Abstract
Differences in internal dose of nicotine and tobacco-derived carcinogens among ethnic/racial groups have been observed. In this study, we explicitly examined the relationships between genetic ancestries (genome-wide average) and 19 tobacco-derived biomarkers in smokers from 3 admixed groups in the Multiethnic Cohort Study (1993–present), namely, African ancestry in African Americans (n = 362), Amerindian ancestry in Latinos (n = 437), and Asian and Native Hawaiian ancestries in Native Hawaiians (n = 300). After multiple comparison adjustment, both African and Asian ancestries were significantly related to a greater level of free cotinine; African ancestry was also significantly related to lower cotinine glucuronidation (P's < 0.00156). The predicted decrease in cotinine glucuronidation was 8.6% (P = 4.5 × 10−6) per a 20% increase in African ancestry. Follow-up admixture mapping revealed that African ancestry in a 12-Mb region on chromosome 4q was related to lower cotinine glucuronidation (P's < 2.7 × 10−7, smallest P = 1.5 × 10−9), although this is the same region reported in our previous genome-wide association study. Our results implicate a genetic ancestral component in the observed ethnic/racial variation in nicotine metabolism. Further studies are needed to identify the underlying genetic variation that could potentially be ethnic/racial specific.
Keywords: association study, blacks, health disparity, Hispanics, nicotine addiction, Pacific Islanders
Considerable differences exist among US racial/ethnic groups in the risk of lung cancer associated with cigarette smoking after accounting for self-reported dose and duration (1, 2). Differences in exposure and in response to tobacco smoke constituents, including nicotine and carcinogens, have been repeatedly cited as possible factors underlying the ethnic/racial differences in lung cancer (3–6). For example, past studies have shown that nicotine uptake per cigarette was greater in African Americans (7, 8) when compared with whites; cigarettes per day-adjusted nicotine equivalents were lower in Asians compared with Native Hawaiians or whites (9); and slower clearance of serum cotinine and lower cotinine glucuronidation were observed for African American than for white smokers (8, 10, 11). These differences suggest that genetic ancestry (resulting from ethnic-specific genetic polymorphisms) and/or ethnic-specific “environmental” factors (such as dietary habits, lifestyle, medication history, etc.) may play a role in the metabolism of nicotine and tobacco smoke-derived carcinogens. Dissecting the contribution of genetic ancestral components from that of environmental factors may lead to discovery of genetic variants influencing the metabolism of tobacco smoke ingredients and, potentially, a better understanding of the disparity in lung cancer risk.
In this study, we examined the relationships between genetic ancestry and metabolic biomarkers of nicotine and the potent tobacco smoke-derived carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) (12), among smokers from 3 admixed populations in the Multiethnic Cohort (MEC) Study, where the gradients in genetic ancestry provide the necessary variation required for testing its effect. We also took advantage of the wide range of questionnaire information in the MEC Study on “environmental” factors, such as lifestyle, diet, and medical history, and adjusted our analysis for possible confounders. Important associations between genetic ancestry and biomarkers were followed up with an admixture mapping scan (13) to identify genomic regions harboring ancestry-specific variants that are related to the traits. An admixture scan can be more powerful than genome-wide association studies (GWAS) in detecting ancestry-specific associations because of a reduced number of statistical tests being performed (13).
Our results confirmed that genetic ancestry could explain part of the variation in nicotine metabolism, although no new genetic variant was found, in comparison with our recently published genome-wide association study (14).
METHODS
Study population
The MEC Study includes over 215,000 participants aged 45–75 years at recruitment from 5 racial/ethnic groups (African Americans, Japanese Americans, Native Hawaiians, Latinos, and European Americans) in Hawaii and California. The cohort was assembled in 1993–1996; a self-administered, 26-page questionnaire was used to obtain extensive information on demographics, medical and reproductive histories, medication use, physical activity, diet, and supplement use. Blood and urine samples were collected mostly between 2002 and 2005 from all willing MEC participants still living in Hawaii or southern California. All MEC members providing biospecimens gave informed consent. Sample collection and all other procedures were approved by the institutional review boards of the University of Hawaii and the University of Southern California. Additional details are provided elsewhere (15). Identification of incident cancer cases is by regular linkage with the Hawaii, Los Angeles County, and California Surveillance, Epidemiology, and End Results registries. Cancer-free smokers in the 3 admixed groups (African Americans, Native Hawaiians, and Latinos) with an available urine and blood sample and who reported having smoked cigarettes in the previous 2 weeks before specimen collection were included in this study.
