Abstract
Charcot–Marie–Tooth (CMT) disease is a genetically heterogeneous group of peripheral neuropathies. Mutations in several aminoacyl-tRNA synthetase (ARS) genes have been implicated in inherited CMT disease. There are 12 reported CMT-causing mutations dispersed throughout the primary sequence of the human glycyl-tRNA synthetase (GARS). While there is strong genetic evidence linking GARS mutations to CMT disease, the molecular pathology underlying the neuromuscular and sensory phenotypes is still not fully understood. In particular, it is unclear whether the mutations result in a toxic gain of function, a partial loss of activity related to translation, or a combination of these mechanisms. We identified a zebrafish allele of gars (garss266). Homozygous mutant embryos carry a C->A transversion, that changes a threonine to a lysine, in a residue next to a CMT-associated human mutation. We show that the neuromuscular phenotype observed in animals homozygous for T209K Gars (T130K in GARS) is due to a loss of dimerization of the mutated protein. Furthermore, we show that the loss of function, dimer-deficient and human disease-associated G319R Gars (G240R in GARS) mutant protein is unable to rescue the above phenotype. Finally, we demonstrate that another human disease-associated mutant G605R Gars (G526 in GARS) dimerizes with the remaining wild-type protein in animals heterozygous for the T209K Gars and reduces the function enough to elicit a neuromuscular phenotype. Our data indicate that dimerization is required for the dominant neurotoxicity of disease-associated GARS mutations and provide a rapid, tractable model for studying newly identified GARS variants for a role in human disease.
Introduction
Charcot–Marie–Tooth (CMT) disease is a genetically heterogeneous group of peripheral neuropathies characterized by impaired sensation and progressive muscle atrophy in the upper and lower extremities. Pathophysiological studies separate CMT disease into two major categories: type I (CMT1 or demyelinating CMT disease) and type II (CMT2 or axonal CMT disease). The estimated prevalence of CMT disease is 1 in 2500 individuals worldwide (1). There is an overlap between CMT2 and a similar group of clinical syndromes classified as distal hereditary motor neuropathy or distal spinal muscular atrophy (dSMA) (2). To date, over 1000 different mutations have been discovered in 80 disease-associated genes. Identification of the genetic causes of CMT elucidated conserved mechanisms that explain the myelin degeneration in CMT1 and axon degeneration in CMT2. Disruption of the myelin structure through mutation of proteins produced by Schwann cells is a recurring mechanism in CMT1. Defects associated with vesicle trafficking (3), mitochondrial morphology (4) and cytoskeletal integrity (5,6) have been implicated in multiple axonal forms of CMT. Interestingly, mutations in six genes encoding enzymes indispensable for protein synthesis [aminoacyl-tRNA synthetase (ARSs)] have been implicated in axonal CMT2 (7–11).
The canonical activity of ARSs is to charge amino acids onto their cognate tRNA molecules in the cytoplasm and mitochondria (12,13), the first step of protein translation, to maintain the fidelity of the genetic code. There are 37 ARS genes in the mammalian nuclear genome that code for 17 cytoplasmic and 17 mitochondrial enzymes. In three cases, a single gene encodes both cytoplasmic and mitochondrial forms of the protein by using alternatively spliced exons or alternative initiation codons, such as the glycyl-tRNA synthetase (GARS) (14). Human GARS functions as a homodimer with the monomer unit having 685 residues and is composed of an N-terminal appended WHEP-TRS domain, a catalytic core and a C-terminal anticodon binding domain (15). The catalytic domain contains the characteristic three conserved motifs of class II tRNA synthetases. There are 12 reported CMT-causing mutations dispersed throughout the primary sequence of human GARS (16,17). Subsequently, mutations in the tyrosyl-(YARS), alanyl-(AARS), histidyl-(HARS) and methionyl (MARS)—tRNA synthetases have been implicated in inherited CMT disease with an axonal pathology (7–11).
Three mouse models of GARS mutations were generated and have been used extensively to study CMT2D onset and progression (18,19). Two of those cause dominant phenotypes due to missense mutations in Gars (GarsNmf249 and GarsC201R) and share pathological features with the human disease. The GarsNmf249 allele causes reduced body weight, loss of larger diameter axons, partial or complete denervation of neuromuscular junctions (NMJs), and lethality by 8 weeks of age (18). The GarsC201R allele, identified in an N-ethyl-N-nitrosourea (ENU) mutagenesis screen, is considerably less severe. Those mice exhibit impaired grip strength, reduced weight and show a trend toward smaller axon diameters (19). Homozygous GarsC201R mice are sub-viable and homozygous GarsNmf249 are embryonic lethal. The embryonic lethality observed in these Gars alleles is probably not related to the neuropathy phenotype, as GARS is necessary for translation in every cell, and loss of function in non-neurological tissue is a likely cause of the embryonic lethality. Moreover, mice heterozygous for a complete null allele (GarsXM256) do not present any phenotype, ruling out haploinsufficiency (18).
A Drosophila gars mutant was identified in a forward genetic screen for dendrite and axon formation and arborization, using the mosaic analysis with a repressible cell marker system. The Drosophila mutation causes a missense change equivalent to a P98L in the human protein. The homozygous phenotype in flies is probably due to impaired translation and it was proposed that the axons and the dendrites have a greater sensitivity to reduced protein synthesis (20).
Finally, structural studies in the human protein have shown that many of the mutations alter dimer formation that is essential for tRNA-charging activity, such as L129P and G240R (17). Peripheral nerves with long, large-diameter axons may be especially vulnerable to decreased activity because of their unusual transport and metabolic demands, suggesting that a reduction of tRNA-charging function could result in neuropathy. Consistent with this, the majority of disease-causing mutations in GARS cause reduced tRNA charging in cell-free assays, which roughly correlates with a decreased ability to support viability in yeast cells deleted for GRS1, the ortholog of GARS (16). Furthermore, some mutations alter the subcellular distribution of GARS in transfected neurons, suggesting a possible loss of function at the cellular level through mislocalization, even if the charging activity is preserved (7,16). While there is strong genetic evidence linking GARS mutations to CMT disease, the molecular pathology that underlies the progression of the neuromuscular and sensory phenotypes is still not fully understood. In particular, it is unclear whether the mutations result in a toxic gain of function, a partial loss of activity related to translation, or a combination of these non-mutually exclusive mechanisms.
The zebrafish is an excellent organism for modeling neuromuscular phenotypes. It is amenable to forward and reverse genetics, and embryos can be obtained in large numbers, are transparent during early developmental stages, and have a simple nervous system. During a forward genetic screen (21), we identified an allele of gars (garss266). Homozygous mutant embryos carry a C->A transversion, leading to a non-conservative substitution within gars that changes a threonine to a lysine at residue 209, which is equivalent to the T130K in the human protein and resides next to a previously studied human mutation (L129P). We show that T209K affects the homodimerization of the GARS protein. Maternally deposited protein and mRNA support the development of embryos up to 48 h post-fertilization (hpf). However, garsT209K/T209K embryos and larvae are underdeveloped with a smaller body axis, exhibit denervated NMJs and a severe muscular atrophy, become progressively immotile and have a cardiac phenotype. Cardiac contractility and blood flow initiate as in wild-type animals but, due to impaired cardiac valve formation, the flow stops and the blood regurgitates between the two heart chambers. Since T209K exists only as a monomer, this ablates the ability to aminoacylate tRNA, thus inhibiting protein translation in mutant embryos. Heterozygous garsT209K/+ embryos and adults do not show any phenotype, are fertile, and can live more than 2 years with no swimming or other behavioral phenotypes. Based on this, we hypothesized that dimerization is required for dominant GARS-associated neuropathy. We modeled several known human mutations in zebrafish to test if garss266 can be used as the first vertebrate animal model for studying CMT-associated human mutations. The garsT209K/T209K phenotype was transiently rescued by overexpressing the wild-type and C236R zebrafish gars (which is equivalent to the C201R mouse mutation). However, overexpression of a human mutation that reduces, but does not ablate dimerization (G319R; equivalent to G240R in GARS) did not improve the phenotype. Finally, overexpressing G605R gars (equivalent to G526R in GARS, a human loss-of-function mutation that does not affect dimerization) in zebrafish embryos heterozygous for T209K gars resulted in a neuromuscular phenotype.
Altogether, these data suggest that: (i) the neuromuscular phenotype observed in animals homozygous for T209K is due to a loss of dimerization of the mutated protein; (ii) the loss of function and dimer-deficient G240R mutant protein is unable to rescue the above phenotype due to the loss-of-function nature of this mutant protein and (iii) G526R gars dimerizes with the remaining wild-type protein in animals heterozygous for T130K gars and reduces the function enough to elicit a neuromuscular phenotype. Our data indicate that dimerization is required for the dominant neurotoxicity of disease-associated GARS mutations and provide a rapid, tractable model for studying newly identified variants for a role in human disease.
