Abstract
Autophagy is a lysosomal degradation mechanism that is essential for cell survival, differentiation, development, and homeostasis. Autophagy protects cells from various stresses, including protecting normal cells from harmful metabolic conditions, and cancer cells from chemotherapeutics. In the current study, a cytarabine arabinoside (Ara-C)-sensitive U937 leukemia cell line and an Ara-C-resistant U937 (U937/AR) cell line were assessed for baseline autophagy activity by investigating the LC3-I conversion to LC3-II, performing EGFP-LC3 puncta, an acidic autophagolysosome assay, and measuring the expression of various autophagy-related genes. The results demonstrated significantly higher autophagic activity in the U937/AR cells compared with the U937 cells, when the cells were cultured with or without serum. Furthermore, an increase in the autophagic activity in starved U937/AR cells was demonstrated, compared with that in the starved U937 cells. Administration of an autophagy inhibitor demonstrated no change in cell death in the two cell lines when cultured with serum, however, it induced cell death regardless of the Ara-C sensitivity when the cell lines were cultured without serum. In addition, the U937 cells demonstrated an Ara-C resistance when cultured without serum. Co-treatment with Ara-C and the autophagy inhibitor significantly induced cell death in the U937/AR and Ara-C-sensitive U937 cells. In conclusion, autophagy serves an important role in protecting U937 cells from Ara-C and in the development of Ara-C resistance. Inhibition of autophagy combined with the Ara-C treatment in the U937 cells augmented the anti-leukemic effect of Ara-C and overcame Ara-C resistance, suggesting that autophagy may be an important therapeutic target to further improve the treatment outcome in patients with acute myeloid leukemia.
Keywords: autophagy, Ara-C-resistant, U937, acute myeloid leukemia, treatment
Introduction
Autophagy refers to any lysosomal degradation pathway and is essential for survival, differentiation, development and homeostasis (1). During autophagy, an autophagosome, a double-membraned vesicle sequestering cytoplasmic materials, and a lysosome, fuse together to form an autolysosome (2). The two principal physiological roles of autophagy are to maintain synthesis of macromolecules and ATP, and to eliminate defective or abnormal proteins and organelles. These functions allow cells to survive various metabolic stresses, such as nutritional and growth factor depletion, or hypoxia, and be protected from infection, neurodegenerative diseases, genomic instabilities or tumor initiation (3). Various pathological conditions are associated with defects in autophagy, as it serves an important role in homeostasis (1). Autophagy has been considered a tumor suppressor pathway (4–6) following the establishment of an association between autophagy and cancer in 1999, when the autophagy-related gene (ATG) beclin-1 was identified as a candidate tumor suppressor (7). Numerous genetic links have been identified between defects in autophagy and cancer. Tumor suppressor genes involved in the upstream inhibition of mammalian target of rapamycin (mTOR) signaling, including phosphatase and tensin homolog (4), tuberous sclerosis 1, and tuberous sclerosis 2 (5), stimulated autophagy. By contrast, TOR-activating oncogene products, such as Ras, phosphoinositide 3-kinase (PI3K) and protein kinase B (Akt), inhibit autophagy (6). Furthermore, a previous study hypothesized that chemoradiotherapy induces the accumulation of autophagosomes in various cancer cell lines and eliminates cancer cells by induction of caspase-independent autophagic and caspase-dependent cell death (8). However, autophagy may promote survival of cancer cells during chemotherapy and serve a role in chemoresistance (9). Therefore, inhibiting autophagy by targeting certain autophagy-related (ATG) genes may accelerate, rather than prevent cell death (10).
Previous studies in the field of hematology have primarily focused on elucidating the role of autophagy in chronic myeloid, chronic lymphoid and acute promyelocytic leukemias, and autophagy was associated with resistance to tyrosine kinase inhibitors, histone deacetylase inhibitors and hypomethylating agents (11–14). However, to the best of our knowledge, no previous study has investigated the possible association between autophagy and chemoresistance to cytosine arabinoside (1-β-d-arabinofuranosylcytosine; Ara-C) in acute myeloid leukemia (AML). Ara-C is a nucleoside analog used to induce complete remission or for post-remission therapy in AML (15). Ara-C acts as an anti-metabolite and induces cell death by competing with pyrimidine for incorporation into replicative DNA, which inhibits DNA polymerase activity and leads to chain elongation termination. Due to the important role of Ara-C in AML treatment, acquired resistance to Ara-C is one of the multiple factors leading to AML persistence or relapse. Therefore, novel therapeutic strategies are required to overcome the Ara-C resistance in myeloid leukemia cells, which is hypothesized to develop through various mechanisms. In the present study, the role of autophagy in myeloid leukemic cell lines was assessed according to Ara-C sensitivity, and whether inhibiting autophagy would overcome Ara-C resistance was investigated.
