Abstract
The present study performed an integral analysis of the gene expression and DNA methylation profile of pilocytic astrocytomas (PAs). Weighted gene co-expression network analysis (WGCNA) was also performed to examine and identify the genes correlated to PAs, to identify candidate therapeutic targets for the treatment of PAs. The DNA methylation profile and gene expression profile were downloaded from the Gene Expression Omnibus database. Following screening of the differentially expressed genes (DEGs) and differentially methylated regions (DMRs), respectively, integrated analysis of the DEGs and DMRs was performed to detect their correlation. Subsequently, the WGCNA algorithm was applied to identify the significant modules and construct the co-expression network associated with PAs. Furthermore, Gene Ontology enrichment analysis of the associated genes was performed using the Database for Annotation, Visualization and Integrated Discovery. A total number of 2,259 DEGs and 235 DMRs were screened out. Integrated analysis revealed that 30 DEGs were DMRs with prominent negative correlation (cor=−0.82; P=0.02). Based on the DEGs, the gene co-expression network was constructed, and nine network modules associated with PAs were identified. The functional analysis results showed that genes relevant to PAs were closely associated with cell differentiation modulation. The screened PA-associated genes were significantly different at the expression and methylation levels. These genes may be used as reliable candidate target genes for the treatment of PAs.
Keywords: pilocytic astrocytomas, differentially expressed genes, differentially methylated regions, weighted gene co-expression network analysis, immune function
Introduction
Pilocytic astrocytoma (PA) is one of the most common types of tumor to emerge in the central nervous system, and occur predominantly in childhood and adolescence. PAs are typically relatively circumscribed astrocytomas, which most frequently develop in the cerebellar hemispheres and in midline structures, including the hypothalamus and optic tract (1). PAs are known as benign tumors, as they are generally indolent with slow growing lesions, and have favorable prognosis (2,3). The survival of rate of >80% of patients with PAs is >10 years following surgical intervention, with or without radiation therapy (4,5).
DNA methylation is a biological mechanism, which is important in several cellular processes. Aberrant DNA methylation of CpG islands, particularly the CpG island shores and first exons, may lead to altered gene expression in human cancer (6,7), resulting in either global hypomethylation or hypermethylation. Gene expression and methylation may be positively and negatively correlated. Previous integrated analysis of DNA methylation and gene expression profiles has become a favorable method in investigating disease pathogenesis, particularly in cancer (8,9).
However, the molecular mechanism underlying the invasion and development of PAs remains to be elucidated. In the present study, the gene expression and DNA methylation profiles were investigated together. The identified differentially expressed genes (DEGs), differentially methylated regions (DMRs) and associated functions may be useful for the elucidation of the underlying mechanisms of PA, providing novel insights and potential therapeutic strategies for the treatment of PAs.
Materials and methods
Microarray data
The microarray data, GSE44684 and GSE44971 (10), were downloaded from the Gene Expression Omnibus (GEO) database (11,12) (http://www.ncbi.nlm.nih.gov/geo/). The GSE44684 data were methylation microarray data based on the GPL13534 platform (Infinium Human Methylation 450 Bead Chips; Illumina, San Diego, CA, USA), which included 67 samples (61 PA samples and six normal cerebellar samples). The GSE44971 data were of an mRNA expression microarray based on the [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array GPL570 platform, comprising 58 samples (49 PA samples and nine normal cerebellar samples). In addition, the clinical features of the data were summarized (Table I), indicating that the patients with PAs were generally younger.
Table I.
Clinical features of the methylation and expression data from the Gene Expression Omnibus database.
| Feature | Methylation data
|
Expression data
|
||||
|---|---|---|---|---|---|---|
| Control group | PA group | Control group | PA group | |||
| Data | GSE44684 | GSE44971 | ||||
| Sample size (n) | 6 | 61 | 9 | 49 | ||
| Gender (male/female) | 3/3 | 36/25 | 6/3 | 26/23 | ||
| Age (years) | 1.5–87 | 0.5–22 | 0–26 | 2–26 | ||
PA, pilocytic astrocytoma.
Data preprocessing
Expression value background calibration and data normalization were performed on the raw expression profile data, which were in the CEL format, using the Affy Package (13) in R (www.r-project.org). The preprocessing process included raw data format transition, missing value interpolation, background calibration (MAS rule) and data quantile normalization (14). The digital transformation, background signal calibration and normalization were performed on the primary methylation signal data using the Genome Studio Methylation Module with R (15) to obtain the chromosome coordinates and the methylation β value in the methylation signal profile.