Biomarker assays
For the biomarkers included in this study, well-established and analytically validated methods exist for their quantitation. All metabolic biomarkers were measured in first morning or overnight urine and were expressed in concentration. To adjust for possible differences in dilution between morning and overnight urine, we quantified and included urinary creatinine concentration as an adjustment variable in data analysis.
Nicotine is primarily metabolized by 3 pathways (Web Figure 1 available at http://aje.oxfordjournals.org/) and converted to 1) cotinine by cytochrome P450 2A6 (CYP2A6)-catalyzed C-oxidation; 2) nicotine N-oxide by flavin monooxygenase-catalyzed N-oxidation; and 3) nicotine glucuronide by 5′-diphospho-glucuronosyltransferase (UGT)-catalyzed conjugation (16). Cotinine is further metabolized to trans-3′-hydroxycotinine (3HC) primarily by CYP2A6. Cotinine and 3HC can be converted to cotinine-glucuronide and 3HC-glucuronide by UGTs. Analysis of the urinary concentrations of total (the sum of free and the glucuronide conjugates) and free nicotine, cotinine, and 3HC was carried out by liquid chromatography-tandem mass spectrometry, as described previously (17). The average percent differences from 115 duplicates across plates were as follows: 3HC, 6.8%; total 3HC, 8.7%; cotinine, 6.0%; total cotinine, 5.4%; nicotine, 9.7%; total nicotine, 11.1%; and nicotine N-oxide, 7.5% (17).
The tobacco-specific lung carcinogen, NNK, can be metabolized by carbonyl reduction to 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL). Both NNK and NNAL are carcinogenic after bioactivation (18). An important detoxification pathway for NNK is through glucuronidation of NNAL by UGTs (19) (Web Figure 2). NNAL and NNAL-glucuronides were determined as previously described with slight modifications (20).
Ancestry estimation
Study subjects were genotyped on the Human 1M-Duo BeadChip array (Illumina, San Diego, California), and genotypes of about 1,089,000 autosomal single-nucleotide polymorphisms for 364 African Americans, 311 Native Hawaiians, and 453 Latinos remained after strict and standard quality-control procedures for GWAS, as described elsewhere (14). We further excluded 2 African Americans, 5 Native Hawaiians, and 11 Latinos who appeared as outliers from their self-reported ethnic groups on the basis of principal components derived from GWAS data.
Genome-wide global African, East Asian, Amerindian, European, and Hawaiian ancestries were estimated with the program ADMIXTURE (21). For easy interpretation of ADMIXTURE results, we included reference populations genotyped on the Illumina 650 array from the Human Genome Diversity Panel (22): Africans (Yoruba, Mandenka, and San); East Asians (Han Chinese and Japanese in Tokyo); Europeans (Basque, French, northern Italians, and Sardinians); and Native Americans (Mayans and Pima Indians). No public reference panel for Native Hawaiians is available, so we separately genotyped on the Human 1M-Duo BeadChip arrays 159 Native Hawaiians in the MEC Study who reported being >75% Hawaiian. Such individuals have been shown to be the best possible substitutes for ancestral Native Hawaiians (23). We followed standard GWAS quality-control procedures for this data set and retained about 1,088,000 autosomal single-nucleotide polymorphisms. Combining the different data sources resulted in a set of about 617,000 markers common to all groups. Among them, 90,700 independent markers (multiple R2 < 0.1) were selected and used in ADMIXTURE.
Local ancestry estimation for the 3 admixed groups was performed separately with LAMP-ANC (Local Ancestry in Admixed Populations-Ancestral), version 2.5 (24). For the 617,000 markers common to all groups, locus-specific ancestral allele frequencies (required parameter) were derived from the above-mentioned reference populations. LAMP-ANC selected unrelated (r2 < 0.1) ancestry informative markers from the 617,000 markers: 185,386 for African Americans; 127,610 for Latinos; and 113,922 for Native Hawaiians. Prior ancestral composition and proportions (required) were set to the estimates derived from a previous MEC analysis (23): 20% European and 80% African for African Americans; 60% European and 40% Amerindian for Latinos because the African ancestral proportion was low in this population (<4% on average); and 27% European, 46% Hawaiian, and 27% East Asian for Native Hawaiians.
We compared the global ancestry estimated by ADMIXTURE and the genome-wide average of locus-specific ancestry estimated by LAMP-ANC for the 4 ancestries of interest, namely, African ancestry in African Americans, Amerindian ancestry in Latinos, and Asian and Native Hawaiian ancestries in Native Hawaiians. Eleven subjects with individual ancestry differences > 0.1 (5 Latinos, 6 Native Hawaiians) were removed. The correlation between global ancestry and average locus-specific ancestry was greater than 0.99 for all 4 ancestries after the removal. A total of 362 African Americans, 300 Native Hawaiians, and 437 Latinos were retained for analysis.
