Abstract
The silkworm baculovirus expression system is widely used to produce recombinant proteins. Several strategies for constructing recombinant viruses that contain foreign genes have been reported. Here, we developed a novel defective-rescue BmNPV Bacmid (reBmBac) expression system. A CopyControl origin of replication was introduced into the viral genome to facilitate its genetic manipulation in Escherichia coli and to ensure the preparation of large amounts of high quality reBmBac DNA as well as high quality recombinant baculoviruses. The ORF1629, cathepsin and chitinase genes were partially deleted or rendered defective to improve the efficiency of recombinant baculovirus generation and the expression of foreign genes. The system was validated by the successful expression of luciferase reporter gene and porcine interferon γ. This system can be used to produce batches of recombinant baculoviruses and target proteins rapidly and efficiently in silkworms.
Introduction
Since its inception more than 30 years ago, the baculovirus expression vector system (BEVS) has been widely used to express heterologous foreign proteins [1–3]. Autographa californica nucleopolyhedrovirus (AcMNPV)-Sf9 and Bombyx mori nucleopolyhedrovirus (BmNPV)-silkworm are two typical BEVSs [4–6]. The BmNPV-silkworm offers several advantages in comparison with the AcMNPV-Sf9 system [7].
Baculoviruses are a group of large viruses with circular double-stranded DNA genomes of 88–153 kb [8], which makes it laborious to manipulate and generate recombinant baculoviruses. Therefore, several construction strategies have been developed to increase the efficiency of recombinant baculovirus generation. To generate the recombinant AcMNPV, Patel et al. have developed a recombinant baculovirus shuttle strategy in yeast with yeast autonomous replicating sequences (ARS) and centromere (CEN) sequences [9]. An AcMNPV baculovirus shuttle vector (Bacmid) that can be manipulated in Escherichia coli (E. coli) as a plasmid has also been reported [10, 11]. This Bacmid is commercially available as Bac-to-Bac BEVS. And also Kitts et al. have developed a BacPAK6 expression system [12] in which several Bsu36I sites have been introduced into the AcMNPV genome to allow preparation of linearized viral DNA with essential gene deficiency for cotransfection. Based on these typical strategies, several defective baculovirus expression system have been engineered. Je et al. have reported an AcMNPV Bacmid system that lack a portion of the essential ORF1629 gene [13]. Jones et al. have also reported the inactivation of this essential gene to improve baculovirus recombination [14]. Furthermore, multigene expression has been achieved by some groups [15, 16].
The BmNPV, as another important baculovirus using for foreign gene expression in cells and silkworms, has been researched deeply. There are also several strategies for generation of recombinant BmNPV just as AcMNPV baculovirus expression system. The Bm-Bac-to-Bac system has been established by Park’s group [17]. Wu et al. have developed the BmBacPAK6 system [18] based on the AcMNPV BacPAK6 system. Some defective baculovirus systems have also been established [19, 20]. A BmNPV expression system using the mating-assisted genetically integrated cloning (MAGIC) strategy has also been developed [21].
These methods for the generation of recombinant AcMNPV and BmNPV have been applied for a long period. The Bac-to-Bac and BacPAK baculovirus expression systems are the most widely used among these expression systems [22].
Baculoviruses have a biphasic life cycle in which two distinct forms of the virus spread throughout tissues and among individuals, which results in two forms of virions, namely a budded virion (BV) and an occlusion derived virion (ODV) [23]. Some virus encoded proteins involved in ODV structure are not required to maintain virus cell to cell infectivity or virus replication. [24–26]. For example, the polyhedrin and p10 genes are not involved in within host virus infection or replication, and thus their promoters are used to drive the expression of foreign proteins [16]. The products of some virus genes, such as cathepsin (cat) and chitinase (chi), can impede foreign gene expression, and hence foreign gene expression efficiency increases in the absence of these two genes [27–29].
In this study, we developed a novel defective-rescue BmNPV Bacmid (reBmBac) expression system to provide an effective and simple strategy for constructing recombinant baculoviruses. The reBmBac was established by using convenient tools, such as phage the λ Red recombinase system, the FLP/FRT (FLP recombinase recognition target) recombinase system [30], the rpsL-neo counter-resistance system [31, 32] and antibiotic resistance genes. The cat and chi genes in the reBmBac genome were inactivated [29] and replaced by an E. coli CopyControl origin of replication [33] to make the baculovirus viral genome DNA easy to manipulate and prepare in E. coli. The ORF1629 gene is essential to the virus life cycle because it encodes the viral capsid-associated protein [34]. This essential gene was stably and partially deleted in E. coli to construct the defective-rescue bacmid and ensure the efficiency and purity of the recombinant baculoviruses obtained through cotransfection [12]. In addition, a general manipulation method for any gene site in the baculovirus genome was established. The reporter gene luciferase (luc) as quality control and porcine gene interferon γ (PoIFN-γ) were successfully expressed by using this system.
Materials and Methods
E. coli DH10B was obtained from Invitrogen (Carlsbad, CA, USA). E. coli BW25113/pKD46 and DH10B/pCP20 were obtained from the Molecular, Cellular and Developmental Biology Department, Kline Biology Tower 830, Yale University. The pGL3-Basic vector and luciferase assay system were obtained from Promega (Madison, WI, USA). The CopyControl pCC1BAC Vector (containing CopyControl origin) and E. coli TransforMax EPI300 were obtained from Epicentre (Madison, WI, USA). DH10Bac/pMON7124 (carrying a tetracycline resistance gene), transfer vector pVL1393 and Lipofectin were obtained from Invitrogen. Low-melting agarose was obtained from Sigma-Aldrich (St. Louis, MO, USA). Anti-PoIFN-γ antibody (Catalogue Number: AB10624) was obtained from Millipore (Boston, MA, USA), and goat anti rabbit pAb-HRP (Code No. 458) was obtained from MBL (Japan). Standard recombinant porcine IFN-γ (Catalogue Number: 985-PI-050) was obtained from R&D SYSTEMS (Minnesota, MN, USA).
Construction of the gene-targeting vector
The pCC-chi-cat vector (GenBank accession number: KU749549) was used to construct BmBac via homologous recombination with BmNPV. Homologous targeting arms were amplified (the primers for fusion PCR, namely Bm-chi-F, Bm-chi-R, Bm-cat-F, and Bm-cat-R, are listed in S1 Table) to remove the cathepsin (GeneID: 1724490) and chitinase (GeneID: 1724489) genes and were inserted into pUC19 via EcoRI/HindIII digestion. A fragment that contained the CopyControl E. coli origin of replication and a chlR gene that was purified from the pCC1BAC vector using SalI digestion was located between the arms.
The pPolh-1629 vector (GenBank accession number: KU749550) was used to construct the defective BmBac lacking a portion of the essential ORF1629 gene. The homologous arms were amplified (the primers for fusion PCR, namely Bm-1629-F, Bm-1629-R, Bm-polh-F and Bm-polh-R are listed in S1 Table) from the BmNPV genome and inserted into the pMD18-T simple vector. The tetracycline resistance (tetR) gene, which served as an antibiotic selectable marker, was amplified (tet-F, tet-R) from the pMON7124 plasmid and inserted between the arms by XhoI digestion.
