecular masses ranging from 40 to 180 kDa (3, 4). Cytoplasmic mRNPs, by contrast, are predominated by three proteins of 52, 76, and 120 kDa (Figure 1a) (3, 5–7). The 76-kDa protein had been previously identified as PABP (now known as PABPC1) (6), and the 52-kDa protein was later found to be Y-box protein (YBX1), which nonspecifically coats mRNAs along their entire length by binding to the sugar-phosphate backbone (8, 9). To date, however, the identity of the 120-kDa protein remains unresolved.
Figure 1.
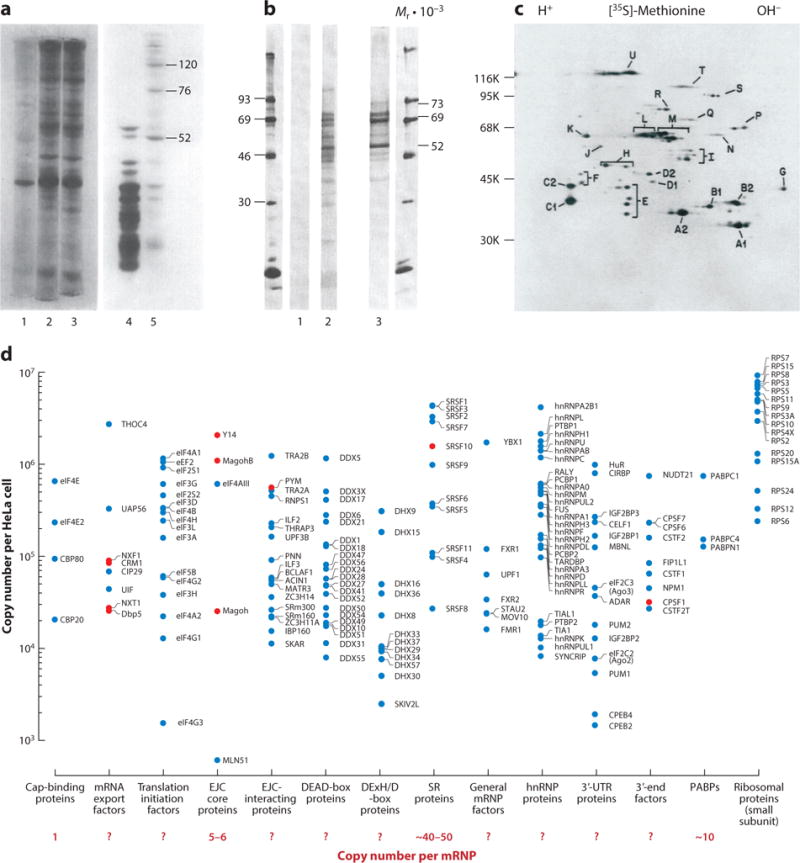
Identification and relative abundance of mRNP components. (a) Oligo-dT-cellulose-selected nuclear hnRNP or cytoplasmic mRNP proteins from [3H]-uridine pulse-labeled HeLa cells. (Lanes 1–3) hnRNP proteins recovered from the unbound fraction (lane 1) or from elution with 25% formamide (lane 2) or with 50% formamide (lane 3). (Lanes 4–5) mRNP proteins in the unbound fraction (lane 4) (mainly ribosomal subunits) or eluted with 50% formamide (lane 5). Panel adapted with permission from plate I in Reference 3. (b) Proteins from oligo-dT-cellulose-selected polyribosomal RNPs from human KB cells labeled with [35S]-methionine without (lane 1) or with (lane 2) short-wave UV irradiation. Oligo-dT-selected proteins are as in lane 2, except the radioactive signal was transferred from [3H]-adenosine- and [3H]-uridine-labeled RNAs after UV cross-linking. Panel modified with permission from Reference 10. (c) [35S]-labeled HeLa hnRNP particle proteins coimmunopurified with 4F4 monoclonal antibody (anti–hnRNP C) and resolved by two-dimensional electrophoresis. Panel modified under a Creative Commons license from Reference 14. (d) Copy numbers of mRNA interactome proteins in HeLa cells. HeLa cell copy number data are from Reference 21, and mRNA interactome data are from Reference 49. The interactome proteins are grouped into categories as shown below the x axis. Some proteins that are absent from the HeLa interactome (red dots) are included for better representation within a category. Shown under each protein category are the estimated copy numbers of its members within an mRNP assembled on an average human mRNA with eight exons and seven introns. EJC copy number is based on ~80% EJC occupancy on human mRNAs (35, 36). The SR protein copy number is based on stoichiometric amounts of SR and SR-like proteins associated with the EJC (36). The number of PABP molecules per mRNA is based on a single PABP occupying ~27 adenosines (144) on an average polyA tail of ×250 nt. Abbreviations: EJC, exon junction complex; hnRNP, heterogeneous nuclear ribonucleoprotein; mRNA, messenger RNA; mRNP, mRNA-containing RNP; PABP, polyadenosine (polyA)-binding protein.
In 1980, the development of a simple yet robust RNA–protein photocrosslinking approach yielded a major breakthrough in the unambiguous definition of mRNP proteins (10). In this approach, proteins directly interacting with RNAs within intact cells were cross-linked to the RNA in vivo by short-wave (~254-nm) UV-light activation of nucleobases. This allowed for specific purification of covalently cross-linked proteins from various cell fractions via polyA tail–oligo-dT hybridization under protein-denaturing conditions (0.5% SDS). The initial study revealed three major bands of 52-, 69-, and 73-kDa apparent molecular mass that cross-linked to cytoplasmic mRNPs in human KB cells (Figure 1b) (10). Subsequent work from other groups reported that proteins cross-linking to nuclear polyA+ RNA were identical to the previously observed hnRNP particle subunits (11–13). Another major advance came with development of monoclonal antibodies against individual hnRNP/mRNP proteins. The Dreyfuss lab (14) accomplished this advance by inoculating mice with the entire mix of oligo-dT-purified, cross-linked proteins from nuclear and cytoplasmic fractions and creating individual hybridomas. Proteins purified via the resulting monoclonals, each of which recognized a different hnRNP, yielded nearly identical patterns of ~24 hnRNP polypeptides when separated on two-dimensional polyacrylamide gels (Figure 1c). On the basis of their relative abundance and gel migration, these proteins were termed hnRNP A1–U.
SR Proteins
Several years later, a different monoclonal antibody (mAb104) that was raised against Xenopus germinal vesicle (nucleus) proteins kick-started the discovery of a second major family of nuclear and cytoplasmic mRNP components—the SR proteins. This antibody decorated transcriptionally active loops of amphibian lampbrush chromosomes and stained nuclear bodies containing small nuclear RNPs (snRNPs) (15). As suggested by these localization patterns, proteins recognized by mAb104, now known as SR proteins (SRSF1 through 12) for their distinctive arginine–serine dipeptide–rich domains, coordinate splicing with transcription (16, 17). With the exception of SC35/SRSF2, all canonical SR proteins shuttle between the nucleus and the cytoplasm, thereby connecting mRNA synthesis events with downstream mRNA utilization events (18, 19). In addition to the 12 canonical SR proteins, numerous other SR-like proteins with diverse functions in splicing, mRNA 3′-end formation, export, translation, and degradation have been recognized (18, 20).
Cap-Binding Proteins
Recent quantitative mass spectrometry analyses have revealed that PABPC1, YBX1, and many hnRNP and SR proteins are highly abundant in most cells (Figure 1d) (21, 22). Because these proteins also bind the majority of mRNAs, they were among the earliest mRNP components identified. Another key class of ubiquitous mRNP proteins consists of proteins that bind the m7G cap characteristic of Pol II transcripts. Polypeptides that specifically cross-link to and/or affinity-purify with the cap structure were first identified from the cytoplasmic fraction (eIF4E) (23, 24) and later from the nuclear fraction [cap-binding complex (CBC) proteins CBP80 and CBP20] (25, 26).
Splicing-Dependent Proteins
All of the above proteins bind mRNAs by recognizing either specific structures (i.e., the m7G cap, the sugar-phosphate backbone, or the polyA tail) or short-sequence motifs. Another key set of mRNP proteins are those left behind by the splicing machinery and thus mark the sites of previous pre-mRNA processing events. The first hints that splicing may affect mRNP composition came from functional analyses of mRNA export and nonsense-mediated decay (NMD). One study from the Reed lab (27) showed that an RNP generated by splicing in vitro migrated differently on native gels and, when injected into a Xenopus oocyte nucleus, was exported more rapidly than an otherwise identical RNP assembled on an intronless transcript. Other functional data indicated that an intron located downstream of a stop codon was a potent enhancer of NMD (28–30). Together, these data stimulated a series of biochemical experiments leading to discovery of the EJC (31–34). Late during the splicing process, a highly conserved set of proteins assemble into a stable complex centered ~24 nt upstream of the exon–exon junctions. In human cells, EJCs decorate the majority of spliced exons (35, 36) and provide a binding platform for more dynamically interacting proteins that link splicing to other nuclear and cytoplasmic processes. Although EJCs bind in a sequence-nonspecific manner, they are incredibly stable once assembled. This vice grip results from the DEAD-box protein eIF4III (DDX48) being locked in its RNA-bound conformation by its cofactors Y14 and Magoh, which prevent completion of eIF4AIII’s ATPase cycle (37, 38).
In addition to the EJC and its associated proteins, ~45 other proteins purify exclusively with in vitro–spliced mRNPs (39). Among these proteins are two other highly abundant DEAD-box proteins, DDX3 and DDX5. As with eIF4AIII, splicing-dependent recruitment of DDX3 and DDX5 also requires the presence of at least 20 nt upstream of the exon junction. Could these proteins nucleate eIF4AIII-independent EJCs? In support of this possibility, eIF4AIII-containing EJCs are undetectable on ~20% of exons within the human transcriptome (35, 36), and some spliced mRNP-specific proteins can be recruited independently of eIF4AIII (40). Interestingly, like eIF4AIII, both DDX3 and DDX5 have been implicated in diverse processes such as transcription, splicing, mRNA export, localization, translation, and decay. Consistent with this finding, and similar to eIF4AIII, DDX3 and DDX5 are associated with subcytoplasmically localized mRNAs in highly polarized cells such as neurons.