DEG and DMR screening
To investigate the differentials between the PA samples and normal control samples, the Significance Analysis of Microarrays method (16) was used to identify the DEGs and for circumventing false positive results by estimating the false discovery rate (FDR) (17,18), which occurred in the multiple tests. The FDRs were estimated using the permutation algorithm. In addition, the fold change (FC) of the expression values between the PA group and control group were calculated. DEGs were identified as those meeting the criteria of FDR <0.05 and |log2FC| >1.
The DMRs between the PA samples and normal cerebellar samples were screened out using a paired t-test conducted using R. FDR <0.05 and |log2FC| >0.585 were set as the cut-off criteria. The corresponding genes and CpG islands of the DMRs were annotated, according to the region correspondence information in GPL13534. As a result, the DMRs, together with the reference genes within it and regions with CpG islands, were obtained.
Chromosome distribution analysis of the DEGs and DMRs
The genes associated with the identified DMRs were located to 23 human chromosomes in the hg19 human genome, according to the corresponding differentially methylated sites. The quantity of the located genes on each chromosome were summarized to calculate the distribution rules of the DMRs on each chromosome, and a chromosome map was then produced on the basis of the distribution information using Circos 0.67 (circos.ca/software/download/circos).
Integrated analysis of DNA methylation and gene expression
The genes, which were identified as being DEGs and reference genes associated with DMRs were screened out. The associations between the methylation and expression levels were measured by the application of Pearson's correlation analysis on the datasets, and Pearson's correlation coefficient was calculated to indicate the correlation using R. The methylation and expression levels of each gene were applied to the rectangular coordinate system and plotted by R.
Construction of gene co-expression networks based on clinical features
The gene co-expression networks were constructed using the weighted gene co-expression network analysis (WGCNA 1.41; labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages) (19) algorithm, which is a typical systematic biological algorithm for constructing gene co-expression networks. Correlation networks are constructed on the basis of high throughput mRNA microarray data in a number of steps Firstly, the network was required to have a scale-free topology. The identification of the gene co-expression matrix and adjacency function formed by the gene network was then performed, following which the coefficient of variation of different nodes were calculated, and gene set modules associated with disease were identified. Finally, the association between modules and the PA disease phenotype were determined. The outlier sample was validated and eliminated.
Functional enrichment of the DEGs
To interpret the biological function in which the screened DEGs were involved, functional analysis was performed by Gene Ontology (GO) enrichment with the Database for Annotation, Visualization and Integrated Discovery (DAVID; david.abcc.ncifcrf.gov) (20) software. This was achieved by identifying the biological processes in which the DEGs were involved in. The cut-off threshold was set as P<0.05.
Results
DEG and DMR screening
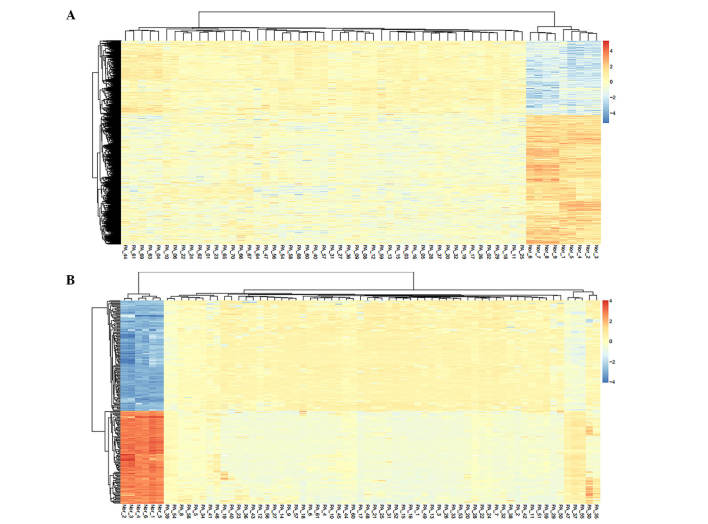
In the present study, the normalized expression data and methylation profile were further mined to identify the DEGs and DMRs. A total number of 2,259 DEGs and 235 DMRs were screened out, of which 123 reference genes to all DMRs were identified, according to the corresponding annotation information. The hierarchical clustering heat maps of the DEGs and DMRs are shown in Fig. 1. The results revealed that the DEGs and DMRs were able to distinguish well between the PA samples and normal control samples.
