Abstract
The ff94 force field that is commonly associated with the AMBER simulation package is one of the most widely used parameter sets for biomolecular simulation. After a decade of extensive use and testing, limitations in this force field, such as over stabilization of α-helices, were reported by us and other researchers. This led to a number of attempts to improve these parameters, resulting in a variety of “AMBER” force fields and significant difficulty in determining which should be used for a particular application. We show that several of these continue to suffer from inadequate balance between different secondary structure elements. In addition, the approach used in most of these studies neglected to account for the existence in AMBER of two sets of backbone φ/ψ dihedral terms. This led to parameter sets that provide unreasonable conformational preferences for glycine. We report here an effort to improve the φ/ψ dihedral terms in the ff99 energy function. Dihedral term parameters are based on fitting the energies of multiple conformations of glycine and alanine tetrapeptides from high level ab-initio quantum mechanical calculations. The new parameters for backbone dihedrals replace those in the existing ff99 force field. This parameter set, which we denote ff99SB, achieves a better balance of secondary structure elements as judged by improved distribution of backbone dihedrals for glycine and alanine with respect to PDB survey data. It also accomplishes improved agreement with published experimental data for conformational preferences of short alanine peptides, and better accord with experimental NMR relaxation data of test protein systems.
Keywords: trialanine, dihedral parameters, molecular dynamics, molecular mechanics, decoy analysis, NMR order parameters, α-helix
Introduction
Molecular modeling studies of biologically important molecules usually require the evaluation of potential energies for alternate conformations. The most accurate approach would encompass the use of the quantum mechanical (QM) wave function. Unfortunately, due to the large sizes of biological macromolecules, such calculations are extremely time consuming and can only be applied to a limited number of conformations. Therefore, most simulation projects employ classical molecular mechanics (MM) energy functions. Due to the need to evaluate these functions a large number of times during the simulation, these functions are relatively simple and utilize many adjustable empirical parameters. These are most often obtained by fitting to data from experiments or from high level quantum mechanical calculations. The energy function together with the set of empirical parameters is known as a force field.
In spite of persistent yet slow emergence of force fields that explicitly account for charge polarization1, the fixed charge additive force fields remain popular due to computational efficiency. For a more in-depth overview of current force fields and trends in force field development, the reader is referred to recent review articles2–4. In this report, we focus on the existing AMBER fixed charge additive force fields.
The “Cornell et al” force field5 (denoted ff94 in AMBER) has been the most widely used with the AMBER suite of programs6,7 since its publication over a decade ago. It introduced the set of parameters for all-atom simulations suitable for protein simulations in the condensed phase, largely inspired by the OPLS potentials8. Some characteristic features of ff94 include fixed partial charges on atom centers, explicit use of all hydrogen atoms, no specific functional form for hydrogen bonding, and dihedral parameters fit to relative quantum-mechanical (QM) energies of alternate rotamers of small molecules. In particular, the protein φ/ψ dihedrals have specific rotational parameters that affect relative energies of alternate backbone conformations. These were fit to optimize agreement with QM relative energies for several conformations of glycine and alanine. The partial atomic charges were derived by fitting the (gas phase) electrostatic potential calculated at the Hartree-Fock 6–31G* level. This approach intentionally “overpolarizes” bond dipoles as present in the gas phase, such that the resulting charge distribution approximates that occurring in aqueous condensed phase9.
Due to limited computational resources at the time, dihedral parameters were fit to a small number of low-energy conformations of glycine and alanine dipeptides. A possible limitation of using dipeptides is that their gas phase energy surfaces do not have a local minimum in the α-helical region, which occurs with high frequency in protein structures. This was addressed in subsequent modifications of ff94, such as ff9610 and, more recently, ff9911. In ff96, identical parameters were used for φ and ψ terms, which were empirically adjusted to reproduce the energy difference between extended and constrained α-helical energies for alanine tetrapeptide. ff99 reflects another attempt at refitting backbone dihedral parameters by including eleven representative structures of alanine tetrapeptide along with the alanine dipeptides. During this process, however, a new problem was introduced in the way ff96/ff99 φ/ψ dihedral parameters were optimized (discussed in detail below), resulting in incorrect conformational preferences for glycine. In addition to changes in backbone dihedral parameters, ff99 also generalized and extended atom types to compounds beyond amino and nucleic acids.
Even though both ff99 and, particularly, ff94 parameter sets have been successfully used for many years, improvement in conformational sampling due to increased computer power and algorithmic advances revealed that both force fields over-stabilize α-helical peptide conformations12,13. On the other hand, the changes introduced in the less frequently used ff96 were observed to overestimate β-strand propensity14–17. Because the backbone dihedral parameters are shared by all amino acids, regardless of the type of sidechain, their cumulative effect is most likely responsible for the bias towards the specific secondary structure. This recently motivated us12,18 and others13,19 to revisit backbone dihedral parametrization for ff94/ff99. Garcia and Sanbonmatsu13 simply zeroed the torsion potential for φ and ψ (although as we explain below, only some of the φ/ψ terms were removed) and noted improved agreement with experimental helix-coil parameters. More recently, Sorin and Pande19,20 modified ff99 by replacing its φ dihedral parameters with the ones from ff94. This improved the agreement with experimental kinetic and thermodynamic measurements for folding of two 21-residue α-helical peptides. Our own modification of ff99 published previously18 specifically addressed strong helical bias and, in this respect, succeeded to achieve better balance of major secondary structures.
All these previous modifications were aimed at correcting an apparent problem encountered with a specific system. The heuristic approaches that were employed might have improved that specific weakness but a more systematic revision was clearly necessary to quantify the problem and address the issue with generality and transferability in mind.
Much of the confusion and suboptimal performance of recent AMBER protein force field variants may have been caused by a somewhat non-intuitive parameterization of protein backbone dihedral angles. In AMBER, each dihedral profile is defined by a set of four atoms. The set of atoms used to define φ and ψ for glycine is as expected, following φ and ψ along the main chain (φ = C-N-Cα-C, ψ = N-Cα-C-N). Importantly, for other amino acids that have a side chain, an additional set of dihedrals also influences rotation about the φ/ψ bonds connecting the Cα atom to the amide C and N atoms. This extra set of terms corresponds to dihedral angles branched out to the Cβ carbon, which in this work we identify as φ′ = C-N-Cα-Cβ and ψ′ = Cβ-Cα-C-N.
This definition of dihedrals was originally implemented in ff94, with φ and ψ fit independently to glycine data and φ′ and ψ′ used to adjust the behavior for alanine. Later modifications of ff94, such as ff96, ff99, Garcia’s13, Pande’s19 and ours12,18 only changed the first set of φ/ψ terms, using them to adjust backbone preferences for alanine. Thus the new ϕ and ψ parameters were fit in the presence of the ff94 alanine-based φ′ and ψ′, and will only give their intended behavior when they accompany these φ′ and ψ′ dihedrals. Importantly, φ′ and ψ′ are not present in glycine. For example, ff99 modified the backbone φ and ψ terms in order to reproduce relative energies for alanine. However, the alanine residue also had the φ′ and ψ′ dihedral parameters present during the φ/ψ modification. These optimizations may perform well for non-glycine residues that possess the φ′/ψ′ terms. However, when these modified backbone dihedral parameters are applied to glycine, the result has little physical justification since they were fit in the presence of φ′ and ψ′. A similar inconsistency is present in Garcia’s modification; only the φ and ψ terms were zeroed, thus while glycine has no backbone dihedral potential all other amino acids retain the ff94 φ′ and ψ′ terms which were fit in the presence of the glycine-based φ and ψ terms and thus have no meaning when applied without them.
We show below that these modifications result in unreasonable sampling of dihedral space for several post-ff94 AMBER force fields. These problems are in addition to the overstabilization of helical conformations present in ff94 and ff99. AMBER and other popular force fields (e.g. CHARMM22 and GROMOS96 variants and OPLS-AA) were previously compared21,22 in terms of conformational sampling of blocked glycine and alanine dipeptide and noted to disagree to various degrees (particularly for glycine21) with simulations employing combined QM/MM force field as well as with statistical analysis of high resolution protein crystal structures. In another study, all force fields were also shown to perform very differently with respect to a number of properties of trialanine compared to NMR and infrared observables23.
We also show that our reparametrization of backbone dihedral parameters improves conformational preferences for typical secondary structures. It should be noted that this work does not attempt to create yet another variant of AMBER force field, but strives to improve the existing ff94 and ff99 force fields as well as specifically address the problem with glycine sampling. Thus rather than contributing to the diverging set of AMBER force fields, we extend the evolution started with ff94, followed by ff99 and subsequently refined in this report. This is also reflected by naming this modified force field ff99SB. We attempted to maximally improve the methodology that we used to derive our new parameters while still following the same general philosophy as was used with ff94/ff99.
A different approach was taken by Duan et al24, who recently introduced a more extensive modification of ff94/ff99 (called ff03), in which a fundamentally different concept to derivation of partial atomic charges was used. Instead of relying on the HF/6–31G* approach to provide aqueous-phase charges, a low-dielectric continuum model corresponding to an organic solvent environment was included directly in the QM calculation of the dihedral parameters and electrostatic potential (from which the charges are obtained). Due to these differences, ff03 should be considered a distinct force field model rather than extension of previous AMBER force fields.