A Polyadenylation-Dependent Mark
Another process affecting mRNP composition is 3′-end formation. Akin to EJC deposition upstream of exon–exon junctions, the nucleolar 32-kDa phosphoprotein nucleophosmin 1 (NPM1) becomes associated in a sequence-independent manner with the region ~10 nt upstream of the polyA signal (AAUAAA) as a consequence of polyadenylation (41). Previously implicated in ribosome biogenesis, centrosome duplication, DNA repair, and cellular stress responses, NPM1 can also act as either an activating oncogene or a tumor suppressor, depending on protein level and cell type. Because its depletion from HeLa cells causes hyperadenylation and nuclear mRNA retention (42), NPM1 deposition during the polyadenylation process may function to limit polyA tail length and enable mRNAs to properly engage the nuclear export machinery. Whether NPM1 binds polyadenylated RNAs stably enough to accompany them to the cytoplasm and affect downstream posttranscriptional processes is currently unknown, but there is evidence to suggest that NPM1 is a nucleocytoplasmic shuttling protein (43).
Promoter-Driven mRNP Composition?
Several studies have now demonstrated promoter-dependent effects on downstream mRNA metabolism. Promoter-driven changes to mRNP composition most readily explain these effects. Back-to-back papers published in 2011 first documented promoter-dependent modulation of cytoplasmic mRNA decay in Saccharomyces cerevisiae. One paper described dramatic destabilization of SWI5 and CLB2 mRNAs at the onset of metaphase (44). This stability switch requires promoter-dependent deposition of the mitotic exit kinase Dbf2p on both mRNAs during transcription. Once in the cytoplasm, interaction with a second kinase, Dbf20p, enables decay of the Dbf2p-bound mRNAs in response to appropriate cell cycle cues. The second paper described a similar decay-enhancing effect mediated by the transcription factor Rap1p (45). In this case, however, the exact mRNP compositional change is not known. More recently, yeast promoter sequences have been shown to direct both mRNA localization and translation during starvation (46), and in mammalian cells, the translation elongation factor eEF1A facilitates transcription, nuclear export, and stabilization of HSP70 mRNA during the heat shock response (47). The rate at which such observations are now being published suggests that these first examples of promoter-driven mRNP compositional changes may be just the tip of the iceberg.
Updated Catalogs Identify Old and New Components
Despite the tremendous progress chronicled above, which spanned four decades of work by hundreds of researchers, the catalog of mRNA-binding proteins was far from complete. Only in the past 3 years, with the application of new high-throughput methodologies to the now-classic technique of in vivo UV cross-linking, has a complete compendium begun to be assembled. In an approach termed mRNA interactome capture, proteins that UV-cross-link to either natural or modified (4-thio-uridine or 6-thio-guanosine) ribonucleosides incorporated into polyadenylated RNAs in vivo are isolated using oligo-dT chromatography and are subsequently identified by highly sensitive state-of-the-art mass spectrometry techniques. Multiple such studies in mammalian and yeast cells have identified a high-confidence list of ~800 proteins that directly crosslink to polyadenylated RNAs (48–50). For many proteins previously annotated as RBPs solely on the basis of sequence similarity to known RBPs, these studies provided the first direct evidence for their in vivo mRNA-binding activity. More importantly, more than 200 new proteins were identified as high-confidence RBPs. Several of these have no known RBP-like signatures, so they likely represent new RBP classes. Intriguing examples include kinases, proteins previously annotated as DNA-binding factors, protein prolyl isomerases, and numerous housekeeping proteins and enzymes. A future challenge is to map their RNA interaction sites and determine which, if any, are of functional significance. Below, we discuss recent insights into three different classes of mRNA-interacting proteins. One of these is a previously well-known family of RBPs, whereas the other two are newly emerging.
DHX and DDX Proteins
The first group encompasses the DExH/D-box (DHX) and DEAD-box (DDX) proteins, which comprise large families (15 and 37 members in humans, respectively) of highly abundant, ATP-dependent RBPs (Figure 1d) (51). Due to their sequence similarity to known DNA unwinding enzymes, both groups are commonly referred to as RNA helicases. The term RNA helicase brings to mind an RNA translocation activity that would actively disrupt RNA secondary structures or stable RNA–protein interactions. To date, however, only a few members of the DHX class have convincingly shown an ability to translocate on RNA. Examples include Prp43 (DHX15) and vaccinia virus helicase NPH-II (52, 53), both of which require a 3′ stretch of single-stranded RNA (ssRNA) as an initial landing pad and can then dislodge either a complementary RNA strand or a tightly bound RBP by traveling in a 3′-to-5′ direction. Thus, these proteins likely do function as active RNP remodelers. In contrast, DDX proteins generally have no requirement for an ssRNA landing pad and exhibit little or no directionality of unwinding. Furthermore, observing unwinding activity in vitro generally requires huge excesses of protein over the double-stranded RNA substrate. These characteristics suggest that any unwinding activity by DDX proteins results from passive capture of transiently appearing ssRNA rather than active strand displacement. We propose that DDX proteins as a whole are not RNA helicases. Instead, these proteins more likely function as structural RNP components that toggle between ATP-bound and ATP-free states to respectively bind and release ssRNA. As exemplified by eIF4AIII, this nucleotide binding can be modulated by companion proteins to either increase or decrease the DDX protein’s affinity for RNA. Because their helicase domain interacts solely with the sugar-phosphate backbone, DDX proteins are perfect for use as sequence-independent RNA clamps that either provide structural stability for synthetic history-dependent marks (e.g., the EJC) or serve as companions to proteins that recognize short-sequence motifs, which are in themselves insufficient to ensure tight binding (e.g., SR proteins). The high cellular abundance of many DDX proteins (Figure 1d) and their stable association with large RNP assemblies such as neuronal transport granules (54) further support this idea of a structural rather than enzymatic role for these key mRNP components.
Small Molecule–Sensing RNA-Binding Proteins
The second group consists of mRNP proteins whose ability to bind RNA is sensitive to the concentration of a small ligand or metabolic intermediate. The founding member of this class is cytosolic aconitase, the enzyme that catalyzes isomerization of citrate to isocitrate in the tricarboxylic acid cycle. This isomerization activity requires an active-site iron–sulfur cluster that exists only when intracellular iron levels are high. In iron-depleted cells, cluster loss converts cytosolic aconitase into an RBP (also known as iron regulatory protein 1) that binds to iron regulatory elements and modulates both translation and mRNA decay of iron homeostasis transcripts (55, 56). Other examples of housekeeping enzymes known to “moonlight” as RBPs include glyceraldehyde-3-phosphate dehydrogenase, isocitrate dehydrogenase, and all three enzymes of the thymidylate cycle. Hentze & Preiss (57) recently suggested that these scattered reports of enzymes functioning as ligand-dependent mRNA regulatory factors herald the existence of much more extensive RNA, enzyme, and metabolite posttranscriptional regulatory networks. Lending strong support to this idea is the recent identification of 46 enzymes as members of the human mRNA interactome (48, 49). The existence of diverse metabolite-sensing RBPs is further supported by the recent discovery of a feedback loop coupling monounsaturated fatty acid levels with translational control of stearoyl-CoA desaturase 1 (SCD) mRNA, a binding target of the RBP Musashi (58). A conformational change induced upon binding of long-chain ω-9 fatty acids to Musashi’s N-terminal RNA recognition motif disfavors RNA binding, which is presumably required for efficient SCD translation. This example illustrates that nonenzymatic RBPs can also sense small-molecule ligand concentrations to modulate gene expression.
Base Modification–Specific RNA-Binding Proteins
The third group consists of proteins that “write,” “read,” and “erase” nucleotide modifications. Whereas stable RNAs such as ribosomal RNAs, transfer RNAs, and small nuclear RNAs have long been known to contain modified nucleobases, selective and reversible modification of mRNA bases has come to be appreciated as a prevalent and important posttranscriptional regulatory mechanism only in the past couple of years. So far, the best-studied base modifications in mRNA are N6-methyladenosine (m6A), 5-methylcytosine (m5C), and pseudouridine (Ψ) (59–61). Thus, another novel class of mRNP proteins consists of those that modify RNA bases (writers), recognize these modifications to modulate gene expression (readers), or remove the modifications (erasers) to reverse the effects. Indeed, writers for m6A (the METTL3 complex) and Ψ (Pus1 and Pus3 proteins) and a reader for m6A (YTHDFC2) were found in the mRNA interactome prior to elucidation of their functions (48, 49). Thus, other writer, reader, and eraser proteins have most likely already been catalogued in the mRNA interactomes and simply await functional discovery.
Protein Stoichiometries and Dynamics
From the work described above, we now have long lists of mRNA-interacting proteins. RNA-seq and quantitative proteomics have also given us fairly accurate per cell copy number estimates for many mRNAs and proteins (21, 22). The next challenge will be to determine the number of individual protein molecules bound per mRNA molecule as well as individual protein dynamics. Both types of information are crucial for developing more sophisticated structural models and understanding the mechanisms involved in mRNP structural evolution as an mRNA moves through its life cycle. For example, structural proteins involved in packaging and compacting mRNA so that the resultant mRNA can efficiently exit the nucleus and find its intended destination in the cytoplasm would be expected to have high stoichiometries and low dissociation rates. The EJC core proteins eIF4AIII, Y14, and Magoh exemplify this class. Resident at ~80% of exon–exon junctions, EJCs constitute regularly spaced stable anchor points throughout the coding regions of most mRNAs (35, 36). Because the average gene is interrupted by seven to eight introns, we can estimate that there are five to six EJCs per generic mRNA (Figure 1d). We also know that numerous SR and SR-like proteins copurify with EJC proteins, some with even higher stoichiometries than EJC cores (36). As a group, stably bound SR proteins outnumber EJC cores by more than eight to one, suggesting that the average mRNA carries 40–50 SR and SR-like protein molecules. This high stoichiometry is consistent with a recently proposed role for these proteins in packaging long stretches of spliced exons into a form strongly resistant to nuclease digestion (36). In contrast, the previously proposed EJC core factor MLN51 is ~1,000-fold less abundant in HeLa cells than the other three core proteins (21) and is ~60-fold less abundant than EJC cores in purified samples from HEK cells (36). Consistent with numerous in vitro EJC assembly and structural studies, these new data rule out MLN51 as an essential EJC or mRNP structural component, but they may suggest that whatever MLN51 protein is present in cells is tightly bound to a subset of EJC cores. Future challenges are to determine which subset this is and how it relates to MLN51’s biological role.