Figure 1.
Hierarchical clustering heat maps of (A) expression values of differentially expressed genes. Red, high expression levels; blue, low expression level; yellow, relative middle expression level. (B) Methylation signal of differentially methylated regions. Red, hypermethylation; blue, hypomethylation; yellow, relative normal methylation.
Chromosome distribution analysis of DEGs and DMRs
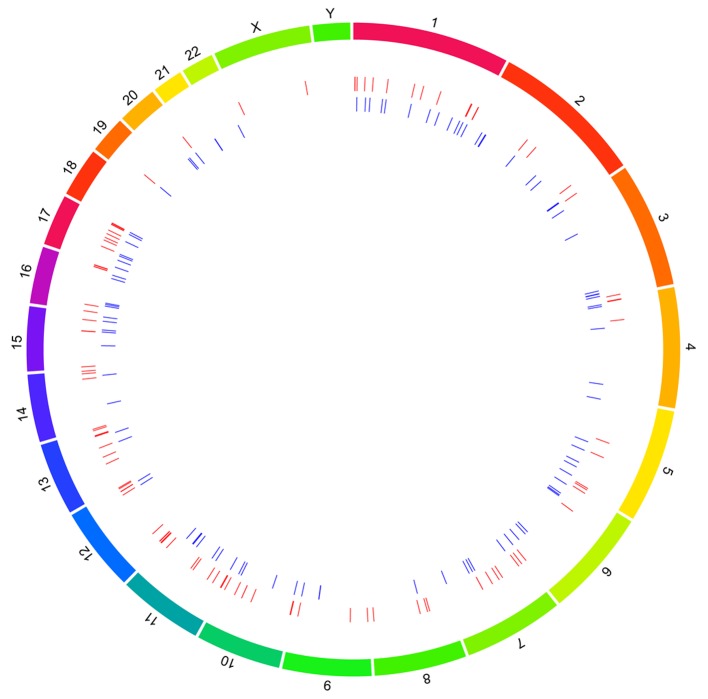
A total of 123 reference genes located to the DMRs were identified by comparing the methylation profiles of the PA samples and normal cerebellar samples. In total, 235 DMRs were located to 23 human chromosomes. Of these, chromosome 1 contained the highest number of DMRs (29 regions), followed by chromosome 16 (20 regions). The upregulated and down-regulated DMRs distributed on each chromosome are shown in Fig. 2.
Figure 2.
Distribution of upregulated and downregulated differentially methylated regions. The outer ring represents chromosomes; the inner circle represents methylated regions. The red indicates hypermethylation and blue indicates hypomethylation levels.
Integrated analyses of DNA methylation and gene expression
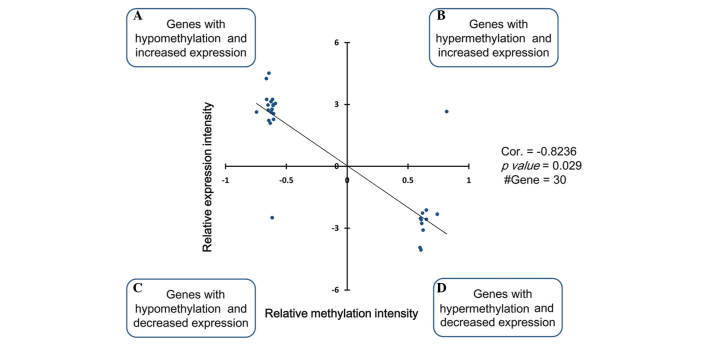
A total of 30 genes were screened out to evaluate the correlation between methylation and expression as a negative correlation was observed, indicating these genes were affected by abnormal methylation. As a result, there was a marked negative correlation (cor=−0.82; P=0.029) between the expression level and the methylation level (Fig. 3).
Figure 3.
Correlation between DNA methylation and gene expression. (A–D) The levels of expression and methylation of the genes in the respective regions of the graph.