The pRN-FRT vector (GenBank accession number: KU749551) contained the counter-resistance gene rpsL-neo (GenBank: GU084141.1), which was synthesized by Genscript Corp. (Nanjing, China) and flanked by a mutant FRT site. The FRT-rpsL-neo element was inserted into the pMD18 vector.
The transfer plasmid pVL1393-luc was used to deliver the luc+ gene into reBmBac, and it achieved rescue with a complete ORF1629 gene via homologous recombination in vivo during cotransfection. The luc+ gene was removed from the pGL3-Basic vector by BglII/XbaI digestion and was then transferred to a BamHI/XbaI-digested pVL1393 vector. The transfer plasmid pVL1393-PoIFN-γ was used to deliver the PoIFN-γ gene (NCBI Reference Sequence: NM_213948.1) [35] into reBmBac and to rescue the defective viral DNA. The porcine IFN-γ gene was amplified (primers for PCR, IFN-F and IFN-R are listed in S1 Table) and inserted into the pVL1393 vector by BamHI/EcoRI digestion.
Manipulation of the baculovirus genome in E. coli
The homologous recombination mediated by phage λ red recombinase and the elimination of the FRT-flanked fragment by FLP recombinase were performed according to Wanner et al., as described briefly below [30]. For the recombination, the targeting element with homologous arms was isolated by digestion or PCR from the targeting vector and purified using gel electrophoresis. A 1 μg sample of the targeting element was transferred into electrocompetent cells (E. coli BW25113 with phage λ Red recombinase) using an ECM 630 Electro Cell Manipulator with a voltage of 15 kV/CM according to the manufacturer’s instructions (BTX Instrument Division of Genetronics, Inc.). Positive recombinant clones were selected after culture with the appropriate antibiotic. The positive clones were further screened by PCR with primers that spanned the viral genome sequence and targeted element sequence. The BmBac vector with the rpsL-neo gene was transferred into electrocompetent cells (E. coli DH10B with FLP recombinase) to eliminate the FRT-flanked fragment (the rpsL-neo counter-selection gene). The counter-selection gene was removed with FLP recombinase. The positive clones were selected on strR antibiotic plates after culture. The positive clones were also further screened by PCR.
A large amount of high-quality reBmBac DNA was induced by L-arabinose [36] and prepared according to the CopyControl BAC cloning kit instructions (Epicentre) in E. coli DH10B. Purified DNA was resuspended in 100 μL of TE buffer, and the concentration was measured with a UV-Vis Spectrophotometer (NanoDrop 2000, Thermo).
Cell culture and virus amplification in cells
The Bombyx mori-derived cell line, Bm5, was cultured in TC100 insect cell culture medium (Applichem, Darmstadt, Germany) with 10% fetal bovine serum (FBS, Gibco, USA) at 27°C according to published procedures [37]. For the manipulation of transfection or cotransfection, Bm5 cells were cultured in 6-well plates at a constant cell density of 1x106 cells per well for 12 h using TC100 medium with FBS. Then cells were washed twice in TC100 medium without FBS and added with the mixture of transfection or cotransfection. After incubation for 4–6 h, the FBS were added into the medium for culture.
For virus amplification or expression in cells, the cells were infected at an MOI of 0.1 (multiplicity of infection of 0.1 p.f.u. per cell) for 1–2 h.
Cotransfection of the transfer vector and reBmBac for recombinant virus with foreign genes
The Bm5 cells were cultured as description. For transfection, 0.5 μg of reBmBac DNA and 1–2 μg of transfer vector DNA were mixed with 6 μL of Lipofectin in a total volume of 60 μL and incubated at room temperature for 15–30 min [38]. Cotransfection was performed with the mixture according to the Lipofectin transfection instruction manual (Invitrogen). The supernatants of cells transfected with recombinant virus was collected after culturing for 5 days at 25–27°C and stored at 4°C.
Expression of foreign genes in silkworm larvae and pupae
Fifth instar silkworm larvae or pupae were injected with recombinant viruses (approximately 105 PFU) between the abdominal knobs on the backside. Silkworm larvae and pupae were reared or incubated at 25–27°C and 65% humidity for 108–120 h. Larval haemolymph was collected by cutting the prolegs, and 1-phenyl-2-thiourea was added at final concentration of 0.1 μM to prevent melanization. Larval haemolymph and pupae were stored at -20°C for subsequent assays.
A plaque assay was performed to test the recombination efficiency using conventional methods [37]. The expression level of luciferase in 50 μg of protein lysate was assayed with a Luciferase Assay kit (Promega). The amount of protein in the lysate was measured by the Bradford method [39]. The expression of recombinant porcine IFN-γ was detected by western blotting according to the Protein Blotting Guide (Bio-Rad). The antiviral activity of recombinant porcine IFNs was assayed in a GFP-reduction assay with recombinant vesicular stomatitis virus (VSV-GFP) [40].
Results
Constructing BmBac (BmNPV shuttle vector) by inserting of the CopyControl origin of replication
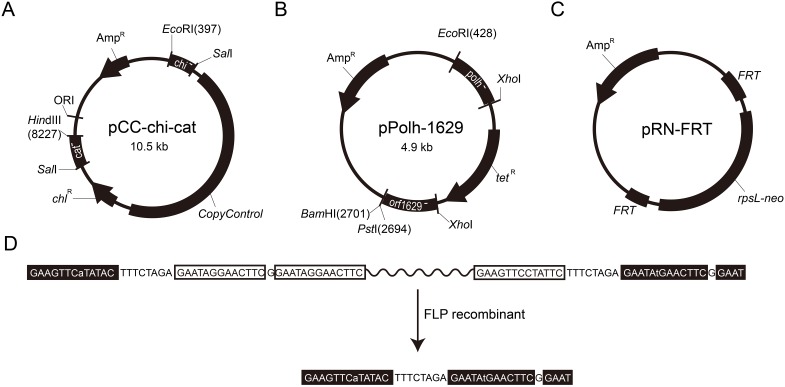
The CopyControl origin of replication in E. coli (which is formed by E. coli F factor-based partitioning, a single-copy origin of replication [41] and an inducible oriV high-copy origin of replication [42]) was chosen to construct the BmNPV shuttle vector (BmBac) which can be manipulated in E. coli. The pCC-chi-cat vector was constructed to insert this origin into the BmNPV genome and replace the cat and chi genes. Homologous arms of the cat and chi genes were present in the vector. The CopyControl origin of replication and a chloramphenicol resistance (chlR) selectable marker gene were placed between the arms. Fig 1A shows the structure of the pCC-chi-cat vector.
Fig 1. Structures of the gene-targeting vectors.
(A) The pCC-chi-cat vector was used to remove the chi and cat genes and insert the CopyControl origin. (B) The pPolh-1629 vector was used to induce a defect in the ORF1629. (C) The pRN-FRT vector contained the rpsL-neo counter-selection gene and several mutant FRT sites. (D) FRT sequence with one mutant site was recognized and reacted using FLP recombinase. However, FRT sequences with two mutant sites were inactivated. Sequences in filled boxes are mutants, and sequences in empty boxes are wild-type.