At the opposite end of the spectrum from mRNP structural components are transiently interacting proteins that alter the structure. Likely in this class are the enzymes that chemically modify either mRNA (e.g., modified base writers and erasers; nucleases) or mRNP structural proteins (e.g., SR protein kinases). Other expected transient interactors are remodeling factors such as bona fide RNA translocases [e.g., the superfamily 1 (SF1) RNA helicase Upf1] and the EJC disassembler PYM. Because many of these likely transient factors have been captured by UV cross-linking as mRNA-interacting proteins, it is tempting to think of them as stable mRNP components. But it is important to keep in mind that most cross-linking experiments provide little or no information about dynamics. That is, through the formation of covalent bonds, cross-linking turns transient interactions into permanent interactions. Thus, UV cross-linkability reveals only that the protein of interest is in direct contact with RNA bases in a geometry that is favorable for forming a covalent bond in the presence of UV light. It gives no information as to RNA–protein interaction lifetime. What are now needed are methodologies that can accurately measure the association and dissociation dynamics of individual mRNP components, preferably on a transcriptome-wide scale. Only with this information will it be possible to obtain the complete picture of how mRNP structure evolves through the whole of an mRNA life cycle.
Alternative mRNA Parts Impact mRNP Composition and Function
In addition to the protein parts list, a complete picture of mRNP structure and dynamics must also take into account the variability of the underlying template: the mRNA. Mammalian cells are estimated to contain anywhere between ~22,000 and ~500,000 mRNAs that range from ~500 to ~10,000 nt in length and between 1 and ~7,000 in copies per cell (62, 63). Whereas many alternative mRNA isoforms encode alternative protein isoforms, others encode the same protein but differ only in the attached untranslated regions (UTRs). This particular set of mRNPs, where alternative UTRs can dramatically change the cis-element landscape and mRNP protein composition—and, in so doing, the localization, translation, and decay of the bound mRNA—are discussed further below.
Alternative 3′ UTRs
It has long been recognized that 3′ UTRs are hotbeds of cis-acting regulatory elements. Chief among these are sequence elements such as adenylate–uridylate (AU)-rich elements and Pumilio protein (PUF), hnRNPs, and miRNA-binding sites. Also notable are complex secondary structures recognized by double-stranded RBPs such as Staufen1 and Staufen2. Although the average 3′-UTR length had been supposed early on to be 150–1,000 nt in metazoans, work over the past several years from many labs has revealed pervasive alternative polyadenylation in numerous systems, resulting in a significantly wider 3′-UTR length range (reviewed in Reference 64). In humans, for example, 3′-UTR lengths are much longer in the brain and shorter in testis when compared with undifferentiated embryonic stem cells (65). Indeed, in brain tissue, many 3′ UTRs exceed 20,000 nt (66). These 3′-UTR length differences result from coordinated alternative polyadenylation across the entire mRNA population as cells go from less to more differentiated states. Given the high prevalence of regulatory elements in 3′ UTRs, gain or loss of 3′-UTR sequence elements via alternative polyadenylation leads to differential recruitment of machineries that regulate mRNA localization, translation, and/or degradation to drive cell fate. For example, 3′-UTR shortening leads to loss of let7-binding sites in IGF2BP1/IMP1 proto-oncogene mRNA in several cancer cell lines, promoting higher protein expression and cellular transformation (67). Thus, 3′-UTR shortening during transformation is now recognized as a key contributor to dysregulated protein expression during cancer. Conversely, 3′-UTR lengthening during brain development likely signals a general shift from largely transcriptional to largely posttranscriptional regulation of gene expression as newly born neurons mature into huge postmitotic cells with highly specialized subcellular compartments.
Alternative 5′ UTRs
Alternative 5′ UTRs are also common within the human transcriptome. Roughly one-third (~6,000) of human genes are estimated to use alternative transcription start sites to create distinct mRNA isoforms, and a significant fraction of these are tissue specific (68). Although some alternative transcription start sites change the translation start site and, therefore, the N terminus of the encoded protein, many others affect only the 5′ UTR. For example, the brain-derived neurotropic factor (BDNF) gene utilizes at least 10 different transcription start sites to generate alternative first exons. Each unique noncoding first exon is spliced to the single protein-coding exon common to all isoforms (69). Although detailed molecular mechanisms have yet to be elucidated, these unique 5′ UTRs are thought to play determinative roles in generating distinct cell-type and subcellular BDNF expression patterns during neuronal development (70). Akin to 3′ UTRs, 5′ UTRs can occasionally contain cis elements that operate via trans-factor recruitment (71). Alternative inclusion/exclusion of these elements can thereby regulate mRNA expression. In S. cerevisiae, transcripts with alternative 5′ UTRs but otherwise identical coding sequences result in >100-fold differences in translation efficiency (72), underscoring the potential of alternative 5′ UTRs to impact mRNP expression.
Alternatively Spliced Internal Exons
In addition to alternative 5′ and 3′ termini, alternative splicing (AS) within UTRs can alter the cis-regulatory element landscape and, hence, mRNP composition. In a recent study, mRNA isoforms generated by ~30% of AS events were found to impact regulatory cis elements in either 5′ UTRs (short upstream ORFs, internal ribosome entry sites) or 3′ UTRs (miRNA-binding sites; Alu elements), leading to differential isoform association with polyribosomes (73). Finally, by introducing an upstream ORF into the 5′ UTR or a premature translation termination codon into the main ORF, many AS events simply redefine the 3′-UTR boundary without significantly altering overall mRNP composition. These altered 3′-UTR boundaries often trigger rapid mRNP disassembly and mRNA degradation via the NMD pathway (see below) (74). In humans, several RBPs employ AS linked to NMD to regulate their own expression (75), or to cross-regulate expression of dozens of other RBPs (76). Furthermore, estimates suggest that nearly one-fifth (17–21%) of all AS events conserved between humans and mouse can alter 3′-UTR boundaries (77). Thus, AS of internal exons is a widespread gene regulatory mechanism with major implications for mRNP composition and fate beyond any obvious changes to the sequence of the encoded protein.
FITTING THE PARTS TOGETHER
Cotranscriptional Assembly and Packaging
As with any complicated outfit, dressing and undressing mRNAs require some order to the addition and removal of individual factors. That is, during mRNP assembly, outerwear addition cannot precede underwear addition, and vice versa for mRNA disassembly. Thus, a major question with regard to mRNP structure and assembly is: Which steps must be sequential and which are nonsequential? Within cells, mRNP assembly begins the moment the nascent transcript emerges from the elongating Pol II. Factors loaded early then begin the process of coupling transcription and pre-mRNA processing to downstream mRNP assembly and postassembly steps. The term coupling generally means enhancement of one process (e.g., mRNA export) by another (e.g., pre-mRNA splicing). This phenomenon has been the subject of many excellent reviews detailing the original observations of functional coupling between myriad steps in eukaryotic gene expression (78, 79), so we do not discuss it in detail here. We instead lay out the general principles underlying this phenomenon by highlighting specific examples. As may be expected, the molecular bases of many sequential and nonsequential coupled events are changes to mRNP structure and composition.
All coupling events fall into one of two broad categories: contemporaneous and sequential. Contemporaneous coupling generally involves cooperative binding between factors that, without the other, would bind more slowly or less tightly. Thus, the factors reinforce each other, making both processes more efficient. A good example of contemporaneous coupling is the interaction between spliceosome assembly factors bound to the 3′-most intron and the cleavage and polyadenylation machinery assembling on a downstream polyA signal. Multiple cooperative interactions [e.g., U2 snRNP–CPSF (80) and U2AF65–polyA polymerase (81)] underlie reciprocal enhancement of pre-mRNA splicing and 3′-end cleavage and polyadenylation.
Sequential coupling, by contrast, requires that some binding or covalent modification event happen first; the stable product then serves as a recognition site for later interacting factors. This type of coupling is often referred to as recruitment; for example, factor X recruits factor Y. Factor X does so by providing a binding surface for factor Y, which binds subsequent to factor X. Most steps during mRNA processing are linked to downstream processes via such sequential coupling. One example is the Pol II C-terminal domain (CTD), wherein its numerous heptad repeats undergo dynamic phosphorylation and cis/trans proline isomerization as the polymerase progresses through initiation, elongation, and termination. These CTD chemical modifications create sequential landing pads for numerous RNA biogenesis factors and mRNP components (Figure 2) (82). These include the mRNA capping complex, the transcription and export (TREX) complex, U1 snRNP, the core splicing factor U2AF65, SR proteins, and the cleavage and polyadenylation machinery. The many effects of nuclear EJC deposition on cytoplasmic mRNA utilization nicely illustrate the principle that sequential coupling need not be limited to a single cellular compartment. As discussed in detail below, there are now many examples of mRNP factors loaded in the nucleus that alter mRNA utilization and metabolism in the cytoplasm.
Figure 2.
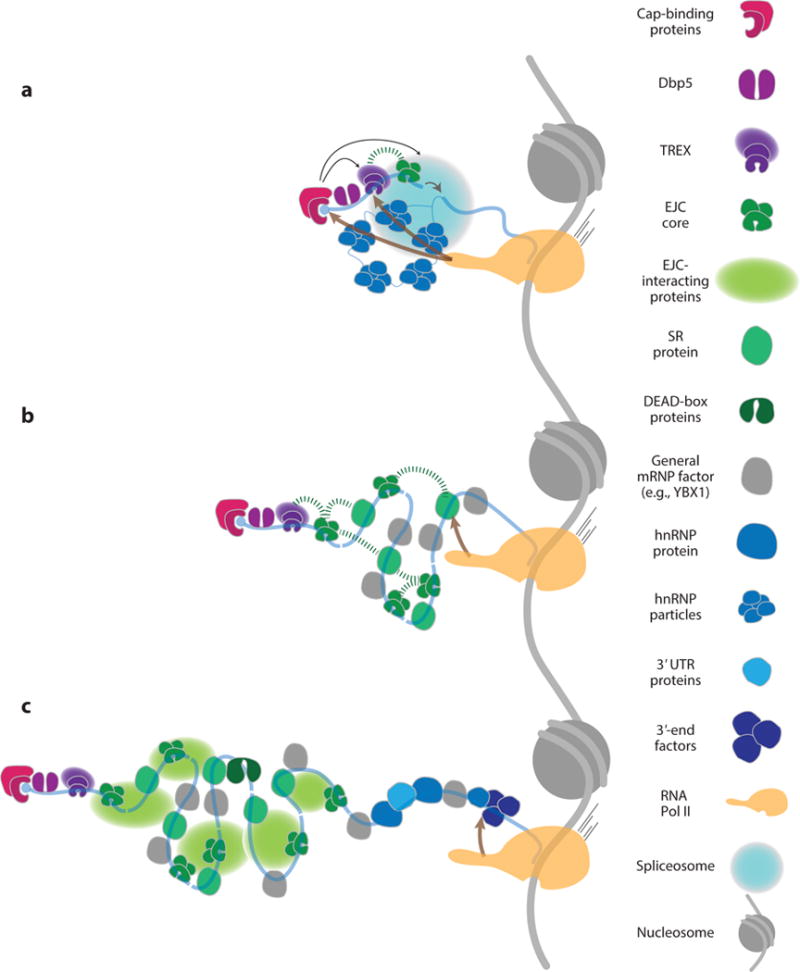
Cotranscriptional assembly and compaction of mRNPs. (a) Early steps in cotranscriptional mRNA processing and mRNP assembly. Nascent RNA is represented by blue lines (exons, thick lines; introns, thin lines). Indicated are sequential coupling events mediated by Pol II CTD (brown arrows), coupling events promoted by CBC (black arrows), and interaction between EJC and TREX that may compact the nascent mRNP (thick green dashed line). Individual components are as shown on the right. (b) EJC interactions with self, SR proteins, or TREX complexes that drive nascent mRNP compaction are indicated by thick green dashed lines. Continued sequential coupling action of Pol II CTD is also depicted as in panel a. All individual components are as in the legend on the right. (c) EJC interactions that drive compaction of internal exon mRNPs are now shown as light green spheres that represent several EJC-interacting proteins. The cotranscriptional assembly of 3′-UTR RNP is also shown. Abbreviations: CBC, cap-binding complex; CTD; C-terminal domain; EJC, exon junction complex; mRNP, messenger RNA–containing ribonucleoprotein; Pol II, RNA polymerase II; RNP, ribonucleoprotein; UTR, untranslated region.