AMBER force fields are not the only ones undergoing such evolutionary changes brought about by discovery of limitations through improved conformational sampling capabilities. Similar efforts have recently been reported by MacKerell et al25, improving the backbone parameters in the CHARMM force field, specifically removing the bias toward π-helical peptide conformations26,27. In this case, the CHARMM22 alanine dipeptide dihedral energy surface was corrected using a grid-based difference map (CMAP) to achieve an almost perfect match with LMP2/cc-pVQZ(-g) energy surface. Likewise, dihedral terms in the original OPLS-AA force field28 were also reparametrized to improve the agreement with high level QM data29.
Methods
Optimization of backbone dihedral parameters
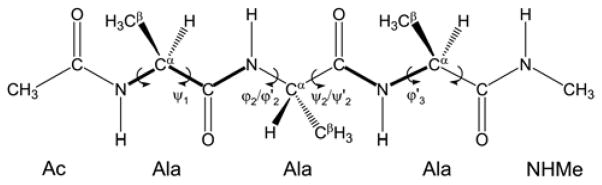
To better understand the procedure we used for obtaining a new set of backbone dihedral parameters, we first outline the general procedure originally used in ff945. Even though it may at first seem more straightforward to have a single set of backbone φ/ψ parameters for all amino acids, the different nature of glycine (no Cβ carbon), motivated the following approach: there is indeed a single set of backbone φ/ψ parameters for glycine (defined as φ =C-N-Cα-C, ψ =N-Cα-C-N), but any other amino acid has an additional set of φ/ψ parameters which are added to the glycine ones (here defined as φ′ = C-N-Cα-Cβ, ψ′ = Cβ-Cα-C-N). In other words, for any non-glycine amino acid, the dihedral energy is the sum of dihedral energies calculated for φ/ψ and φ′/ψ′. Note however, that φ′/ψ′ is calculated for a dihedral angle shifted by ~120° because that is the offset of the two torsions (as follows from their definitions using different sets of four atoms). Figure 1 demonstrates the definition of these two sets of dihedral angles on Ala3 tetrapeptide.
Figure 1.
Blocked alanine tetrapeptide (three alanine residues but four peptide bonds) used in the optimization procedure. Note the definition of dihedral angles: φ =C-N-Cα-C, ψ =N-Cα-C-N, φ′ = C-N-Cα-Cβ, ψ′ = Cβ-Cα-C-N. There are three φ/ψ pairs and three additional φ′/ψ′ pairs in this tetrapeptide. Glycine tetrapeptide would only have the φ/ψ dihedrals due to the absence of Cβ carbon. As an example, ψ1 and φ′3 are shown in bold.
The glycine φ/ψ parameters were optimized first, usually based on the best reasonable fit of QM and MM energies for a set of glycine conformers. These are total potential energies for the molecule, where backbone dihedral parameters are the adjustable variables in the optimization. For example, if we optimized the absolute difference of QM and MM energies for glycine tetrapeptide (Gly3), we would minimize the absolute error (ae) between the MM and QM energies (Equation 1):
| (1) |
where EQM(i) and EMM(i) correspond to the QM and MM energies respectively for i-th glycine tetrapeptide conformer and N is the number of all glycine conformers. In our case, MM energy for a given conformer is given by the AMBER energy function (see ff945):
| (2) |
where the dihedral energy term is:
| (3) |
Vn is dihedral force constant (amplitude), n is dihedral periodicity, and γn is a phase of the dihedral angle θ (which would be either φ or ψ for backbone dihedral terms). The Fourier series in Edihedral is approximated using a small number of terms. Our choice of three terms in the expansion is consistent with the AMBER philosophy of using dihedral terms that can be physically rationalized 30 and yields a reasonable number of terms per single dihedral (six) that need to be optimized.
Dihedral energy parameters in Equation 3 corresponding to backbone φ/ψ dihedrals (i.e. Vn and γn) are the adjustable parameters. We seek values of these parameters that minimize the function in Equation 1. Once the glycine parameters are optimized, they are held fixed and used in a second round of fitting where φ′/ψ′ parameters are optimized. This is carried out by fitting the parameters to best reproduce QM and MM energies for a set of alanine tetrapeptide (Ala3) conformations. This second round of φ′/ψ′ optimization was initially misunderstood by us and by others who modified the original ff94. The use of a single round fitting in which the glycine φ/ψ dihedral parameters were modified to improve agreement for alanine data where the ‘real’ φ′/ψ′ alanine parameters were untouched led to incorrect parametrization (primarily for glycine) and overall confusion about the effect of dihedral parameters on the backbone conformation.
In the following, we describe the specifics of this general procedure that were employed for the present study. All optimizations were carried out with blocked (acetyl and N-methylamine groups at the N- and C-termini, respectively) glycine and alanine tetrapeptides (Gly3, Ala3) as shown in Figure 1. A set of Gly3 and Ala3 tetrapeptide conformers was chosen to represent local minima on the ff94 energy surface (ff99 was not used here due to apparent flaws in the ff99 energy function for glycine). Because the 6-dimensional dihedral space of tetrapeptides is too large to be represented exhaustively (as would be possible for simple 2-dimensional surface of dipeptides), we chose to optimize the relative depths of local minima on the energy surface, because these are most relevant to the thermodynamic stability of alternate secondary structure types. Each point in this dihedral space corresponds to a specific tetrapeptide conformation. Local minima were identified through a stochastic search in φ/ψ dihedral space followed by local minimization.
All conformations corresponding to the proposed minima were optimized at the Hartree-Fock level with 6–31G* basis set. After discarding the conformers that converged to the same geometry, we ended up with 28 glycine and 51 alanine tetrapeptide conformers (with a maximum range ff94 energy cutoff of 10 kcal/mol). The energies of these conformations were calculated using higher level (gas phase) QM LMP2/cc-pVTZ(-f) in Jaguar31 employing the methodology described by Beachy32, who showed that inclusion of electron correlation (in this case using LMP2) is important and has a significant effect on relative energies of peptide conformations. Tables of all Gly3/Ala3 conformers and their QM energies are provided in the supplementary material (Tables S1 and S2). Note that we only used these energies to refit backbone dihedral parameters, and no change was made to the ff94 partial atomic charges.
The optimization of dihedral parameters was done in two steps. First, the φ/ψ torsion parameters for Gly3 were optimized as follows. We used an exhaustive grid search in 12-dimensional dihedral parameter space: three amplitudes (V1, V2, V3) and three phases (γ1, γ2, γ3) for each φ and ψ. Amplitudes were systematically varied between 0 and 2 kcal/mol with a step of 0.2, phases were set to either 0 or π radians. Thus the total number of grid points in the initial search was 116 x 26 (over 100 million).
As the initial grid spacing was 0.2 kcal/mol for amplitudes (Vn), that would be the maximum precision of optimized parameters. To improve this, we employed ‘grid focusing’ by repeating the grid search with finer grid spacing centered on the parameters resulting from the initial course grid. Our refined grid had a spacing of 0.01 kcal/mol for amplitudes, thus that is the ultimate precision of our optimized grid parameters.
The function we used for optimization is somewhat more complicated than that shown in Equation 1. Since the zero of the MM energy function is arbitrary, the optimization should be performed using energy differences between alternate conformations. This was achieved in ff99 by setting one of the energies (the lowest one) to zero, and all other energies were assigned values relative to this “zero energy” reference. However, we observed that the optimized parameter set changes depending on which conformer’s energy is used as a reference. Therefore the function we optimized was calculated as an average of QM and MM energy differences with each conformer’s energy set as a reference in turn. This gives an average absolute error (aae) defined as follows:
| (4) |
where is the QM energy of conformer j with conformer i as a reference, and is the MM energy of conformer j with conformer i as a reference, N is number of conformers, which is 28 for Gly3. Another possibility to define a function for optimization is to concentrate on what the maximum absolute error (mae) might be when we consider all individual QM and MM differences and, again, take into account that any conformer may serve as a reference “zero energy”. This would give a function of the following form:
| (5) |
Here, we would search for such set of dihedral parameters that give the smallest mae. As it turns out, there are many sets of dihedral parameters with very similar aae. However, inspecting mae reveals that this parameter is much more sensitive to different parameter sets and therefore the selection of the final set was based on this error estimate. In general, small values of mae always correlate with small values of aae, such that the best parameter set picked based on mae will also have one of the lowest aae values.
For Gly3 optimization, an additional constraint was introduced, requiring the first phase for φ (i.e. γ1 for φ) to be 0. This was necessary to obtain physically reasonable dihedral functions with energy barrier located at φ ≈ 0° on the glycine Ramachandran map. Even though glycine parameter sets without this constraint achieved slightly better fit of QM and MM data, the resulting dihedral energy function would have a physically unreasonable minimum in the φ ≈ 0° region of Ramachandran map (where we also did not have any QM data). This arises from our use of only local minima to train the dihedral correction terms.