Statistical analysis
We analyzed the following 19 biomarkers (Web Table 1): total nicotine equivalents (sum of total cotinine, nicotine, 3HC, and nicotine N-oxide) as a measure of internal dose of tobacco smoke exposure; free nicotine; free cotinine; free 3HC; nicotine N-oxide; and total NNAL as a biomarker of total NNK uptake (18, 25, 26); percentages of free nicotine, nicotine-glucuronide, nicotine C-oxidation products (total cotinine + total 3HC), and nicotine N-oxide among total nicotine equivalents; nicotine, cotinine, and 3HC glucuronidation [(total − free)/total]; percentages of free cotinine, cotinine-glucuronide, and total 3HC among nicotine C-oxidation products; 2 measures of CYP2A6 activity for comparison with the literature (9), total 3HC/total cotinine and total 3HC/free cotinine; and NNAL glucuronidation [(total − free)/total NNAL] as a biomarker for detoxification of NNK.
Linear regression was performed separately in the 3 admixed groups to assess correlation between these traits and global African ancestry in African Americans, Amerindian ancestry in Latinos (adjusted for African ancestry), and Asian and Hawaiian ancestry in Native Hawaiians. Other ancestries were not included in modeling because of their low proportions (<1% on average). We adjusted for age at urine collection, sex, creatinine (log transformed), total nicotine equivalents (log transformed) where appropriate, self-reported numbers of cigarettes smoked per day during the last 2 weeks before urine collection, and other potential confounders (see below). Transformation (log or cube root; Web Table 1) was applied to reduce skewness and heavy tails in the distribution for some traits. For total NNAL and NNAL glucuronidation, we also adjusted for batch of NNAL assays. For the others, additionally controlling for batch did not change the relations materially.
The potential confounders included were body mass index, physical activity, ethanol drinking (0, ≤15, >15 g/day), caffeine intake (≤100, >100–300, >300–500, >500 mg/day), and education (<8th grade, high school, some college, college degree, and graduate degree) as a surrogate for socioeconomic status. The following variables were also considered but were not strong confounders and were not included in the regression analysis: use of estrogen in any form in women at urine collection (yes/no); ever use of birth control pills for ≥1 month (yes/no); ever use of aspirin 2 or more times per week for ≥1 month (yes/no); menopausal status; red meat consumption; total intake (from food and supplement use) of vitamins A, B6, B12, C, D, and E; and β carotene, calcium, folate, niacin, riboflavin, and thiamin. We checked residual plots to make sure the assumptions for linear regression were roughly met.
For significant associations between global ancestries and biomarkers, admixture mapping was performed by regressing biomarkers on local ancestry and adjusting for similar covariates as above plus global ancestries in the corresponding admixed groups.
The statistical significance threshold for analysis of global ancestry was 0.05/8/4 = 0.00156, accounting for 4 ancestries and 8 independent measured phenotypes. For admixture scans, we used the number of independent ancestry informative markers selected by LAMP-ANC in Bonferroni correction within each admixed group. The resulting significance threshold was 2.7 × 10−7, 3.9 × 10−7, and 4.4 × 10−7 for African Americans, Latinos, and Native Hawaiians, respectively. SAS, version 9.3 (SAS Institute, Inc., Cary, North Carolina), and R (http://www.r-project.org/) were used for data analysis. Two-sided tests were used.
RESULTS
Basic characteristics of the smokers in this study are shown in Table 1. The mean self-reported smoking dose at urine collection was lowest in Latinos (9.4 cigarettes smoked per day), followed by African Americans (11.2 cigarettes smoked per day), and Native Hawaiians (15.2 cigarettes smoked per day), although total nicotine equivalents were highest in African Americans. Caffeine and red meat consumption were much greater in Native Hawaiians and Latinos compared with African Americans, whereas alcohol intake was the lowest in Native Hawaiians compared with African Americans and Latinos. Average body mass index, physical activity, and smoking duration were similar among ethnic groups. The mean African ancestry was 77.5% (range, 29.3%–100%) in African Americans. The average Amerindian ancestry was 39.6% (range, 0%–80.3%) in Latinos. In Native Hawaiians, the average Asian and Hawaiian ancestry was 22.0% (range, 0%–100%) and 46.8% (range, 0%–100%), respectively.
Table 1.