Given that sequential coupling requires an ordered process, what is the order of mRNP assembly? Because formation of the m7G cap is essentially complete by the time nascent Pol II transcripts are 20–30 nt long (83, 84), and this is the sole feature recognized by the nuclear CBC, mRNP assembly likely initiates with CBC acquisition. CBC then promotes TREX complex acquisition in the cap-proximal region (85) and enhances cotranscriptional pre-mRNA splicing by recruiting several spliceosomal components (Figure 2a) (86, 87). Although CBC binding is not necessary for EJC deposition, it does promote stable association of several proteins that specifically bind spliced mRNA in vitro (e.g., ILF2, THRAP3) (Figure 2a) (39). It is not currently known, however, whether these proteins associate only with the first exon in vivo or whether they also bind later internal exons.
The packaging of internal exons likely involves both sequential and contemporaneous interactions between SR proteins and EJC cores (Figure 2b). Most SR proteins function as splicing activators that help recruit spliceosomes to internal exons via simultaneous and direct interactions with pre-mRNA splicing enhancer elements and the core splicing machinery (18). Thus, the initial loading of SR proteins likely precedes EJC assembly [which occurs within catalytically activated spliceosomes (88)], but once deposited, EJCs seem to stabilize SR protein binding. Even after extensive RNase digestion, all 12 SR proteins and several other SR-like proteins copurify with EJC core factors, and knockdown of eIF4AIII decreases SR protein cross-linking to polyA+ RNA (36). Furthermore, these EJC–SR interactions protect long (80–130-nt) stretches of spliced mRNA from nuclease digestion, suggesting that EJCs and SR proteins collaborate to condense and package spliced exons into a highly nuclease-resistant form.
In mammals, the average intron is ~10 times longer than the average exon. Therefore, the bulk of the RNA emerging from Pol II is intronic, and these sequences are likely packaged co-transcriptionally by hnRNP proteins. Such a role is best understood for hnRNP C, described in early studies as a core component of hnRNP particles. A heterotetramer of hnRNP C1 and C2 proteins (present in a three-to-one ratio) packages intronic sequences into particles with uniform size and shape. The existence of such particles on nascent transcripts in vivo was recently bolstered by a CLIP-seq study demonstrating direct contact between the RNA recognition motif domains of hnRNP C monomers and uridine-rich tracts found at regular intervals of ~165 nt and 300 nt within human introns (89). These new findings are in striking consonance with previous biochemical analyses of both in vivo isolated and in vitro assembled hnRNP C particles that wrap similar RNA lengths (90). Such hnRNP C binding patterns are detected on more than half (~55%) of human introns. hnRNP C proteins thus constitute a major hnRNP particle core that other hnRNP proteins, whose roles in RNA packaging are less well defined, could interact with to form larger hnRNP assemblies with more diverse structures and sequence specificities. Pre-mRNA packaging by hnRNP proteins likely functions to condense long intronic regions, thereby hiding cryptic splice and polyadenylation signals while facilitating constitutive splicing through three-dimensional juxtaposition of bona fide splice sites. Alternative packaging may expose or tuck away alternative exons.
In addition to introns, many hnRNP proteins assemble onto 3′ UTRs (see below). Thus, as with other nucleus-acquired mRNP components, 3′ UTR–bound hnRNPs can travel with mRNAs to the cytoplasm and influence downstream mRNA metabolism. Because many hnRNP proteins also harbor their own nuclear export signals, they may also play roles in facilitating or regulating mRNA export. Finally, the newly synthesized polyA tail is bound by multiple copies of nuclear polyA-binding protein (PABPN1), whose acquisition is also thought to facilitate nuclear export.
A small yet significant fraction of mammalian transcripts has either no introns (e.g., c-Jun and c-Fos) or no introns within the ORF (e.g., BDNF and Arc). How are these mRNAs packaged? It has long been known that SRSF1, SRSF3, and SRSF7 can bind to and facilitate export of intronless mRNAs (91, 92). These proteins could be recruited either via their interaction with the Pol II CTD or via direct interaction with methylated DNA. Another dual-specificity RNA- and DNA-binding protein is YBX1, which serves as both a transcription factor and a sequence-independent RNA packager. In vitro YBX1 assembles into stable and uniform particles that wrap and compact RNA (9). In specialized cells from diverse systems (e.g., neurons, Xenopus oocytes), YBX1 is highly enriched in packaged and silenced mRNPs (93). Given its high abundance in purified mRNPs (Figure 1a,b), YBX1 likely also binds spliced mRNAs. The latter association, however, may be limited to EJC-free stretches, as YBX1 does not specifically copurify with EJCs and their attendant SR proteins (36).
What if mRNAs are not quickly and efficiently packaged? If left unpackaged, newly transcribed RNA can hybridize with the unpaired template DNA in the transcription bubble. Such RNA–DNA hybrids formed in the wake of elongating polymerase are known as R-loops and are observed when cells are depleted of TREX components or SRSF1 (94, 95). The unpaired sense strand thereby exposed and stabilized by an R-loop can serve as a recombination hot spot, resulting in transcription elongation defects, high recombination rates, and genomic instability. Interestingly, genomic instability occurring upon SRSF1 depletion can be rescued by overexpression of RNPS1 (96), another SR-like protein. This suggests that multiple redundant RNA–protein interactions contribute to cotranscriptional mRNA packaging. Notably, cells depleted of EJC core proteins also display genomic instability (97), but whether R-loop formation also underlies this phenotype is not yet known. Importantly, several splicing factors, including hnRNP C, have also been discovered as the largest group of functionally related proteins necessary for genomic stability (98). Thus, by quickly capturing the RNA emerging from elongating Pol II, various mRNP and hnRNP components also serve to maintain genome integrity.
mRNP Domains
What is the overall shape/structure of a fully processed and packaged mRNP? To date, such structural insight is available only for the Balbiani ring (BR) mRNPs from Chironomus tentans salivary glands (99). These mRNPs assemble on exceptionally long (~30-kb) and abundant mRNAs, making them particularly amenable to electron microscopic ultrastructural analyses in salivary gland nuclei. As a BR mRNA emerges from Pol II, cotranscriptional recruitment of SR and hnRNP proteins leads to the formation of a coiled fibril. This fibril is tightly packed into a ribbon-like structure that is ultimately bent to form a croissant-shaped mRNP ~50 nm in diameter and ~15 nm thick (100). Remarkably, BR mRNA thus packaged is compacted 100–200-fold in comparison with its fully stretched linear form. What is currently unknown, however, is whether the packaging and compaction processes at work on these gargantuan mRNAs are also at work on normal-sized metazoan mRNAs. An electron microscopic analysis of S. cerevisiae mRNPs revealed ~5-nm-wide structures of varying length (15–30 nm) wherein mRNAs were calculated to be only 10-fold compacted (101). However, S. cerevisiae mRNPs contain neither EJCs nor SR proteins. Thus, mammalian mRNPs constitute an area desperately in need of structural analysis.
In the absence of definitive structural information, what can we surmise about overall mRNP architecture? The tremendous amount of in situ mRNA-binding data now available for mammalian RBPs enabled us to analyze the distribution of CLIP tags for several RBPs across all annotated 5′ UTRs, coding sequences (CDS), and 3′ UTRs (Figure 3a). This analysis suggests to us that spliced mammalian mRNPs assemble into three compositionally distinct domains around first, internal, and last exons, which loosely correspond to the 5′ UTR, ORF, and 3′ UTR, respectively (Figure 3b). The 5′ domain consists of the first exon, which is usually coincident with the 5′ UTR. As may be predicted for the ribosome-landing pad, this 5′ domain is comparatively protein free (102). Because the vast majority of introns in mammalian mRNAs interrupt the protein-coding region, EJCs and their partner SR proteins are disproportionately positioned over ORFs and specifically depleted from 3′ UTRs. The coding exons thus form the CDS domain, wherein the RNA is compacted by EJC and SR proteins. Also enriched here are proteins that regulate translation by engaging elongating ribosomes (e.g., FMR1 and Lin28). Lastly, the 3′ domain consists of the 3′ UTR, which is generally also the last exon. The 3′ domain is highly enriched for mRNA-bound hnRNP proteins (e.g., hnRNP U, FUS/TLS, and PTB) as well as many localization, translation, and decay factors, including HuR, RISC (also known as Ago), PUF, and IGF2BPs (also known as zipcode-binding proteins). Such a domain arrangement would have the advantage of leaving the 5′ and 3′ ends of the mRNA accessible to engage and regulate the translation machinery while compacting the CDS to protect the ORF and facilitate mRNP movement around the cell. Much future work is required to assess the validity of this model and determine its exact functional consequences.
Figure 3.