Gene co-expression networks
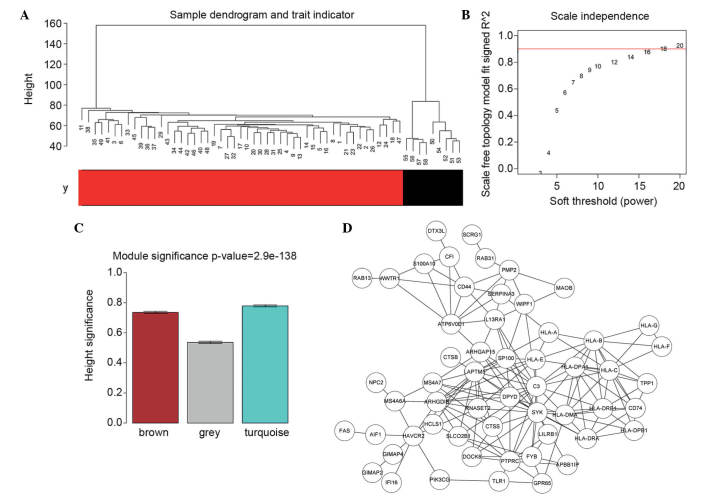
In order to validate the outlier in the samples, a sample hierarchical clustering dendrogram was plotted (Fig. 4), with the states of each sample labeled. As shown in Fig. 4A, the PA and normal samples had their own integral polymerism, in which no sample outliers were present. Thus, all the PA samples were incorporated in the analysis.
Figure 4.
Gene co-expression network construction based on the weighted gene co-expression network analysis method. (A) Hierarchical clustering dendrogram plot and corresponding sample information. Red represents pilocytic astrocytoma samples and black represents control cerebellar samples. (B) Plot of the weighting parameter β of the adjacency matrix. The horizontal axis represents the weighting parameter (β) and the vertical axis represents the corresponding square of correlation coefficient (R2) for log(K) and log(p(K)) in the network. The red line is the standard line of R2=0.9. (C) Modules highly correlation with disease status. Different colors represent different network modules. The vertical axis represents the correlation and the error bars represent the confidence interval for correlation between the gene modules and disease status. (D) Differentially expressed gene co-expression network of the modules possessing the highest correlation with disease.
In order to meet the preconditions of scale-free network distribution, the β value weighting parameter of the adjacency matrix was optimized for setting the ranges of network construction parameters, and for calculating the scale-free distribution of the topology matrix. The β value ranges were set as 1–20, and the corresponding model statistics were calculated for graph construction (Fig. 4B). The higher the R2 value, the closer the network is to an ideal scale-free system. The β value of 18, at which the R2 reached 0.9 for the first time was selected.
Initially, the dissimilarity coefficient between genes was calculated to obtain the hierarchical clustering dendrogram. Subsequently, the minimum number of genes in each gene network was set as 30, in accordance to the standard of the Dynamic Branch Cut method (21). Following determination of the gene module using the Dynamic Branch Cut method, the eigengenes of each module were calculated successively, followed by cluster analysis on each module, in which the close modules were merged into a new module. The correlation coefficient between the eigengenes of each module and the disease status were calculated. For the disease state variable (y), y=0 indicated a normal group and y=1 indicated a PA group). As shown in Fig. 4C, the following three modules showed a high degree of correlation with disease status: Brown (959 DEGs), grey (238 DEGs) and turquoise (1,062 DEGs). The genes in the turquoise module possessed the highest correlation with disease. Subsequently, the correlation coefficients of the genes in the turquoise module were output, and 148 DEGs, the R2 values of which were all >0.9 m were selected to construct the co-expression network. The co-expression network is shown in Fig. 4D, and the WIPF1, IFI16, GIMAP2 genes in the network possessed three common features: i) significant differential expression; ii) involved with DMRs exhibiting overexpression and hypomethylation; iii) closely associated with disease.
Functional enrichment of DEGs
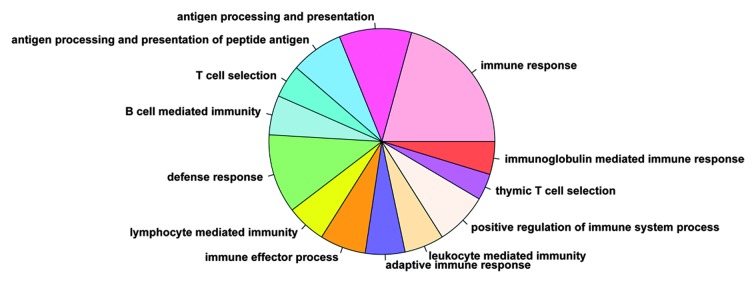
To determine the functional features of the screened DEGs, the PA-associated genes that were involved in the gene co-expression network construction were uploaded to DAVID to analyze the biological functional processes. As shown in Fig. 5, 13 significantly correlated biological functions were identified, of which immune response was the most enriched process, and the remaining processes were all associated with the immune system function.