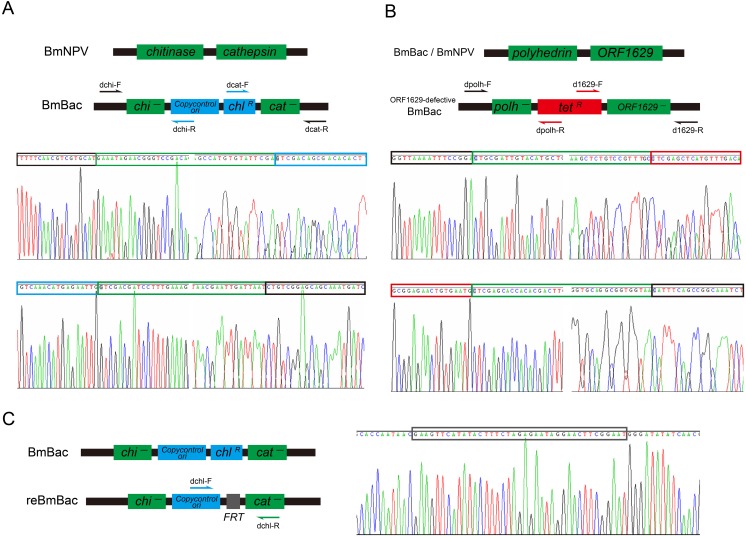
The CopyControl origin was inserted into BmNPV genomic DNA to inactivate the cat and chi genes. A 7.9 kb fragment (chlR-ori) was obtained from the pCC-chi-cat vector by EcoRI/HindIII digestion and was then purified using gel electrophoresis. A mixture of the purified fragment (1 μg) and BmNPV DNA (2 μg) was transformed by electroporation into E. coli containing Red λ recombinase. The chlR transformants contained a recombinant BmNPV in which the chi and cat genes were deleted and replaced by the origin fragment. Positive clones were identified and screened by a PCR assay. A fragment spanning the viral genome sequence and targeting vector sequence was amplified with the primers, dchi-F, dchi-R, dcat-F and dcat-R, which are listed in S1 Table. Positive clones were further confirmed by DNA sequencing (Fig 2A). The recombinant BmNPV shuttle vector was named BmBac.
Fig 2. DNA sequencing of recombinant sites in BmBac and reBmBac.
(A) The deletion of chi and cat genes in BmBac DNA was analyzed by PCR and sequencing as the schematic shown. (B) The defective ORF1629 was analyzed as schematic shown. (C) The deletion of tetR gene in reBmBac was analyzed as schematic shown. In BmBac DNA, the length of PCR product was different with that in reBmBac DNA.
BmBac DNA was extracted by the alkaline lysis method and transfected into Bm5 cells to confirm that the shuttle vector could replicate in both E. coli and insect cells. The extracted BmBac DNA was then transformed back into E. coli for the following study. The results showed that the CopyControl origin was successfully inserted into baculovirus DNA and that BmBac could reproduce in Bm cells.
The effectiveness of the CopyControl origin induction was examined to test the BmBac vector. Selected BmBac DNA was transformed into E. coli TransforMax EPI300, which carries an inducible trfA gene. L-arabinose (0.01%) was added to the E. coli host culture medium to induce DNA replication [36]. Both induced and uninduced BmBac DNA was purified and quantified using agarose gel electrophoresis. At least 5 μg of BmBac DNA was extracted from 3 mL of induced E. coli cultures, but only 0.3 μg of DNA was extracted from uninduced cultures. These results demonstrated that induction improved the DNA yield by at least tenfold.
Construction of an ORF1629-defective BmBac
The ORF1629 gene, which is located downstream of the polyhedrin gene, was partly deleted in our system on the basis of BacPAK and flashBAC strategies. Baculoviruses that lack a portion of the essential ORF1629 gene cannot replicate in insect cells [12]. Therefore, the defective ORF1629 gene can be rescued only during homologous recombination with an intact ORF1629, which allows the recombinant baculovirus to replicate in insect cells, when the foreign gene is inserted downstream of the polyhedrin gene promoter. This feature allows for more efficient acquisition of recombinant baculoviruses [12]. The pPolh-1629 vector was constructed to knock out the function of this gene. This vector contained two homologous arms of the polyhedrin and ORF1629 genes, with a tetracycline resistance gene inserted between the arms as a selectable marker (Fig 1B).
A portion of the essential ORF1629 gene was deleted from the BmBac DNA. A 2.3 kb tetR element fragment that was flanked by homologous arms of the partial polyhedrin gene and ORF1629 was obtained from pPolh-1629 via PstI/EcoRI digestion and purification. DNA (1 μg) of the fragment was used to electroporate BW25113 electrocompetent cells that contained the BmBac and phage λ Red recombinase. The tetR and chlR transformants were selected on antibiotic-containing plates. The partial polyhedrin gene and ORF1629 gene were deleted and replaced with the tetR gene via homologous recombination. The defective BmBac was also identified and screened by a PCR assay (primers d1629-F, d1629-R, dpolh-F and dpolh-R in S1 Table) and further confirmed by DNA sequencing (Fig 2B).
BmBac DNA without an essential gene replicates in E. coli cells but not insect cells. The cotransfection of reBmBac alone was performed in Bm5 cells and showed no sign of infection. However, cotransfection of the mixture containing the above-mentioned BmBac DNA and the pVL1393 transfer vector was successful. The PCR and cotransfection results confirmed that the essential ORF1629 gene-defective BmBac had been constructed successfully.
Deletion of the excessive resistance gene and construction of reBmBac
We separately introduced the chlR and tetR genes in the ORF1629 gene-defective BmBac vector in combination with the CopyControl origin of replication and partially deleted ORF1629. The tetR gene would be replaced during cotransfection, and it was not present in the recombinant baculoviruses, but the chlR gene could not be replaced. One antibiotic resistance gene is sufficient for genetic manipulation in E. coli. Thus the chlR gene should also be deleted to ensure environmental biosafety. The strategy for deleting the chlR gene involved the use of the FLP/FRT recombinase system combined with the rpsL-neo counter-selection marker. The rpsL-neo gene (GenBank: GU084141.1) flanked by FRT sites was synthesized and inserted into the pMD18 vector to construct the pRN-FRT vector (Fig 1C). The FRT sites used for this step have been optimized for high efficiency and stable knockout [43, 44]. The remaining FRT site was inactivated by FLP recombinase after recombination (Fig 1D).
The FRT-rpsL-neo element fragment was flanked by chlR deletion homologous arms by using PCR (primers, Re-chl-F and Re-chl-R, in S1 Table) and was purified through gel electrophoresis. A 1 μg sample of the fragment was transformed into BW25113 electrocompetent cells that contained the ORF1629-defective BmBac and phage λ Red recombinase. The tetR and kanR (kanamycin resistance) transformants were selected on antibiotic-containing plates. The chlR gene was replaced with the FRT-rpsL-neo fragment by homologous recombination. The positive plasmid was transformed into electrocompetent DH10B cells that contained FLP recombinase. The rpsL-neo counter-resistance gene was removed through FLP recombination, and the streptomycin antibiotic resistance of the DH10B cells was restored. Positive clones that lacked the chlR and rpsL-neo genes were selected on tetR and strR (streptomycin resistance) antibiotic-containing plates. The ORF1629 gene-defective BmBac with only one antibiotic resistance gene (tetR) was screened with a PCR assay (primers, dchl-F and dchl-R, in S1 Table) and further confirmed using DNA sequencing (Fig 2C). The deletion of chi and cat genes, introduction of CopyControl ori and the defective ORF1629 gene were verified again by using the PCR analyze and sequencing as described before. This construct was named reBmBac (GenBank accession number: KU749552).