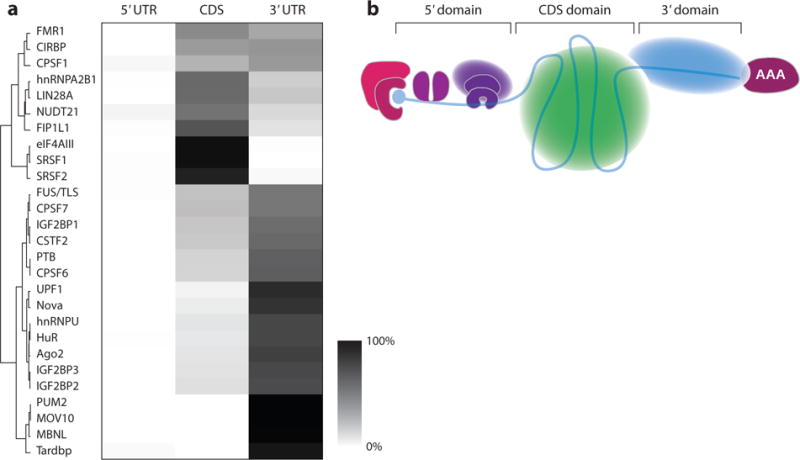
Major domains of spliced mammalian mRNPs. (a) Heat map showing percent of binding sites for given RBPs in the 5′ UTR, CDS, or 3′ UTR. The lightest shade indicates no binding, and the darkest shade indicates 100% binding in a given region. Abbreviations: CDS, coding sequence; mRNP, mRNA-containing ribonucleoprotein; UTR, untranslated region. (b) Schematic of the major hypothetical domains of spliced mRNPs: RNA (blue line), internal exon interactome (green sphere), and the 3′-UTR interactome (light blue sphere). Other shapes are as in Figure 2.
mRNP REMODELING DURING EXPORT AND TRANSLATION
Dynamics and Remodeling During Export
Once biogenesis and packaging are complete, the mRNP is properly dressed for nucleocytoplasmic export. The best-understood mRNA export pathway to date involves NPC transit mediated by the export receptor heterodimer NXF1–NXT1 (also known as TAP–p15) (reviewed in Reference 103). In the nucleus, NXF1 and NXT1 are major mRNP outerwear components, recruited by multiple underwear adaptors including the TREX components Aly/REF and UAP56-interacting factor (UIF); the SR proteins SRSF1, SRSF3, and SRSF7; the DDX protein Dbp5; and cleavage and polyadenylation specificity factor 6. These diverse recruitment sites span all three mRNP domains and suggest that individual mRNPs likely associate with multiple NXF1–NXT1 molecules. Multiple parallel and redundant NXF1–NXT1 recruitment mechanisms on the same mRNP could make export more robust and enhance its kinetics, as has been observed for spliced versus unspliced mRNAs (104). Furthermore, the presence of export adaptors in all three mRNP domains (first, internal, and last exons) suggests that NXF1-draped mRNPs can traverse the NPC in several possible orientations. This would contrast with BR mRNPs, which have been reported to emerge 5′ end first and engage ribosomes while still in transit through the pore (99). At the opposite extreme, many localized mRNAs are specifically sequestered away from the translation machinery until they reach their intended subcellular destination. mRNAs of this type have no apparent need to emerge 5′ end first, so they could exit in another orientation.
The existence of multiple NXF1 recruitment mechanisms may also allow for specific export control of particular mRNP cohorts (also known as regulons, sets of mRNAs jointly regulated by one or more RBPs) (105). An example is NXF1 recruitment to alternative mRNA export (ALREX) elements embedded within RNA sequences encoding the signal peptides of proteins targeted to the endoplasmic reticulum and mitochondria (106). Although its underwear adaptors are not yet known, this pathway is functionally distinct from the well-characterized splicing-dependent export pathway. Another distinct mRNA export pathway employs the karyopherin CRM1 instead of NXF1, and a minority of messages specifically outfit themselves with CRM1 to escape the nucleus. Examples are mRNAs that contain AU-rich elements and employ HuR (a protein that binds AU-rich elements) as an export adaptor (107), as well as mRNAs encoding several cell cycle regulators (e.g., cyclins CycD1 and CycE1) that bind to eIF4E in the nucleus to serve as a CRM1 adaptor (108). Another recently described mRNP export pathway bypasses NPCs altogether and delivers ultralarge mRNPs to the cytoplasm via nuclear envelope budding (109). Currently, which export adaptor underwear and receptor outerwear direct mRNPs to this pathway are completely unknown.
During transit through the central NPC channel (~10 nm diameter), the exceptionally large BR mRNPs undergo dramatic structural rearrangements. Electron micrographs of in-transit BR mRNPs show their ribbon structures unfolding into elongated fibrils (~25-nm-wide and ~135-nm-long rods) to fit through the pore (99, 100). Unlike these gigantic mRNPs, however, yeast mRNPs (5 nm wide and 20–30 nm long) (101) can easily fit through the pore without major structural perturbation. Thus, average mRNPs may require only minimal remodeling to transit through the pore. In support of this idea, single actin mRNPs transit very rapidly through the NPC, and unlike BR mRNPs, long-lived transit intermediates are rarely observed (110, 111).
Upon reaching the cytoplasmic side of the pore, mRNPs must be stripped of their export receptor outerwear to prevent backsliding into the nucleus (Figure 4a). Key players in this process are the DDX protein Dbp5 (DDX19) and its interaction partner Gle1 (103). Both proteins are nucleocytoplasmic shuttling proteins, and Dbp5 can be observed in association with BR mRNPs during both their cotranscriptional assembly and their exploration of the nucleoplasm. As proposed above for other DDX proteins, Dbp5 likely functions as a stable nuclear mRNP component that, in its closed, ATP-bound state, helps anchor other factors (e.g., export adaptors and receptors) to the mRNA. Upon reaching the cytoplasm, an interaction between Gle1, its small-molecule cofactor inositol hexakisphosphate (IP6), and nucleoporins within the pore cytoplasmic fibrils facilitates conversion of Dbp5 to its open, ADP-bound state (112). The resultant conformational change both decreases Dbp5’s affinity for RNA and promotes displacement of NXF1 and NAB2 (the yeast nuclear PABP) from the mRNP (113). The exact mechanical details of how this displacement works, however, have yet to be elucidated (114).
Figure 4.
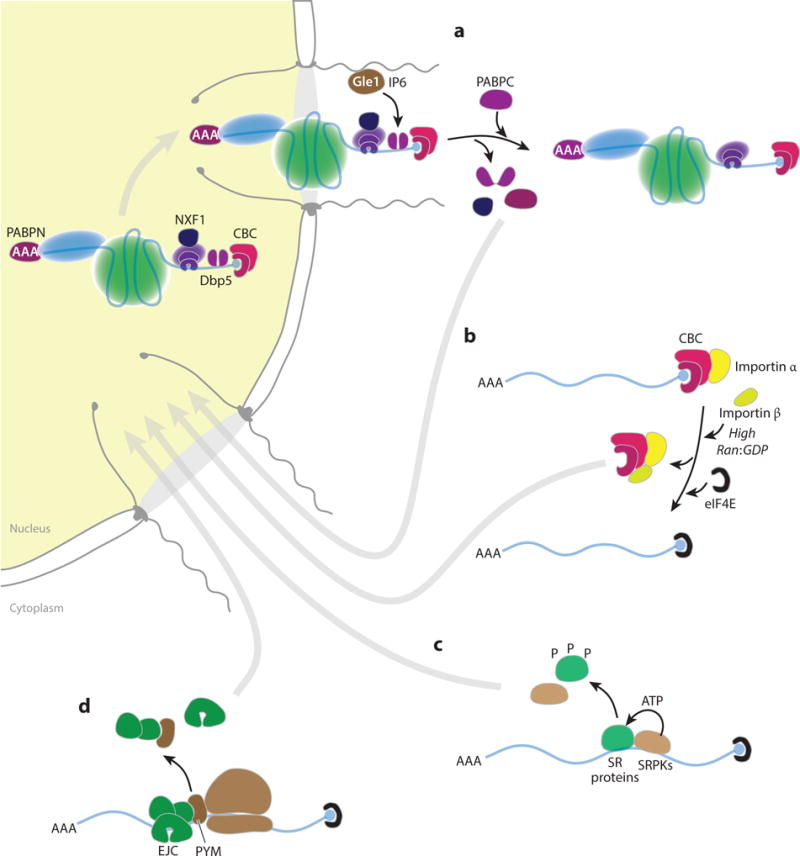
Mechanisms of mRNP remodeling in the cytoplasm. (a) mRNP remodeling during mRNA export that is catalyzed by Dbp5 at the cytoplasmic face of the NPC. Molecules represented by each individual shape are either labeled or are as in Figure 2. (b) Active mechanism of nuclear CBC exchange with the cytoplasmic cap-binding protein eIF4E. (c) Phosphorylation (P) and mRNA release of SR proteins mediated by the SRPKs. (d) EJC disassembly during translation by the ribosome-bound PYM protein. Abbreviations: CBC, cap-binding complex; EJC, exon junction complex; mRNA, messenger RNA; mRNP, mRNA-containing ribonucleoprotein; NPC, nuclear pore complex; PABP, polyadenosine (polyA)-binding protein; SRPK, SR-specific protein kinase.
Mass Action Versus Active Remodeling
Once the mRNP enters the cytoplasm, the next wardrobe change begins. Depending on their strength of interaction, replacement of nuclear mRNP components with cytoplasmic ones involves either passive or active remodeling. Passive remodeling is driven by mass action and occurs upon stochastic dissociation of less stably bound nuclear proteins; the low concentrations of these factors in the cytoplasm disfavor their rebinding. The newly exposed binding sites are then free to interact with proteins that are more abundant in the cytoplasm. That native mRNPs can be purified by oligo-dT hybridization to the polyA tail suggests that PABPs fall into this category of dynamically associating mRNP proteins. Similarly, eIF4E can readily associate with the 5′-m7G cap once the nuclear CBC proteins clear off. The CBC’s grip on the cap is loosened when high cytoplasmic Ran-GDP levels promote karyopherin importin-β association with CBC–importin-α complexes. A resultant structural change in CBP20 decreases its affinity for cap (115, 116). As the CBC–importin-α/β complex departs, the vacated cap can immediately be occupied by eIF4E, which is present at much higher concentrations in the cytoplasm than is CBC (Figure 4b). Exactly when this exchange occurs, however, may depend on the first or “pioneer” round of translation. Enrichment of CBP80 and not eIF4E in translationally repressed neuronal mRNPs suggests that CBC dissociation requires translational activation (117). Consistent with this suggestion is that the CBC can serve as an initiation factor for early rounds of translation, so its departure may require active mRNP remodeling by the ribosome (118).
Active remodeling events require the expenditure of energy, usually in the form of nucleoside triphosphate (NTP) hydrolysis. Because scanning of the small (40S) subunit along the 5′ UTR and translocation of 80S ribosomes through the ORF require much NTP hydrolysis, the first round of translation can be viewed as one big active remodeling event. During this process, every protein must necessarily be stripped away to offer the naked mRNA to the decoding center of the ribosome. Although the highly stable EJCs found on the 5′ UTR and ORF could be an impediment to ribosome translocation, the ribosome carries a key to the EJC’s lock. Positioned near the prow of the 40S subunit, the specific EJC disassembly factor PYM is perfectly poised to displace EJCs as the first ribosome moves down the mRNA. Interaction between PYM and Magoh weakens association between Magoh–Y14 and eIF4AIII (119, 120), allowing eIF4AIII to complete its ATPase cycle, release both the mRNA and ADP, and thus complete EJC disassembly (Figure 4c). Removal of SR proteins also requires active remodeling, in this case mediated by SR-specific protein kinases (SRPKs) (Figure 4c) that phosphorylate serine residues within the RS domain. This phosphorylation decreases SR proteins’ affinity for RNA, leading to their dissociation and reimport into the nucleus (121). What is not yet known, however, is whether SRPK action precedes or is coincident with the first round of translation.