Once the Gly3 parameter set was obtained, the same procedure was repeated with 51 Ala3 conformers, and parameter sets corresponding to φ′/ψ′ dihedrals were obtained. While the resulting set of parameters appeared satisfactory, a more detailed analysis of errors revealed that Ala3 conformer number 16 was repeatedly responsible for the largest fitting errors (as measured by aae or mae). This conformer also falls into the unusual region of the Ramachandran map (see Figure 2 below), therefore it was excluded from the optimization of Ala3 dihedral parameters.
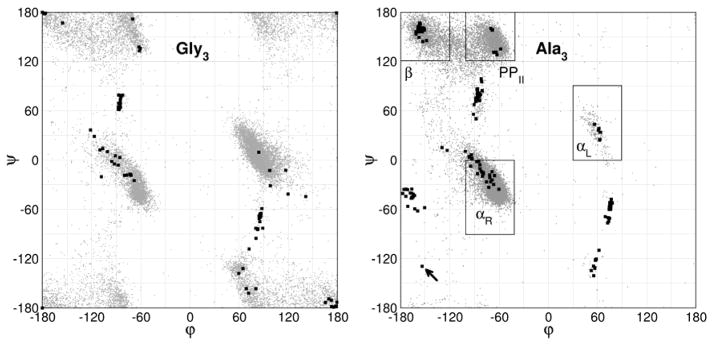
Figure 2.
Ramachandran plot of 28 Gly3 (left) and 51 Ala3 (right) conformers as obtained from QM geometry optimization of structures that were acquired using stochastic search with MM energies. All three dihedral pairs are shown for each conformer (black squares). The gray points in the background are φ/ψ values of glycine and alanine residues collected from a subset of the PDB. Typical secondary structure regions are outlined by boxes in the Ala3 plot (see Methods for additional details). Ala3 conformer number 16 (outlier) in the lower left of Ala3 plot is designated by an arrow.
The resulting parameters in the form suitable for use with AMBER are provided in supplementary material (Table S3).
Simulations in explicit water
All molecular dynamics simulations for Gly3 and Ala3 tetrapeptides were carried out with the sander module in AMBER87 using several different force fields as discussed in the main text: ff945, ff9911, Garcia’s modification of ff9413 with C-N-Cα-C and N-Cα-C-N terms zeroed (denoted ff94gs in the text), Pande’s modified ff9919 with C-N-Cα-C term replaced by the one from ff94 (denoted ff99ϕ), ff0324 (as present in AMBER8 distribution) and ff99SB developed as described above. The time step was 2 fs, and all bonds involving hydrogen were constrained by SHAKE with a tolerance of 10−4Å. Glycine and alanine tetrapeptide systems were solvated by approximately 520 TIP3P33 water molecules in a periodic box. Simulations were carried out in the NPT ensemble at 300 K. A cutoff of 8 Å was used for nonbonded interactions, and long-range electrostatic interactions were treated with the particle mesh Ewald (PME) method34.
Replica exchange molecular dynamics (REMD) simulations35, as implemented in AMBER8, were run for Ala3 using explicit water solvation and the ff99SB and ff03 force fields. REMD simulations were set up under similar conditions as the corresponding standard MD runs. Ala3 was solvated in a truncated octahedral box using 595 TIP3P water molecules. 26 replicas were used to span the temperatures range of 267 K to 571 K. Temperatures were optimized to give a uniform exchange acceptance ratio of ~30%. Exchange between neighboring temperatures was attempted every picosecond and each REMD simulation was run for ~30,000 exchange attempts (30 ns). Due to the high temperatures of some replicas, the NVT ensemble was employed.
Calculation of NMR order parameters
Hen egg white lysozyme (PDB code 6LYT36) and ubiquitin (PDB code 1UBQ37,38) simulations were run under the same conditions as the short peptides described above, but with truncated octahedral periodic boundary conditions including 4350 and 3300 water molecules respectively. Equilibration was done in multiple steps, with positional restraints on all heavy atoms in the crystal structure gradually released from 2 to 0 kcal/mol-Å2. The production run trajectories were 30 ns long. These trajectories were used to calculate the N-H internuclear vector autocorrelation functions with ptraj module. Order parameters (S2) were obtained from a plateau region of autocorrelation functions39. Specifically, the autocorrelation function was calculated up to time of half of trajectory length and the mean of last 5 ns was taken as S2. Standard deviation of the mean was used to plot the error bars. Since autocorrelation functions for a few backbone amides typically do not converge well (meaning that they do not show clear plateau), the mean will be a less reliable estimate of S2 for these amides, which will be reflected by larger standard deviations (i.e. larger error bars).
Dihedral plots and free energy surfaces
A Ramachandran plot showing the distribution of glycine or alanine φ/ψ angles in protein crystal structures was constructed based on a subset of proteins from a PDB database, with high resolution (<1.6 Å), small R-factor (<0.25) and less than 20% sequence homology40,41. Dihedral plots based on this statistical survey of the PDB provide experimental indication of which values of backbone torsions are commonly found in proteins.
Gly3/Ala3 simulations were used to produce free energy surfaces (or potential of mean force, PMF, maps) for backbone dihedral angles, assuming that φ/ψ are represented in the ensembles according to a Boltzmann distribution. First, the values of all φ/ψ dihedral pairs over the production portions of simulations were collected. Then, 2-dimensional φ/ψ normalized histograms were constructed from these values and converted to free energies using equation 6:
| (6) |
where Ni is the population of a particular histogram bin for specified values of φ/ψ, and N0 is the most populated bin. Thus the global free energy minimum always has a value of 0 kcal/mol.
The φ/ψ histograms were also used to evaluate populations in different regions of secondary structures. The definitions of the four principal regions were as follows: right-handed α-helix (αR): (φ, ψ) ≈ (−70±30°, −45±45°), left handed α-helix (αL): +60±30°/+45±45°, poly-proline II (PPII): −70±30°, +150±30°, and extended β-strand conformation (β): −150±30°, +150±30°. Most ranges are ±30° except from the ψ range of αR and αL, which were slightly wider (±45°) to capture the shift in these regions that resulted from using the ff94gs parameter set. The number of structures in individual regions were summed and divided by the total number of structures to obtain population fractions. Relative fractions are also reported; these differ in that the populations in the secondary structure basins were normalized by the total number of structures in just the four regions.
The convergence of Ala3 simulations was estimated by comparing the first and second half of the trajectories in terms of differences of population fractions. The fractions were calculated independently for the two parts of the trajectory and the resulting values are given as averages, with deviations reflecting the differences of the two independent calculations.
Decoy analysis
Similar to work that we previously published12 for ff94 and ff99, we used “decoy analysis” to test the capability of the force field to identify the experimentally determined native structure as that with the lowest potential energy. Three systems were used for decoy analysis: trpzip242 (SWTWENGKWTWK-NH2), Baldwin type43 alanine based α-helical peptide44 (Ace-GGG(KAAAA)3K-NH2) and trpcage miniprotein45 (NLYIQWLKDGGPSSGRPPPS). The decoys for the three systems were generated by a number of unrestrained as well as forced MD simulations at 300K as described previously12. The sets cover structures generated in simulations started from various initial structures, such as native, extended, and multiple unfolded or partially folded structures from intermediary stages of folding trajectories. The decoy ensembles thus contain local and global structural variation with a greater diversity of structures than could be obtained from a single simulation. The sizes of the three decoy sets are as follows: 177,000 for trpzip2, 70,000 for the helical peptide, and 118,000 for trpcage. The energies of these decoy sets were analyzed with different force fields using a Generalized Born solvation model46 implemented in AMBER. For each force field, the potential energy and RMSD from the experimentally determined structure were plotted for each of the decoy structures. The expectation for a “correct” force field is that the lowest energies are obtained for structures with low RMSDs, i.e. for native folds.
Because many different structures can have similar RMSD values, and because different force fields may favor slightly different conformations in each RMSD range, we performed a further simplification of the data. Since we are primarily interested in structures with low energies, we only plot one energy value, which is the average of the 20 lowest energies in that particular RMSD bin. This results in a curve that outlines the lowest energies sampled for the RMSD range (“lowest energy profile”), which can be more easily interpreted as being representative of the underlying energy surface for the force field. This approach has several limitations, including neglect of entropic contributions to the actual free energy, and also uses an implicit solvation model which may have its own effects on the stability of different conformations. Nevertheless, this methodology is useful for fast qualitative screening of many variations of a force field on larger peptides.
Results and Discussion
The unique conformations of 28 Gly3 and 51 Ala3 that resulted from QM optimization are shown in Figure 2. In general, these conformers overlap the regions outlined by PDB survey data. There are several distinct clusters that fall within the regions of typical protein secondary structures, such as right handed (αR) and left handed (αL) α-helix, polyproline II (PPII) and extended β-strand conformations.