Materials and methods
Reagents and antibodies
Ara-C was purchased from Sigma-Aldrich (St. Louis, MO, USA), and was dissolved in distilled water and stored as a 100 mM stock solution. Hydroxychloroquine (HCQ) was purchased from Myung In Pharmaceutical Company, Ltd. (Seoul, South Korea) and dissolved in dimethyl sulfoxide (Sigma-Aldrich). Bafilomycin A1 (Ba-A1) was purchased from Sigma-Aldrich. Fetal bovine serum (FBS) and Gibco RPMI 1640 medium containing 50 U/ml and 50 µg/ml streptomycin were purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Rabbit polyclonal antibodies against human microtubule-associated protein 1 light chain 3 (LC3; cat. no. NB100-2220; dilution, 1:1,000), beclin-1 (cat. no. NB500-249; dilution, 1:1,000), ATG9A (cat. no. NB110-56893; dilution, 1:1,000), ATG7 (NB110-74811) and mouse anti-human p62 (cat. no. H0008878; dilution, 1:1,000) were purchased from Novus Biologicals LLC (Littleton, CO, USA). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit (cat. no. 7074; dilution, 1:3,000) and horse anti-mouse (cat. no. 7072; dilution, 1:3,000) IgG secondary antibodies were obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA). The Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit was purchased from BD Pharmingen (San Diego, CA, USA). All other chemicals were from Sigma-Aldrich. The GFP-LC3 plasmid was kindly provided by Dr Kim (Division of Pulmonology, Yonsei University College of Medicine, Seoul, South Korea).
Cell culture
The human leukemia cell line, U937, was obtained from the American Type Culture Collection (Rockville, MD, USA). The Ara-C-resistant U937 (U937/AR) cell line was established in our laboratory by exposing parental U937 cells to stepwise increasing concentrations of Ara-C (1 nM – 2 mM, usually at intervals of 10 passages) in complete RPMI 1640 medium supplemented with 10% heat-inactivated FBS and 100 U/ml penicillin. Resistant sublines were grown for longer than 6 months. To maintain exponential growth, cells were seeded at 1×105 cells/ml and passaged every 3 days.
Cell death assay
U937 and U937/AR cells were cultured on 12-well plates (Corning Incorporated, Corning, NY, USA) at 2×105 cells/well in medium containing RPMI 1640, 10% FBS and 100 U/ml penicillin and streptomycin for 24, 48 and 72 h. Cultured cells were washed with phosphate-buffered saline (PBS; Gibco; Thermo Fisher Scientific, Inc.) and incubated in 100 µl 1X binding buffer containing 5 µl Annexin V-FITC, and the nuclei were counterstained with 10 µl propidium iodide according the kit manufacturer's instructions. The percentage of apoptotic cells was determined using a FACSCalibur flow cytometer and analyzed using CellQuest version 3.3 software (BD Immunocytometry Systems, San Jose, CA, USA).
Western blot analysis
U937 and U937/AR cells were cultured in 12-well plates at 2×105 cells/well in RPMI 1640 medium with or without 10% FBS for 48 h. Cells were lysed in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 120 mM NaCl, 20 mM NaF, 1 mM EDTA, 5 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid, 15 mM sodium pyrophosphate, 1 mM benzamidine, 0.1 mM phenylmethylsulfonyl fluoride, and 1% Nonidet P-40, and this was briefly sonicated. The mitochondrial and cytosolic fractions were obtained using the QIA88 ProteoExtract Cytosolic/Mitochondria Fractionation kit (Oncogene Research Products, La Jolla, CA, USA). Protein yields were quantified using a detergent-compatible protein assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according to the manufacturer's instructions. Equivalent amounts of protein (10 mg) were boiled for 10 min and separated by SDS-PAGE (15% for LC3; 12% for beclin-1 and p62; and 10% for ATG9A and ATG7). Proteins were subsequently transferred to nitrocellulose membranes (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA). Following blocking with 0.05% Tris-buffered saline with Tween 20 (TBST) and 5% bovine serum albumin solution, or 5% skim milk, for 1 h at room temperature, blots were incubated with LC3, beclin-1, ATG9A, ATG7 and p62 primary antibodies overnight at 4°C. Blots were washed four times for 5 min with TBST and incubated for 1 h at room temperature with HRP-conjugated anti-rabbit or anti-mouse IgG secondary antibodies (1:3,000). Subsequent to washing with TBST, the reactive proteins were visualized using the enhanced chemiluminescence reagent (Amersham; GE Healthcare Life Sciences). The density of the protein bands on the membrane were scanned and analyzed with ImageJ (imagej.nih.gov/ij/).
EGFP-LC3 puncta assay
U937 and U937/AR cell suspensions (2×106 cells) were immediately electroporated with pEGFP-LC3 cDNA using the Nucleofector™ 2b system (program T-20; Lonza Cologne GmbH, Cologne, Germany) according to the manufacturer's instructions. Immediately following electroporation, cells were resuspended in complete medium and incubated at 37°C in a humidified 5% CO2 incubator. pEGFP vector transfection was performed as control. After 24 h, cells were rinsed and resuspended in serum-free medium and incubated at 37°C for 48 h. Cultured cells were centrifuged at 800 × g onto glass slides. Fluorescence images were analyzed using an LSM 700 laser-scanning confocal microscope (Zeiss GmbH, Jena, Germany). The GFP-LC3 puncta in a single cell were manually counted using confocal microscopy. For each group, 50 cells were randomly selected to estimate the average number of GFP-LC3 puncta per cell. Data presented are one representative experiment of a minimum of three independent repeats.