Once the 5′-UTR and ORF domains have been cleared of proteins by transit of the first ribosome, do the underlying RNA sequences remain forever unclothed? Almost assuredly not. A single-molecule analysis of β-actin mRNAs in neurons has revealed that ribosome-colocalizing mRNAs initially in a dormant “masked” state rapidly shift to an “unmasked” state in response to neuronal activity. This unmasked state is characterized by new local protein synthesis and an increased mRNA detectability by fluorescent oligo hybridization. The ability of protease treatment to mimic this unmasking suggests that the normal process involves stripping of RBPs from mRNAs. Interestingly, the activity-induced unmasking is reversed upon deactivation of synaptic activity, reverting mRNAs back to a masked state (122). What cytoplasmic factors replace the previously shed proteins upon remasking remain to be identified, although the highly abundant YBX1 protein would be an obvious top candidate. This process may be similar to what happens during stress granule assembly, wherein translationally active mRNAs are assembled into a repressive masked state via acquisition of stress granule assembly proteins such as TIAR/TIA1 (123).
LARGE mRNP ASSEMBLIES AND mRNP LOCALIZATION
A Variety of mRNP Assemblies
Given that transit through the NPC is likely the predominant means of mRNA export in most cells, and that the central channel allows for transport only of smaller solid objects with diameters less than 39 nm, most mRNPs are likely exported as singletons. A burgeoning literature using fluorescently tagged mRNAs in living cells or single-molecule FISH (fluorescence in situ hybridization) in fixed samples supports this view (124, 125). Nonetheless, much larger assemblies also exist. For example, both electron microscopic and biochemical fractionation data from brain tissue indicate the existence of extremely large mRNA-containing complexes that likely contain multiple mRNA molecules (126). These so-called neuronal granules may act as bundled telecommunication signals to allow efficient transport of multiple different mRNA species along neurites, thereby ensuring that mRNAs encoding all necessary components for proper modulation of synaptic strength arrive within a single information packet (127). Furthermore, recent data from the Sossin lab (128) indicate that at least some dendritic mRNAs are transported as stably paused polysomes that can be rapidly reactivated in response to synaptic activity. Although attractive, however, all these ideas await rigorous testing to determine their validity and/or generality.
Neuronal granules are just one example of the large cytoplasmic mRNA assemblage observed in diverse cell types (123). Others include processing bodies (P bodies), stress granules, and germ granules. P bodies contain high levels of RNA degradation factors, so they are thought to be major sites of mRNA degradation (123). Stress granules are transient assemblies that form when cells experience negative conditions such as oxidative and hyperosmotic stress (123). They are thought to serve as sequestration sites for mRNAs that have been transiently removed from the translationally active pool, thus both protecting bulk cellular mRNAs from degradation and freeing up the translation machinery to synthesize stress response proteins. Once the stress has been alleviated, translation of the sequestered mRNAs is reactivated and stress granules dissipate. Other granules include polar granules (Drosophila), P granules (Caenorhabditis elegans), and chromatoid bodies (mammals), which are all different names for mRNA and PIWI-interacting RNA–containing material that accumulates around early germ cell nuclei and is thought to suppress transposon activity in these nuclei (129).
Granules or Droplets?
The word granule derives from the Latin word grānulum (meaning “small grain”) and thus connotes a small, solid object. Recent work, however, indicates that most RNA-containing assemblages currently dubbed granules or bodies are more likely droplets resulting from liquid–liquid phase separation (as in a lava lamp). Evidence for this idea includes the spherical shape of many RNA-containing assemblages, the ability of smaller spheres to fuse with one another to form larger spheres, and their tendency to “flow” when placed in a streaming fluid (130, 131). Phase transitions occur when the members of one set of molecules have higher affinity for one another than for molecules in the surrounding medium. In comparison to a solid, liquid-phase transitions are readily driven by heteropolymeric interactions and can be easily reversed with surfactants, making them highly dynamic. Furthermore, it is much easier for solutes to move in and out of liquids than in and out of solids. Thus, dissolving a liquid can occur much more rapidly than dissolving a solid, which is limited by solute access only to the solid–liquid interface. For these reasons, liquid droplets make much more sense than solids for the construction of dynamic structures required for biological regulation.
A general molecular feature that drives biopolymer phase transitions is multiple weak interactions between inherently unstructured domains or low-complexity sequences. Such domains are highly enriched in mRNP proteins (49). These domains are also sites of dynamic posttranslational modification, which likely serves to modulate the affinity of the unstructured domains for one another. But if this affinity becomes too strong, then liquids turn into solid aggregates that are much more difficult to dissolve. Such aggregates are often observed in proteinopathy disorders such as neurodegenerative diseases. So, the flip side of an ability to form useful liquid-phase transitions is the tendency of these phases to solidify. Most of the mechanistic work to date on phase transitions involving RNPs has focused on roles played by RBP low-complexity sequences in nucleating phase transitions. What have not yet been explored, however, are direct interactions between RNA molecules. Do RNAs merely serve as scaffolds for binding proteins that drive the phase transitions, or can RNAs play more active roles by making direct intermolecular contacts?
END-OF-LIFE mRNP DISASSEMBLY
All mRNPs have a limited life span: The half-life of the average mammalian mRNA, and hence mRNP, is ~8 h. However, half-life range spans from a few minutes to well over 24 h. Regardless, once an mRNA’s time is up, its protein attire must be completely removed to enable efficient turnover. Mutations that prevent or slow this undressing process result in accumulation of mRNA decay intermediates and decreased cell fitness. Excellent recent reviews cover the overall processes of both general and regulated mRNA decay (132) and how these pathways are intertwined with mRNA translation (133), so they are not covered here. What we focus on instead is how proteins are disassembled from mRNPs to make RNA available to degradative enzymes, providing a particularly well studied example of an active mRNP remodeler that facilitates translation-dependent mRNA decay.
As discussed above, 5′ UTRs are relatively devoid of stably bound proteins, and all CDS-bound proteins are necessarily stripped away by ribosome transit. Therefore, the domain most in need of disassembly prior to mRNA decay is the 3′ UTR. One protein with such a function is the SF1 RNA helicase, Upf1, which is a central player in the NMD pathway. It is also a bona fide RNA translocase that can harness the energy of ATP hydrolysis to travel along ssRNA in a 5′-to-3′ direction (134). When mRNAs are endonucleolytically cleaved as a consequence of premature termination codon recognition, Upf1’s helicase activity is necessary to disassemble proteins from the 3′ fragment so that exonucleases can access and degrade the underlying RNA (135). In vivo impairment of Upf1’s ATPase activity leads to accumulation of these mRNA degradation intermediates in P bodies along with several NMD and mRNP proteins. Although it had been widely presumed that Upf1 was recruited to an NMD target only upon recognition of a premature termination codon, new data suggest that Upf1 can interact with mRNAs across their entire length even prior to translation (136, 137). Other recent data indicate that Mov10, another SF1 helicase that also localizes to P bodies, cross-links on some mRNA 3′ UTRs in close vicinity to Upf1, and may assist Upf1 in the clearance of proteins from degrading mRNPs (138). However, both proteins also cross-link to a large subset of mRNAs in a mutually exclusive manner (138). These observations highlight yet-to-be-revealed complexities that underlie substrate specificity and cooperativity between RNP remodelers.
If Upf1 is present in mRNPs prior to premature termination codon recognition, how is it maintained in an inactive state until needed? The answer here is a pair of autoinhibitory interactions, one of which is disrupted upon physical interaction with another NMD factor and the other upon phosphorylation by an NMD-specific kinase (139). Once activated, Upf1 blocks eIF3 recruitment to inhibit new translation of NMD-targeted mRNPs (140). Because translation initiation machineries occupy both the mRNA cap and the polyA tail, they antagonize mRNA decapping and deadenylation. Blockade of their assembly by Upf1-mediated events may thus provide greater opportunity for decapping enzymes and deadenylases to access their substrates and begin mRNA degradation. This process could be further enhanced by Upf1 interactions with the general mRNA degradation machineries.
End-of-life mRNP disassembly may not be the sole domain of processive helicases. Dhh1, the yeast homolog of DDX6, is believed to function primarily as a translation repressor, which it can achieve even when defective for ATPase activity (141, 142). This mutant, however, accumulates in P bodies along with mRNA decay machinery (143). This finding suggests that one function of Dhh1 could be to evict itself and other interacting proteins from dying mRNPs to allow degradation to proceed. Such functions may be widespread among other DDX proteins, many of which stably associate with translationally repressed mRNPs.
Many questions with regard to final mRNP disassembly remain to be addressed: What factors function to dislodge stably bound 3′-UTR proteins during general mRNA decay? What is the disassembly process for inherently unstable mRNAs, such as those encoding inflammatory proteins (e.g., interleukin-2) or proto-oncogenes (e.g., c-Myc)? What about mRNP disassembly during miRNA- or m6A-mediated decay? Is the same set of remodelers used by all pathways, or are some remodelers unique to individual pathways? Clearly, there is still much to be done before we can fully understand how the last mRNP proteins are cast off so they can start the “fashion show” anew with nascent mRNAs in need of dressing in the nucleus.
SUMMARY POINTS.
Early research identified abundant mRNP proteins and provided the technical and conceptual foundation essential to obtain the comprehensive mRNP parts list available today.
More than 1,000 proteins ranging over four orders of magnitude in abundance interact with mRNAs to form mRNPs. Within mRNPs, proteins bind with different stabilities and stoichiometries, although these remain largely unknown for most components.
Alternate mRNA parts, namely alternate 5′-UTR, ORF, and 3′-UTR sequences, are major contributors to compositional and functional mRNP diversity.
mRNPs assemble cotranscriptionally with many steps coupled via contemporaneous and sequential interactions between processing factors and nascent mRNP components.
Cotranscriptional interactions (both contemporaneous and sequential) lead to immediate packaging of nascent mRNA into mRNPs. This rapid packaging is essential for genome integrity and cell viability.