Following the procedure described in Methods, we optimized a set of dihedral parameters (Table I). The root mean square difference (RMSD) between the QM and MM energies for glycine tetrapeptide using these parameters is 1.17 kcal/mol. The average absolute error and maximum error are 0.93 and 2.71 kcal/mol, respectively. Alanine tetrapeptide calculations used the glycine φ/ψ backbone parameters as well as the φ′/ψ′ dihedral terms. In this case, the root mean square difference, absolute average error and maximum error are 1.31, 1.05 and 3.43 kcal/mol. This is comparable to similar fitting errors obtained in ff9911 and ff0324. For comparison, the values of backbone dihedral parameters for many other variants of AMBER force field are given in the supplementary information (Table S4). It should be noted again that RMSD, absolute average error (aae) and maximum absolute error (mae) are calculated such that each conformer in turn is set as a “zero energy” reference, and thus all pairs of energy differences are used in the average. The sensitivity of errors (RMSD, aae, mae) to the choice of reference can be demonstrated by calculating these errors for all 51 Ala3 conformers with a reference conformation fixed to each conformer in turn. Such evaluation of errors produces best RMSD of 0.94 if conformer #31 is a reference and worst RMSD of 1.97 for conformer #45 as a reference. Similarly, depending on the choice of conformer as a reference, aae ranges between 0.77 and 1.82, and mae between 1.74 and 3.43 kcal/mol. Thus, averaging over all pair wise differences should provide a more stringent error estimation (e.g. mae is always the largest of all pair wise differences when using all pairs) and yields an independence of results with respect to which conformer is chosen as a reference.
Table I.
Optimized dihedral parameters: amplitudes (V) and phases (γ) for backbone dihedral angles. The φ/ψ terms are always used by all residues, φ′/ψ′ are used by all residues except glycine. Phases are in radians and amplitudes are in kcal/mol.
| V1 | γ1 | V2 | γ2 | V3 | γ3 | ||
|---|---|---|---|---|---|---|---|
| φ | C-N–Cα–C | 0.00 | 0 | 0.27 | 0 | 0.42 | 0 |
| ψ | N-Cα–C–N | 0.45 | π | 1.58 | π | 0.55 | π |
| φ′ | C-N–Cα–Cβ | 2.00 | 0 | 2.00 | 0 | 0.40 | 0 |
| ψ′ | N-C–Cα–Cβ | 0.20 | 0 | 0.20 | 0 | 0.40 | 0 |
Force field validation
Explicit water simulations of Gly3/Ala3
We performed molecular dynamics simulations on the same systems we used for parameter optimization, i.e. Gly3 and Ala3 peptides, with different AMBER force fields. All simulations were fully solvated with explicit water and extended to ~80ns. Histogram analysis was used to calculate relative free energies of different conformers with respect to φ/ψ backbone angles. These free energy (PMF) surfaces are shown in Figure 3 for both Gly3 and Ala3.
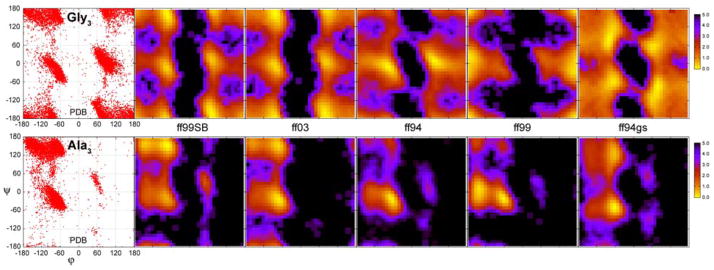
Figure 3.
Free energy φ/ψ maps for Gly3 (top row) and Ala3 (bottom row) from 80ns simulations with explicit TIP3P water. The energies are color coded from 0 up to 5 kcal/mol. PDB survey data is represented by a simple Ramachandran plot. Individual force fields are designated as: ff99SB (this work), ff0324, ff945, ff9911 and ff94gs13.
The correspondence of the distributions obtained from PDB and simulations has been used extensively in force field validation and development21,24–26. This is the basis for our first test and qualitatively evaluates our dihedral parameter set. PDB survey data are presented here as simple Ramachandran maps, not as free energy surfaces. One might expect that the φ/ψ regions sampled in crystal structures should be also accessible to solvated Gly3/Ala3 peptides. However, it would be wrong to assume that the relative populations of φ/ψ angles in static crystal structures should match populations in small tetrapeptides solvated in water. Not only are the two environments very different but also populations in crystal structures will certainly be biased by occurrence of common secondary structures, such as α-helices and β-sheets. Furthermore, there is no obvious choice for temperature associated with the PDB distribution, which would make any free energy calculation difficult.
We first discuss the plots for Gly3. The plots obtained from the Gly3 simulations are symmetric with respect to the origin, while alanine plots are not due to the presence of the chiral center on the alanine α-carbon. A certain amount of asymmetry in the glycine PDB data arises from the influence of chiral centers in other residues, which are not present in the Gly3 simulations.
The analysis of glycine conformations in the PDB data reveals that both right- and left-handed α-helical regions are densely populated. PPII and extended β-strand regions are also found with high frequency. One should expect similar conformational preferences resulting from MD simulations (Figure 3). This is largely true for ff99SB and ff03 and to a lesser extent for ff94 (which is missing PPII and β, and has strong bias favoring α). However, ff99 and ff94gs both show unexpected patterns for glycine conformations, sampling regions which are not represented in PDB survey data. As we noted before, these likely arise from lack of treatment of both sets of backbone parameters (φ/ψ and φ′/ψ′). The ff99 glycine parameters were fit using alanine data and favor conformations that show no population in the PBD data (φ = −180° and ψ = 0°). The ff94gs surface is also in significant disagreement with PDB data, with a surface that has minima and low barrier heights that appear highly dissimilar to the PDB data and to any of the force fields that do include φ and ψ terms. This may artificially increase the glycine conformational transition rates. The plots for ff96, ff99ϕ as well as the plots obtained from simulations that used our previous modification of dihedral parameters12,18 are shown in Figure S6 (supplementary material) and, similarly to ff99 and ff94gs, exhibit incorrect sampling for glycine dihedrals due to the lack of explicit treatment of both sets of backbone parameters.
Alanine free energy maps for the different force fields (Figure 3) show more similarity to each other, with major regions represented by right handed α-helix, PPII and extended β conformations. Once again, differences in the relative free energies of the basins are readily apparent in the different force fields. While ff99SB and ff03 show a reasonable balance between αR and PPII, ff94 and ff99 show clear bias in favor of αR. ff99 also shows the same tendency to adopt too-low (close to −180°) values of φ that was seen in its corresponding Gly3 data. ff94gs samples both αR and PPII, but little population of extended β is present; αR is also shifted low in ψ. The αL conformation (with a positive backbone φ value) is sampled most in ff99SB and least in ff03. This conformation occurs in several types of β-turns47, particularly those found in β-hairpins, and thus reproducing αL region may be important for modeling β-hairpins.
In order to better interpret such significant differences in relative populations of various secondary structure elements, we turned to available experimental data that provide more direct evidence concerning the dominant structure of short alanine dipeptides in aqueous solution. All of these observations have important implications for the varied conformational states of proteins and therefore should be captured in the force field if it should ever be used for the description of unfolded proteins or for protein folding studies.
A recent two-dimensional infrared spectroscopy study of alanine dipeptide in aqueous solution48 estimated backbone dihedral angles in the range (−70±25°, +120±25°), corresponding to PPII-like conformation. Another structural study of alanine dipeptide using 13C NMR49 confirmed the presence of PPII conformation, but also suggests that a mixture of PPII and αR is more likely in water. The dominance of PPII in trialanine was reiterated by two-dimensional vibrational spectroscopy studies of Woutersen & Hamm50,51 and later modified by the same authors to include around 20% of αR apart from PPII 52. The experimental studies of Schweitzer-Stenner & Eker on trialanines53,54 and tetraalanines55 in water using polarized Raman, FT-IR and VCD spectroscopy confirm the dominance of PPII conformation in tetraalanines, while a 50:50 mixture of PPII and extended β-strand-like conformations was observed for trialanines. Other studies56–58, mostly from Kallenbach and colleagues, have confirmed that short alanine peptides form predominantly PPII conformation, which is in temperature dependent equilibrium with extended β-strand conformations. Taken together, these experimental findings suggest that short alanine peptides are found predominantly in PPII-like structures with varying degree of extended β-strand, and possibly smaller fractions of α-helical conformations.
We analyzed the Ala3 simulation data obtained from each force field in terms of fractional population of local conformational basins corresponding to the four prevalent secondary structure elements. The frequent transitions between PPII, αR and β regions result in relatively rapid thermalization of these basins during the simulations. However, the αL region is separated from the other basins by a higher barrier near φ =0, resulting in much less frequent transitions from other regions (see Figure S5 in the supplementary information for further details). To ensure that the relative populations in that region were determined reliably, we ran replica exchange molecular dynamics (REMD) for ff99SB and ff03, which were the two force fields that provided conformational ensembles most consistent with the PDB data. All results are summarized in Table II. Comparison of standard and replica exchange molecular dynamics for ff99SB and ff03 demonstrates that relative populations in the four regions are essentially unchanged, which indicates that the results are well converged and that the differences do not arise from poor sampling.
Table II.