Results
The s266 mutant allele destabilizes the dimerization of Gars protein
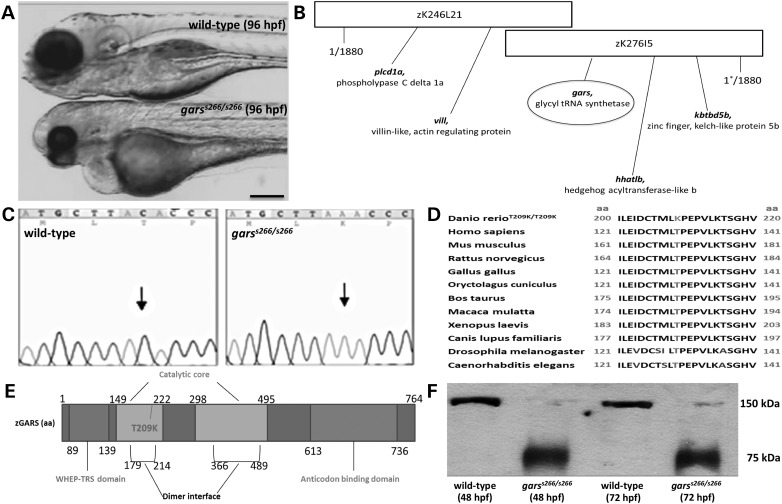
We identified a recessive lethal mutation, s266, with retrograde blood flow between the two heart chambers in a forward genetic screen in zebrafish (21). Homozygous mutant embryos exhibit pericardial edema and are underdeveloped with unconsumed yolk and smaller head and body axis (Fig. 1A). They become progressively immotile starting at 48 hpf, but survive up to 120 hpf. To identify the s266 gene, we employed a positional cloning strategy. Bulk segregant analysis placed the s266 locus on zebrafish chromosome 24. Fine mapping with 1880 diploid mutant embryos positioned the s266 locus to a 0.01 cM genomic region using markers z17203 and z9321 and subsequently new markers we developed within two partially overlapping BACs: DKEY-246L21 and DKEY-276I5 (Fig. 1B). We sequenced all genes in the defined region and found a single-nucleotide transversion (C to A) in the gars gene, which is predicted to cause a substitution of a threonine with lysine (T209K, equivalent to a T130K in human GARS) (Fig. 1C and D). Importantly, zebrafish have a single gars gene on chromosome 24 (ZDB-GENE-030131-9174) encoding both the mitochondrial and cytoplasmic Gars, as in Humans. T209 is inside the catalytic domain at the dimer interface of the protein (Fig. 1E), suggesting that it could affect homodimerization. Indeed, native western blot analysis from garss266/s266 mutant embryo lysates demonstrated the presence of only monomeric Gars proteins (Fig. 1F).
Figure 1.
Identification and cloning of a novel gars allele. (A) Bright-field images of wild-type (A, upper), garss266/s266 mutant (A, lower) at 96 hpf. garss266/s266 embryos are delayed in development. They exhibit smaller head, eyes, body axis and a pericardial edema (B) The mutation was mapped on linkage group 24 (BAC clone DKEY-276I5) using 1880 mutant embryos. (C) s266 embryos carry a C->A transversion, leading to a substitution within the gars gene that changes a threonine to a lysine at residue 209. The gars sequence is shown at the region of the mutation from a wild-type (left panel) and a homozygous mutant (right panel). (D) The amino acid change is in a highly conserved region of GARS protein throughout species (conserved amino acids are in black; the altered lysine is highlighted). (E) gars is a 17-exon gene encoding 764-amino acid protein with two catalytic domains. The T209 mutant corresponds to the T130K in the human protein and is inside the first catalytic domain and in the dimer interface domain. (F) Using a specific human anti-GARS antibody to protein extracts from garss266/s266 and wild-type embryos at 48 and 72 hpf on a native western blot, we showed that T130K interferes with the dimerization of the protein. Scale bar: 100 μm.
The T209K Gars mutation reduces yeast cell viability
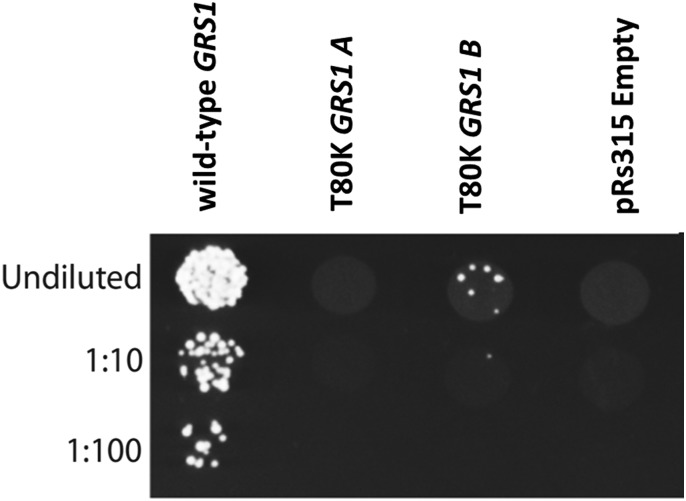
To determine if the T209K amino-acid substitution alters the function of the Gars protein, we employed a yeast complementation assay (Fig. 2). Yeast complementation assays have been used to identify loss-of-function properties of human GARS mutations (7,16). To assess the functional consequences of T209K gars, we modeled this mutation in the yeast ortholog GRS1 (residue T209 in zebrafish GARS is equivalent to residue T80 in yeast GRS1 and T130 in human GARS) and tested for the ability to support yeast cell growth compared with wild-type GRS1 or to a vector with no GRS1 gene (‘Empty’). A previously validated haploid yeast strain with the endogenous GRS1 gene deleted and a vector that expresses wild-type GRS1 and URA3 to maintain viability (22) was transformed with a separate vector harboring a LEU2 selection marker and either no insert or a wild-type or T80K GRS1 allele. Yeast were then selected on media containing 5-fluoroorotic acid (5-FOA), which is toxic to yeast cells expressing URA3 and thus selects for cells that have spontaneously lost the maintenance vector. Only yeast cells expressing a functional GRS1 allele from the LEU2-bearing vector will survive on 5-FOA. The wild-type GRS1 expression vector sustained yeast viability, while the empty vector was unable to complement the knockout allele, consistent with GRS1 being an essential gene (Fig. 2) (22). Yeast expressing T80K GRS1 demonstrate minimal survival on 5-FOA media (Fig. 2). These results indicate that T209K gars is a loss-of-function allele.
Figure 2.
T80K GRS1 fails to support yeast cell growth. Haploid Δgrs1 yeast strains were transformed with a vector containing no insert (pRS315 Empty) or an insert to express wild-type or T80K GRS1 (equivalent to the T209K, s266 zebrafish mutation, T130K in human GARS). Cultures resulting from each transformation were spotted undiluted and diluted (1:10 and 1:100) on plates containing 0.1% 5-FOA. Experiments were performed using two independently generated T80K GRS1 expression constructs (A and B).
S266 larvae exhibit partial innervation and degeneration of fast muscle fibers in the developing skeletal muscles
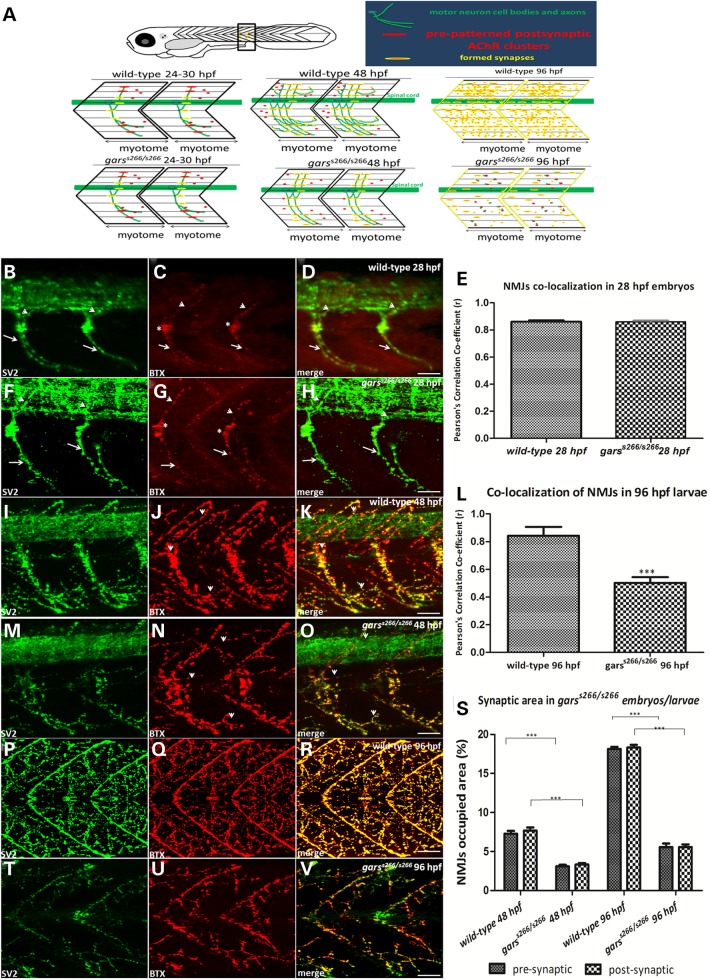
To form a functional muscular unit, motor neurons grow out from the spinal cord to establish contact with muscle fibers of the corresponding somite. Zebrafish have three types of primary motor neurons (PMNs), the caudal primary (CaP), the middle primary (MiP) and the rostral primary. Between 16 and 24 hpf the axon of each PMN grows along the common pathway, lateral to the notochord, to the choice point, pauses for several hours and grows across the medial surface of one myotomal muscle (23,24). Axons subsequently grow along the lateral myosepta and branch extensively to innervate more lateral muscle fibers. At the same time, pre-patterned acetylcholine receptor (AChR) clusters at the choice point are present in almost all medially located muscle fibers. Each muscle fiber has one cluster situated in the middle of the myotome (Fig. 3C, asterisks). Between 20 and 28 hpf, each primary axon extends into its appropriate territory of the myotome and the synapses are formed by overlapping of the axon and the AChR clusters (Fig. 3D, arrows and arrowheads). We examined garss266/s266 homozygous mutants (n = 8 embryos at 28 hpf) for development and the formation of NMJs. Embryos were co-stained with the synaptic-vesicle 2 (SV2) antibody marking the pre-synaptic motor nerve ends and a-Bungarotoxin (aBTX), binding to the post-synaptic acetylcholine receptors, AChRs. At 28 hpf s266 mutants do not exhibit a phenotype and we could not detect any differences within the clutch. The primary axons (CaP and MiP) are developing and projecting at the center of the myotome normally (Fig. 3F) and the pre-patterned AChRs are clustering, too (Fig. 3G, merge in Fig. 3H). Robust overlapping NMJs are formed without any difference in the co-localization of SV2 and aBTX as shown by the Pearson’s correlation co-efficiency analysis (Fig. 3E).