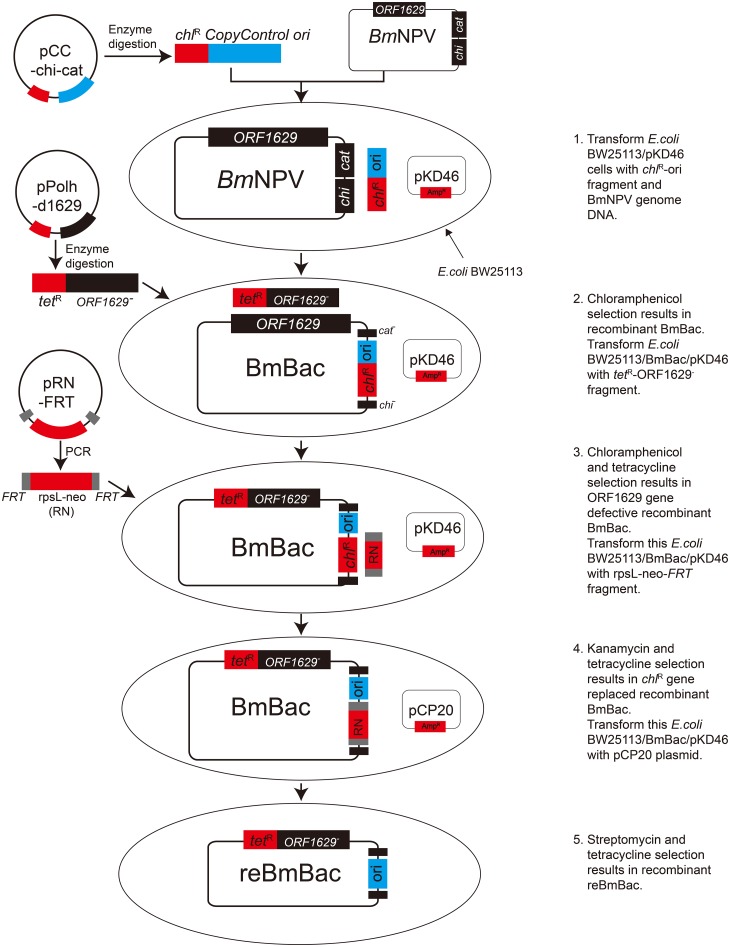
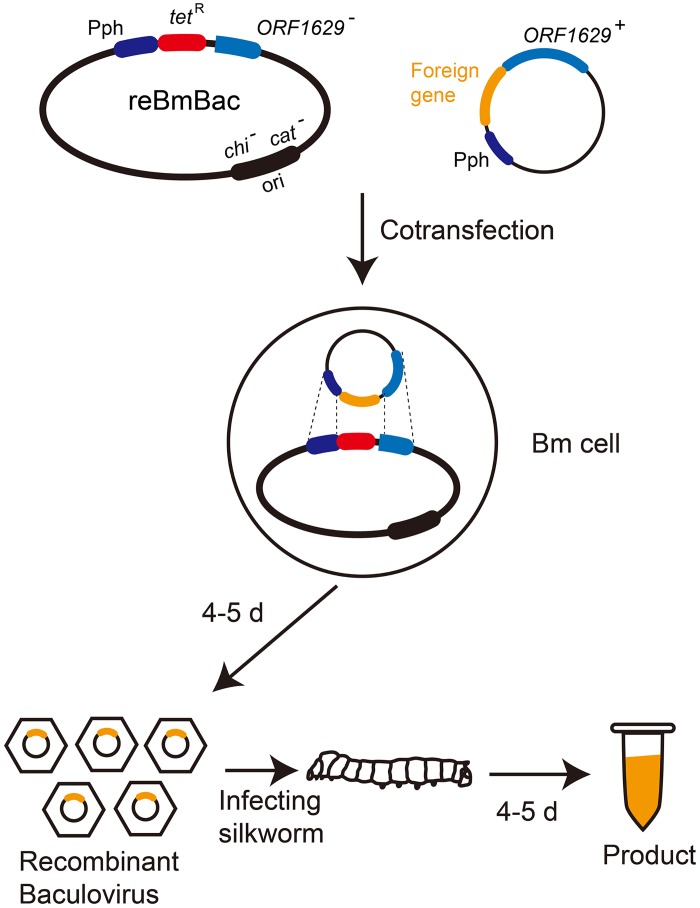
A novel reBmBac expression system was developed though the steps described above. Fig 3 illustrates an overview of this system (with reference to Airenne et al. [45]).
Fig 3. Overview of the novel reBmBac expression system (refer to Airenne et al.).
In BmBac DNA, the chi and cat genes was replaced by the Copycontrol origin and a chlR gene. Then the essential ORF1629 gene was partially deleted by using tetR gene. The chlR gene was removed by using FLP/FRT system. The finally acquired bacmid DNA was reBmBac.
Luciferase expression
The luciferase gene is one of the most sensitive reporter genes, and it is remarkably easy to detect. Therefore, this gene was used to test the reBmBac expression system. The luciferase gene was obtained from the pGL3-Basic vector by using BglII/XbaI digestion and was transferred into the BamHI/XbaI-digested pVL1393 vector to construct the pVL1393-luc vector. A mixture of pVL1393-luc and reBmBac DNA was cotransfected into Bm5 cells. The supernatant, which contained recombinant BmNPV (reBm-luc), was collected after 4–5 days of incubation, and it was used to infect silkworms. A 50 μg sample of protein from lysed cells or larval haemolymph was used in the luciferase assay. The luminescence of the cotransfected cells was approximately 4.86±0.47×106 RLU (relative light unit), and the luminescence of silkworm larval haemolymph was approximately 3.42±0.52×108 RLU. The background luminescence of the cell culture and larval haemolymph from luc-negative virus-infected samples was 150–300 RLU (S1 Fig).
We continuously amplified the recombinant reBm-luc from cotransfected viral stocks in Bm5 cells for three rounds, and infected silkworms to verify the purity of the recombinant baculovirus. If the recombinant BmNPV is not pure, the expression levels of luciferase in cells and larvae will reduced to 10% or even less in the first two rounds of amplification. Here, the luciferase expression in silkworms was not significantly different from that of the original viral stocks. This result indicated that the stable deletion of ORF1629 and the use of CopyControl ori ensured the efficiency of recombinant BmNPV in cotransfection. It also showed that the viral stock acquired during cotransfection was pure to be directly used for viral amplification and protein expression. There was scarcely any wild baculovirus mixing in the target recombinant virus. S1A Fig shows the luminescence data from cells and larvae in the three rounds of amplification and expression. Therefore, the recombinant virus acquired in cotransfection was pure and with stably expression level of luciferase.
Twenty-four reBm-luc plaques were isolated and used to infect Bm5 cells cultured in 24-well plates to measure the recombination efficiency. Viral supernatants were collected from 24 dishes after 108–120 h of incubation. Groups of ten silkworm larvae were injected with the supernatant samples. Luciferase activity was detected in each of the 24 samples by a luminescence assay. The luminescence of 50 μg of protein from infected larval haemolymph samples that had been lysed ranged from 5×107 to 5×108 RLU (S1 Fig). All 24 plaques were successful recombinant BmNPVs that contained the luciferase gene. The purity of the recombinant virus harvested via cotransfection approached 100%.