Characteristics of the Study Population in the Multiethnic Cohort Study, 1993-Present
| Characteristic | African Americans (n = 362) |
Native Hawaiians (n = 300) |
Latinos (n = 437) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | Mean (SD) | No. | % | Mean (SD) | No. | % | Mean (SD) | |
| Female | 252 | 69.6 | 191 | 63.7 | 207 | 47.4 | |||
| Education | |||||||||
| <8th grade | 3.0 | 2.7 | 31.2 | ||||||
| 8th–12th grade | 35.6 | 51.3 | 31.4 | ||||||
| Some college | 41.2 | 33.6 | 28.6 | ||||||
| College degree | 11.2 | 7.4 | 4.4 | ||||||
| Graduate school | 9.0 | 5.0 | 4.4 | ||||||
| Age at urine collection, years | 64.7 (7.5) | 61.3 (7.0) | 65.5 (6.4) | ||||||
| Body mass indexa | 27.8 (5.8) | 28.0 (5.8) | 27.1 (4.8) | ||||||
| Cigarettes per day at urine collection | 11.2 (7.3) | 15.2 (9.8) | 9.4 (7.4) | ||||||
| Total nicotine equivalents, nmol/mL | 56.1 (41.9) | 37.7 (28.0) | 41.8 (34.6) | ||||||
| Smoking duration, years | 28.2 (10.1) | 29.3 (9.1) | 29.8 (10.8) | ||||||
| Physical activityb | 1.6 (0.3) | 1.6 (0.4) | 1.7 (0.4) | ||||||
| Caffeine intake, mg/day | 188 (160) | 250 (180) | 265 (189) | ||||||
| Ethanol intake, g/day | 16.0 (36.2) | 12.9 (38.7) | 16.8 (44.6) | ||||||
| Red meat consumption, g/day | 72.4 (60.8) | 88.8 (60.8) | 98.3 (84.3) | ||||||
| Used estrogen at urine collection (among women) | 52 | 20.6 | 28 | 14.7 | 41 | 19.8 | |||
| Ever used birth control pills (among women) | 143 | 56.7 | 114 | 59.7 | 105 | 50.7 | |||
| Proportion of ancestry, % | |||||||||
| African | 77.5 (14.1) | 1.0 (1.9) | 3.7 (2.4) | ||||||
| Amerindian | 1.0 (1.8) | 0.4 (0.9) | 39.6 (13.6) | ||||||
| European | 20.8 (13.9) | 29.8 (26.6) | 55.4 (13.9) | ||||||
| East Asian | 0.4 (0.8) | 22.0 (24.8) | 0.9 (1.8) | ||||||
| Hawaiian | 0.3 (0.4) | 46.8 (24.5) | 0.4 (0.5) | ||||||
Abbreviation: SD, standard deviation.
a Expressed as weight (kg)/height (m)2.
b Metabolic equivalents, calculated as [(no. of hours sleeping × 0.91) + (no. of hours sitting × 1.0) + (no. of hours in light activities × 2.4) + (no. of hours in moderate activities × 4.0) + (no. of hours in vigorous activity × 7.2)]/24.
The medians of nicotine C-oxidation proportions among total nicotine equivalents (i.e., transformation to cotinine and its metabolites) were 81%, 81%, and 73% in African Americans, Latinos, and Native Hawaiians, respectively, consistent with a previous observation that the majority of nicotine is cleared through the C-oxidation pathway (27). The majority of nicotine C-oxidation products are in the form of 3HC or 3HC-glucuronide; the median proportions of total 3HC were 68%, 64%, and 56% in African Americans, Latinos, and Native Hawaiians, respectively, whereas free cotinine and cotinine-glucuronide comprised <21% and <25% of C-oxidation products on average in all groups. There were considerable differences in the distribution of the biomarkers among ethnic groups based on nonparametric tests (P's < 0.0001, Kruskal-Wallis test) (Web Table 2).
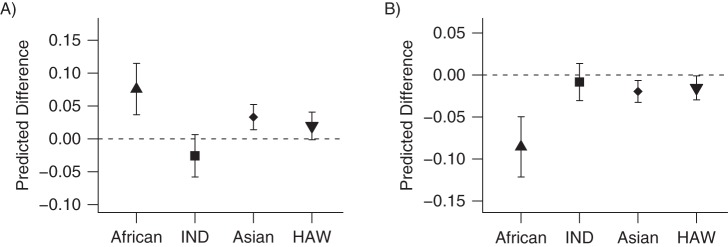
After adjustment for age at urine collection, sex, total nicotine equivalents, cigarettes smoked per day, and other potential confounders, 4 traits were significantly (P < 0.00156) related to African ancestry in African Americans, 2 of which were also related to Asian ancestry in Native Hawaiians (Table 2; Figure 1). The free cotinine level (concentration or relative percentage among nicotine C-oxidation products) was higher with increased African and Asian ancestries. On a log scale, the percentage of free cotinine among C-oxidation products was predicted to increase 0.13 (95% confidence interval (CI): 0.053, 0.21) with a 20% increase in African ancestry (P = 0.0012) or, equivalently, the predicted ratio of change for this percentage was 1.14 (95% CI: 1.05, 1.24) between 2 persons with a 20% difference in African ancestry. Similarly, a 20% increase in Asian ancestry was associated with a predicted ratio of change of 1.08 (95% CI: 1.04, 1.13) (P = 5.0 × 10−4) or an 8% increase. Because of the cube root transformation used, it was not easy to produce a meaningful prediction of change in the free cotinine level. African ancestry was also related to lower cotinine glucuronidation, among total cotinine or among nicotine C-oxidation products. The predicted decrease for cotinine glucuronidation was 8.6% (95% CI: 5.0, 12.1) (P = 4.5 × 10−6) per 20% increase in African ancestry.