Figure 3.
Abnormal NMJs are observed in garss266/s266 mutant larvae. (A). Cartoon summary of neuromuscular synaptogenesis in wild-type zebrafish (A, top view) 24–30, 48 and 96 hpf. For each stage a side view of two myotomal segments are shown. In each panel, green structures represent motor neuron cell bodies and axons; red structures represent AChR clusters, and yellow represents overlap of pre- and post-synaptic structures and thus a synapse. At the appropriate stage, the phenotype of garss266/s266 mutant is shown. At 24–30 hpf in wild-type and mutant embryos, motor axons have reached the dorsal and ventral edges of the myotome, and AChRs are clustered beneath synaptic vesicle clusters. Non-synaptic AChRs are present along the myosepta between adjacent myotomes. At 48 hpf in wild-type zebrafish, motor axons have turned and have grown along the rostral lateral myosepta, and axon branches are present in all muscle fiber layers. Synapses form along all axon branches, and synaptic vesicle clusters and AChR clusters are well co-localized (yellow). However, in garss266/s266 embryos pre- and post-synaptic areas are smaller (bottom). By 96 hpf in wild-type zebrafish, motor axon branching and synapse formation has increased and the entire length of the rostral lateral myosepta is innervated (yellow in the myosepta). In mutant embryos at 96 hpf, synapse number is reduced in the myotome and not properly localized (neuromuscular synapses are labeled with antibodies against SV2 (green, pre-synaptic vesicles) and aBTX (red, post-synaptic AChRs). Images are oriented so that left is rostral and top is dorsal. (B–H) At 28 hpf the PMN of the embryo projected straight axons dorsally (MiP axons, arrowheads) and ventrally (CaP axons, arrows) and robustly overlapped with NMJs. (E) Quantification of co-localization of SV2 and aBTX in 28 hpf garss266/s266 (n = 8) and wild-type (n = 15) embryos showed no difference at this stage. However, as the mutant embryos develop they showed a progressive loss of NMJs resulting to a severe muscular atrophy. (I–O) Projections of confocal z-stacks at the level of hindgut extension (I: 62 μm, O: 70 μm). In 48 hpf, mutant embryos (n = 13) had reduced synaptic occupied areas, compared with the wild-type embryos (n = 9). (L) Quantification of Pearson’s correlation (r) of pre- and post-synaptic regions of wild-type (n = 10) and garss266/s266 (n = 14) 96 hpf larvae. (P–R) Projections of confocal z-stacks at the level of hindgut extension (R: 64 μm) of 96 hpf wild-type and mutant embryos. During the development of garss266/s266 denervated post-synaptic regions were observed, as only the 5.5% ± 0.8% of synaptic area was present in mutants (T–V) larvae (W: 63 μm). These measurements reflected differences in larval muscle innervation, explaining the immotility of garss266/s266-deficient larvae. (S) Quantification of synaptic occupied areas in the two somites of the region of hindgut between wild-type (n = 9) and garss266/s266 (n = 13) 48 hpf embryos and wild-type (n = 15) and garss266/s266 (n = 20) at 96 hpf larvae. (L and S) mutants had significantly lower synaptic occupied area when compared with siblings and wild-types (**P < 0.0001, unpaired t-test). Scale bars: 25 μm. Agarose sections width 150 μm.
In contrast, starting at 48 hpf (Fig. 3M–O) and more profound at 96 hpf (Fig. 3T–V) we observed a progressive reduction of NMJs in the developing muscle. Quantifications of pre- and post-synaptic areas at 48 and 96 hpf showed that garss266/s266 larvae had lower volumes of both SV2 and aBTX (Fig. 3S). Particularly, at 48 hpf wild-type embryos had NMJs occupied area of 7.5% ± 0.398% as a percentage of the total area of two somites at the level of the hindgut extension, in comparison with the 3.3% ± 0.765% of garss266/s266. At 96 hpf wild-type larvae had NMJs occupied area of 18% ± 0.255% in comparison with 5.5% % ± 0.255% of garss266/s266 larvae. NMJs occupied area were not significantly altered between wild-type and siblings larvae (data not shown). The reduction of the muscle innervation is more pronounced in the fast muscle fibers in the center of the myotomes, since the NMJs are located at the edge of slow muscle fibers and form chevron-shaped lines at the boundary of body segments (Fig. 3R). Moreover, co-localization of SV2 and aBTX was significantly reduced by 1.6-fold (Fig. 3L). In summary, T209K causes NMJ defects in homozygous mutant embryos by arresting innervation. The primary motor axons entered the muscle and branched properly, while the nerve terminal completely overlaps the post-synaptic receptor field on the muscle. However, from 48 hpf and onwards the innervation of the muscle is significantly decreased, causing the observed paralysis of garss266/s266 larvae at 96 hpf. Those data agree with the GarsNmf249/+ mice that exhibit denervated or partially innervated NMJs from P36 to P37, but not earlier (18). A possible explanation of normal NMJ development during early stages is maternal contribution of wild-type mRNA and protein (Supplementary Material, Fig. S1A and B).
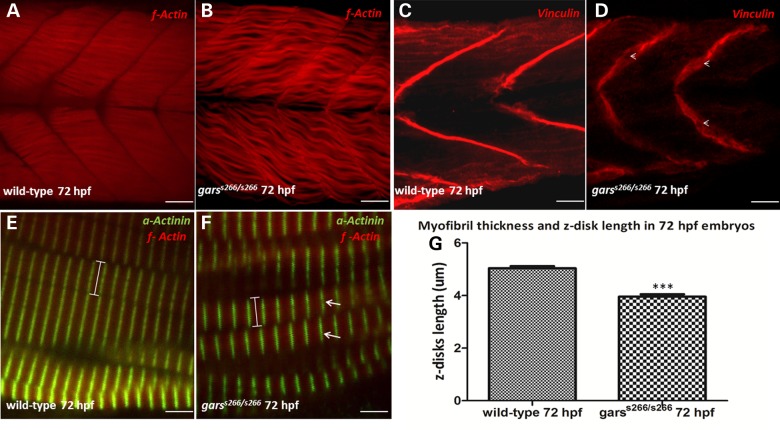
Since GARS mutations are also associated with distal spinal muscular atrophy V, we examined the development and formation of skeletal muscles in garss266/s266 larvae by immunostaining with sarcomeric α-Actinin that stains the z-disks and phalloidin that specifically recognizes f-actin of fast muscle fibers. We observed that the myofibrils of homozygous mutant larvae lack the normally aligned arrangement to the chevron-shape somatic semi-segment. Moreover, they lost their integrity and tight formation (Fig. 4B) and were thinner compared with the myofibrils of wild-type larvae (Fig. 4A). The thickness of the myofibrils is reduced from 5 μm ± 0.08 (average ± SE, n = 10) in wild-type animals to 3 μm ± 0.07 [average ± standard error (SEM) of the mean (SEM), n = 11] in mutant animals (Fig. 4G). The formation of this looser muscle structure in homozygous mutants affects the striation of the sarcomeres, since in garss266/s266 larvae myofibrils are discontinuously striated (Fig. 4F, arrows). Finally, we investigated the mechanical stability of the muscles by immunostaining with Vinculin, a cytoskeletal protein associated with cell–cell and cell–matrix junctions. Vinculin normally localizes to the myotendinous junctions (MTJs) (25). In garss266/s266 embryos, Vinculin stains discontinuously in MTJs with gaps at regions where the actin filaments have retracted (Fig. 4D, arrowheads). In addition, we observed larger angle and more U-shaped like somites. Heterozygous larvae showed no neuromuscular alterations (Supplementary Material, Fig. S2A–F).