Fractions (percentages) of the four secondary structures in Ala3 MD simulations in explicit water. See Methods for definitions of secondary structure regions. Numbers in parentheses are relative fractions, only considering populations within the four regions.
| force field | Region
|
|||
|---|---|---|---|---|
| PPII (%) | β (%) | αR (%) | αL (%) | |
| ff99SB | 38.1±1.2 (51.9±1.7) | 20.1±0.7 (27.4±0.5) | 13.0±1.6 (17.8±2.6) | 2.1±0.9 (2.8±1.2) |
| ff99SB (REMD) | 36.9±0.1 (50.1±0.3) | 20.5±0.8 (27.9±1.0) | 12.7±0.5 (17.2±0.7) | 3.4±0.1 (4.6±0.1) |
| ff03 | 28.0±0.4 (37.8±0.4) | 12.4±0.5 (16.7±0.5) | 33.6±0.5 (45.4±0.9) | 0.0±0.0 (0.0±0.0) |
| ff03 (REMD) | 28.6±0.6 (38.2±0.6) | 12.6±0.4 (16.9±0.5) | 33.4±0.7 (44.7±1.1) | 0.1±0.0 (0.1±0.0) |
| ff94 | 2.7±0.5 (3.5±0.7) | 0.8±0.1 (1.1±0.2) | 72.0±0.5 (94.6±0.6) | 0.5±0.2 (0.7±0.3) |
| ff99 | 1.0±0.0 (2.5±0.1) | 2.4±0.3 (6.3±0.8) | 35.8±0.4 (90.8±0.5) | 0.1±0.1 (0.3±0.3) |
| ff94gs | 20.7±3.1 (30±6.0) | 2.3±0.3 (3.4±0.7) | 46.3±6.8 (66.0±6.5) | 0.4±0.2 (0.6±0.2) |
| ff99ϕ | 1.6±0.7 (2.2±0.9) | 0.4±0.2 (0.5±0.2) | 68.8±0.5 (96.6±0.6) | 0.4±0.4 (1.2±1.2) |
Simulations with ff99SB produce ensembles that are largely PPII (38%) with significant fraction of β (20%) and lower population of αR (13%). Compared to other force fields, this appears to be in best agreement with experimental observations, in that PPII is the most favorable conformation with lower presence of extended β and even lower fraction of α-helical structures.
ff94 and ff99 show clear preference towards αR (72% and 36% respectively), with the relative fractions of αR as compared to the other basins being over 90% for both force fields. The lower absolute fraction in ff99 (36%) arises from the unusually high population of structures around (−150°, 0°) which do not correspond to any of the four canonical secondary structure basins and are also much less populated in other force fields. Overall, the data from ff94 and ff99 are both at variance with experimental observations.
ff03 behaves most similarly to ff99SB, but it may still be slightly over stabilizing α-helices as the most populated basin is the αR conformation (34%). It is also somewhat unsettling that ff03 samples very poorly the αL region that is important for β-turn conformations. We obtained no sampling of αL in standard 80ns long MD simulations (0.0% fraction) indicating that this region is at least 5 kcal/mol (this is based on our cutoff for free energy plots, see Figure 3) less stable than the most populated αR region. However, REMD simulations do sample αL region with 0.06% population fraction. The sampling is insufficient to make a reliable estimate (see φ versus time plots in Figure S5 of supplementary material) of free energy difference between αR and αL, but based on the populations in those regions, αL is at least 3.8 kcal/mol higher than αR. Even though ff99SB only adopts 2–3% population in that region, it is still the largest compared to other force fields.
Simulations with ff99ϕ result in essentially the same behavior as was obtained for ff94, with the relative basin populations overwhelmingly dominated by αR conformations (97 vs. 95% for ff94, see relative fractions of individual secondary structures in Table II and free energy surfaces in Figure S6 of supplementary material). Thus, at least at the level of glycine/alanine tetrapeptide, the small change of a single φ term in ff99ϕ does not seem to result in significantly different or improved behavior.
Decoy analysis of longer peptides
A critical property of the energy function is whether it can distinguish the native state such that it corresponds to the lowest (free) energy. We previously employed “decoy analysis”12 as a rapid test of how well the energy function identifies the native structure. By plotting the potential energy against RMSD from the native structure one can readily see if the native state generally has lowest energies. This has the advantage that energy profiles for multiple force fields can be compared without obtaining converged ensembles for each peptide/force field combination. As a result of neglecting entropy and the use of an implicit water model, however, the results do not provide quantitative stability data for any of the peptides simulated.
We show the “lowest profile” (see Methods) of energies plotted versus RMSD from the native structure (Figure 4) with the expectation to see lowest potential energies (including solvation free energy) for small RMSD values, reflecting that the native basin indeed can be distinguished based merely on the energy function. We performed decoy screening using several peptides that have been experimentally demonstrated to adopt stable secondary and/or tertiary structures. Trpzip2 represents an exceptionally well-defined β-hairpin42, stabilized by cross-strand pairs of indole rings. The Baldwin type alanine-based peptide44 was designed to adopt α-helix. Lastly, the trpcage mini-protein45 has a mixture of secondary structure elements (α, 310 and polyproline).
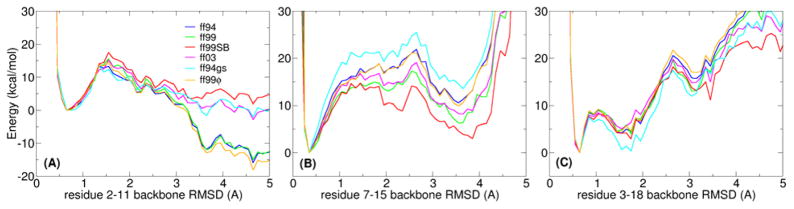
Figure 4.
“Lowest energy” profiles for three prototypical decoy systems, each tested with six AMBER force field variants (ff94, ff99, ff99SB, ff03, ff94gs, ff99ϕ): (A) trpzip2, which is known to adopt a β-hairpin, (B) a Baldwin-type sequence as a representative of an α-helix, and (C) trpcage as a representative of mixed secondary structures. RMSD values are calculated using the experimentally determined structure as a reference. Ideally, a force field should show lowest energies for the lowest RMSD values.
We previously used decoy analysis of the trpzip2 β-hairpin to demonstrate the extreme helical bias of the ff94/ff99 force fields12, showing that non-native α-helical decoys had the lowest energies and were also the most populated in simulations. Therefore it comes as no surprise that the present decoy plots again clearly reveal the inadequacy of the ff94 and ff99 force fields to identify the native state of the hairpin. Indeed the pronounced minimum around 4–5 Å (Figure 4A) represents a large set of helical structures. Consistent with our observations from the tetrapeptide simulations, ff99ϕ behaves much like ff94. The native trpzip2 structure scores as lowest in potential energy only with ff99SB. ff03 also performs much better than ff94/ff99 but, as we noted before, may still be slightly too favorable for helices. ff94gs behaves similarly to ff03.
On the other hand, all tested force fields successfully score the α-conformation of the Baldwin helix with lowest energy. As expected, the energy gap between the helical conformation and those with larger RMSD values is greater for the force fields with strong helical bias (ff94, ff99, ff99ϕ). Even though ff94gs performed almost as well as ff99SB for trpzip2, it scores the Baldwin helix even more favorably than the ff94/ff99 force fields that are known to over stabilize α-helix conformations. Likewise, all of the force fields correctly identify the trpcage native fold.
Overall, both ff99SB and ff03 perform well in all three decoy cases. ff99ϕ follows very closely the behavior of ff94. Since ff94gs and ff99ϕ have not been used as extensively as the original AMBER force fields upon which they are based (ff94/ff99), and did not perform well on the tetrapeptide and decoy screens, we will not consider them in further validation.
Comparison with NMR parameters for lysozyme and ubiquitin
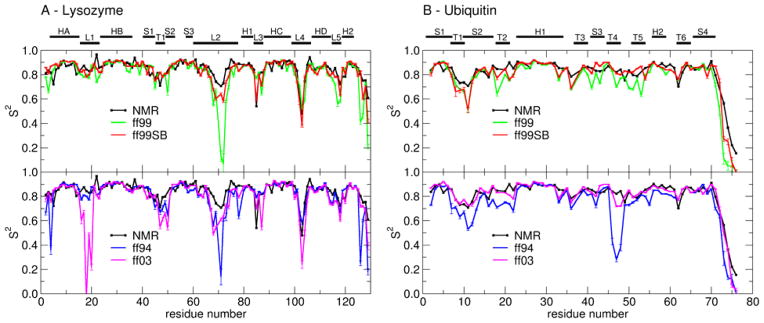
Hen egg white lysozyme has become a standard for evaluating the quality of force fields by comparing internal dynamics parameters calculated from MD simulations to NMR relaxation experiments59–63. The degree of backbone flexibility is specified by experimentally derived order parameters S2, which correspond to amide bond N-H librational motion. Lower S2 values reflect increased backbone flexibility. We generated 30ns MD simulations of lysozyme with explicit water using four different parameter sets: ff94, ff99, ff99SB and ff03. Order parameters calculated from these simulations are compared to experimentally derived values in Figure 5A. The root mean square difference and the correlation coefficient between experimental and calculated S2 are given in Table III. It can be seen that the agreement of ff99SB derived values with the experiment is very good, with the best correlation to experimental data and only half of the root mean square difference (RMSD) compared to all other force fields.