Transmission electron microscopy (TEM)
For TEM evaluation, U937 and U937/AR cells were seeded in 12-well plates at 2×105 cells/well and cultured with or without FBS for 48 h at 37°C. Following incubation, the cells were collected and fixed for 2 h at 4°C with ice-cold 2% glutaraldehyde-paraformaldehyde (EMD Millipore, Billerica, MA, USA) and post-fixed at 4°C with 1% OsO4 (KeyGen Biotech Co. Ltd.; dissolved in 0.1 M PBS) for 2 h and dehydrated through serial dilutions of ethanol (50, 70, 90 and 100% for 15 min and then three times at 100%) and infiltrated with propylene oxide (Sigma-Aldrich) prior to embedding in epoxy resin using a Poly/Bed 812 kit (Polysciences, Inc., Warrington, PA, USA) at 65°C electron microscope oven (TD-700; Dosaka EM, Kyoto, Japan) for 24 h. The embedded sections were cut into sections of 250–250 nm for staining with toluidine blue (Sigma-Aldrich) and observation under a light microscope and into ultrathin sections (70 nm) were double-stained with 7% uranyl acetate for 20 min and lead citrate (Thermo Fisher Scientific, Inc.) for 10 min for contrast staining. The sections were cut with an ultramicrotome (Leica EM UC7; Leica Microsystems GmbH, Wetzlar, Germany) and mounted on copper and nickel grids. Sections were viewed using a JEM-1011 system (JEOL USA, Inc., Peabody, MA, USA) at an acceleration voltage of 80 kV (Camera Megaview, Soft Imaging System, Berlin, Germany). Photoshop (Adobe Systems, Inc., San Jose, CA, USA) was used to further quantify the TEM images.
Statistical analysis
The statistical analysis was performed using the two-tailed Student's t-test. Unless otherwise indicated, data are expressed as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Sensitivity of U937 and U937/AR leukemia cell lines to Ara-C
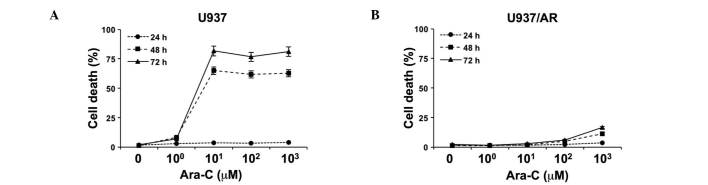
The relative sensitivity of U937 and U937/AR to Ara-C was assessed in vitro as a baseline study. Following treatment with logarithmically scaled concentrations of Ara-C (100–103 µM), cell death was assessed using an Annexin V binding assay, combined with flow cytometry. As demonstrated in Fig. 1A, cell death was induced in a time-dependent manner in the Ara-C U937 cells. Ara-C (101 µM) induced cell death in 65.0±1.0 and 81.7±0.8% of U937 cells after 48 and 72 h, respectively. The U937/AR cells were resistant to Ara-C and the maximum level of cell apoptosis observed was 16.6±0.2% at 72 h after treatment with the highest concentration of Ara-C (Fig. 1B).
Figure 1.
Induction of cell death by treating leukemia cell lines with Ara-C. (A) Ara-C-sensitive U937 cells demonstrated time-dependent induction of cell death. (B) U937/AR cells were highly resistant to Ara-C. Ara-C, cytarabine arabinoside; AR, Ara-C-resistant.
Status of autophagy in U937 and U937/AR cells according to the culture conditions
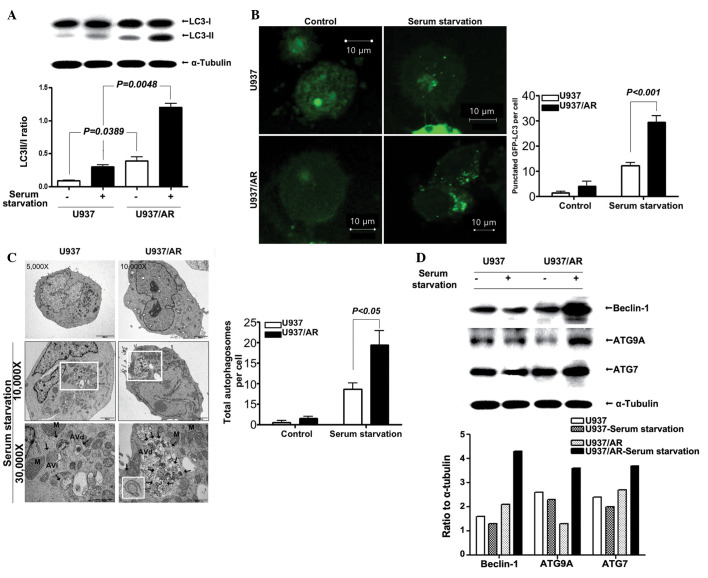
Following standard cell culture conditions (media supplemented with FBS) and serum starvation, the status of autophagy was assessed. Western blot analysis results demonstrated that the LC3-II/I ratio in the U937/AR cells was significantly higher compared with that of the Ara-C sensitive U937 cells, under the standard culturing condition with FBS (0.39±0.08 vs. 0.09±0.01; P=0.0389; Fig. 2A) and under serum starvation (1.21±0.08 vs. 0.30±0.04; P=0.0048; Fig. 2A). The fractional increases in the LC3-II/I ratio under serum starvation were ~3-fold in the two cell lines and thus, were not observed to be significantly different (P>0.05).
Figure 2.
The status of autophagy in U937 and U937/AR cells according to the culturing conditions. (A) Assay for conversion of LC3-I to LC3-II showed a significantly higher LC3-II/LC3-I ratio in U937/AR cells compared to that in U937 cells cultured with or without serum. (B) The number of GFP-LC3 puncta as assessed by the EGFP-LC3 puncta assay. (C) The number of autophagosomes observed by transmission electron microscopy are significantly larger in the U937/AR cells compared to the U937 cells when cultured without serum. (D) Autophagy related genes are highly expressed in U937/AR cells when cultured without serum. AR, cytarabine arabinoside-resistant; LC3, microtubule-associated protein 1 light chain 3; GFP, green fluorescent protein; AVd, late autolysosomes; AVi, early autophagosomes; ATG9A, autophagy-related protein 9A.