These results demonstrated that the reBmBac expression system could be used to express foreign genes successfully. The recombinant BmNPV in the cotransfection viral stock could be used directly for expression.
Expression of recombinant porcine interferon-γ
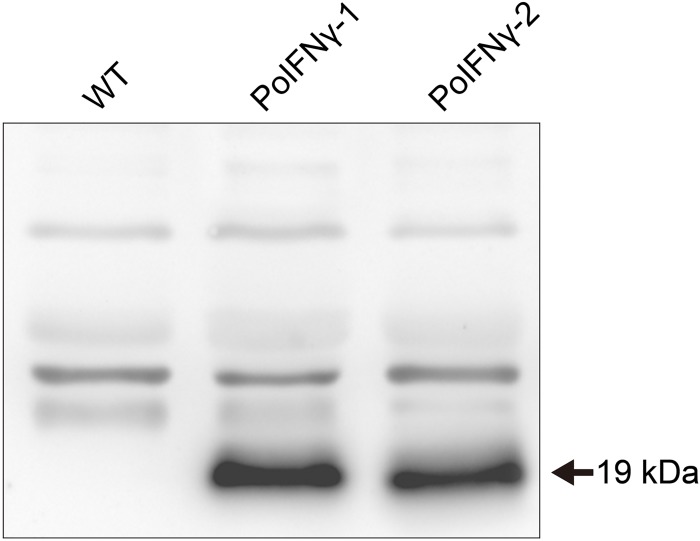
PoIFN-γ gene expression was tested to further examine the utility of the reBmBac expression system. First, the PoIFN-γ gene was cloned into the transfer vector pVL1393. Second, the gene was inserted into the viral genome to construct recombinant reBm-PoIFN-γ via homologous recombination. The recombinant reBm-PoIFN-γ virus was injected into larvae or pupae. The expression product of porcine IFN-γ was analysed by western blotting (Fig 4) with an anti-PoIFN-γ antibody (Millipore, USA). Fig 4 shows that an approximately 19 kDa protein band that reacted with the anti-PoIFN-γ antibody was detected in the samples. No corresponding immunoreactive protein was detected in the negative control sample from larval haemolymph infected with WT BmNPV. Therefore, these results indicate that porcine IFN-γ was successfully expressed in silkworms.
Fig 4. Western blot analyses of PoIFN-γ expression in silkworm larvae.
The PoIFNγ-1 sample is the larval haemolymph infected with one of 24 plaques of recombinant virus stock, and the PoIFNγ-2 sample is the larval haemolymph infected with the recombinant virus acquired during cotransfection. PoIFN-γ proteins were detected in PoIFNγ-1 and PoIFNγ-2 samples as an approximately 19 kDa band. No corresponding immunoreactive protein was detected in wild-type (WT) samples.
The antiviral activity assay indicated that the reBm-PoIFN-γ product exhibited antiviral activity that exceeded 1 x 106 IU/mL in haemolymph. Twenty-four plaques were also isolated and used to infect silkworms. The antiviral activity assay demonstrated that the activity of these plaque samples were approximately 6.2 ± 1.3×105 to 2.4 ± 0.7×106 IU/mL of haemolymph (S2 Fig). The highest antiviral activity was 2.4 ± 0.7x106 IU/mL, which represented a twofold improvement in antiviral activity in comparison with the cotransfected sample. The activity of PoIFN-γ in one mililiter of haemolymph from the best sample was equal to that of 271.4 ± 81.5 μg standard recombinant porcine IFN-γ (R&D Systems, USA).
Discussion
The above data demonstrated the successful development of the novel defective-rescue reBmBac expression system. Recombinant BmNPV viral stocks were obtained within 4–5 days with this system, and recombinant proteins expressed in silkworm larvae or pupae were harvested after an additional 4–5 days (Fig 5). This system is remarkably efficient. Many experiments have shown that the expression level of a heterologous protein in five silkworm larvae or pupae is approximately equal to the expression levels of fermentation products from 1 L of sf9-AcMNPV cells [7]. Therefore, this reBmBac-silkworm expression system is a rapid and highly efficient system for producing large amounts of foreign protein.
Fig 5. Schematic for the generation of recombinant viruses and expression of interested foreign genes.
Recombinant viruses containing foreign genes were generated 4–5 days after cotransfection. The recombinant protein was harvested 4–5 days after silkworm infection.
The BmNPV-silkworm expression system has been reported that it is a powerful and potential system for foreign gene expression. Scientists have long attempted to develop more convenient methods for generation of recombinant BmNPV and high-level expression of recombinant proteins in silkworm. The recombinant baculovirus generation strategies of BmNPV are similar to those corresponding ones of AcMNPV [46]. The Bac-to-Bac system and the BacPAK system are two typical and commonly used baculovirus expression system. A foreign gene can be inserted into BmNPV baculovirus DNA in E. coli by using the Bm-Bac-to-Bac system, which is a convenient and easy-to-used expression vector for recombinant baculovirus construction and expression [17]. There were studies shown that the recombinant Bacmid DNA or special E. coli containing Bacmid could be used to inject the silkworm directly for expression, but the expression efficiency and level were not ideal [17, 47]. The Bac-to-Bac system is widely used in the BEVS expression fields, which benefit from the convenience and stabilization of its operation in E. coli. However, this system is also relatively inconvenient for the heterologous expression of many genes because each gene must be manipulated independently in E. coli. In the BmBacPAK system, there are several Bsu36I sites in baculovirus DNA [48]. The Bsu36I digested baculovirus DNA is linear and lacks part of ORF1629. Incomplete enzyme digestion dramatically reduces the ratio of recombinant virus. In comparison with these two BEVSs, the reBmBac expression system combines their advantages, i.e., convenient viral genome DNA preparation in E. coli and easy recombinant baculovirus production via cotransfection. The stable deletion of the essential ORF1629 gene in E. coli beforehand made the digestion step could be omitted and ensured the efficiency and purity of recombinant baculoviruses that were obtained by cotransfection. In addition, some disadvantages of these two BEVSs were avoided. It always takes at least two weeks to produce one recombinant baculovirus using Bac-to-Bac BEVS, whereas it takes only approximately five days when using reBmBac BEVS to produce various recombinant baculoviruses that contain different genes of interest at the same time. The batch production of different baculoviruses can be achieved conveniently via one-step cotransfection, and thus, a large amount of time is saved. It is much more convenient to prepare a large amount of high-quality parental viral genome DNA by using the reBmBac BEVS rather than the BmBacPAK system because of the CopyControl ori [33]. Besides, the ORF1629-defective Bacmid bBpGOZA expression system is another easy-to-use system [19]. Our reBmBac system has two major improvements relative to this system. First, the fragment replaced during homologous recombination in our reBmBac system is only a 1.2 kb tetR gene. In contrast, the fragment in bBpGOZA is more than 6 kb, and it contains a miniF replicon and a kanR gene. Second, the CopyControl origin in our system facilitates the large-scale preparation of high-quality reBmBac DNA. Our results over the course of many experiments indicated that the high quality of reBmBac DNA and transfer plasmid DNA are key factors for achieving good recombination results. We observed a greater than fivefold difference in the luciferase expression level and the expression levels of other proteins of interest depending on the quality of the DNA (S2 Table). The quality of well-preserved (fresh or store at -20°C) and bad-preserved (freeze-thawed for five rounds) transfer vectors are different. And the quality of parent reBmBac viral DNA, extracted from E. coli by induction or non-induction methods, are different. Over many protein expression trials in this reBmBac system, we found that when we used the same amount but different quality of pVL1393-luc transfer vector DNA to mix with the same tube of reBmBac DNA and to cotransfect cells, the luminescence from the cells and larvae could reach a tenfold difference. When we use the same amount but different quality of reBmBac DNA to mix with the same tube of well-preserved pVL1393-luc vector and to carry out the cotransfection, the luminescence from cells and larvae were at least fivefold different.