Table 2.
Predicted Differences in Metabolic Traits Per 20% Increase in Ancestries for Statistically Significant Relations (P < 0.00156) From Linear Regression With Adjustment for Lifestyle Confounding Factors in the Multiethnic Cohort Study, 1993-Present
| Trait | Basic Adjustmenta |
Additional Adjustmentb |
||||
|---|---|---|---|---|---|---|
| Difference | 95% CI | P Value | Difference | 95% CI | P Value | |
| African ancestry | ||||||
| Free cotinine, nmol/mLc | 0.057 | 0.021, 0.094 | 0.0023 | 0.076 | 0.037, 0.12 | 1.8 × 10−4 |
| Free cotinine among C-oxidation productsd | 0.11 | 0.037, 0.19 | 0.004 | 0.13 | 0.053, 0.21 | 0.0012 |
| % cotinine glucuronidatione | −7.4 | −10.6, −4.1 | 1.3 × 10−5 | −8.6 | −12.1, −5.0 | 4.5 × 10−6 |
| % cotinine glucuronidation among C-oxidation productsf | −2.5 | −4.2, −0.9 | 0.003 | −3.1 | −4.9, −1.3 | 0.0011 |
| Asian ancestry | ||||||
| Free cotinine, nmol/mLc | 0.033 | 0.015, 0.052 | 0.0004 | 0.033 | 0.014, 0.052 | 8.8 × 10−4 |
| Free cotinine among C-oxidation productsd | 0.08 | 0.04, 0.12 | 1.3 × 10−4 | 0.080 | 0.034, 0.12 | 5.0 × 10−4 |
Abbreviation: CI, confidence interval.
a Basic adjustment for age at urine collection, sex, creatinine (log transformed), total nicotine equivalents (log transformed), and self-reported numbers of cigarettes smoked per day during the last 2 weeks before urine collection.
b Additional adjustment for body mass index, physical activity, ethanol drinking, caffeine intake, and education.
c Differences shown are for cube root-transformed values.
d Differences shown are for log[free cotinine/(total cotinine + total trans-3′-hydroxycotinine)].
e Calculated as [cotinine-glucuronide/total cotinine].
f Calculated as [cotinine-glucuronide/(total cotinine + total trans-3′-hydroxycotinine)].
Figure 1.
Plots of predicted differences per 20% increase in 4 ancestries in the Multiethnic Cohort Study (1993-present) after adjustment for lifestyle factors for A) free cotinine (cube root) and B) cotinine glucuronidation. The vertical bars represent 95% confidence intervals for predicted differences. HAW, Native Hawaiian ancestry; IND, Amerindian ancestry.
No other relationships between the biomarkers and global ancestries, including all those for Amerindian ancestry in Latinos and Hawaiian ancestry in Native Hawaiians, passed the threshold of statistical significance; however, several suggestive relations (P's < 0.05) may be of interest, given that our sample sizes were relatively small, and are shown in Web Table 3. The relationships did not differ significantly by total nicotine equivalents or levels of cigarettes smoked per day (data not shown). The results did not change in sensitivity analyses when prevalent lung cancer and other major cancer cases (n = 40) were removed. Parameter estimates changed >25% for some relationships with and without adjustment for lifestyle factors (Table 2), suggesting that these factors may affect nicotine metabolism directly or indirectly. We list these suggestive relations (P's < 0.05) in Web Table 4 for further interest. Results additionally adjusting for estrogen use at urine collection and birth control pill use are in Web Table 5.