LC3 expression was assessed using the GFP-LC3 puncta assay. The results demonstrated a significant increase in the number of GFP-LC3 puncta per cell in the U937/AR cells compared with the U937 cells, regardless of culturing conditions (4.0±2.0 vs. 1.5±0.5 puncta per cell with FBS; P=0.0239 or 29.5±2.5 vs. 12.2±1.2 puncta per cell without FBS; P<0.001; Fig. 2B). Furthermore, the total number of autophagosomes per cell was counted using TEM (Fig. 2C), and the results were similar to those from western blot and GFP-LC3 puncta assays. As demonstrated in Fig. 2C, the number of autophagosomes significantly increased when cells were cultured without FBS. The number of autophagosomes in U937/AR cells cultured without FBS was significantly higher than that in the U937 cells (19.3±3.5 vs. 8.7±1.5; P<0.05; Fig. 2C).
In addition, the expression of several autophagy-associated genes was determined. As demonstrated in Fig. 2D, the increase in beclin-1, ATG9A, ATG7 and p62/SQSTM1 protein expression levels was more prominent in the U937/AR cells (ratio to α-tubulin, 4.3, 2.6 and 3.7, respectively) compared with the U937 cells (ratio to α-tubulin, 1.3, 2.3 and 2.0, respectively) when cultured without FBS.
Effect of autophagy inhibitors in U937 and U937/AR cells cultured without serum
Following cell treatment with or without serum, the mechanism of action of the autophagy inhibitors affecting apoptosis was investigated. The U937 and U937/AR cells were treated with two autophagy inhibitors separately, Ba-A1 (2 nM) and HCQ (20 µg/ml) for 48 h. Untreated cell lines grow when cultured with FBS, but autophagic activity is required to prolong cell survival when cultured without FBS. As demonstrated in Fig. 3, the autophagy inhibitors induced cell death in <25% of the cells cultured with serum. In the cultures with FBS, the autophagy inhibitors increased cell death in the U937/AR cells compared with the U937 cells as follows: Ba-A1, 6.2±0.5 vs. 1.6±0.4% (P<0.05); HCQ, 23.0±3.0 vs. 11.7±1.6% (P<0.05). These results were consistent with the status of autophagy in the U937 and U937/AR cells treated with FBS (Fig. 2).
Figure 3.
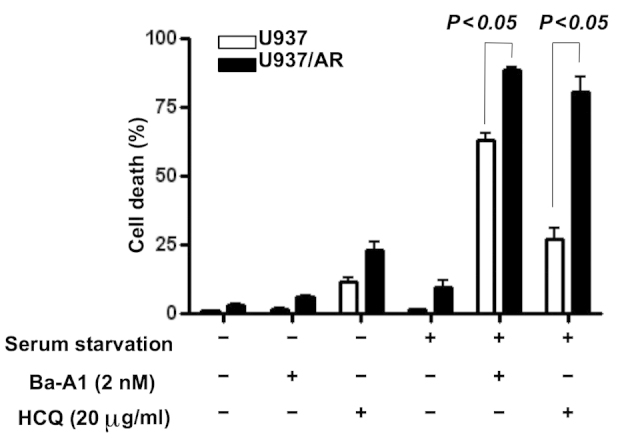
The effect of autophagy inhibitors on inducing cell death in U937 and U937/AR cells according to the culturing conditions. The effects of the autophagy inhibitors, Ba-A1 and HCQ, on inducing cell death were minimal following culturing with serum. However, in the U937 cells cultured without serum, treatment with the autophagy inhibitors induced cell death regardless of Ara-C resistance. AR, cytarabine arabinoside-resistant; Ba-A1, bafilomycin-A1; HCQ, hydrochloroquine.
Furthermore, in a comparison of FBS-cultured and serum starved cells, the autophagy inhibitors induced cell death in a significantly higher percentage of serum starved cells, compared with the FBS-treated cells as follows: Ba-A1, 62.9±2.7 vs. 1.6±0.4% in U937 (P<0.05) and 88.7±0.9 vs. 6.2±0.5% in U937/AR (P<0.05); HCQ, 27.0±3.9 vs. 11.7±1.6% in U937 (P<0.05) and 80.8±5.0 vs. 23.0±3.0% in U937/AR (P<0.05). In addition, the difference in cell death between U937 and U937/AR induced by the autophagy inhibitors was consistent and unaffected by culture conditions or the type of autophagy inhibitor used.
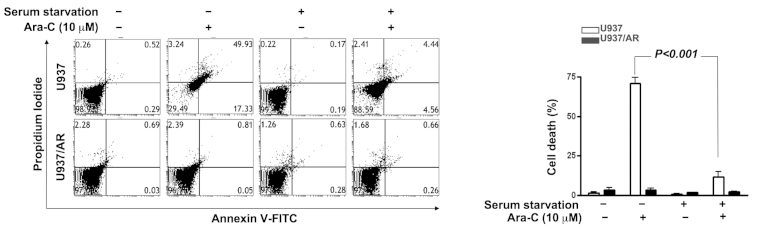
Induction of Ara-C resistance by culture condition, and overcoming Ara-C resistance by co-treatment with Ara-C and autophagy inhibitors
The association of autophagic activity and Ara-C resistance was investigated. As demonstrated in Fig. 4, U937/AR cells were Ara-C resistant in all culture conditions. However, Ara-C treatment induced cell death in the U937 cell line with FBS. The Ara-C-sensitive U937 cells acquired Ara-C resistance upon culturing without FBS (Fig. 4; cell death rate, 70.9±3.8% with FBS vs. 11.9±3.0% without FBS; P<0.001).