The expression tests of luciferase and PoIFN-γ revealed that the purity of the recombinant viruses acquired by cotransfection was sufficient to directly infect silkworms for expression. The amount of recombinant virus from one cotransfection was sufficient to infect at least 5,000 silkworm larvae or pupae. The expression was sufficiently high to perform functional research on the expressed protein. Furthermore, the three continuous rounds of viral reproduction demonstrated that the efficiency and purity of recombinant BmNPV were sufficient for virus amplification and protein expression. The plaque assays of reBm-luc and reBm-PoIFN-γ showed that the reBmBac system allows 100% recombinant BmNPV formation. Combining the three rounds of amplification test, the direct expression in silkworms obviates the need for further virus plaque purification assay. Therefore, the time required from expression plasmid production to harvesting a large amount recombinant protein is only 10 days, in theory. Obtaining recombinant virus requires only a simple cotransfection, which could be performed in batch mode and would be suitable for the expression of different foreign proteins after batches. In general, we performed 8–12 cotransfections in two 6-well cell culture plates (6 x 35 mm) for one batch and used the luciferase reporter to evaluate the recombination and expression efficiency in silkworms. We were confident that a group of recombinations and gene expression trial in the silkworms was successful when the luminescence of the cell lysate from the cotransfection reached 2 x 106 RLU or greater, and the larval haemolymph achieved 5 x 107 RLU or greater. These features are beneficial for the rapid and efficient expression of foreign proteins.
The three rounds of amplification and infection of reBm-luc in cells and silkworms showed that the luciferase expression level was stable by using this reBmBac system. We have used the uninduced reBmBac DNA to generate the luciferase gene recombinant BmNPV. The luminescence reduced dozens-fold in the first round of amplification and expression in larvae, which indicated that the recombinant BmNPV acquired in cotransfection is not pure and not suitable for directly expressing in larvae. The same phenomenon sometimes appears in the using of BacPAK6 system. Here, the stable expression data in the three rounds indicated that the reBm-luc acquired in cotransfection was pure and could be used for expression directly. In the plaque assay of reBm-luc, all of the screened virus stocks drive positive expression of luciferase. But the expression levels of them were steadily different. This is because of the reBm-luc acquired in cotransfection is a mixture of various types of recombinant viruses, while the screened virus stock is single clone recombinant virus in theory. Therefore, the expression level of luciferase of 24 plaque stocks were higher or lower than that of contransfection virus stock. And the plaque assay of reBm-PoIFN-γ also verified this result. Even so, the several virus stocks that drive higher expression levels could be selected for further researches or large-scale expression of target genes. Generally, the expression levels of these optimized recombinant virus stocks could be at least twofold higher than that of cotransfection stock. This optimization of expression level is recommended step for further large-scale producing.
The introduction of the CopyControl replicon facilitates the generation of large-plasmid DNA in E. coli. Wild [33] and the CopyControl BAC cloning kit instructions indicate that the copy number of the vectors, containing CopyControl origin, depends on the vectors’ size. The copy number for Bacmids could increase by 10- to 20-fold upon induction. The reBmBac DNA yield was similar in our system. The oriV origin of replication should be silent during genetic manipulations in baculovirus genome. The single-copy replicon allows for the autonomous replication and stable segregation of plasmids at a low number, which facilitates positive clone selection. The high-copy oriV origin of replication is induced when it is necessary to prepare Bacmid DNA for cotransfection. High-quality reBmBac DNA, which can be prepared easily on a large scale, is one of the key factors for successful homologous recombination. The above mentioned results showed that the quantity of reBmBac DNA extracted from 3 mL of overnight culture medium was sufficient for performing at least 10 cotransfections.
Recombinant BmNPV that contained foreign genes contained no antibiotic resistance gene in our system, which provides improved environmental biosafety. The tetR gene was replaced with a foreign gene during cotransfection. The chlR gene was deleted with the FLP/FRT system and the rpsL-neo counter-resistance selection system. The deletion of the chlR gene was also an example of the genetic manipulation of viral genes or the introduction of foreign genes into the other baculoviral genome sites in E. coli. This makes the multigene expression can also be achieved by using our reBmBac expression system as the using of other published system in silkworms [49]. The FRT site used here is a mutant site [43, 44]. The remaining FRT sequence is inactivated after recombination, which is favourable for ensuring the stability of the viral genome.
Conclusions
A novel defective-rescue reBmBac expression system was successfully developed. The examples of luciferase and PoIFN-γ expression showed that foreign genes can be successfully expressed with this system. These results show that the novel reBmBac silkworm expression system is excellent and has the following advantages: (i) This system allows for simple and rapid generation of recombinant BmNPVs carrying foreign genes. Because the defective ORF1629 and the introduction of the CopyControl origin of replication, the purity of the recombinant virus harvested during cotransfection approaches 100%. The luciferase expression level could be used as the quality control for the cotransfection and expression efficiency of foreign genes in larvae. The amount and quality of recombinant viral stock is sufficient for various research applications. Plaque purification can be used to further improve the expression level but is not a necessary step. (ii) This system facilitates the batch generation of recombinant BmNPV with various foreign genes, thus allowing for the mass production of proteins of interest. (iii) No antibiotic resistance gene is present in recombinant BmNPV, and thus the biosafety of this system is ensured.
Supporting Information
(XLSX)
The luminescence of 50 μg of proteins lysed from cells and larval haemolymph indicates the expression of luciferase. (A) Luciferase expression in cells that were consecutively passaged for three rounds is stable. The three rounds of expression in larval haemolymph were not obviously different from the original viral stocks. The Cells-Control and Larvae-Control were the cells and larvae samples which were infected with non-luc recombinant BmNPV. (B) Luciferase expression of 24 plaques samples in silkworm was detected, and the luminescence of the best sample achieved 5.1 ± 1.0×108 RLU/50 μg protein. This result indicates that the expression of foreign protein was doubled compared with the cotransfection sample. The “M-C” bar was the mock infected control sample. The “N-C” bar was the non-luciferase recombinant BmNPV infected control sample.
(TIF)
The product of reBm-PoIFN-γ (cotransfection viral stock) exhibited antiviral activity that exceeded 1 x 106 IU/mL haemolymph (“co-t” bar). 24 plaque viral stocks were screened using plaque assay and used to infect silkworms. The antiviral activity of the best sample was 2.4 ± 0.7x106 IU/mL, which exhibited a twofold improvement in antiviral activity compared with the cotransfection sample. The commercial positive control (“c-1” bar) sample was the standard recombinant porcine IFN-γ (R&D Systems, USA). The standard sample was reconstituted at 50 μg/mL and its antiviral activity was about 4.5 x 106 IU/mL. The negative control (“c-2” bar) sample was the non-interferon recombinant BmNPV infected larvae and it shown no antiviral activity. The activity of PoIFN-γ in one milliliter haemolymph of the best sample (in our system) was equal to that of 271.4 ± 81.5 ug standard control sample.