Figure 5.
Order parameters (S2) derived from experiment (black line) and calculated from MD simulations in explicit water with different force field parameter sets: ff99 (green), ff99SB (red), ff94 (blue) and ff03 (magenta). Error bars reflect the convergence of calculated S2 values. (A) Lysozyme. Secondary structures of lysozyme are labeled: helix A (HA: residues 4–15), loop 1 (L1: 16–23), helix B (HB: 24–36), strand 1 (S1: 41–45), turn 1 (T1: 46–49), strand 2 (S2: 50–53), strand 3 (S3: 58–60), long loop 2 (L2: 61–78), 310 helix 1 (H1: 80–84), loop 3 (L3: 85–89), helix C (HC: 89–99), loop 4 (L4: 100–107), helix D (HD: 108–115), loop 5 (L5: 116–119) and 310 helix 2 (H2: 120–124). (B) Ubiquitin. Secondary structure elements in ubiquitin: strand 1 (S1: residues 1–7), turn 1 (7–10), strand 2 (S2: 10–17), turn 2 (T2: 18–21), helix 1 (H1: 23–34), turn 3 (T3: 37–40), strand 3 (S3: 41–44), turn 4 (T4: 45–48), turn 5 (T5: 51–54), helix 2 (H2: 56–59), turn 6 (T6: 62–65) and strand 4 (S4: 66–71). Differences are described in the text.
Table III.
Correlation of experimental and calculated order parameters (S2) for several AMBER force fields.
| ff94 | ff99 | ff99SB | ff03 | |
|---|---|---|---|---|
|
|
||||
| lysozyme
|
||||
| RMSD | 0.11 | 0.12 | 0.06 | 0.13 |
| Ra | 0.61 | 0.62 | 0.83 | 0.62 |
|
| ||||
| ubiquitin
|
||||
| RMSD | 0.13 | 0.10 | 0.07 | 0.05 |
| R | 0.83 | 0.93 | 0.95 | 0.96 |
linear correlation coefficient
ff99SB describes well the increased flexibility of residues in loops L2 and L4 as well as residue 84 in L3 (which is not described well by other force fields). As has been noted previously64, molecular mechanics force fields perform most poorly in the loop regions, usually exaggerating the flexibility of loop residues. This is confirmed by our simulations where the most obvious disagreements with experimental order parameters occur in loops. Some of these discrepancies might be explained by a particular bias/weakness of the specific force field that was revealed in our model system studies. For example, we demonstrated that ff03 did not sample the αL region of φ/ψ space for alanine tetrapeptide. The pronounced failure of ff03 to match experimental data in the L1 loop (with simulated S2 values much too low) may be a result of the αL conformation that is adopted by several L1 residues in the crystal structure. The local instability in this conformation may give rise to increased structural dynamics in the ff03 simulations. Interestingly, significantly lower order parameters specifically in regions with glycines occupying the αL conformation were also noted for CHARMM22 force field by its developers26.
We also showed that ff94/ff99 are both biased towards helical structures, lacking sampling in PPII and β regions. This is likely the cause for increased flexibility in the middle of the long L2 loop, where several residues occupy the extended β-like conformation. This problem may be exacerbated in ff99 due to the presence of two glycines; we showed above that the glycine free energy surface in ff99 is highly distorted. Overall, many of the problematic regions are found to contain glycines, further supporting our assertion of the inadequacy of glycine parameters in many AMBER force field variants.
We repeated the same type of analysis for another protein, human ubiquitin. This is another small and well characterized protein of 76 residues with a known crystal structure37,38, for which NMR relaxation parameters were also determined experimentally65. Based on the crystal structure, the C-terminal region extends away from and makes few contacts with the remainder. This is reflected by very low S2 parameters for the four terminal residues. Again, Figure 5B graphically compares the ability of the four force fields to reproduce experimental S2 parameters. The quantitative indicators for the agreement with experimental data is given in Table III. For this system, all four AMBER force fields work reasonably well, with no large discrepancies with the exception of the region near turn T4 when using ff94. All force fields but ff03 exaggerate flexibility of turn T1 connecting strands S1 and S2. Overall, ff03 performs best, even though ff99SB still provides slight improvement over ff94/ff99. Examination of trajectories did not reveal any specific structural differences; rather the variations appear to arise from the magnitude of fluctuations about similar average structures.
Overall, these simulations serve as a good example of sensitivity of internal dynamics, as measured by NMR order parameters, to particular details of the molecular mechanics force field. Also, it is clear from these two sets of simulations that the quality of agreement to experimental relaxation data is somewhat system dependent. All force fields achieved better agreement with S2 values for ubiquitin than for lysozyme.
Conclusions
We have shown that several variants of AMBER force field have deficiencies arising from inadequate backbone dihedral term parametrization. The original ff94 force field introduced backbone dihedral treatment to be able to reproduce relative energies of selected glycine and alanine dipeptide conformers. Due to limited computational resources at the time, only a few conformers were included in the fitting procedure. We have shown here and previously that ff94 strongly favors helical conformations. More recent modifications of ff94 attempted to rectify the problem, but only partially succeeded. All of them failed to recognize that not just one, but two sets of dihedral parameters control backbone preferences in ff94. The first set is optimized solely for glycine. Any other amino acid will use both glycine set of parameters and a second set of dihedral parameters which are usually parametrized using alanine. Prior modifications of ff94, such as ff96, ff99, ff94gs, ff99ϕ, and our previous modification of ff9918 only changed the first set of dihedrals, effectively overwriting glycine dihedral parameters with the ones intended for alanine, unaware that there existed a set of alanine dihedral parameters that do not use the traditional definition of φ and ψ. This resulted in incorrect parametrization of glycine. Our data demonstrate (Figure 3, Figure S6) that these parameter sets result in conformational ensembles for glycine peptides that are in significant disagreement with both the PDB data and with force fields that explicitly include glycine in the fitting process. We therefore recommend against using these parameter sets for protein simulations.
The modification of ff94/ff99 force field presented in this work aims to rectify the problems caused by misunderstanding of parametrization procedure, but also endeavors to improve the parametrization initially done in ff94. This was primarily achieved through two courses of action. First, we used a large number of glycine and alanine tetrapeptide local minima instead of a small number of dipeptide conformations when fitting QM and MM energies. We believe that, unlike the dipeptide, tetrapeptide dihedral space bears closer resemblance to that found in proteins, and also has a local minimum for the α-helical conformation, permitting direct fitting to avoid over stabilization that was observed in earlier variants. Second, we used a procedure that avoids biasing the outcome of the dihedral parameters optimization with respect to a single a priori chosen reference conformation.
The resulting parameter set was shown to result in better balance of prevalent secondary structures, such as PPII, α-helix and extended/β-strand in simulations of short glycine and alanine peptides in water. This was supported by better agreement of dihedral angle regions sampled in the simulations with regions generated from PDB survey data. Here, the improvement for glycine residue is most noticeable, where some of the force fields produced free energy surfaces that bore no resemblance to those observed for real proteins. The relative sampling of the typical secondary structure regions in simulations of alanine tetrapeptide using the new force field is in good agreement with experimental data which used various techniques to infer the prevalent states of short alanine peptides. Beyond short glycine and alanine peptides, the qualitative performance of the new parameters was tested on several larger systems representing typical secondary structures: trpzip2 β-hairpin, Baldwin type α-helix and the trpcage miniprotein. ff99SB correctly identified native structure as that with the lowest energy in all three cases, which was not observed for any of the other force field variants. Finally, we performed long explicit water simulations of lysozyme and ubiquitin and tested the performance of different force field variants by comparing relaxation order parameters calculated from simulations to experimental NMR values. Once again, the newly derived parameters performed the best. The changes we introduced with ff99SB modification are extremely minimal and therefore excellent compatibility should be preserved with existing parameters developed to be compatible with ff94/ff99. This is in contrast to the ff03 protein parameters, for which no compatible parameters currently exist for small molecules and most importantly nucleic acids. It is therefore unclear whether ff03 could be used for simulations of these complexes (which would include potentially incompatible charge models).
As usual, caution should be taken when any force fields are used beyond their range of validity. For example, one can expect a substantial effect of solvation model used. Implicit solvation models, such as Generalized Born, may shift the balance between different secondary structure elements. If, for example, PPII is indeed stabilized by specific interactions with explicit water as recently suggested57,66–68, such stabilization is not possible with implicit solvents, and thus structural preference may change. In fact, we recently reported that GB solvation models increase α-helical propensity69. Different approaches in force field development might be needed to account for these effects, and force fields should likely be tested with the particular characteristics (and perhaps idiosyncrasies) of these models in mind.