Figure 4.
Induction of Ara-C resistance in U937 cells cultured without serum. Following culturing of Ara-C sensitive U937 cells without serum, the U937 cells gained Ara-C resistance. The percentage of cell death by Ara-C treatment alone was significantly decreased with serum starvation. Ara-C, cytarabine arabinoside; AR, Ara-C-resistant; FITC, fluorescein isothiocyanate.
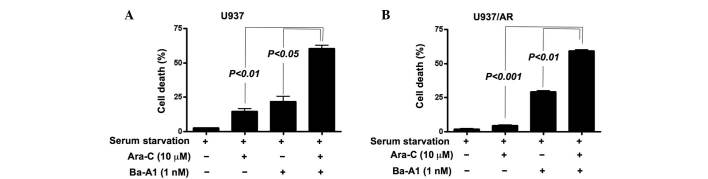
Cell death was then assessed in the U937 and U937/AR cell lines following culturing without FBS and co-treatment with Ara-C and Ba-A1. To evaluate whether the co-treatment had an additive or synergistic effect on inducing cell death, a lower concentration of Ba-A1 (1 nM) was used to induce moderate cell death. Resistance to Ara-C in Ara-C-sensitive U937 cells was demonstrated upon serum starved culture and co-treatment with Ba-A1 and Ara-C (cell death rate, 14.8±2.5% for Ara-C monotherapy vs. 60.5±3.2% for Ara-C and Ba-A1; P<0.01; Fig. 5A). Furthermore, the acquired Ara-C resistance was of higher significance in the U937/AR cells (cell death rate, 4.5±0.8% for Ara-C monotherapy vs. 59.2±1.2% for Ara-C and Ba-A1; P<0.001; Fig. 5B).
Figure 5.
The effect of co-treatment with Ara-C and autophagy inhibitors on inducing cell death in U937 and U937/AR cells. (A) U937 cells and (B) U937/AR cells were co-treated with Ara-C and the autophagy inhibitor Ba-A1. Co-treated cells demonstrated a significantly higher percentage of cell death compared with the untreated or Ara-C-treated groups. Ara-C, cytarabine arabinoside; AR, Ara-C-resistant; Ba-A1, bafilomycin-A1.
Discussion
Autophagy is a catabolic process that recycles intracellular components and selectively eliminates organelles for regulation and maintenance of quality control (1). The first step of autophagy is the formation of an isolation membrane and phagophore, the expansion of the membrane. The edges of the phagophore fuse to form an autophagosome, which further fuses with a lysosome to make an autolysosome, while the inner membrane and materials are degraded (2). Physiologically, autophagy defends against various forms of metabolic stress, including nutrient deprivation, growth factor depletion and hypoxia. Autophagic degradation generates free amino acids and fatty acids used by the tricarboxylic acid cycle to maintain cellular ATP production (3). Autophagy performs cellular functions to eliminate defective proteins or organelles, prevent abnormal protein aggregate accumulation and remove intracellular pathogens. In addition, previous studies demonstrated that autophagy may act as a guardian of the genome to limit DNA damage and chromosomal instability (16–18). This role may be associated with the preventive effects of autophagy against the initiation and progression of cancer. By contrast, defective autophagy has been proposed as a contributor to various human diseases. For example, patients with neurodegenerative diseases, such as Alzheimer's disease, Parkinson's and Huntington's diseases, have abnormal accumulation of autophagosomes in the brain (19,20), and germline or somatic mutations in genes, such as dynactin subunit p150 glued or ceroid-lipofuscinosis neuronal 3, have been associated with neurologic diseases (21). In addition, the liver, muscles and heart may be damaged by defective autophagy, and several inherited diseases, such as Danon or Pompe diseases, demonstrate the role of defective autophagy in their pathogenesis (22–24).
Abnormalities in the autophagic process may present in cancer. Liang et al (7) demonstrated that the ATG gene, beclin-1, was a candidate tumor suppressor gene in 1999, and previous studies have indicated that defective autophagy is closely associated with the initiation or progression of cancer (25–27). Certain mutations in genes affecting autophagy, such as beclin-1, Akt, PI3K, p53 and Bcl-2, serve a role in the pathogenesis of malignant lymphoma and breast, ovarian and prostate cancer (1).
Autophagy protects against cell starvation and hypoxia, which are the hallmarks of the tumor microenvironment. A previous study suggested that autophagy serves an important role in the chemoresistance of cancer to therapeutics that typically induce apoptosis (9). Numerous clinical trials evaluating autophagy in various solid cancers, including breast (28), lung (29), melanoma (30), rectal/colon (31), renal cell carcinoma (32), prostate (33) and pancreatic cancer (34), have demonstrated inconsistent results regarding the anticancer effect of autophagy manipulation (35). Certain previous studies demonstrated that autophagic cell death observed during chemotherapy acted as an anticancer machinery (8,36), however other studies suggested that autophagy prevents apoptosis of cancer cells from chemotherapy (9,37). These inconsistent results may be due to the dynamic nature of autophagy and the diversity of molecules or organelles targeted by it. Autophagy may target tumor-initiating proteins developed in normal cells, therefore suppressing tumor activity. In addition, cancer cells may preserve themselves using autophagy during chemotherapy, thus promoting cancer cell survival.