(TIF)
(DOC)
(DOC)
Acknowledgments
This work was supported by the National High-tech R&D Program of China (“863” Program, No.2011AA100603, http://www.863.gov.cn), the National Program of Key Basic Research project of China (“973” Program, 2012CB114600, http://www.973.gov.cn) and the National Natural Sciences Foundation of China (No. 31200275, http://www.nsfc.gov.cn).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National High-Tech R&D Program of China (“863” Program, No. 2011AA100603, http://www.863.gov.cn), the National Program of Key Basic Research project of China (“973” Program, 2012CB114600, http://www.973.gov.cn) and the National Natural Sciences Foundation of China (No. 31200275, http://www.nsfc.gov.cn). ZZ received all of the fundings.
References
- 1.Luckow VA. Baculovirus systems for the expression of human gene products. Curr Opin Biotechnol. 1993;4(5):564–72. 10.1016/0958-1669(93)90078-B . [DOI] [PubMed] [Google Scholar]
- 2.Merrington CL, Bailey MJ, Possee RD. Manipulation of baculovirus vectors. Mol Biotechnol. 1997;8(3):283–97. 10.1007/BF02760782 . [DOI] [PubMed] [Google Scholar]
- 3.Felberbaum RS. The baculovirus expression vector system: A commercial manufacturing platform for viral vaccines and gene therapy vectors. Biotechnol J. 2015;10(5):702–14. 10.1002/biot.201400438 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith GE, Summers MD, Fraser MJ. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol. 1983;3(12):2156–65. 10.1128/MCB.3.12.2156 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reis U, Blum B, von Specht BU, Domdey H, Collins J. Antibody production in silkworm cells and silkworm larvae infected with a dual recombinant Bombyx mori nuclear polyhedrosis virus. Biotechnology (N Y). 1992;10(8):910–2. 10.1038/nbt0892-910 . [DOI] [PubMed] [Google Scholar]
- 6.O'Reilly DR, Miller LK, Luckow VA. Baculovirus expression vectors: a laboratory manual: Oxford University Press; 1994. [Google Scholar]
- 7.Usami A, Ishiyama S, Enomoto C, Okazaki H, Higuchi K, Ikeda M, et al. Comparison of recombinant protein expression in a baculovirus system in insect cells (Sf9) and silkworm. J Biochem. 2011;149(2):219–27. 10.1093/jb/mvq138 . [DOI] [PubMed] [Google Scholar]
- 8.Burgess S. Molecular weights of Lepidopteran baculovirus DNAs: derivation by electron microscopy. J Gen Virol. 1977;37(3):501–10. 10.1099/0022-1317-37-3-501 [DOI] [Google Scholar]
- 9.Patel G, Nasmyth K, Jones N. A new method for the isolation of recombinant baculovirus. Nucleic Acids Res. 1992;20(1):97–104. 10.1093/nar/20.1.97 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luckow VA. Cloning and expression of heterologous genes in insect cells with baculovirus vectors. Recombinant DNA technology and applications. 1991;97:97–152. [Google Scholar]
- 11.Luckow VA, Lee SC, Barry GF, Olins PO. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol. 1993;67(8):4566–79. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitts PA, Possee RD. A method for producing recombinant baculovirus expression vectors at high frequency. BioTechniques. 1993;14(5):810–7. . [PubMed] [Google Scholar]
- 13.Je YH, Chang JH, Choi JY, Roh JY, Jin BR, O'Reilly DR, et al. A defective viral genome maintained in Escherichia coli for the generation of baculovirus expression vectors. Biotechnol Lett. 2001;23(8):575–82. [Google Scholar]
- 14.Zhao Y, Chapman DA, Jones IM. Improving baculovirus recombination. Nucleic Acids Res. 2003;31(2):E6- . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noad RJ, Stewart M, Boyce M, Celma CC, Willison KR, Roy P. Multigene expression of protein complexes by iterative modification of genomic Bacmid DNA. BMC Mol Biol. 2009;10:87 10.1186/1471-2199-10-87 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol. 2004;22(12):1583–7. 10.1038/nbt1036 . [DOI] [PubMed] [Google Scholar]
- 17.Motohashi T, Shimojima T, Fukagawa T, Maenaka K, Park EY. Efficient large-scale protein production of larvae and pupae of silkworm by Bombyx mori nuclear polyhedrosis virus bacmid system. Biochem Biophys Res Commun. 2005;326(3):564–9. 10.1016/j.bbrc.2004.11.060 . [DOI] [PubMed] [Google Scholar]
- 18.Wu X, Kamei K, Sato H, Sato SI, Takano R, Ichida M, et al. High-level expression of human acidic fibroblast growth factor and basic fibroblast growth factor in silkworm (Bombyx mori L.) using recombinant baculovirus. Protein Expr Purif. 2001;21(1):192–200. 10.1006/prep.2000.1358 . [DOI] [PubMed] [Google Scholar]
- 19.Je YH, Chang JH, Kim MH, Roh JY, Jin BR, O'Reilly DR. The use of defective Bombyx mori nucleopolyhedrovirus genomes maintained in Escherichia coli for the rapid generation of occlusion-positive and occlusion-negative expression vectors. Biotechnol Lett. 2001;23(21):1809–17. [Google Scholar]
- 20.Possee RD, Hitchman RB, Richards KS, Mann SG, Siaterli E, Nixon CP, et al. Generation of baculovirus vectors for the high-throughput production of proteins in insect cells. Biotechnol Bioeng. 2008;101(6):1115–22. 10.1002/bit.22002 . [DOI] [PubMed] [Google Scholar]
- 21.Yao LG, Liu ZC, Zhang XM, Kan YC, Zhou JJ. A highly efficient method for the generation of a recombinant Bombyx mori nuclear-polyhedrosis-virus Bacmid and large-scale expression of foreign proteins in silkworm (B. mori) larvae. Biotechnol Appl Biochem. 2007;48(Pt 1):45–53. 10.1042/BA20070017 . [DOI] [PubMed] [Google Scholar]
- 22.Jarvis DL. Baculovirus-insect cell expression systems. Methods Enzymol. 2009;463:191–222. 10.1016/S0076-6879(09)63014-7 . [DOI] [PubMed] [Google Scholar]
- 23.Blissard GW, Rohrmann GF. Baculovirus diversity and molecular biology. Annu Rev Entomol. 1990;35(1):127–55. [DOI] [PubMed] [Google Scholar]
- 24.Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202(2):586–605. 10.1006/viro.1994.1380 . [DOI] [PubMed] [Google Scholar]
- 25.Ahrens CH, Russell RL, Funk CJ, Evans JT, Harwood SH, Rohrmann GF. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology. 1997;229(2):381–99. 10.1006/viro.1997.8448 . [DOI] [PubMed] [Google Scholar]
- 26.Gomi S, Majima K, Maeda S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Virol. 1999;80 (Pt 5):1323–37. 10.1099/0022-1317-80-5-1323 . [DOI] [PubMed] [Google Scholar]
- 27.Hawtin RE, Zarkowska T, Arnold K, Thomas CJ, Gooday GW, King LA, et al. Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology. 1997;238(2):243–53. 10.1006/viro.1997.8816 . [DOI] [PubMed] [Google Scholar]