Force field development is a complex process in which many approximations are typically required. In the present case, conformational energies from high-level gas-phase QM calculations were used to fit dihedral parameters for a force field in which the partial charges should more closely represent those in aqueous solution. Additionally, the large difference in the size of systems that are used to train biomolecular force fields and those for which they are intended to be used results in a slow maturation period, often with deficiencies becoming apparent only after extensive use. For this reason, we extensively used these parameters on a variety of systems not discussed here before making this report. For example, the improved glycine parametrization was important in our recent simulations of HIV-1 protease70, where glycine rich “flaps” play an essential role in protease dynamics. Another study69 reported a hybrid replica exchange method, where ff99SB was used to test the methodology based on conformational sampling of polyalanine peptides of varying lengths. Our recent simulations71 of fragments of the villin headpiece with ff99SB also achieved good agreement with experimental trends72 in calculated J-coupling constants and helical propensities. These studies have supplied further evidence that the ff99SB parameter set provides a reasonable parameter set for protein simulations, in the context of an efficient, additive molecular mechanics model.
Supplementary Material
Acknowledgments
The authors thank Alex MacKerell for useful discussions concerning order parameters and Angel Garcia and Vijay Pande for helpful discussions and providing details for their AMBER force field modifications. Supercomputer time at NCSA (NCSA MCA02N028 to CS and MCA05S010 to AER) and financial support (CS) from the National Institutes of Health (NIH GM6167803) and Department of Energy (Contract DE-AC02-98CH10886) are gratefully acknowledged. C.S. is a Cottrell Scholar of Research Corporation.
References
- 1.Rick SW, Stuart SJ. Potentials and algorithms for incorporating polarizability in computer simulations. Reviews in Computational Chemistry. 2002;18:89–146. [Google Scholar]
- 2.Ponder JW, Case DA. Force fields for protein simulations. Protein Simulations. Volume 66. Advances in Protein Chemistry. 2003:27. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- 3.Mackerell AD. Empirical force fields for biological macromolecules: Overview and issues. Journal of Computational Chemistry. 2004;25(13):1584–1604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 4.Jorgensen WL, Tirado-Rives J. Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(19):6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second Generation Force Field For the Simulation of Proteins, Nucleic Acids, and Organic Molecules. Journal of the American Chemical Society. 1995;117(19):5179–5197. [Google Scholar]
- 6.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, Debolt S, Ferguson D, Seibel G, Kollman P. Amber, a Package of Computer-Programs for Applying Molecular Mechanics, Normal-Mode Analysis, Molecular-Dynamics and Free-Energy Calculations to Simulate the Structural and Energetic Properties of Molecules. Computer Physics Communications. 1995;91(1–3):1–41. [Google Scholar]
- 7.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. Journal of Computational Chemistry. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jorgensen WL, Tiradorives J. The Opls Potential Functions for Proteins - Energy Minimizations for Crystals of Cyclic-Peptides and Crambin. Journal of the American Chemical Society. 1988;110(6):1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 9.Kuyper LF, Hunter RN, Ashton D, Merz KM, Kollman PA. Free-Energy Calculations on the Relative Solvation Free-Energies of Benzene, Anisole, and 1,2,3-Trimethoxybenzene - Theoretical and Experimental-Analysis of Aromatic Methoxy Solvation. Journal of Physical Chemistry. 1991;95(17):6661–6666. [Google Scholar]
- 10.Kollman P, Dixon R, Cornell W, Fox T, Chipot C, Pohorille A. The development/application of the “minimalist” organic/biochemical molecular mechanic force field using a combination of ab initio calculations and experimental data. In: van Gunsteren WF, Weiner PK, Wilkinson AJ, editors. Computer Simulations of Biomolecular Systems. Vol. 3. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 83–96. [Google Scholar]
- 11.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? Journal of Computational Chemistry. 2000;21(12):1049–1074. [Google Scholar]
- 12.Okur A, Strockbine B, Hornak V, Simmerling C. Using PC clusters to evaluate the transferability of molecular mechanics force fields for proteins. Journal of Computational Chemistry. 2003;24(1):21–31. doi: 10.1002/jcc.10184. [DOI] [PubMed] [Google Scholar]
- 13.Garcia AE, Sanbonmatsu KY. alpha-Helical stabilization by side chain shielding of backbone hydrogen bonds. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(5):2782–2787. doi: 10.1073/pnas.042496899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamiya N, Higo J, Nakamura H. Conformational transition states of a beta-hairpin peptide between the ordered and disordered conformations in explicit water. Protein Science. 2002;11(10):2297–2307. doi: 10.1110/ps.0213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higo J, Ito N, Kuroda M, Ono S, Nakajima N, Nakamura H. Energy landscape of a peptide consisting of alpha-helix, 3(10)-helix, beta-turn, beta-hairpin, and other disordered conformations. Protein Science. 2001;10(6):1160–1171. doi: 10.1110/ps.44901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono S, Nakajima N, Higo J, Nakamura H. Peptide free-energy profile is strongly dependent on the force field: Comparison of C96 and AMBER95. Journal of Computational Chemistry. 2000;21(9):748–762. [Google Scholar]
- 17.Wang L, Duan Y, Shortle R, Imperiali B, Kollman PA. Study of the stability and unfolding mechanism of BBA1 by molecular dynamics simulations at different temperatures. Protein Science. 1999;8(6):1292–1304. doi: 10.1110/ps.8.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simmerling C, Strockbine B, Roitberg AE. All-atom structure prediction and folding simulations of a stable protein. Journal of the American Chemical Society. 2002;124(38):11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 19.Sorin EJ, Pande VS. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophysical Journal. 2005;88(4):2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorin EJ, Pande VS. Empirical force-field assessment: The interplay between backbone torsions and noncovalent term scaling. Journal of Computational Chemistry. 2005;26(7):682–690. doi: 10.1002/jcc.20208. [DOI] [PubMed] [Google Scholar]
- 21.Hu H, Elstner M, Hermans J. Comparison of a QM/MM force field and molecular mechanics force fields in simulations of alanine and glycine “dipeptides” (Ace-Ala-Nme and Ace-Gly-Nme) in water in relation to the problem of modeling the unfolded peptide backbone in solution. Proteins-Structure Function and Genetics. 2003;50(3):451–463. doi: 10.1002/prot.10279. [DOI] [PubMed] [Google Scholar]
- 22.Zaman MH, Shen MY, Berry RS, Freed KF, Sosnick TR. Investigations into sequence and conformational dependence of backbone entropy, inter-basin dynamics and the flory isolated-pair hypothesis for peptides. Journal of Molecular Biology. 2003;331(3):693–711. doi: 10.1016/s0022-2836(03)00765-4. [DOI] [PubMed] [Google Scholar]
- 23.Mu YG, Kosov DS, Stock G. Conformational dynamics of trialanine in water. 2. Comparison of AMBER, CHARMM, GROMOS, and OPLS force fields to NMR and infrared experiments. Journal of Physical Chemistry B. 2003;107(21):5064–5073. [Google Scholar]
- 24.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong GM, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang JM, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. Journal of Computational Chemistry. 2003;24(16):1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 25.MacKerell AD, Feig M, Brooks CL. Improved treatment of the protein backbone in empirical force fields. Journal of the American Chemical Society. 2004;126(3):698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 26.Mackerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. Journal of Computational Chemistry. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 27.Feig M, MacKerell AD, Brooks CL. Force field influence on the observation of pi-helical protein structures in molecular dynamics simulations. Journal of Physical Chemistry B. 2003;107(12):2831–2836. [Google Scholar]
- 28.Jorgensen WL, Maxwell DS, TiradoRives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. Journal of the American Chemical Society. 1996;118(45):11225–11236. [Google Scholar]
- 29.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. Journal of Physical Chemistry B. 2001;105(28):6474–6487. [Google Scholar]
- 30.Bowen JP, Allinger NL. Molecular Mechanics: The Art and Science of Parametrization. In: Lipkowitz KB, Boyd DB, editors. Reviews in Computational Chemistry. Vol. 2. New York: VCH Publishers, Inc; 1991. pp. 81–98. [Google Scholar]
- 31.Jaguar. version 4.1. Portland, Oregon: Schrodinger, Inc; 2000. [Google Scholar]
- 32.Beachy MD, Chasman D, Murphy RB, Halgren TA, Friesner RA. Accurate ab initio quantum chemical determination of the relative energetics of peptide conformations and assessment of empirical force fields. Journal of the American Chemical Society. 1997;119(25):5908–5920. [Google Scholar]