The myeloid leukemias are a heterogeneous group of diseases characterized by neoplastic cells that infiltrate the blood, bone marrow and other tissues of the hematopoietic system. In previous studies, induction of autophagy was demonstrated to be important for the death of leukemic cells (11–14,38), and the induction of autophagy by various drugs, such as imatinib mesylate, arsenic trioxide, everolimus, brevinin-2R and eupalinin A, was attempted. Autophagy may be an important pathway of cell death during various chemotherapeutic modalities, and manipulation of autophagy may be a useful clinical application for targeting multidrug-resistant leukemia. Although triggering autophagy may be a potential therapeutic strategy to overcome drug resistance, inhibiting autophagy may be another therapeutic strategy to improve the outcome of anticancer treatments. For example, autophagy acts as a prosurvival mechanism and contributes to drug resistance in various types of leukemia (38). By contrast, inhibition of autophagy was documented to enhance the therapeutic benefit of tyrosine kinase inhibitors in Philadelphia (Ph)-positive leukemias, and the tumor anti-leukemic effect of the histone deacetylase inhibitor, SAHA, was augmented by co-treatment with an autophagy inhibitor (11). Overcoming drug resistance by manipulating autophagy was primarily attempted for Ph-positive types of leukemia, such as chronic myeloid leukemia, Ph-positive acute lymphoblastic leukemia and acute promyelocytic leukemia (39,40). However, the role of autophagy in association with drug resistance in AML remains unclear.
In the current study, the status of autophagy in AML cell lines was assessed according to the resistance against Ara-C, a chemotherapeutic agent used to induce remission. In addition, an attempt was made to overcome the Ara-C resistance by combination treatment with an autophagy inhibitor and Ara-C. As demonstrated in Fig. 2, specific characteristics of autophagy in the U937 and U937/AR cell lines were identified, including LC3-I-to-LC3-II conversion (Fig. 2A), formation of EGFP-LC3 puncta (Fig. 2B) and acidic autophagolysosomes (Fig. 2C). The three assays demonstrated a consistent highly active autophagic status in the U937/AR cells compared with the U937 cells. To verify the autophagic activity at the molecular level, the expression of autophagy-associated molecules was investigated. Following autophagy initiation by the ATG1-ATG13 protein complex, which is activated by the absence of signaling of the nutrient-sensing kinase mTOR, class III PI3K-beclin-1 complexes promote formation of the isolation membrane. Elongation of the isolation membrane is then mediated by two ubiquitin-like conjugation systems as follows: i) ATG7 and ATG10 act to conjugate ATG5 to ATG12; ii) the ATG5-ATG12 conjugate acts with ATG7 and ATG3 to conjugate ATG8 to phosphatidylethanolamine in the membrane of the growing autophagosome (41). In addition, the expression of beclin-1, ATG9A, ATG7 and p62/SQSTM1 was determined. The autophagy-associated molecules were expressed in similar patterns as observed in the LC3-I-to-LC3-II conversion, EGFP-LC3 puncta and autophagosome assays. The results of the current study demonstrated that the U937/AR cells had higher levels of autophagic activity compared with the U937 cells, and that autophagic activity increased after Ara-C treatment, thus the ability of an autophagy inhibitor to overcome Ara-C resistance in the U937/AR cells and to improve the antileukemic effect of Ara-C in the U937 cells was assessed. As the Ara-C-resistant U937/AR cell line was established by exposing parental U937 cells to increasing concentrations of Ara-C in culture media with FBS, the increasing autophagic activity in U937/AR cells may be induced by Ara-C. Ara-C is an anti-metabolite and other anti-metabolites, such as 5-fluorouracil or 6-thioguanine have been demonstrated to induce autophagy (42,43). The current study demonstrated that the U937/AR cells had a higher level of autophagic activity than U937 cells when the cells were treated with Ara-C (Fig. 2). The augmented autophagic activity in Ara-C-treated U937/AR cells may be one of the mechanisms leading to the U937/AR cells resistance to higher concentrations of Ara-C with longer treatment durations (Fig. 1B). The evidence of the anti-apoptotic role of autophagy in the U937 cells during metabolic stress was supported by the results demonstrated when U937 cells were cultured with autophagy inhibitors under starvation (Fig. 3). As U937 is a leukemia cell line, serum starvation alone cannot induce sufficient cell death, however, when the cell lines were cultured without FBS and treated with autophagy inhibitors, sufficient cell death was induced. Furthermore, culturing the cells without FBS may transform Ara-C sensitive U937 cells to Ara-C resistant U937 cells with no further manipulations (Fig. 4). Although the underlying mechanism of this effect was not fully investigated, the maintenance of autophagic activity may be associated with the acquisition of Ara-C resistance, even in the absence of Ara-C. Combination treatment with a low dose of the autophagy inhibitor and Ara-C improved the anti-leukemic activity in the U937 cells (Fig. 5A) and overcame Ara-C resistance in the U937/AR cells.