For the 6 relations observed in the global ancestry analysis (4 for African, 2 for Asian ancestry), we conducted admixture mapping in the corresponding admixed populations, namely, African Americans or Native Hawaiians. We found a significant signal only between local African ancestry on chromosome 4q and cotinine glucuronidation (Web Figure 3). P values for association in this 12-Mb region from 62,418,658 to 74,039,463 base pairs (bp) were less than 2.7 × 10−7, which is the significance threshold for admixture mapping in African Americans after Bonferroni correction. The top region spans from 66,071,716 to 66,454,906 bp (383 kilobases), corresponding to a 5.4% (95% CI: 3.7, 7.0) predicted decrease in cotinine glucuronidation per a 20% increase in local African ancestry (P = 1.5 × 10−9). This is the same region reported in our genome-wide association study of cotinine glucuronidation (14). After adjustment for the genotyped rs294777 (69,682,471 bp), one of the top hits from the GWAS results, the strength of association at the top region from admixture mapping was substantially reduced (P = 0.59), indicating that they represent the same signal. rs294777 is intronic to UGT2B10, the gene encoding the enzyme catalyzing nicotine and cotinine glucuronidation. It is worth noting that, in the model including rs294777, global African ancestry was still significantly related to cotinine glucuronidation (P = 0.0001), suggesting that more genetic variation influences this trait. A similar extent of association was seen at the same region for cotinine glucuronidation among nicotine C-oxidation products (smallest P = 1.7 × 10−8).
DISCUSSION
Building upon previous reports of ethnic/racial differences in tobacco smoke-related metabolism, we took an in-depth look at the hypothesis that genetic ancestries are related to nicotine metabolism and internal dose of a tobacco smoke carcinogen. Our results, with adjustment for an array of important “environmental” and lifestyle factors, demonstrated that African and Asian ancestries were related to a higher free cotinine level and that African ancestry was related to lower cotinine glucuronidation. A follow-up admixture scan revealed a 12-Mb region possibly containing African ancestry-specific variants on chromosome 4q related to cotinine glucuronidation. Although the region, which includes UGT2B10, was the same one identified in our previous genome-wide association study (14), these results indicate the existence of ancestry-specific genetic factors underlying the ethnic/racial variation in nicotine metabolism.
We showed that Asian ancestry was associated with a higher cotinine level. Few past studies focused on East Asians. Previous reports suggested that CYP2A6 activity (9) and nicotine equivalents (adjusted for cigarettes smoked per day) were lower (9, 28) in East Asians, compared with whites. In this study, East Asian ancestry was only suggestively related to lower CYP2A6 activity (P = 0.008) (Web Table 3), perhaps because of the stringent significance level used and a relatively small sample size.
The findings that African ancestry was related to a higher level of free cotinine and a lower level of cotinine glucuronidation are consistent with previous reports (8, 10, 11). In humans, the majority of nicotine is metabolized to cotinine through C-oxidation quickly by CYP2A6; cotinine is further metabolized by CYP2A6 to 3HC at a relatively slower rate (29–31) or converted to cotinine-glucuronide primarily by UGT2B10 (11). Higher free cotinine levels could be due to faster conversion into cotinine from nicotine, a lower cotinine glucuronidation rate, and/or slower conversion of cotinine to 3HC. Past studies suggested that higher African ancestry correlates with reduced CYP2A6 enzymatic activity that affects more of the conversion from cotinine to 3HC than from nicotine to cotinine, because of a higher prevalence of slow-metabolizing CYP2A6 alleles in this population (32), which results in a higher free cotinine level (31). In our data, the evidence was weak that African ancestry was related to lower CYP2A6 activity (P = 0.07) (Web Table 3) or total nicotine C-oxidation products (P = 0.15). Therefore, it is likely that the correlation of higher cotinine with increasing African ancestry was mainly driven by lower cotinine glucuronidation. However, because of our small sample size, we cannot rule out the possibility that both mechanisms (i.e., lower cotinine glucuronidation and lower CYP2A6 activity) contributed to the correlation between higher cotinine and increasing African ancestry.
The role of lower cotinine glucuronidation or high free cotinine with regard to lung cancer risk is not clear, and there have not been enough incident lung cancer cases so far (n = 12) in our cohort study to address this question, even though an association between higher free cotinine and lung cancer risk has been suggested (33). UGT2B10 that catalyzes cotinine glucuronidation is also involved in detoxification/glucuronidation of the lung-cancer carcinogen NNAL. However, we did not observe an important relation between African ancestry and NNAL glucuronidation, although the percentages of glucuronidation for NNAL and cotinine were moderately correlated (ρ = 0.43; P < 0.0001). This suggests that other UGTs (UGT2B7, UGT2B17, etc.) in the NNAL detoxification pathway (Web Figure 2), but not in cotinine glucuronidation, play an important role.