Nonrandom distribution of mRNP proteins on first, internal, and last exons (loosely corresponding to 5′ UTRs, ORFs, and 3′ UTRs) organizes mRNPs into three distinct domains.
mRNPs undergo remodeling as they cross the nuclear pore to achieve unidirectional export into the cytoplasm. Subsequent remodeling during translation leads to a major overhaul in mRNP composition.
Active mechanisms displace mRNA protective structures and disassemble RNPs to expose the RNA polymer for degradation.
FUTURE ISSUES.
A major future challenge is to assign molecular functions to the hundreds of newly identified mRNA-interacting proteins. We also need to define upstream regulators and downstream targets of mRNA-interacting proteins to precisely place them within gene regulatory circuits.
What differentiates mRNPs from long noncoding RNPs, given that both contain long, polyadenylated Pol II transcripts?
Numerous RBPs, including the EJC, SR, and hnRNP proteins, are involved in virtually every step in mRNA biogenesis and metabolism. An important future goal is to understand their precise contribution to or function during each step.
Transcriptome-wide RBP-binding studies are identifying huge numbers of RBP-interaction sites. A major challenge for elucidating RNP protein function will be to distinguish the subset of binding sites that are functional from those that are of little or no consequence.
Researchers need to develop new methodologies that can accurately measure, preferably on a transcriptome-wide scale, the dynamics of individual mRNP component interactions.
Another important future goal is to define the precise composition and dynamics of a single mRNP as it passes through its various life stages. High-resolution structures of single mRNPs are also currently lacking.
Principles of assembly, function, and dynamics of large mRNP aggregates/assemblies need to be established.
A detailed mechanistic understanding of RNA remodeling activities during mRNP expression and degradation will be essential to reveal new regulators of posttranscriptional gene expression.
Acknowledgments
The authors thank Thoru Pederson for stimulating discussions and invaluable insights into the early years of mRNP research. G.S. acknowledges support from The Ohio State University in the form of institutional funds. M.J.M. is supported by a grant from the National Institute of Health (GM53007) and is a Howard Hughes Medical Institute Investigator. G.W.Y. and G.P. are funded by grants from the National Institute of Health (HG004659, NS075449, and U54HG007005) and the California Institute for Regenerative Medicine (RB4-06045 and TR3-05676). G.P. also acknowledges support by the National Science Foundation in the form of a Graduate Research Fellowship (DGE-1144086). G.W.Y. is an Alfred P. Sloan Research Fellow.
Glossary
- Heterogeneous nuclear ribonucleoproteins (hnRNPs)
a large family of sequence-specific RBPs that complex with both introns and exons and affect multiple posttranscriptional events
- Pre-mRNA
the precursor messenger RNA transcribed from the genome that encompasses exons and introns
- CLIP-seq
an approach that combines UV cross-linking of RBPs, immunoprecipitation, and high-throughput sequencing for studies of RNA–protein interactions
- mRNA interactome
collection of all proteins that are directly in contact with and can be covalently cross-linked to mRNA with UV
- SR proteins
a family of RBPs rich in arginine–serine dipeptides that bind degenerate sequences in and around exons to promote spliceosome assembly on pre-mRNAs and facilitate mRNA export, translation, and decay
- Nonsense-mediated decay (NMD)
a translation-dependent mRNA decay pathway that identifies and rapidly degrades mRNAs in which translation termination is aberrant as on nonsense codons
- DHX proteins
RecA homology domain–containing proteins with conserved DEAH or DExH motifs that integrate ATP binding and hydrolysis with RNA-binding and translocation activity
- DDX proteins
RecA homology domain–containing proteins characterized by conserved DEAD amino acid motif that couples ATP binding and hydrolysis with RNA-binding activity
- Spliceosome
the large dynamic machine that assembles from >100 proteins and 5 RNAs on intronic sequences and catalyzes intron excision and exon ligation
- Pol II CTD
C-terminal domain of RNA polymerase II with 25–52 tandem copies of serine- and proline-rich heptads
- TREX
a multisubunit protein complex that contains transcription elongation THO complex proteins and components of mRNA export machinery
- R-loops
RNA–DNA hybrid structures formed via base pairing of nascent RNA with the unpaired template DNA in the transcription bubble
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Guramrit Singh, Email: singh.734@osu.edu.
Melissa J. Moore, Email: melissa.moore@umassmed.edu.
LITERATURE CITED
- 1.Gall JG. On the submicroscopic structure of chromosomes. Brookhaven Symp Biol. 1956;8:17–32. [PubMed] [Google Scholar]
- 2.Dreyfuss G. Structure and function of nuclear and cytoplasmic ribonucleoprotein particles. Annu Rev Cell Biol. 1986;2:459–98. doi: 10.1146/annurev.cb.02.110186.002331. [DOI] [PubMed] [Google Scholar]
- 3.Kumar A, Pederson T. Comparison of proteins bound to heterogeneous nuclear RNA and messenger RNA in HeLa cells. J Mol Biol. 1975;96:353–65. doi: 10.1016/0022-2836(75)90165-5. [DOI] [PubMed] [Google Scholar]
- 4.Beyer AL, Christensen ME, Walker BW, LeStourgeon WM. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977;11:127–38. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- 5.Barrieux A, Ingraham HA, David DN, Rosenfeld MG. Isolation of messenger-like ribonucleoproteins. Biochemistry. 1975;14:1815–21. doi: 10.1021/bi00680a002. [DOI] [PubMed] [Google Scholar]
- 6.Blobel G. Protein tightly bound to globin mRNA. Biochem Biophys Res Commun. 1972;47:88–95. doi: 10.1016/s0006-291x(72)80014-7. [DOI] [PubMed] [Google Scholar]
- 7.Bryan RN, Hayashi M. Two proteins are bound to most species of polysomal mRNA. Nat New Biol. 1973;244:271–74. doi: 10.1038/newbio244271a0. [DOI] [PubMed] [Google Scholar]
- 8.Evdokimova VM, Wei CL, Sitikov AS, Simonenko PN, Lazarev OA, et al. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J Biol Chem. 1995;270:3186–92. doi: 10.1074/jbc.270.7.3186. [DOI] [PubMed] [Google Scholar]
- 9.Skabkin MA, Kiselyova OI, Chernov KG, Sorokin AV, Dubrovin EV, et al. Structural organization of mRNA complexes with major core mRNP protein YB-1. Nucleic Acids Res. 2004;32:5621–35. doi: 10.1093/nar/gkh889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagenmakers AJ, Reinders RJ, van Venrooij WJ. Cross-linking of mRNA to proteins by irradiation of intact cells with ultraviolet light. Eur J Biochem. 1980;112:323–30. doi: 10.1111/j.1432-1033.1980.tb07207.x. [DOI] [PubMed] [Google Scholar]
- 11.Dreyfuss G, Choi YD, Adam SA. Characterization of heterogeneous nuclear RNA–protein complexes in vivo with monoclonal antibodies. Mol Cell Biol. 1984;4:1104–14. doi: 10.1128/mcb.4.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayrand S, Setyono B, Greenberg JR, Pederson T. Structure of nuclear ribonucleoprotein: identification of proteins in contact with poly(A)+ heterogeneous nuclear RNA in living HeLa cells. J Cell Biol. 1981;90:380–84. doi: 10.1083/jcb.90.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Eekelen CA, Riemen T, van Venrooij WJ. Specificity in the interaction of hnRNA and mRNA with proteins as revealed by in vivo cross linking. FEBS Lett. 1981;130:223–26. doi: 10.1016/0014-5793(81)81125-8. [DOI] [PubMed] [Google Scholar]
- 14.Pinol-Roma S, Choi YD, Matunis MJ, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–27. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 15.Roth MB, Murphy C, Gall JG. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–23. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–96. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–47. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 18.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 19.Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boucher L, Ouzounis CA, Enright AJ, Blencowe BJ. A genome-wide survey of RS domain proteins. RNA. 2001;7:1693–701. [PMC free article] [PubMed] [Google Scholar]
- 21.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 23.Sonenberg N, Morgan MA, Merrick WC, Shatkin AJ. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. PNAS. 1978;75:4843–47. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonenberg N, Rupprecht KM, Hecht SM, Shatkin AJ. Eukaryotic mRNA cap binding protein: purification by affinity chromatography on sepharose-coupled m7GDP. PNAS. 1979;76:4345–49. doi: 10.1073/pnas.76.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patzelt E, Blaas D, Kuechler E. CAP binding proteins associated with the nucleus. Nucleic Acids Res. 1983;11:5821–35. doi: 10.1093/nar/11.17.5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozen F, Sonenberg N. Identification of nuclear cap specific proteins in HeLa cells. Nucleic Acids Res. 1987;15:6489–500. doi: 10.1093/nar/15.16.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo MJ, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. PNAS. 1999;96:14937–42. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–99. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 29.Carter MS, Li S, Wilkinson MF. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–75. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–83. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, et al. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–82. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 32.Kim VN, Dreyfuss G. Nuclear mRNA binding proteins couple pre-mRNA splicing and post-splicing events. Mol Cells. 2001;12:1–10. [PubMed] [Google Scholar]
- 33.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000;19:6860–69. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev. 2000;14:1098–108. [PMC free article] [PubMed] [Google Scholar]
- 35.Sauliere J, Murigneux V, Wang Z, Marquenet E, Barbosa I, et al. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19:1124–31. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- 36.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, et al. The cellular EJC interactome reveals higher-order mRNP structure and an EJC–SR protein nexus. Cell. 2012;151:750–64. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–69. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 38.Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346–51. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 39.Merz C, Urlaub H, Will CL, Lührmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–28. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, Krainer AR. Splicing remodels messenger ribonucleoprotein architecture via eIF4A3-dependent and -independent recruitment of exon junction complex components. PNAS. 2007;104:11574–79. doi: 10.1073/pnas.0704946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palaniswamy V, Moraes KC, Wilusz CJ, Wilusz J. Nucleophosmin is selectively deposited on mRNA during polyadenylation. Nat Struct Mol Biol. 2006;13:429–35. doi: 10.1038/nsmb1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagawa F, Ibrahim H, Morrison AL, Wilusz CJ, Wilusz J. Nucleophosmin deposition during mRNA 3′ end processing influences poly(A) tail length. EMBO J. 2011;30:3994–4005. doi: 10.1038/emboj.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okuwaki M. The structure and functions of NPM1/nucleophsmin/B23, a multifunctional nucleolar acidic protein. J Biochem. 2008;143:441–448. doi: 10.1093/jb/mvm222. [DOI] [PubMed] [Google Scholar]
- 44.Trcek T, Larson DR, Moldon A, Query CC, Singer RH. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell. 2011;147:1484–97. doi: 10.1016/j.cell.2011.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M. Promoter elements regulate cytoplasmic mRNA decay. Cell. 2011;147:1473–83. doi: 10.1016/j.cell.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Zid BM, O’Shea EK. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature. 2014;514:117–21. doi: 10.1038/nature13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vera M, Pani B, Griffiths LA, Muchardt C, Abbott CM, et al. The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. eLife. 2014;3:e03164. doi: 10.7554/eLife.03164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–90. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 49.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 50.Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–33. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–51. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–25. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–25. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 55.Hentze MW, Argos P. Homology between IRE-BP, a regulatory RNA-binding protein, aconitase, and isopropylmalate isomerase. Nucleic Acids Res. 1991;19:1739–40. doi: 10.1093/nar/19.8.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rouault TA, Stout CD, Kaptain S, Harford JB, Klausner RD. Structural relationship between an iron-regulated RNA-binding protein (IRE-BP) and aconitase: functional implications. Cell. 1991;64:881–83. doi: 10.1016/0092-8674(91)90312-m. [DOI] [PubMed] [Google Scholar]