Figure 4.
garss266/s266 mutants exhibit structural defects in muscles and somites. (A and B) Confocal analysis in 100 μm agarose sections of 72 hpf wild-type and garss266/s266 larvae stained with filamentous Actin (wt n = 30, garss266/s266 n = 25), (C and D) with Vinculin, staining the chevron shaped-septa along the anterior–posterior axis (wt n = 16, garss266/s266 n = 15) and (E and F) with sarcomeric α-Actinin (wt n = 10, garss266/s266 n = 11). In garss266/s266 mutants (B and F) the myofibrils lacked the aligned arrangement to the chevron-shape somatic semi-segment and lost their integrity and tight formation, comparing with wild-type (A and E) embryos. In garss266/s266 embryos (F) myofiblis were discontinuously striated (arrows). (G) The sarcomeres in mutant embryos were 30% shorter, compared with wild-type when measuring the length of z-disks (α-actinin), (wt n = 10, garss266/s266 n = 11 in C and D). Vinculin staining showed that garss266/s266 larvae do not exhibit the typical chevron pattern in septa, but a U-shaped, more fainted signal and a discontinuous formation with gaps (arrowheads, D). Arrows (F) are indicating z-disk and arrowheads (D) the discontinuous septa. (G) The myofibril thickness and z-disk length was significantly reduced in mutants when compared with wild-type (**P < 0.001, unpaired t-test). Scale bars are 25 μm in (A–D) and 5 μm in (E and F).
Finally, we performed touch-evoke response assays at three developmental stages (62, 72 and 96 hpf) on wild-type and garss266/s266 animals to assess for muscle weakness (26). Wild-type embryos responded to mechanosensory stimuli with rapid and vigorous swimming in a straight line (Supplementary Material, Fig. S3A–D). In contrast, homozygous mutants tended to weakly flex and move for only a short distance (Supplementary Material, Fig. S3E–H). Statistical analysis of the captured videos showed that the time interval for swimming of a fixed distance (1.1 cm) was significantly increased up to 5-fold for mutant compared with wild-type embryos (Supplementary Material, Fig. S3K). However, at this developmental stage garss266/s266 embryos did not have any structural dystrophies in the skeletal muscles, as exhibited at later stages. (Supplementary Material, Fig. S3J compare with Supplementary Material, Fig. S3I). This suggests that the increased interval time for swimming was a result of a muscular weakness.
S266 larvae exhibit immature AV cushion formation and pericardial edema
The s266 mutant allele causes a recessive cardiac phenotype with pericardial edema and blood stasis by 96 hpf. Blood circulation initiates but from 72 hpf is gradually reduced and then at the level of the bulbus arteriosus stops, with the blood regurgitating between the two chambers of the heart. All mutant larvae die by 5 dpf due to the lack of blood circulation similar to most mutant lines with cardiac defects from the forward genetic screen (21) or the silent heart mutant line (sih) where a mutation in the cardiac troponin T results to a non-contractile myocardium (27). We, therefore, aimed to examine myocardial function and the morphology of the AV cushion. Bright-field videos of the heart showed that garss266/s266 animals had no difference in myocardial function compared with wild-type or sibling larvae and exhibit an unaltered heart rate (Supplementary Material, Fig. S4A). To establish whether cardiac valve morphogenesis was affected, we used the transgenic lines Tg(kdrl:EGFP)s843 to mark endothelial cells and the wnt signaling reporter Tg(7xTCF-Xla.Siam:nslCherry)ia5. In these lines endocardial and valve cells can be visualized. Confocal analysis of hearts showed that AV canal endocardial cells in mutant larvae acquired the cuboidal shape. However, analysis of Tg(7xTCF-Xla.Siam:nslCherry)ia5 showed a dramatic decrease in the amount of TCF:cherry positive, mesenchymal looking endocardial valve cells in the AV cushions (Supplementary Material, Fig. S4B arrows in Supplementary Material, Fig. S4J and K, compare with Supplementary Material, Fig. S4D and E and Movie 2A and B). Taken together, these results indicated that at early developmental stages AV valve differentiation occurs normally, but at later stages impaired Gars function prevents formation of functional cardiac valves.
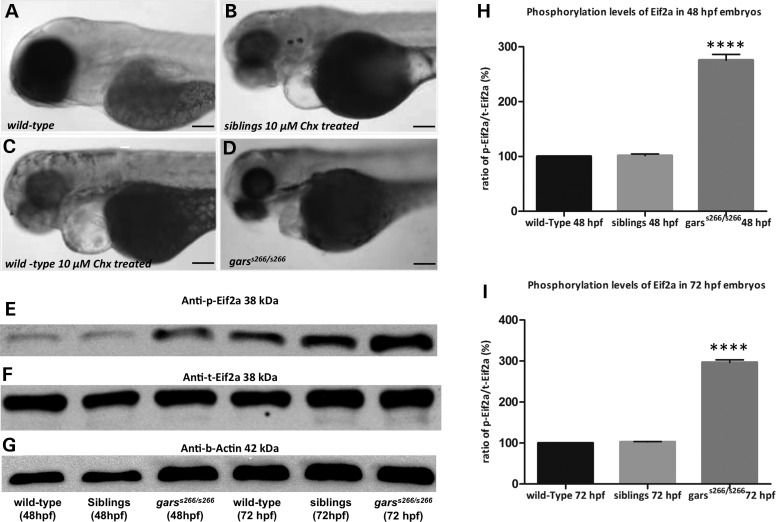
T209k is a loss-of-function variant of Gars protein
The canonical function of GARS is to catalyze the ligation of glycine to cognate tRNAs, resulting in glycyl-tRNA complexes necessary for protein translation. To analyze whether the observed phenotypes in s266 mutants are caused by defective protein synthesis, we chemically inhibited protein synthesis between 24 and 96 hpf by the addition of cycloheximide (Chx) to wild-type and siblings embryos. Treatment of wild-type animals with Chx (Fig. 5B and C) led to a phenotype similar to garss266/s266 (Fig. 5D) with treated embryos being underdeveloped with smaller body axis and having a smaller head and decreased swimming capability (Supplementary Material, Movie S3). Chx-treated larvae also developed a progressive neuromuscular phenotype with a significant reduction in the NMJs occupied area (Supplementary Material, Fig. S5A and B and quantification Supplementary Material, Fig. S5C) and reduced myofibril thickness and shorter z-disks in the skeletal muscles (Supplementary Material, Fig. S5D and E and quantification in Supplementary Material, Fig. S5F). These findings are consistent with the s266 phenotype being caused by impaired translation. Since T209K Gars exists only as a monomer, it would be predicted to ablate aminoacylation of tRNA-Gly molecules. Furthermore, this could result in increased levels of uncharged tRNA and phosphorylation of Eif2α (Fig. 5E and quantification in Fig. 5I). Thus, T209K demonstrates loss-of-function characteristics, which result in impaired protein translation and the observed phenotype.
Figure 5.
Eif2a phosphorylation is induced in garss266/s266 embryos. (A–D) Bright-field images of wild-type, siblings and mutant larvae at 72 hpf, after treatments with Chx. Wild-type larvae (n = 60) and siblings (n = 40) treated with 10 μm Chx phenocopied the garss266/s266 phenotype. (A) Wild-type untreated control and (D) garss266/s266 untreated embryo. (E) Western blot analysis showed that the levels of Eif2a phosporylation is increased up to 2.8-folds in homozygous mutants at 48 hpf (H) and up to 3.2-folds at 72 hpf (I), compared with wild-type embryos. All mutants were significantly altered comparing with wild-type (****P < 0.00001, one-way ANOVA). The quantification of Eif2a protein levels was performed with the ImageJ software. Scale bars: 50 μm.