Previous studies have emphasized the role of autophagy in AML. One study demonstrated that overexpression of melanoma differentiation-associated gene-7/interleukin-24 inhibited autophagy and strongly augmented the anti-leukemia activity in an AML cell line and in a mouse model of leukemia (44). Another study demonstrated that bone-morphogenetic protein 4, a member of the TGF-β super-family, has a role in promoting chemoresistance through the activation of autophagy and subsequent inhibition of apoptosis in leukemic cells (45). S100A8, a member of the S100 calcium-binding protein family, has also been proposed to be involved in the development of chemoresistance in leukemic cells by regulating autophagy, and has been suggested as a novel target for improving leukemia therapy (12,46).
In conclusion, the role of autophagy as a protector against cellular stress may be important for the survival of AML cells when treated with chemotherapeutics. Chemoresistant AML cells demonstrated increased autophagic activity that augmented the anti-leukemic activity of Ara-C and overcame Ara-C resistance in vitro. Prior to validating the clinical benefit of autophagy inhibition, the genetic and epigenetic mechanisms of autophagy that protect leukemic cells and promote resistance to chemotherapeutics should be further investigated.
Acknowledgments
The current study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant no. NRF-2012R1A1A2 009114).
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: Implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: The PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz RA, Fernández G, Kotulska K, Jóźwiak S. Tuberous sclerosis complex: Advances in diagnosis, genetics, and management. J Am Acad Dermatol. 2007;57:189–202. doi: 10.1016/j.jaad.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Ligresti G, Militello L, Steelman LS, Cavallaro A, Basile F, Nicoletti F, Stivala F, McCubrey JA, Libra M. PIK3CA mutations in human solid tumors: Role in sensitivity to various therapeutic approaches. Cell Cycle. 2009;8:1352–1358. doi: 10.4161/cc.8.9.8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 8.Notte A, Leclere L, Michiels C. Autophagy as a mediator of chemotherapy-induced cell death in cancer. Biochem Pharmacol. 2011;82:427–434. doi: 10.1016/j.bcp.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y, Xie M, Yin X, Livesey KM, Lotze MT, et al. HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia. 2011;25:23–31. doi: 10.1038/leu.2010.225. [DOI] [PubMed] [Google Scholar]
- 10.Pan Y, Gao Y, Chen L, Gao G, Dong H, Yang Y, Dong B, Chen X. Targeting autophagy augments in vitro and in vivo antimyeloma activity of DNA-damaging chemotherapy. Clin Cancer Res. 2011;17:3248–3258. doi: 10.1158/1078-0432.CCR-10-0890. [DOI] [PubMed] [Google Scholar]
- 11.Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, Houghton JA, Huang P, Giles FJ, Cleveland JL. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Yang M, Zhang H, Wang Z, Yu Y, Xie M, Zhao M, Liu L, Cao L. S100A8-targeting siRNA enhances arsenic trioxide-induced myeloid leukemia cell death by down-regulating autophagy. Int J Mol Med. 2012;29:65–72. doi: 10.3892/ijmm.2011.806. [DOI] [PubMed] [Google Scholar]
- 13.Zhu S, Cao L, Yu Y, Yang L, Yang M, Liu K, Huang J, Kang R, Livesey KM, Tang D. Inhibiting autophagy potentiates the anticancer activity of IFN1α/IFNα in chronic myeloid leukemia cells. Autophagy. 2013;9:317–327. doi: 10.4161/auto.22923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amrein L, Soulières D, Johnston JB, Aloyz R. p53 and autophagy contribute to dasatinib resistance in primary CLL lymphocytes. Leuk Res. 2011;35:99–102. doi: 10.1016/j.leukres.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 15.Larson RA. New agents for induction and postremission therapy of acute myeloid leukemia. Leukemia. 2001;15:675–676. doi: 10.1038/sj.leu.2402064. [DOI] [PubMed] [Google Scholar]
- 16.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan KM. p53 and autophagy in cancer: Guardian of the genome meets guardian of the proteome. Eur J Cancer. 2011;47:44–50. doi: 10.1016/j.ejca.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 18.García-Arencibia M, Hochfeld WE, Toh PP, Rubinsztein DC. Autophagy, a guardian against neurodegeneration. Semin Cell Dev Biol. 2010;21:691–698. doi: 10.1016/j.semcdb.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheper W, Nijholt DA, Hoozemans JJ. The unfolded protein response and proteostasis in Alzheimer disease: Preferential activation of autophagy by endoplasmic reticulum stress. Autophagy. 2011;7:910–911. doi: 10.4161/auto.7.8.15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: When the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 21.Puls I, Oh SJ, Sumner CJ, Wallace KE, Floeter MK, Mann EA, Kennedy WR, Wendelschafer-Crabb G, Vortmeyer A, Powers R, et al. Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann Neurol. 2005;57:687–694. doi: 10.1002/ana.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czaja MJ, Ding WX, Donohue TM, Jr, Friedman SL, Kim JS, Komatsu M, Lemasters JJ, Lemoine A, Lin JD, Ou JH, et al. Functions of autophagy in normal and diseased liver. Autophagy. 2013;9:1131–1158. doi: 10.4161/auto.25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katsetos CD, Koutzaki S, Melvin JJ. Mitochondrial dysfunction in neuromuscular disorders. Semin Pediatr Neurol. 2013;20:202–215. doi: 10.1016/j.spen.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Sugimoto S. A novel vacuolar myopathy with dilated cardiomyopathy. Autophagy. 2007;3:638–639. doi: 10.4161/auto.4931. [DOI] [PubMed] [Google Scholar]