- 33.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics. 1983;79(2):926–935. [Google Scholar]
- 34.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N. Log(N) Method for Ewald Sums in Large Systems. Journal of Chemical Physics. 1993;98(12):10089–10092. [Google Scholar]
- 35.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chemical Physics Letters. 1999;314(1–2):141–151. [Google Scholar]
- 36.Young ACM, Dewan JC, Nave C, Tilton RF. Comparison of Radiation-Induced Decay and Structure Refinement from X-Ray Data Collected from Lysozyme Crystals at Low and Ambient-Temperatures. Journal of Applied Crystallography. 1993;26:309–319. [Google Scholar]
- 37.Vijaykumar S, Bugg CE, Cook WJ. Structure of Ubiquitin Refined at 1. 8 a Resolution. Journal of Molecular Biology. 1987;194(3):531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 38.Vijaykumar S, Bugg CE, Wilkinson KD, Cook WJ. 3-Dimensional Structure of Ubiquitin at 2. 8 a Resolution. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(11):3582–3585. doi: 10.1073/pnas.82.11.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic-Resonance Relaxation in Macromolecules 1. Theory and Range of Validity Journal of the American Chemical Society. 1982;104(17):4546–4559. [Google Scholar]
- 40.Wang GL, Dunbrack RL. PISCES: a protein sequence culling server. Bioinformatics. 2003;19(12):1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 41.Wang GL, Dunbrack RL. PISCES: recent improvements to a PDB sequence culling server. Nucleic Acids Research. 2005;33:W94–W98. doi: 10.1093/nar/gki402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cochran AG, Skelton NJ, Starovasnik MA. Tryptophan zippers: Stable, monomeric beta-hairpins. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(10):5578–5583. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marqusee S, Baldwin RL. Helix Stabilization by Glu- … Lys+ Salt Bridges in Short Peptides of Denovo Design. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(24):8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fesinmeyer RM, Peterson ES, Dyer RB, Andersen NH. Studies of helix fraying and solvation using C-13′ isotopomers. Protein Science. 2005;14(9):2324–2332. doi: 10.1110/ps.051510705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neidigh JW, Fesinmeyer RM, Andersen NH. Designing a 20-residue protein. Nature Structural Biology. 2002;9(6):425–430. doi: 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- 46.Hawkins GD, Cramer CJ, Truhlar DG. Pairwise Solute Descreening of Solute Charges From a Dielectric Medium. Chemical Physics Letters. 1995;246(1–2):122–129. [Google Scholar]
- 47.Wilmot CM, Thornton JM. Analysis and Prediction of the Different Types of Beta-Turn in Proteins. Journal of Molecular Biology. 1988;203:221–232. doi: 10.1016/0022-2836(88)90103-9. [DOI] [PubMed] [Google Scholar]
- 48.Kim YS, Wang JP, Hochstrasser RM. Two-dimensional infrared spectroscopy of the alanine dipeptide in aqueous solution. Journal of Physical Chemistry B. 2005;109(15):7511–7521. doi: 10.1021/jp044989d. [DOI] [PubMed] [Google Scholar]
- 49.Mehta MA, Fry EA, Eddy MT, Dedeo MT, Anagnost AE, Long JR. Structure of the alanine dipeptide in condensed phases determined by C-13 NMR. Journal of Physical Chemistry B. 2004;108(9):2777–2780. [Google Scholar]
- 50.Woutersen S, Hamm P. Structure determination of trialanine in water using polarization sensitive two-dimensional vibrational spectroscopy. Journal of Physical Chemistry B. 2000;104(47):11316–11320. [Google Scholar]
- 51.Woutersen S, Hamm P. Isotope-edited two-dimensional vibrational spectroscopy of trialanine in aqueous solution. Journal of Chemical Physics. 2001;114(6):2727–2737. [Google Scholar]
- 52.Woutersen S, Pfister R, Hamm P, Mu YG, Kosov DS, Stock G. Peptide conformational heterogeneity revealed from nonlinear vibrational spectroscopy and molecular-dynamics simulations. Journal of Chemical Physics. 2002;117(14):6833–6840. [Google Scholar]
- 53.Eker F, Cao XL, Nafie L, Schweitzer-Stenner R. Tripeptides adopt stable structures in water. A combined polarized visible Raman, FTIR, and VCD spectroscopy study. Journal of the American Chemical Society. 2002;124(48):14330–14341. doi: 10.1021/ja027381w. [DOI] [PubMed] [Google Scholar]
- 54.Eker F, Griebenow K, Schweitzer-Stenner R. Stable conformations of tripeptides in aqueous solution studied by UV circular dichroism spectroscopy. Journal of the American Chemical Society. 2003;125(27):8178–8185. doi: 10.1021/ja034625j. [DOI] [PubMed] [Google Scholar]
- 55.Schweitzer-Stenner R, Eker F, Griebenow K, Cao XL, Nafie LA. The conformation of tetraalanine in water determined by polarized raman, FT-IR, and VCD spectroscopy. Journal of the American Chemical Society. 2004;126(9):2768–2776. doi: 10.1021/ja039452c. [DOI] [PubMed] [Google Scholar]
- 56.Shi ZS, Chen K, Liu ZG, Ng A, Bracken WC, Kallenbach NR. Polyproline II propensities from GGXGG peptides reveal an anticorrelation with beta-sheet scales. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(50):17964–17968. doi: 10.1073/pnas.0507124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McColl IH, Blanch EW, Hecht L, Kallenbach NR, Barron LD. Vibrational Raman optical activity characterization of poly(L-proline) II helix in alanine oligopeptides. Journal of the American Chemical Society. 2004;126(16):5076–5077. doi: 10.1021/ja049271q. [DOI] [PubMed] [Google Scholar]
- 58.Shi ZS, Olson CA, Rose GD, Baldwin RL, Kallenbach NR. Polyproline II structure in a sequence of seven alanine residues. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(14):9190–9195. doi: 10.1073/pnas.112193999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soares TA, Daura X, Oostenbrink C, Smith LJ, van Gunsteren WF. Validation of the GROMOS force-field parameter set 45A3 against nuclear magnetic resonance data of hen egg lysozyme. Journal of Biomolecular Nmr. 2004;30(4):407–422. doi: 10.1007/s10858-004-5430-1. [DOI] [PubMed] [Google Scholar]
- 60.Stocker U, van Gunsteren WF. Molecular dynamics simulation of hen egg white lysozyme: A test of the GROMOS96 force field against nuclear magnetic resonance data. Proteins-Structure Function and Genetics. 2000;40(1):145–153. [PubMed] [Google Scholar]
- 61.Buck M, Boyd J, Redfield C, Mackenzie DA, Jeenes DJ, Archer DB, Dobson CM. Structural Determinants of Protein Dynamics - Analysis of N-15 Nmr Relaxation Measurements for Main-Chain and Side-Chain Nuclei of Hen Egg-White Lysozyme. Biochemistry. 1995;34(12):4041–4055. doi: 10.1021/bi00012a023. [DOI] [PubMed] [Google Scholar]
- 62.Buck M, Karplus M. Internal and overall peptide group motion in proteins: molecular dynamics simulations for lysozyme compared with results from X-ray and NMR spectroscopy. Journal of the American Chemical Society. 1999;121(41):9645–9658. [Google Scholar]
- 63.Buck M, Bouguet-Bonnet S, Pastor RW, MacKerell AD. Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophysical Journal. 2006;90(4):L36–38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Case DA. Molecular dynamics and NMR spin relaxation in proteins. Accounts of Chemical Research. 2002;35(6):325–331. doi: 10.1021/ar010020l. [DOI] [PubMed] [Google Scholar]
- 65.Tjandra N, Feller SE, Pastor RW, Bax A. Rotational diffusion anisotropy of human ubiquitin from N-15 NMR relaxation. Journal of the American Chemical Society. 1995;117(50):12562–12566. [Google Scholar]
- 66.Fleming PJ, Fitzkee NC, Mezei M, Srinivasan R, Rose GD. A novel method reveals that solvent water favors polyproline II over beta-strand conformation in peptides and unfolded proteins: conditional hydrophobic accessible surface area (CHASA) Protein Science. 2005;14(1):111–118. doi: 10.1110/ps.041047005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mezei M, Fleming PJ, Srinivasan R, Rose GD. Polyproline II helix is the preferred conformation for unfolded polyalanine in water. Proteins-Structure Function and Bioinformatics. 2004;55(3):502–507. doi: 10.1002/prot.20050. [DOI] [PubMed] [Google Scholar]
- 68.Mezei M, Srinivasan R, Fleming PJ, Rose GD. Solvent effect on the conformational preference of poly alanine. Biophysical Journal. 2004;86(1):630A–630A. [Google Scholar]
- 69.Okur A, Wickstrom L, Layten M, Geney R, Song K, Hornak V, Simmerling C. Improved efficiency of replica exchange simulations through use of a hybrid explicit/implicit solvation model. Journal of Chemical Theory and Computation. 2006;2(2):420–433. doi: 10.1021/ct050196z. [DOI] [PubMed] [Google Scholar]
- 70.Hornak V, Okur A, Rizzo RC, Simmerling C. HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(4):915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wickstrom L, Okur A, Song K, Hornak V, Raleigh DP, Simmerling C. The Unfolded State of the Villin Headpiece Helical Subdomain: Computational Studies of the Role of Locally Stabilized Structure. Journal of Molecular Biology. 2006 doi: 10.1016/j.jmb.2006.04.070. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang YF, Rigotti DJ, Fairman R, Raleigh DP. Peptide models provide evidence for significant structure in the denatured state of a rapidly folding protein: The villin headpiece subdomain. Biochemistry. 2004;43(11):3264–3272. doi: 10.1021/bi035652p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.