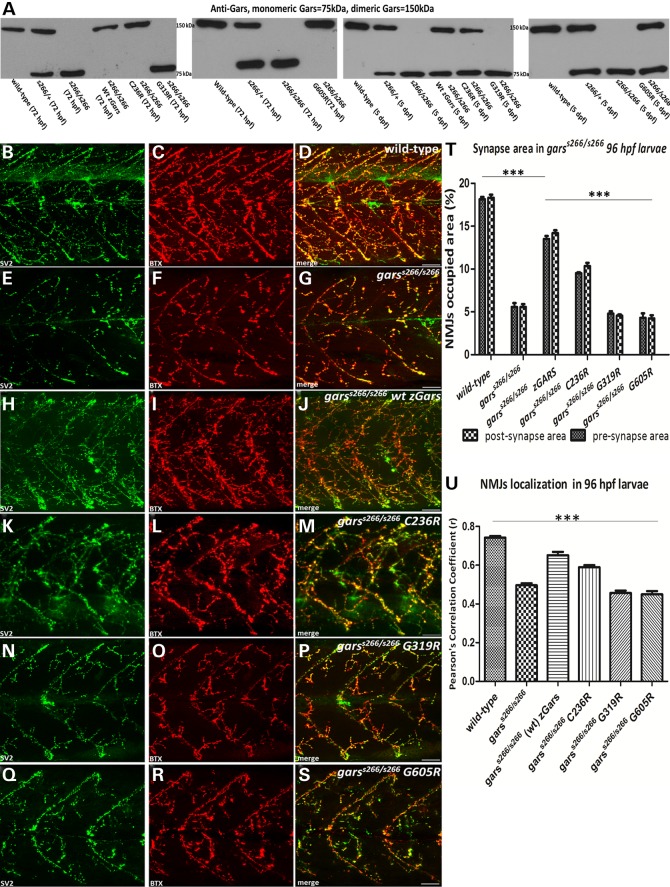
In support of a loss-of-function mechanism, overexpression of wild-type Gars protein improves the motility and extends the life span of mutant embryos by 2 days. garss266/s266 larvae overexpressing wild-type Gars also maintained blood circulation up to 6 dpf (Supplementary Material, Movie S4A) and have a 2-fold increase in pre- and post-synaptic density in NMJs compared with uninjected controls (Fig. 6H–J and quantification in Fig. 6T). This partial rescue is achieved only by the overexpression of a functional protein. Indeed, native western blot analysis from injected embryo lysates showed the presence of the active homodimer form of Gars in garss266/s266 larvae injected with wild-type gars mRNA at 72 hpf and a double band of both forms (monomeric and dimeric) of the protein (Fig. 6A). These data support our conclusion that T209K is a loss-of-function gars allele.
Figure 6.
Functional analysis of C236R, G319R and G605R mutations using garss266/s266 larvae. (A) Native western blot analysis of protein extracts from injected with wild-type, C236R and G605R zGars and uninjected mutant embryos at 72 hpf showed that in injected garss266/s266 embryos, Gars existed in an active, dimer formation, while in the control as a monomer. In contrast, injection with G319R did not result in the formation of heterodimers. (B–D). Projections of confocal z-stacks at the level of hindgut extension of 96 hpf. (E–G) During the development of garss266/s266, denervated post-synaptic regions were observed, as only the 5.5% ± 0.8% of synaptic occupied areas were present in mutant larvae. (H–J) Forty-eight percent of the homozygous mutant larvae injected with wild-type zGars exhibited blood flow (n = 41, not shown) at 6 dpf and showed nearly wild-type levels of synaptic areas at 96 hpf. (K–M) Seventeen percent of the homozygous mutant larvae injected with C236R mRNA exhibit blood flow (n = 128, not shown) at 6 dpf and showed an increase of synaptic areas up to 2-fold at 96 hpf compared with mutant control. (N–P) Homozygous mutant larvae injected with G319R mRNA (n = 35). Injected embryos had denervated muscles with a similar decrease of NMJs occupied areas compared with garss266/s266 uninjected larvae. (Q–S) Homozygous mutant larvae (n = 47) injected with G605R mRNA. Injected embryos had denervated muscles with a similar decrease of NMJs density compared with garss266/s266 uninjected larvae. Although dimers formed in the injected larvae, both mutations rendered the dimer non-functional and as a result embryos died by 5 dpf. (S and T) Quantification of synaptic occupied areas in injected larvae (wild-type n = 10, garss266/s266 n = 14, garss266/s266 wild-type zGars n = 10, garss266/s266 C157R zGars n = 9, garss266/s266 G240R zGars n = 10, garss266/s266 G526R zGars n = 11, ***P < 0.0001, one-way ANOVA). (U) Quantification of co-localization of SV2 and aBTX in injected embryos (wild-typen = 10, garss266/s266 n = 14, garss266/s266 wild-type zGars n = 10, garss266/s266 C236R zGars n = 9, garss266/s266 G319R zGars n = 10, garss266/s266 G605R zGars n = 11, ***P < 0.0001, one-way ANOVA). Scale bars: 25 μm.
In contrast, we did not detect rescue of the phenotype upon overexpressing G319R gars (equivalent to the G240R in human GARS), which reduces dimerization and aminoacylation (17). Overexpression was confirmed by immunoblots at 72 hpf and injected larvae had significantly increased levels of GARS protein (up to 3-fold) compared with uninjected controls. Native western blot analysis from injected mutant embryo lysates demonstrated only the monomeric form of GARS in garss266/s266 injected mutants (Fig. 6A) consistent with previous in vitro data showing impaired dimerization of this protein (17). These data for the first time show in vivo that G240R GARS affects dimerization.
The disease-associated G526R mutation ablates enzyme activity but retains dimerization (16,17) and, as mentioned above, the C201R mouse mutation is a partial loss-of-function allele. We, therefore, tested these two mutant proteins for the ability to rescue the s266 phenotype by modeling them in the Gars protein (G605R and C236R, respectively) and overexpressing them up to 3-fold compared with endogenous levels. C236R does not interfere with dimerization of the GARS protein, but does reduce enzymatic activity in homozygous mice (19). Overexpression of C236R transiently rescues the levels of NMJs occupied area by increasing pre- and post-synaptic areas (Fig. 6K–M) and by maintaining blood circulation up to 6 dpf (Supplementary Material, Movie S4B). Native western blot analysis of injected embryo lysates revealed only the homodimeric form of Gars in garss266/s266 injected larvae at 72 hpf (Fig. 6A). At 5 days a double band indicating the presence of both the monomeric and dimeric forms of the protein was observed (Fig. 6A), and only monomers were detected at 7 dpf (data not shown). A likely explanation for this partial rescue is that C236R is a hypomorphic allele. Indeed, this is consistent with the milder phenotype in heterozygous C201R mice and the partial viability of homozygous C201R mice.
Finally, overexpression of G605R gars, (a functional null allele equivalent to G526R GARS that does not reduce dimerization 7,16,17), failed to rescue the neuromuscular phenotype of garss266/s266, since the pre- and post-synaptic NMJs occupied areas show a decreasing shift to lower values comparing to garss266/s266 larvae (Fig. 6Q–S). In parallel, the garss266/s266 injected larvae could not maintain the blood circulation later than 96 hpf. Previous reports showed that G526R does not affect dimer formation (15). Indeed, on a native western blot analysis from injected embryo lysates, we observed the homodimeric form of Gars (Fig. 6A); however, G526R ablates enzymatic activity by direct blocking of the AMP-binding site (16,17). Importantly, this is the first evidence in a vertebrate model system that G526R causes a loss-of-function effect.
Dominant toxicity is associated with dimerization
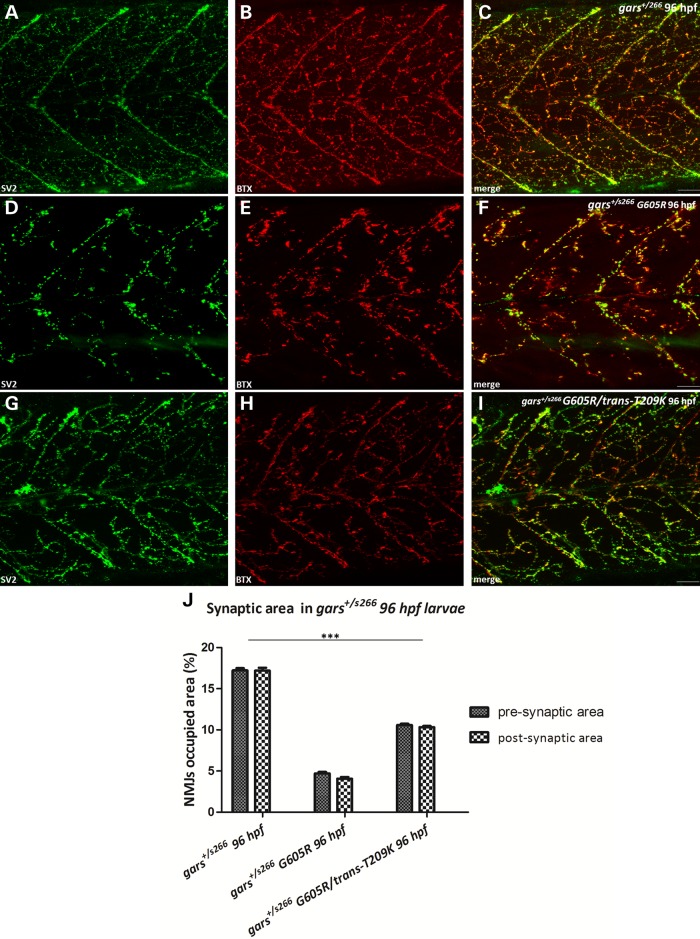
We injected gars capped mRNA to overexpress T209K, G319R and G605R proteins (equivalent to the T130K, G240R and G526R human mutations) in s266 siblings (heterozygous and wild-type embryos). T209K and G319R overexpression had no effect (data not shown), but G605R enhanced the NMJ phenotype. In particular, we observed a severe reduction of pre- and post-synaptic densities in NMJs at similar levels to garss266/s266 larvae (Fig. 7D–F). In addition, injected heterozygous larvae exhibited pericardial edema and reduced blood circulation. To determine if reduced dimerization would improve the phenotype associated with G605R expression we co-injected both mRNAs harboring the T209K and the G605R in trans. Interestingly, this ameliorated the NMJ phenotype by increasing the NMJs occupied area by almost 2-fold (Fig. 7G–I, quantification Fig. 7J). A possible explanation for this improvement is that the toxic potential of G605R is reduced when the dimerization of the protein is reduced by an excess of T209K in trans, which would impair any dimerization with endogenous, wild-type Gars and therefore reduce any dominant-negative effects of G605R.