The limitations of this study include the moderately small sample size in each admixed group. As a result, statistical power for detecting significant associations was relatively low. The Bonferroni correction applied here, accounting for 4 ancestries and 8 traits, was conservative. Although this reduced the chance of false positives, it increased the possibility of missing true but weak associations. We, therefore, present suggestive relations in our Web Material data for further interest. Also, as in all observational studies, even though we carefully adjusted for many possible confounders in the analysis based on general knowledge (e.g., creatinine level to control for differences in overnight or morning urine, education as surrogate for socioeconomic status, estrogen use because estrogen influences CYP2A6 activity (34–36)), some confounding factors may have been overlooked or unmeasured, and a systematic search for specific “environmental” risk factors associated with nicotine metabolism should be carried out separately. One concern may be that the dietary or lifestyle factors used here were obtained from the baseline questionnaire that was administered about 10 years prior to urine collection. Only body mass index was available from follow-up (1–9 years after enrollment) for 84% of the subjects, and the correlation was 0.89 between the 2 measurements. However, it is unlikely that a change in diet or lifestyle could lead to systematic bias in our ancestry analysis, as the change is more likely to be random rather than related to a person's genetic ancestry within ethnic/racial groups. Finally, only a single measurement was available for the biomarkers. Serial measurements would have reduced the effect of intraindividual variability, which is likely to have been nonsystematic and to have lowered statistical power in our study.
In summary, our results support that genetic factors underlie the ethnic/racial variation in nicotine metabolism and that larger studies are needed to further identify these factors. Smoking remains a major preventable cause of lung cancer worldwide. Understanding variations in nicotine metabolism improves our understanding of disparities in smoking behavior and susceptibility to nicotine addiction. It may also help in designing new smoking cessation interventions and in discovery of the mechanisms contributing to ethnic/racial differences in lung cancer risk.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: University of Hawaii Cancer Center, Honolulu, Hawaii (Hansong Wang, Lynne R. Wilkens, Laurence N. Kolonel, Loïc Le Marchand); Department of Preventive Medicine and Norris Comprehensive Cancer Center, Keck School of Medicine, University of Southern California, Los Angeles, California (Sungshim L. Park, Daniel O. Stram, Christopher A. Haiman); and Department of Biochemistry, Molecular Biology, and Biophysics and Masonic Cancer Center, University of Minnesota, Minneapolis, Minnesota (Stephen S. Hecht, Sharon E. Murphy).
This study was funded by the National Institutes of Health (grant P01 CA138338). The Multiethnic Cohort Study was funded by the National Institutes of Health (grants R37 CA54281, R01 CA63464, P01 CA33619, and UM1 CA164973).
Conflict of interest: none declared.
REFERENCES
- 1.Le Marchand L, Wilkens LR, Kolonel LN. Ethnic differences in the lung cancer risk associated with smoking. Cancer Epidemiol Biomarkers Prev. 1992;12:103–107. [PubMed] [Google Scholar]
- 2.Haiman CA, Stram DO, Wilkens LR et al. . Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med. 2006;3544:333–342. [DOI] [PubMed] [Google Scholar]
- 3.Gadgeel SM, Severson RK, Kau Y et al. . Impact of race in lung cancer: analysis of temporal trends from a Surveillance, Epidemiology, and End Results database. Chest. 2001;1201:55–63. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz AG, Swanson GM. Lung carcinoma in African Americans and whites. A population-based study in metropolitan Detroit, Michigan. Cancer. 1997;791:45–52. [DOI] [PubMed] [Google Scholar]
- 5.Stellman SD, Chen Y, Muscat JE et al. . Lung cancer risk in white and black Americans. Ann Epidemiol. 2003;134:294–302. [DOI] [PubMed] [Google Scholar]
- 6.Rubinstein ML, Shiffman S, Rait MA et al. . Race, gender, and nicotine metabolism in adolescent smokers. Nicotine Tob Res. 2013;157:1311–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kandel DB, Hu MC, Schaffran C et al. . Urine nicotine metabolites and smoking behavior in a multiracial/multiethnic national sample of young adults. Am J Epidemiol. 2007;1658:901–910. [DOI] [PubMed] [Google Scholar]
- 8.Pérez-Stable EJ, Herrera B, Jacob P 3rd et al. . Nicotine metabolism and intake in black and white smokers. JAMA. 1998;2802:152–156. [DOI] [PubMed] [Google Scholar]
- 9.Derby KS, Cuthrell K, Caberto C et al. . Nicotine metabolism in three ethnic/racial groups with different risks of lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;1712:3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benowitz NL, Perez-Stable EJ, Fong I et al. . Ethnic differences in N-glucuronidation of nicotine and cotinine. J Pharmacol Exp Ther. 1999;2913:1196–1203. [PubMed] [Google Scholar]