- 25.Edinger AL, Thompson CB. Defective autophagy leads to cancer. Cancer Cell. 2003;4:422–424. doi: 10.1016/S1535-6108(03)00306-4. [DOI] [PubMed] [Google Scholar]
- 26.Karantza-Wadsworth V, White E. Role of autophagy in breast cancer. Autophagy. 2007;3:610–613. doi: 10.4161/auto.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheen JH, Zoncu R, Kim D, Sabatini DM. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell. 2011;19:613–628. doi: 10.1016/j.ccr.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karpathiou G, Sivridis E, Koukourakis M, Mikroulis D, Bouros D, Froudarakis M, Giatromanolaki A. Light-chain 3A autophagic activity and prognostic significance in non-small cell lung carcinomas. Chest. 2010;140:127–134. doi: 10.1378/chest.10-1831. [DOI] [PubMed] [Google Scholar]
- 30.Savaraj N, You M, Wu C, Wangpaichitr M, Kuo MT, Feun LG. Arginine deprivation, autophagy, apoptosis (AAA) for the treatment of melanoma. Curr Mol Med. 2010;10:405–412. doi: 10.2174/156652410791316995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koukourakis MI, Giatromanolaki A, Sivridis E, Pitiakoudis M, Gatter KC, Harris AL. Beclin 1 over- and underexpression in colorectal cancer: Distinct patterns relate to prognosis and tumour hypoxia. Br J Cancer. 2010;103:1209–1214. doi: 10.1038/sj.bjc.6605904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14:90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim RH, Bold RJ, Kung HJ. ADI, autophagy and apoptosis: Metabolic stress as a therapeutic option for prostate cancer. Autophagy. 2009;5:567–568. doi: 10.4161/auto.5.4.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boone BA, Bahary N, Zureikat AH, Moser AJ, Normolle DP, Wu WC, Singhi AD, Bao P, Bartlett DL, Liotta LA, et al. Safety and biologic response of pre-operative autophagy inhibition in combination with gemcitabine in patients with pancreatic adenocarcinoma. Ann Surg Oncol. 2015;22:4402–4410. doi: 10.1245/s10434-015-4566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy JM, Thorburn A. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol Ther. 2011;131:130–141. doi: 10.1016/j.pharmthera.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fanzani A, Zanola A, Rovetta F, Rossi S, Aleo MF. Cisplatin triggers atrophy of skeletal C2C12 myotubes via impairment of Akt signalling pathway and subsequent increment activity of proteasome and autophagy systems. Toxicol Appl Pharmacol. 2011;250:312–321. doi: 10.1016/j.taap.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Zhao D, Yuan H, Yi F, Meng C, Zhu Q. Autophagy prevents doxorubicin-induced apoptosis in osteosarcoma. Mol Med Rep. 2014;9:1975–1981. doi: 10.3892/mmr.2014.2055. [DOI] [PubMed] [Google Scholar]
- 38.Ekiz HA, Can G, Baran Y. Role of autophagy in the progression and suppression of leukemias. Crit Rev Oncol Hematol. 2012;81:275–285. doi: 10.1016/j.critrevonc.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Helgason GV, Karvela M, Holyoake TL. Kill one bird with two stones: Potential efficacy of BCR-ABL and autophagy inhibition in CML. Blood. 2011;118:2035–2043. doi: 10.1182/blood-2011-01-330621. [DOI] [PubMed] [Google Scholar]
- 40.Orfali N, McKenna SL, Cahill MR, Gudas LJ, Mongan NP. Retinoid receptor signaling and autophagy in acute promyelocytic leukemia. Exp Cell Res. 2014;324:1–12. doi: 10.1016/j.yexcr.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watson AS, Mortensen M, Simon AK. Autophagy in the pathogenesis of myelodysplastic syndrome and acute myeloid leukemia. Cell Cycle. 2011;10:1719–1725. doi: 10.4161/cc.10.11.15673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Donovan TR, O'Sullivan GC, McKenna SL. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy. 2011;7:509–524. doi: 10.4161/auto.7.5.15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng X, Yan T, Schupp JE, Seo Y, Kinsella TJ. DNA mismatch repair initiates 6-thioguanine-induced autophagy through p53 activation in human tumor cells. Clin Cancer Res. 2007;13:1315–1321. doi: 10.1158/1078-0432.CCR-06-1517. [DOI] [PubMed] [Google Scholar]
- 44.Yang C, Tong Y, Ni W, Liu J, Xu W, Li L, Liu X, Meng H, Qian W. Inhibition of autophagy induced by overexpression of mda-7/interleukin-24 strongly augments the antileukemia activity in vitro and in vivo. Cancer Gene Ther. 2010;17:109–119. doi: 10.1038/cgt.2009.57. [DOI] [PubMed] [Google Scholar]
- 45.Zhao X, Liu J, Peng M, Liu J, Chen F. BMP4 is involved in the chemoresistance of myeloid leukemia cells through regulating autophagy-apoptosis balance. Cancer Invest. 2013;31:555–562. doi: 10.3109/07357907.2013.834925. [DOI] [PubMed] [Google Scholar]
- 46.Yang M, Zeng P, Kang R, Yu Y, Yang L, Tang D, Cao L. S100A8 contributes to drug resistance by promoting autophagy in leukemia cells. PLoS One. 2014;9:e97242. doi: 10.1371/journal.pone.0097242. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]