Figure 7.
Dominant toxicity is associated with dimerization. (A–I) Projections of confocal z-stacks at the level of hindgut extension (C: 42,57 μm; F: 42,3 μm; I: 31,22 μm) of gars+/s266, gars+/s266 G605R, gars+/s266 G605R/trans-T209K larvae at 96 hpf. (A–C) gars+/s266 uninjected larvae. (D–F) gars+/s266 larvae injected with G605R mRNA (n = 228 injected larvae, 33% of injected larvae gave a s266 phenotype). The expression of G605R caused a dramatic reduction of the pre- and post-synaptic densities in the injected larvae. (G–I) gars+/s266 larvae injected with G605R/trans-T209K mRNA (n = 153 injected larvae). However, trans-expression of T209K mutation ameliorated the observed NMJ phenotype probably due to lower levels of G605R Gars available for dimerization with wild-type Gars. (J) Quantification of synapse density in injected larvae (gars+/s266 n = 11 larvae, gars+/s266 G605R n = 15 larvae, gars+/s266 G605R/trans-T209K n = 19 larvae, ***P < 0.0001, one-way ANOVA). Scale bars: 50 μm.
T130K GARS mislocalizes in neurons
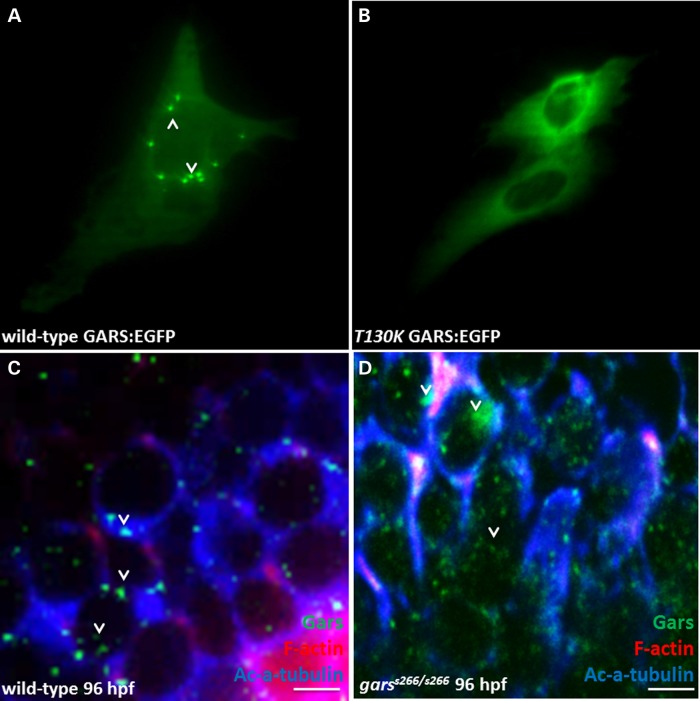
Altered subcellular localization has been reported for mutant forms of GARS (7,16). The affect on localization of the garss266 allele (T209K) was examined by introducing the mutation into the human coding sequence (T130K) in frame with an enhanced green fluorescent protein (EGFP) tag and transfecting constructs into immortalized motor neurons (MN-1). In cells expressing wild-type GARS-EGFP, the protein formed puncta (Fig. 8A, arrows). In contrast, T130K GARS-EGFP was diffusely localized throughout the cytoplasm and did not show any points of concentrated localization. In addition, endogenous zebrafish Gars protein was visualized by staining with a GARS antibody in wild-type and homozygous mutant larvae at 96 hpf and puncta were observed in the cytoplasm of the soma and cell processes of the midbrain. However, the number and the intensity of these puncta were highly variable. In mutant larvae, Gars showed a more diffuse staining pattern and seems to accumulate in the cytoplasm of neuron somata of midbrain (Fig. 8D, arrowheads).
Figure 8.
T209K-Gars has altered sub-cellular localization. (A) Expression of wild-type and mutant GARS-EGFP in mouse motor neurons. The T130K mutation was introduced into the human-coding sequence in frame with a C-terminal EGFP tag, and transfected into MN-1 cells. Cells expressing wild-type GARS-EGFP demonstrated localization of the protein to cytoplasmic puncta (arrowhead). (B) T130K GARS-EGFP appeared diffused throughout the cytoplasm and did not form punctate structures. (C and D) Confocal analysis in agarose sections width 50 μm of 96 hpf wild-type and garss266/s266 larvae stained with filamentous actin (red), anti-Gars (green) and the ac-a-tubulin (blue), a pan-axonal marker (wt n = 12, garss266/s266 n = 11). (C) Endogenous Gars is localized in cytoplasmic puncta of the neurons in the midbrain of wild-type larvae at 96 hpf. (D) In contrast, in mutant larvae Gars showed fainter, diffused staining. Scale bars: 5 μm.
Discussion
GARS is the most well-studied of the ARS enzymes implicated in CMT disease; however, the molecular and cellular mechanisms linking GARS and other ARSs to neuromuscular disorders remain unclear. Mechanistic insights into CMT2D pathology came from cellular and yeast studies (7,16) and from two mouse models (18,19). The broad distribution of CMT-causing mutations on the primary sequence of GARS has long been puzzling. Understanding how GARS mutations cause a highly specific phenotypic peripheral neuropathy (28–31) requires additional insight into mutant forms of GARS found in affected individuals. In this study, we present the first CMT2D-associated zebrafish model, which arose from an ENU-mutagenesis screen. Positional cloning led to the identification of a threonine to lysine change (T209K) in Gars, which corresponds to T130K in human GARS. This residue is highly conserved, resides in the catalytic domain, and is directly adjacent to the diseased-associated human mutation L129P. The T209K mutation causes a progressive, but severe, denervation of muscles with subsequent muscular atrophy of fast muscle fibers and disorganization of the muscle cytoskeleton leading eventually to paralysis and premature death in homozygous animals. In addition, we studied two human and one mouse mutation associated with peripheral neuropathy in our model by overexpressing them via injecting capped mRNA into one-cell stage embryos: C201R, a known hypomorphic mouse allele with partial function in vivo; G240R, a complete loss of function human disease-associated allele that demonstrates decreased dimerization and G526R, a functional null allele that is capable of dimerization (7,15,16). Our results indicate that our zebrafish model can be used to systematically study human disease-associated GARS polymorphisms.
Modeling T209K GARS in the yeast ortholog (GRS1) revealed that it does not complement loss of endogenous wild-type GRS1. In previous studies, modeling human GARS mutations in the yeast revealed that several mutations (e.g. L129P, H418R and G526R) do not complement deletion of wild-type GRS1, whereas others (E71G and G240R) do complement loss of endogenous GRS1. Furthermore, T130K GARS mislocalizes in cultured neurons similar to other GARS mutant proteins that cause CMT disease (7,16). In summary, T209K is a loss-of-function allele that has functional consequences similar to human disease-associated GARS mutations.