Figure 5.
Biological functions of the differentially expressed genes in the co-expression network.
Discussion
In the present study, WGCNA was performed, selecting the appropriate weighting coefficient to meet the scale-free network distribution precondition. This meant that the gene networks/modules were obtained by constructing a hierarchical clustering dendrogram following matrix transformation, which results in the gene network/module construction to be of a soft threshold by transforming the correlation coefficient to a continuous variable. The use of soft threshold, a parameter in the WCGNA algorithm, allows the network to have increased stability and reliability. Subsequently, the genes were clustered using the Dynamic Branch Cut method, and genes with high expression similarity were allocated to the same gene network/module as a whole factor to examine their correlation with disease and each other. Gene function analyses can be performed on the constructed modules based on WGCNA. This analytical method is now widely used in investigations of disease and have been reported to screen multiple reliable target genes for various disease (22,23).
Methylation in CpG islands has been reported to inhibit genomic binding sites of activating transcription factors or other proteins, and shows a high level of association with gene repression (6,24). Integrated analysis, involving the screening of DMRs in PA samples and comparing them with normal samples, was performed in the present study. The results revealed that chromosome 1 contained the maximum ratio of significantly methylated genes, and that the DEGs showed marked negative correlation with the DMRs (cor=−0.82; P=0.029).
In the present study, integrated analysis of the expression profile and DNA methylation profile, and identification of PA-associated genes using the WGCNA method were performed to investigate the pathogenesis of PAs. Previously, several pathways and genes have been reported to be disturbed or modulated in PAs. The mitogen-activated protein kinase (MAPK) signaling pathway is a conserved signaling cascade, which utilizes a series of kinases to transduce signals from the cell membrane to the nucleus, thereby mediating cell growth, cell survival and cell differentiation. It has been reported that BRAF gene fusion leads to MAPK pathway activation in PAs, which is a novel oncogenic fusion gene with diagnostic, prognostic and therapeutic potential (3). The tumor suppressor, A-kinase anchor protein 12 (AKAP12) in human diffuse astrocytomas and PAs is regulated by promoter methylation, and is expressed at a high level in PAs, but at a low level in diffuse astrocytomas (25).
Based on the WGCNA of the PA expression data, three network modules associated with PA were identified and the gene co-expression network were constructed. Functional enrichment analysis showed that 13 significantly correlated biological functions, including immune response and defense response, were associated with the immune system. It has been reported that genes, including HLA-DRa, HLA-DPB1, HLA-DQB1, IgG3, IgGK, FCER1G, A2M, FCRN, IFI-56K, and DAP12 are upregulated in PAs, compared with the normal cerebellum, grade II astrocytomas and oligodendrogliomas, and that all the genes are immune defense-associated genes (1). These results suggested that immunological investigations may be beneficial and offer potential in investigating PAs.
In conclusion, integrated analyses of DNA methylation profiles and gene expression profiles, combined with correlation and functional enrichment analysis may provide novel insights for further investigation of PAs, in target gene screening and in functional process identification.