- 11.Berg JZ, Mason J, Boettcher AJ et al. . Nicotine metabolism in African Americans and European Americans: variation in glucuronidation by ethnicity and UGT2B10 haplotype. J Pharmacol Exp Ther. 2010;3321:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem Res Toxicol. 2008;211:160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith MW, O'Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat Rev Genet. 2005;68:623–632. [DOI] [PubMed] [Google Scholar]
- 14.Patel YM, Stram DO, Wilkens LR et al. . The contribution of common genetic variation to nicotine and cotinine glucuronidation in multiple ethnic/racial populations. Cancer Epidemiol Biomarkers Prev. 2015;241:119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolonel LN, Henderson BE, Hankin JH et al. . A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;1514:346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hukkanen J, Jacob P 3rd, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;571:79–115. [DOI] [PubMed] [Google Scholar]
- 17.Murphy SE, Park SS, Thompson EF et al. . Nicotine N-glucuronidation relative to N-oxidation and C-oxidation and UGT2B10 genotype in five ethnic/racial groups. Carcinogenesis. 2014;3511:2526–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol. 1998;116:559–603. [DOI] [PubMed] [Google Scholar]
- 19.Upadhyaya P, Kenney PMJ, Hochalter JB et al. . Tumorigenicity and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers and metabolites in the A/J mouse. Carcinogenesis. 1999;208:1577–1582. [DOI] [PubMed] [Google Scholar]
- 20.Carmella SG, Ming X, Olvera N et al. . High throughput liquid and gas chromatography-tandem mass spectrometry assays for tobacco-specific nitrosamine and polycyclic aromatic hydrocarbon metabolites associated with lung cancer in smokers. Chem Res Toxicol. 2013;268:1209–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;199:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li JZ, Absher DM, Tang H et al. . Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;3195866:1100–1104. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Haiman CA, Kolonel LN et al. . Self-reported ethnicity, genetic structure and the impact of population stratification in a multiethnic study. Hum Genet. 2010;1282:165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sankararaman S, Sridhar S, Kimmel G et al. . Estimating local ancestry in admixed populations. Am J Hum Genet. 2008;822:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Church TR, Anderson KE, Caporaso NE et al. . A prospectively measured serum biomarker for a tobacco-specific carcinogen and lung cancer in smokers. Cancer Epidemiol Biomarkers Prev. 2009;181:260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan JM, Koh WP, Murphy SE et al. . Urinary levels of tobacco-specific nitrosamine metabolites in relation to lung cancer development in two prospective cohorts of cigarette smokers. Cancer Res. 2009;697:2990–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benowitz NL, Jacob P 3rd, Fong I et al. . Nicotine metabolic profile in man: comparison of cigarette smoking and transdermal nicotine. J Pharmacol Exp Ther. 1994;2681:296–303. [PubMed] [Google Scholar]
- 28.Benowitz NL, Pérez-Stable EJ, Herrera B et al. . Slower metabolism and reduced intake of nicotine from cigarette smoking in Chinese-Americans. J Natl Cancer Inst. 2002;942:108–115. [DOI] [PubMed] [Google Scholar]
- 29.Messina ES, Tyndale RF, Sellers EM. A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. J Pharmacol Exp Ther. 1997;2823:1608–1614. [PubMed] [Google Scholar]
- 30.Nakajima M, Yamamoto T, Nunoya K et al. . Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther. 1996;2772:1010–1015. [PubMed] [Google Scholar]
- 31.Zhu AZ, Renner CC, Hatsukami DK et al. . The ability of plasma cotinine to predict nicotine and carcinogen exposure is altered by differences in CYP2A6: the influence of genetics, race, and sex. Cancer Epidemiol Biomarkers Prev. 2013;224:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haberl M, Anwald B, Klein K et al. . Three haplotypes associated with CYP2A6 phenotypes in Caucasians. Pharmacogenet Genomics. 2005;159:609–624. [DOI] [PubMed] [Google Scholar]
- 33.Boffetta P, Clark S, Shen M et al. . Serum cotinine level as predictor of lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;156:1184–1188. [DOI] [PubMed] [Google Scholar]
- 34.Higashi E, Fukami T, Itoh M et al. . Human CYP2A6 is induced by estrogen via estrogen receptor. Drug Metab Dispos. 2007;3510:1935–1941. [DOI] [PubMed] [Google Scholar]
- 35.Dempsey D, Jacob P 3rd, Benowitz NL. Accelerated metabolism of nicotine and cotinine in pregnant smokers. J Pharmacol Exp Ther. 2002;3012:594–598. [DOI] [PubMed] [Google Scholar]
- 36.Benowitz NL, Lessov-Schlaggar CN, Swan GE et al. . Female sex and oral contraceptive use accelerate nicotine metabolism. Clin Pharmacol Ther. 2006;795:480–488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.