A common pathological mechanism for genetic disorders is a loss of function through altered mRNA and protein levels or enzymatic inactivity. The active form of GARS is a homodimer. Native western blot analysis showed that in mutant embryos/larvae T209K Gars exists only as a monomer compared with heterozygous fish that have both monomeric and dimeric Gars. In addition, when G240R GARS was modeled in the zebrafish gene (G319R gars) and overexpressed in embryos, it failed to promote dimerization of the T209K protein in mutants, while overexpression of C236R Gars induced dimerization with T209K Gars and transiently rescued garss266/s266 larvae. From the structural perspective, it is not surprising that T209K and G319R mutations disrupt dimer formation since these residues are involved in the interaction domains of monomers to form a dimer (17). Since GARS must form a dimer to charge tRNA, the failure of mutant monomers to dimerize and charge tRNA could be the general cause for decreased protein synthesis. Importantly, protein-synthesis inhibition studies on wild-type embryos at 48 and 72 hpf phenocopy s266 fish, suggesting that the cause of the phenotype is the lack of protein synthesis. Further evidence for a loss-of-function effect is that the CMT-disease-relevant NMJ histopathology is rescued by wild-type Gars. Injections with wild-type Gars extended the life span of mutant larvae by 2 days with improved blood circulation and increased NMJs occupied area to near wild-type levels. Our results for the human CMT2D mutation G319R are consistent with this mutation being a loss-of-function allele. Overexpression resulted in the presence of only the monomeric form of GARS and injection of G319R mRNA did not complement the lethality or neuromuscular phenotypes of garss266/s266 larvae. In contrast, overexpression of the C236R gars (equivalent to the C201R mouse Gars mutation) ameliorates the phenotype in the NMJ as it increases the occupied area of NMJs by almost 2-fold (but not to wild-type levels) and extends the lifespan of mutant larvae by 2 days. A possible explanation for the amelioration of the neuromuscular phenotype is that C201R is a hypomorphic (but not functional null) allele. Indeed, in GarsC201R/C201R mice the enzymatic activity is ∼60% decreased (19), but overexpression of the mutant protein may compensate for this reduction of enzymatic activity. Transient overexpression of variants capable of forming dimers with the T209K mutant variant seems to transiently rescue the phenotype, indicating that such a ‘heterodimer’ can be functional. Moreover, even though G526R GARS does not affect dimerization of the enzyme, the protein is not active and as a result the overexpression of G526R in our model did not rescue the NMJ phenotype. Importantly, these findings reveal our in vivo model as relevant for the systematic analysis of CMT-associated human mutations to understand their impact on dimerization. A recent report (32) demonstrated a neomorphic function for mutated forms of GARS binding to the receptor Nrp1 and competing with the VEGF-Nrp1 signaling. Also, a non-canonical function for several ARSs regulating angiogenesis has been demonstrated (33,34).
s266 homozygous zebrafish appear normal up to 48 h post-fertilization with a normal heart rate. This is very likely due to maternally provided mRNA and protein. Only at later stages do embryos show a cardiac valve phenotype. Heterozygous zebrafish appear normal and have no observable phenotype up to 2 years of age at which point they continue to actively swim and remain fertile. We overexpressed the zebrafish equivalents of four GARS mutations (zebrafish T209K, mouse C201R and human G240R and G526R) in clutch-matched wild-type and T209K heterozygous embryos. Overexpression of T209K, C236R and G319R Gars had no effect on the NMJ phenotype. In contrast, expression of G605R had a severe effect on the NMJs of both wild-type and heterozygous larvae at 96 hpf, causing a significant denervation of muscles comparable with homozygous T130K animals. Combined, these findings show that the dominant toxicity of GARS mutations is likely associated with enzyme dimerization. That is, the mutant protein must be able to interact with the remaining wild-type protein to enact the toxic mechanisms (e.g. a dominant-negative effect that would further reduce GARS function well <50%). Indeed, when both G605R and T209K gars mRNA was co-injected to overexpress both mutations in trans, the toxic effect of G605R was reduced. A possible explanation is that T209K reduces dimerization with wild-type, thus reducing the toxic effect of G605R. Therefore, Gars T209K (T130K in GARS) could be a ‘separation of function’ mutation that shows that dimerization between a mutant monomer and a wild-type monomer is required for the autosomal dominant disease; this observation could explain why s266 heterozygotes show no neuropathy. These findings indicate that a dominant-negative effect may be a potential mechanism of GARS-mediated CMT disease and warrant follow-up studies to further test this mechanism.
Materials and Methods
Zebrafish maintenance and breeding
Zebrafish embryos were raised under standard laboratory conditions at 28°C. s266 is a recessive mutation identified during an ENU screen (21). The mutants were anesthetized using a tricaine chemical (tricaine methanesulfonate). The genetic backgrounds used were wild-type Ab strains, s266 carrying the following transgenes Tg(kdrl:EGFP)s843 and Tg(7xTCF-Xla:Siam:nlsCherry)ia5 (35,36). All zebrafish animal work has been approved by the BRFAA ethics committee and the Attica Veterinary Department (EL25BIO001/no. 4739).
Positional cloning of s266
Bulk segregant analysis for s266 was performed and the mutation was mapped between zk246Ls1 (@4 kb, 1 recombinant/1880 mutants tested) and zk276I5 (@80 kb, 1 recombinants/1880 mutants tested) on LG24 near the gars gene. Sequencing of the gars gene in homozygous and heterozygous larvae revealed a C->A transversion, causing a non-conservative threonine to lysine amino acid change at residue 209 in zebrafish (T209K), which is the equivalent for a T130K mutation in the human protein.
Yeast complementation assays
Yeast complementation assays were performed as previously described (7,16). Briefly, mutation-containing oligonucleotides were designed and used with the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA, USA) (per the manufacturer's instructions) to model the zebrafish T209K gars mutation in the yeast ortholog GRS1 in a pDONR221 Gateway entry clone (Invitrogen, Carlsbad, CA, USA). Plasmids were isolated from individual clones and sequenced to confirm mutagenesis and exclude polymerase errors. The T209K GRS1/pDONR221 entry clone was recombined into a Gateway-compatible LEU2-bearing pRS315 destination vector. Resulting clones were purified and digested with BsrGI (New England Biolabs, Ipswich, MA, USA) to confirm recombination. The Δgrs1 haploid yeast strain [harboring a pRS316 maintenance vector to express wild-type GRS1 and URA3 (22) was transformed with wild-type or mutant GRS1 in a LEU2-bearing pRS315 vector and selected on medium lacking uracil and leucine (Teknova, Hollister, CA, USA). For each transformation, two colonies were selected for further analysis. Each colony was grown to saturation in the selection medium for 48 h. Next, 10 μl of undiluted and diluted (1:10 and 1:100) samples from each culture were spotted on plates containing 0.1% 5-FOA complete medium or SD -leu -ura growth medium (Teknova) and incubated at 30°C for 72 h. Yeast cell growth was determined by visual inspection.
Cell culture and protein localization studies
The mouse motor neuron, neuroblastoma fusion cell line (MN-1) was cultured and transfected as previously described (7,37). Briefly, MN-1 cells were grown in a four-well culture slide (BD Biosciences, Bedford, MA, USA) and incubated at 37°C for 24 h. Cells were then rinsed with one-time PBS and transfected with 1.5 μg of purified plasmid DNA or the equivalent volume of water for mock transfections constructs to express wild-type or mutant GARS in-frame with an EGFP tag [using Lipofectamine 2000 (Invitrogen) per the manufacturer's instructions]. Cells were incubated with the OptiMEM/lipofectamine/plasmid solution at 37°C for 4 h. Next, transfection reactions were removed and replaced with normal growth medium and the cells were incubated at 37°C for 48 h. Growth medium was removed and the cells were washed in one-time PBS and then incubated in one-time PBS/0.4% paraformaldehyde for 10 min at room temperature. Cells were washed in one-time PBS, co-stained with 300 nm 4′,6-diamidino-2-phenylindole for 5 min, washed again in one-time PBS and finally coated with pro-long antifade reagent (Invitrogen). The IX71 Inverted Microscope using the cellSens Standard image software (Olympus) was used to obtain images.
Immunohistochemistry
Time-staged zebrafish were fixed overnight at 4°C in 4% PFA. Whole-mount antibody staining was carried out as described previously (21). The antibodies were used in the following concentrations: anti-α-Actinin (1:500), anti-SV2 (Developmental Studies Hybridoma Bank, catalog no. sv2; 1:250), anti-Vinculin (Sigma-Aldrich, catalog no. V9131; 1:400), anti-GARS (Abcam, catalog no. ab42905; 1:3000), Rhodamine Phalloidin (1:500) and aBTX-Alexa Fluor 555 conjugated (1:500). The appropriate secondary antibodies were used with Alexa Fluor 488 (Molecular Probes, catalog no. A-10235; 1:500) or 633 (Molecular Probes, catalog no. A-20170; 1:250).
Confocal microscopy
Imaging was performed using a Leica TCS SP5 inverted confocal microscope. The images were captured with the LAS AF software. Images shown are representatives samples of at least 10 embryos/larvae examined.
Synapses quantification and co-localization
Quantification of pre- and post-synapses were made on confocal scans at three developmental stages of 24, 48 and 96 hpf. Images were obtained from trunk segments located at the level of the yolk-sac extension. Measurements were carried out as described previously (38,39). In particular, to quantify the NMJs we used the ImageJ analysis software to measure the pre- and post-synaptic area (SV2 and BTX, respectively positive area as a percentage of the total area in those two myotomes, Fig. 3A). To study the co-localization of pre- and post-synaptic regions, we quantified the Pearson’s correlation co-efficient using the Bitplane: Imaris 7.7.2. software. Data analysis was made in the Microsoft Excel and GraphPad Prism version 5.01 software.