- 57.Hentze MW, Preiss T. The REM phase of gene regulation. Trends Biochem Sci. 2010;35:423–26. doi: 10.1016/j.tibs.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Clingman CC, Deveau LM, Hay SA, Genga RM, Shandilya SM, et al. Allosteric inhibition of a stem cell RNA-binding protein by an intermediary metabolite. eLife. 2014;3:e02848. doi: 10.7554/eLife.02848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–46. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–33. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014;11:669–72. doi: 10.4161/rna.28829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Islam S, Kjällquist U, Moliner A, Zajac P, Fan JB, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–67. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marinov GK, Williams BA, McCue K, Schroth GP, Gertz J, et al. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 2014;24:496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci. 2013;38:312–20. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27:2380–96. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res. 2013;23:812–25. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mayr C, Bartel DP. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–84. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–35. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–60. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Li Y, Song MG, Kiledjian M. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol Cell Biol. 2008;28:939–48. doi: 10.1128/MCB.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rojas-Duran MF, Gilbert WV. Alternative transcription start site selection leads to large differences in translation activity in yeast. RNA. 2012;18:2299–305. doi: 10.1261/rna.035865.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sterne-Weiler T, Martinez-Nunez RT, Howard JM, Cvitovik I, Katzman S, et al. Frac-seq reveals isoform-specific recruitment to polyribosomes. Genome Res. 2013;23:1615–23. doi: 10.1101/gr.148585.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay: mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013;1829:612–23. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 75.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–93. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 76.Hamid FM, Makeyev EV. Emerging functions of alternative splicing coupled with nonsense-mediated decay. Biochem Soc Trans. 2014;42:1168–73. doi: 10.1042/BST20140066. [DOI] [PubMed] [Google Scholar]
- 77.Bicknell AA, Cenik C, Chua HN, Roth FP, Moore MJ. Introns in UTRs: why we should stop ignoring them. Bioessays. 2012;34:1025–34. doi: 10.1002/bies.201200073. [DOI] [PubMed] [Google Scholar]
- 78.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 79.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 80.Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 81.Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 2000;14:403–13. [PMC free article] [PubMed] [Google Scholar]
- 82.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–37. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jove R, Manley JL. In vitro transcription from the adenovirus 2 major late promoter utilizing templates truncated at promoter-proximal sites. J Biol Chem. 1984;259:8513–21. [PubMed] [Google Scholar]
- 84.Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. PNAS. 1993;90:7923–27. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 86.Pabis M, Neufeld N, Steiner MC, Bojic T, Shav-Tal Y, Neugebauer KM. The nuclear cap-binding complex interacts with the U4/U6 U5 tri-snRNP and promotes spliceosome assembly in mammalian cells. RNA. 2013;19:1054–63. doi: 10.1261/rna.037069.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Görnemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 88.Reichert VL, Le Hir H, Jurica MS, Moore MJ. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev. 2002;16:2778–91. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang M, Rech JE, Northington SJ, Flicker PF, Mayeda A, et al. The C-protein tetramer binds 230 to 240 nucleotides of pre-mRNA and nucleates the assembly of 40S heterogeneous nuclear ribonucleoprotein particles. Mol Cell Biol. 1994;14:518–33. doi: 10.1128/mcb.14.1.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 92.Huang Y, Gattoni R, Stévenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–43. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 93.Tafuri SR, Familari M, Wolffe AP. A mouse Y box protein, MSY1, is associated with paternal mRNA in spermatocytes. J Biol Chem. 1993;268:12213–20. [PubMed] [Google Scholar]
- 94.Domínguez-Sánchez MS, Barroso S, Gómez-González B, Luna R, Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLOS Genet. 2011;7:e1002386. doi: 10.1371/journal.pgen.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–78. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 96.Li X, Niu T, Manley JL. The RNA binding protein RNPS1 alleviates ASF/SF2 depletion-induced genomic instability. RNA. 2007;13:2108–15. doi: 10.1261/rna.734407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, et al. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci. 2010;13:551–58. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–39. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Daneholt B. Packing and delivery of a genetic message. Chromosoma. 2001;110:173–85. doi: 10.1007/s004120000127. [DOI] [PubMed] [Google Scholar]
- 100.Skoglund U, Andersson K, Björkroth B, Lamb MM, Daneholt B. Visualization of the formation and transport of a specific hnRNP particle. Cell. 1983;34:847–55. doi: 10.1016/0092-8674(83)90542-1. [DOI] [PubMed] [Google Scholar]
- 101.Batisse J, Batisse C, Budd A, Bottcher B, Hurt E. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J Biol Chem. 2009;284:34911–17. doi: 10.1074/jbc.M109.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Silverman IM, Li F, Alexander A, Goff L, Trapnell C, et al. RNase-mediated protein footprint sequencing reveals protein-binding sites throughout the human transcriptome. Genome Biol. 2014;15:R3. doi: 10.1186/gb-2014-15-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci. 2009;122:1933–37. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Valencia P, Dias AP, Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. PNAS. 2008;105:3386–91. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–43. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 106.Cenik C, Chua HN, Zhang H, Tarnawsky SP, Akef A, et al. Genome analysis reveals interplay between 5′ UTR introns and nuclear mRNA export for secretory and mitochondrial genes. PLOS Genet. 2011;7:e1001366. doi: 10.1371/journal.pgen.1001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brennan CM, Gallouzi IE, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J Cell Biol. 2000;151:1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Topisirovic I, Siddiqui N, Borden KL. The eukaryotic translation initiation factor 4E (eIF4E) and HuR RNA operons collaboratively regulate the expression of survival and proliferative genes. Cell Cycle. 2009;8:960–61. [PubMed] [Google Scholar]
- 109.Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149:832–46. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grünwald D, Singer RH. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–7. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oeffinger M, Zenklusen D. To the pore and through the pore: a story of mRNA export kinetics. Biochim Biophys Acta. 2012;1819:494–506. doi: 10.1016/j.bbagrm.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–59. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 113.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–54. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 114.Valkov E, Dean JC, Jani D, Kuhlmann SI, Stewart M. Structural basis for the assembly and disassembly of mRNA nuclear export complexes. Biochim Biophys Acta. 2012;1819:578–92. doi: 10.1016/j.bbagrm.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 115.Dias SM, Wilson KF, Rojas KS, Ambrosio AL, Cerione RA. The molecular basis for the regulation of the cap-binding complex by the importins. Nat Struct Mol Biol. 2009;16:930–37. doi: 10.1038/nsmb.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Görlich D, Kraft R, Kostka S, Vogel F, Hartmann E, et al. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 117.Fritzsche R, Karra D, Bennett KL, Ang FY, Heraud-Farlow JE, et al. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 2013;5:1749–62. doi: 10.1016/j.celrep.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 118.Maquat LE, Hwang J, Sato H, Tang Y. CBP80-promoted mRNP rearrangements during the pioneer round of translation, nonsense-mediated mRNA decay, and thereafter. Cold Spring Harb Symp Quant Biol. 2010;75:127–34. doi: 10.1101/sqb.2010.75.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bono F, Ebert J, Unterholzner L, Güttler T, Izaurralde E, Conti E. Molecular insights into the interaction of PYM with the Mago–Y14 core of the exon junction complex. EMBO Rep. 2004;5:304–10. doi: 10.1038/sj.embor.7400091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536–48. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 121.Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. PNAS. 2004;101:9666–70. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Buxbaum AR, Wu B, Singer RH. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014;343:419–22. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–8. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Batish M, van den Bogaard P, Kramer FR, Tyagi S. Neuronal mRNAs travel singly into dendrites. PNAS. 2012;109:4645–50. doi: 10.1073/pnas.1111226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, et al. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science. 2014;343:422–24. doi: 10.1126/science.1239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–96. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 127.Carson JH, Gao Y, Tatavarty V, Levin MK, Korza G, et al. Multiplexed RNA trafficking in oligodendrocytes and neurons. Biochim Biophys Acta. 2008;1779:453–58. doi: 10.1016/j.bbagrm.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Graber TE, Hébert-Seropian S, Khoutorsky A, David A, Yewdell JW, et al. Reactivation of stalled polyribosomes in synaptic plasticity. PNAS. 2013;110:16205–10. doi: 10.1073/pnas.1307747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. RNA granules in germ cells. Cold Spring Harb Perspect Biol. 2011;3:a002774. doi: 10.1101/cshperspect.a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–32. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 131.Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–91. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 132.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–59. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Roy B, Jacobson A. The intimate relationships of mRNA decay and translation. Trends Genet. 2013;29:691–99. doi: 10.1016/j.tig.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–35. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell. 2010;143:938–50. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zünd D, Gruber AR, Zavolan M, Mühlemann O. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol. 2013;20:936–43. doi: 10.1038/nsmb.2635. [DOI] [PubMed] [Google Scholar]
- 137.Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23:1636–50. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, et al. MOV10 is a 5′ to 3′RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell. 2014;54:573–85. doi: 10.1016/j.molcel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 139.Fiorini F, Boudvillain M, Le Hir H. Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res. 2013;41:2404–15. doi: 10.1093/nar/gks1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–27. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carroll JS, Munchel SE, Weis K. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol. 2011;194:527–37. doi: 10.1083/jcb.201007151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sweet T, Kovalak C, Coller J. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLOS Biol. 2012;10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dutta A, Zheng S, Jain D, Cameron CE, Reese JC. Intermolecular interactions within the abundant DEAD-box protein Dhh1 regulate its activity in vivo. J Biol Chem. 2011;286:27454–70. doi: 10.1074/jbc.M111.220251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baer BW, Kornberg RD. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. PNAS. 1980;77:1890–99. doi: 10.1073/pnas.77.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]