References
- 1.Huang H, Hara A, Homma T, Yonekawa Y, Ohgaki H. Altered expression of immune defense genes in pilocytic astrocytomas. J Neuropathol Exp Neurol. 2005;64:891–901. doi: 10.1097/01.jnen.0000183345.19447.8e. [DOI] [PubMed] [Google Scholar]
- 2.Rorive S, Maris C, Debeir O, Sandras F, Vidaud M, Bièche I, Salmon I, Decaestecker C. Exploring the distinctive biological characteristics of pilocytic and low-grade diffuse astrocytomas using microarray gene expression profiles. J Neuropathol Exp Neurol. 2006;65:794–807. doi: 10.1097/01.jnen.0000228203.12292.a1. [DOI] [PubMed] [Google Scholar]
- 3.Jeuken JW, Wesseling P. MAPK pathway activation through BRAF gene fusion in pilocytic astrocytomas; a novel oncogenic fusion gene with diagnostic, prognostic and therapeutic potential. J Pathol. 2010;222:324–328. doi: 10.1002/path.2780. [DOI] [PubMed] [Google Scholar]
- 4.Giannini C, Scheithauer BW, Burger PC, Christensen MR, Wollan PC, Sebo TJ, Forsyth PA, Hayostek CJ. Cellular proliferation in pilocytic and diffuse astrocytomas. J Neuropathol Exp Neurol. 1999;58:46–53. doi: 10.1097/00005072-199901000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Burkhard C, Di Patre PL, Schüler D, Schüler G, Yaşargil MG, Yonekawa Y, Lütolf UM, Kleihues P, Ohgaki H. A population-based study of the incidence and survival rates in patients with pilocytic astrocytoma. J Neurosurg. 2003;98:1170–1174. doi: 10.3171/jns.2003.98.6.1170. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 7.Gopalakrishnan S, Van Emburgh BO, Robertson KD. DNA methylation in development and human disease. Mutat Res. 2008;647:30–38. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Balch C, Montgomery JS, Jeong M, Chung JH, Yan P, Huang TH, Kim S, Nephew KP. Integrated analysis of DNA methylation and gene expression reveals specific signaling pathways associated with platinum resistance in ovarian cancer. BMC Med Genomics. 2009;2:34. doi: 10.1186/1755-8794-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan M, Yan PS, Hartman-Frey C, Chen L, Paik H, Oyer SL, Salisbury JD, Cheng AS, Li L, Abbosh PH, et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res. 2006;66:11954–11966. doi: 10.1158/0008-5472.CAN-06-1666. [DOI] [PubMed] [Google Scholar]
- 10.Lambert SR, Witt H, Hovestadt V, Zucknick M, Kool M, Pearson DM, Korshunov A, Ryzhova M, Ichimura K, Jabado N, et al. Differential expression and methylation of brain developmental genes define location-specific subsets of pilocytic astrocytoma. Acta Neuropathol. 2013;126:291–301. doi: 10.1007/s00401-013-1124-7. [DOI] [PubMed] [Google Scholar]
- 11.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: Mining tens of millions of expression profiles-database and tools update. Nucleic Acids Res. 2007;35(Database issue):D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic. Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 14.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and computational biology solutions using R and Bioconductor. Springer-Verlag; New York, NY: 2005. pp. 397–pp420. [DOI] [Google Scholar]
- 15.Turan N, Ghalwash MF, Katari S, Coutifaris C, Obradovic Z, Sapienza C. DNA methylation differences at growth related genes correlate with birth weight: A molecular signature linked to developmental origins of adult disease? BMC Med Genomics. 2012;5:10. doi: 10.1186/1755-8794-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S. A comprehensive evaluation of SAM, the SAM R-package and a simple modification to improve its performance. BMC Bioinformatics. 2007;8:230. doi: 10.1186/1471-2105-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. 1995;57:289–300. [Google Scholar]
- 18.Benjamini Y. Discovering the false discovery rate. J R Stat Soc Series B Stat Methodol. 2010;72:405–416. doi: 10.1111/j.1467-9868.2010.00746.x. [DOI] [Google Scholar]
- 19.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for. R Bioinformatics. 24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 22.Aggarwal A, Guo DL, Hoshida Y, Yuen ST, Chu KM, So S, Boussioutas A, Chen X, Bowtell D, Aburatani H, et al. Topological and functional discovery in a gene coexpression meta-network of gastric cancer. Cancer Res. 2006;66:232–241. doi: 10.1158/0008-5472.CAN-05-2232. [DOI] [PubMed] [Google Scholar]
- 23.Dewey FE, Perez MV, Wheeler MT, Watt C, Spin J, Langfelder P, Horvath S, Hannenhalli S, Cappola TP, Ashley EA. Gene coexpression network topology of cardiac development, hypertrophy and failure. Circ. Cardiovasc Genet. 2011;4:26–35. doi: 10.1161/CIRCGENETICS.110.941757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 25.Goeppert B, Schmidt CR, Geiselhart L, Dutruel C, Capper D, Renner M, Vogel MN, Zachskorn C, Zinke J, Campos B, et al. Differential expression of the tumor suppressor A-kinase anchor protein 12 in human diffuse and pilocytic astrocytomas is regulated by promoter methylation. J Neuropathol Exp Neurol. 2013;72:933–941. doi: 10.1097/NEN.0b013e3182a59a88. [DOI] [PubMed] [Google Scholar]