- 28.Lee KS, Je YH, Woo SD, Sohn HD, Jin BR. Production of a cellulase in silkworm larvae using a recombinant Bombyx mori nucleopolyhedrovirus lacking the virus-encoded chitinase and cathepsin genes. Biotechnol Lett. 2006;28(9):645–50. 10.1007/s10529-006-0030-7 . [DOI] [PubMed] [Google Scholar]
- 29.Park EY, Abe T, Kato T. Improved expression of fusion protein using a cysteine-protease- and chitinase-deficient Bombyx mori (silkworm) multiple nucleopolyhedrovirus bacmid in silkworm larvae. Biotechnol Appl Biochem. 2008;49:135–40. 10.1042/Ba20070098 WOS:000253080900006. [DOI] [PubMed] [Google Scholar]
- 30.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–5. 10.1073/pnas.120163297 WOS:000087526300074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Muyrers JP, Rientjes J, Stewart AF. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Mol Biol. 2003;4(1):1 10.1186/1471-2199-4-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuntufye HN, Goddeeris BM. Use of lambda Red-mediated recombineering and Cre/lox for generation of markerless chromosomal deletions in avian pathogenic Escherichia coli. FEMS Microbiol Lett. 2011;325(2):140–7. 10.1111/j.1574-6968.2011.02421.x . [DOI] [PubMed] [Google Scholar]
- 33.Wild J, Hradecna Z, Szybalski W. Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Res. 2002;12(9):1434–44. 10.1101/gr.130502 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Possee RD, Sun TP, Howard SC, Ayres MD, Hill-Perkins M, Gearing KL. Nucleotide sequence of the Autographa californica nuclear polyhedrosis 9.4 kbp EcoRI-I and -R (polyhedrin gene) region. Virology. 1991;185(1):229–41. 10.1016/0042-6822(91)90770-C . [DOI] [PubMed] [Google Scholar]
- 35.Dijkmans R, Vandenbroeck K, Beuken E, Billiau A. Sequence of the porcine interferon-gamma (IFN-gamma) gene. Nucleic Acids Res. 1990;18(14):4259 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–30. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Summers MD, Smith GE. A manual of methods for baculovirus vectors and insect cell culture procedures: Texas Agricultural Experiment Station; 1987. [Google Scholar]
- 38.Hartig PC, Cardon MC, Kawanishi CY. Generation of recombinant baculovirus via liposome-mediated transfection. BioTechniques. 1991;11(3):310, 2–3. . [PubMed] [Google Scholar]
- 39.Kruger NJ. The Bradford method for protein quantitation. Methods Mol Biol. 1994;32:9–15. 10.1385/0-89603-268-X:9 . [DOI] [PubMed] [Google Scholar]
- 40.Ge J, Wen Z, Wang X, Hu S, Liu Y, Kong X, et al. Generating vesicular stomatitis virus pseudotype bearing the severe acute respiratory syndrome coronavirus spike envelope glycoprotein for rapid and safe neutralization test or cell-entry assay. Ann N Y Acad Sci. 2006;1081:246–8. 10.1196/annals.1373.030 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim UJ, Birren BW, Slepak T, Mancino V, Boysen C, Kang HL, et al. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34(2):213–8. 10.1006/geno.1996.0268 . [DOI] [PubMed] [Google Scholar]
- 42.Stalker DM, Thomas CM, Helinski DR. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):8–12. 10.1007/BF00338997 . [DOI] [PubMed] [Google Scholar]
- 43.Senecoff JF, Rossmeissl PJ, Cox MM. DNA recognition by the FLP recombinase of the yeast 2 mu plasmid. A mutational analysis of the FLP binding site. J Mol Biol. 1988;201(2):405–21. DNA recognition by the FLP recombinase of the yeast 2 mu plasmid. A mutational. . [DOI] [PubMed] [Google Scholar]
- 44.Lyznik LA, Rao KV, Hodges TK. FLP-mediated recombination of FRT sites in the maize genome. Nucleic Acids Res. 1996;24(19):3784–9. 10.1093/nar/24.19.3784 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Airenne KJ, Peltomaa E, Hytonen VP, Laitinen OH, Yla-Herttuala S. Improved generation of recombinant baculovirus genomes in Escherichia coli. Nucleic Acids Res. 2003;31(17):e101 10.1093/nar/gng102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jarvis DL. Baculovirus-insect cell expression systems. Methods in Enzymol. 2009;463:191–222. 10.1016/S0076-6879(09)63014-7 . [DOI] [PubMed] [Google Scholar]
- 47.Sun J, Yao L, Yao N, Xu H, Jin P, Kan Y. Production of recombinant Bombyx mori nucleopolyhedrovirus in silkworm by intrahaemocoelic injection with invasive diaminopimelate auxotrophic Escherichia coli containing BmNPV–Bacmid. Biotechnol Appl Biochem. 2010;57(3):117–25. 10.1042/BA20100148 [DOI] [PubMed] [Google Scholar]
- 48.Wu X, Yang G, Hu J. A recombinant rescue linearizable BmNPV baculovirus BmBacPAK. China Patent (application No 981109632). 1998. [Google Scholar]
- 49.Yao L, Wang S, Su S, Yao N, He J, Peng L, et al. Construction of a baculovirus-silkworm multigene expression system and its application on producing virus-like particles. PLoS One. 2012;7(3):e32510 10.1371/journal.pone.0032510 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
The luminescence of 50 μg of proteins lysed from cells and larval haemolymph indicates the expression of luciferase. (A) Luciferase expression in cells that were consecutively passaged for three rounds is stable. The three rounds of expression in larval haemolymph were not obviously different from the original viral stocks. The Cells-Control and Larvae-Control were the cells and larvae samples which were infected with non-luc recombinant BmNPV. (B) Luciferase expression of 24 plaques samples in silkworm was detected, and the luminescence of the best sample achieved 5.1 ± 1.0×108 RLU/50 μg protein. This result indicates that the expression of foreign protein was doubled compared with the cotransfection sample. The “M-C” bar was the mock infected control sample. The “N-C” bar was the non-luciferase recombinant BmNPV infected control sample.
(TIF)
The product of reBm-PoIFN-γ (cotransfection viral stock) exhibited antiviral activity that exceeded 1 x 106 IU/mL haemolymph (“co-t” bar). 24 plaque viral stocks were screened using plaque assay and used to infect silkworms. The antiviral activity of the best sample was 2.4 ± 0.7x106 IU/mL, which exhibited a twofold improvement in antiviral activity compared with the cotransfection sample. The commercial positive control (“c-1” bar) sample was the standard recombinant porcine IFN-γ (R&D Systems, USA). The standard sample was reconstituted at 50 μg/mL and its antiviral activity was about 4.5 x 106 IU/mL. The negative control (“c-2” bar) sample was the non-interferon recombinant BmNPV infected larvae and it shown no antiviral activity. The activity of PoIFN-γ in one milliliter haemolymph of the best sample (in our system) was equal to that of 271.4 ± 81.5 ug standard control sample.
(TIF)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.