Touch-evoke response assays and muscular weakness analysis
Touch-evoke response assays were carried out as described before (26). In brief, we captured videos (60 frames/s) of embryos/larvae at stages 62, 72 and 96 hpf and they were analyzed frame-by-frame using the ImageJ analysis software. Using a metric ruler, we marked the start and end points of the swimming bout (1.1 cm). After delivering mechanosensory stimuli to the embryo/larvae by touching the tail with a needle, we measured the time interval of swimming the fixed distance. Data analysis was made in the Microsoft Excel and GraphPad Prism version 5.01 software.
Statistical analysis
Results were obtained from at least three independent experiments and shown as mean ± SEM. Statistical analysis (unpaired t-test) was performed to compare two groups (Figs. 3E, L, S and 4G; Supplementary Material, Figs. S3L, S5Cand S5F). A one-way analysis of variance test was performed for statistical analysis in all other cases by using the GraphPad Prism version 5.01 software (Figs. 5H and I, 6T and U and 7J), (*P < 0.05, **P < 0.01 and ***P < 0.001).
Western blots
Proteins were isolated according to (40) and run on a native and denatured 10% acrylamide gel. Membranes were incubated with the anti-GARS antibody (Abcam, catalog no. ab42905; 1:5000), anti-p-Eif2a (Eukaryotic translation initiation factor 2A, Cell Signaling Technology, catalog no. 9722; 1:250) and total-Eif2α (Cell Signaling Technology, catalog no. 2103; 1:500). Normalization for loading was done by using a monoclonal anti-β-actin (Sigma, catalog no. A5441; 1:2000).
Heart rate measurements
Heart rate [beats per minute (bpm)], analysis of captured videos (60 frames/s) was done as described previously (41). In brief, larvae at 72 hpf (AB, wild-type, n = 10; siblings, n = 11; garss266/s266, n = 11; P > 0.05, ns) were anesthetized for 2 min in 0.04 g/ml tricaine and then placed lateral to the microscope lens for video capture.
In situ hybridization
Whole-mount in situ hybridization experiments with gars antisense probe were performed in different stages embryos and larvae, according to (42).
Site-directed mutagenesis and overexpression of C201R, G240R and G526R in gars
The zebrafish gars open-reading frame was polymerase chain reaction (PCR) amplified using a full-length cDNA clone as a template. PCR-amplified gars was cloned into the pBluescript R vector (Agilent Technologies, catalog no. 212240) using standard methods. Subsequent site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Agilent Technologies, catalog no. 200518). The appropriate C236R-bearing oligos are FW: 5′-GGATGTCAAGAACGGAGAGCGTTTTCGTGCAGACCACCTTC-3′ and REV: 5′-GAAGGTGGTCTGCACGAAAACGCTCTCCGTTCTTGACATCC-3′, the G319R are FW: 5′-GCCTGGAGGCAACATGCAACGCTATTTAAGGCCAGAAACC-3′ and REV: 5′-GGTTTCTGGCCTTAAATAGCGTTGCATGTTGCCTCCAGGC-3′ and the G605R are FW: 5′-TGTGATCGAACCCTCTTTCCGTATCGGGAGGATCATGTAC-3′ and REV: 5′-GTACATGATCCTCCCGATACGGAAAGAGGGTTCGATCACA-3′. Then, we in vitro transcribed the mutagenized gars mRNA (C236R equivalent to the C201R, G319R equivalent to human G240R and G605R equivalent to G526R) using a MEGAscript® T7 Transcription Kit (Ambion, catalog no. AM1333) and we injected the capped-mRNAs at 330pg for mutations C236R and G319R and at 250 pg for G605R mutation in one or two cells in wild-type, siblings and garss266/s266 embryos. The overexpression of Gars was quantified by denaturing immunoblots with a specific anti-GARS antibody (Abcam, catalog no. ab42905; 1:5000).
Supplementary Material
Funding
This work was supported in part by the Human Frontier Science Program Organization (CDA to D.B.); The Greek General Secretariat of Research and Technology, European Social Fund and National Funds (ZfValves, Aristia I grant to D.B.); A.A. is supported by the Muscular Dystrophy Association (MDA294479) and the Fletcher Family Fund. L.B.G. was supported by the National Institutes of Health (Cellular and Molecular Biology Training grant GM007315, Medical Scientist Training grant GM07863 and F30 NRSA NS092238).
Supplementary Material
Acknowledgements
We thank Stamatis Pagakis and the BRFAA BioImaging Unit. N.M. was a PhD student at the Medical School, University of Crete.
Conflict of Interest statement. None declared.
References
- 1.Skre H. (1974) Genetic and clinical aspects of Charcot-Marie-Tooth's disease. Clin. Genet., 6, 98–118. [DOI] [PubMed] [Google Scholar]
- 2.Barisic N., Claeys K.G., Sirotković-Skerlev M., Löfgren A., Nelis E., De Jonghe P., Timmerman V. (2008) Charcot-Marie-Tooth disease: a clinico-genetic confrontation. Ann. Hum. Genet., 72, 416–441. [DOI] [PubMed] [Google Scholar]
- 3.Verhoeven K., De Jonghe P., Coen K., Verpoorten N., Auer-Grumbach M., Kwon J.M., FitzPatrick D., Schmedding E., De Vriendt E., Jacobs A. et al. (2003) Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am. J. Hum. Genet., 72, 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vettori A., Bergamin G., Moro E., Vazza G., Polo G., Tiso N., Argenton F., Mostacciuolo ML (2011) Developmental defects and neuromuscular alterations due to mitofusin 2 gene (MFN2) silencing in zebrafish: a new model for Charcot-Marie-Tooth type 2A neuropathy. Neuromuscul. Disord., 21, 58–67. [DOI] [PubMed] [Google Scholar]
- 5.Evgrafov O.V., Mersiyanova I., Irobi J., Van Den Bosch L., Dierick I., Leung C.L., Schagina O., Verpoorten N., Van Impe K., Fedotov V. et al. (2004) Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat. Genet., 36, 602–606. [DOI] [PubMed] [Google Scholar]
- 6.Mersiyanova I.V., Perepelov A.V., Polyakov A.V., Sitnikov V.F., Dadali E.L., Oparin R.B., Petrin A.N., Evgrafov O.V. (2000) A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. Am. J. Hum. Genet., 67, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonellis A., Lee-Lin S.Q., Wasterlain A., Leo P., Quezado M., Goldfarb L.G., Myung K., Burgess S., Fischbeck KH, Green E.D. (2006) Functional analyses of glycyl-tRNA synthetase mutations suggest a key role for tRNA-charging enzymes in peripheral axons. J. Neurosci., 26, 10397–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordanova A., Irobi J., Thomas F.P., Van Dijck P., Meerschaert K., Dewil M., Dierick I., Jacobs A., De Vriendt E., Guergueltcheva V. et al. (2006) Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat. Genet., 38, 197–202. [DOI] [PubMed] [Google Scholar]
- 9.Latour P., Thauvin-Robinet C., Baudelet-Méry C., Soichot P., Cusin V., Faivre L., Locatelli M.C., Mayençon M., Sarcey A., Broussolle E. et al. (2010) A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. Am. J. Hum. Genet., 86, 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vester A., Velez-Ruiz G., McLaughlin H.M., Lupski J.R., Talbot K., Vance J.M., Züchner S., Roda R.H., Fischbeck K.H. et al. , NISC Comparative Sequencing Program, (2013) A loss-of-function variant in the human histidyl-tRNA synthetase (HARS) gene is neurotoxic in vivo. Hum. Mutat., 34, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez M., McLaughlin H., Houlden H., Guo M., Yo-Tsen L., Hadjivassilious M., Speziani F., Yang X.L., Antonellis A., Reilly M.M. et al. (2013) Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J. Neurol. Neurosurg. Psychiatry, 84, 1247–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibba M., Söll D. (2004) Aminoacyl-tRNAs: setting the limits of the genetic code. Genes Dev., 18, 731–738. [DOI] [PubMed] [Google Scholar]
- 13.Schimmel P. (2008) Development of tRNA synthetases and connection to genetic code and disease. Protein Sci., 17, 1643–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonellis A., Green E.D. (2008) The role of aminoacyl-tRNA synthetases in genetic diseases. Annu. Rev. Genomics Hum. Genet., 9, 87–107. [DOI] [PubMed] [Google Scholar]
- 15.Xie W., Nangle L.A., Zhang W., Schimmel P., Yang X.L. (2007) Long-range structural effects of a Charcot–Marie–Tooth disease-causing mutation in human glycyl-tRNA synthetase. Proc. Natl Acad. Sci. USA, 104, 9976–9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin L.B., Sakaguchi R., McGuigan D., Gonzalez M.A., Searby C., Züchner S., Hou Y.M., Antonellis A. (2014) Impaired function is a common feature of neuropathy-associated glycyl-tRNA synthetase mutations. Hum. Mutat., 35, 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nangle L.A., Zhang W., Xie W., Yang X.L., Schimmel P. (2007) Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc. Natl Acad. Sci. USA, 104, 11239–11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seburn K.L., Nangle L.A., Cox G.A., Schimmel P., Burgess R.W. (2006) An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron, 51, 715–726. [DOI] [PubMed] [Google Scholar]
- 19.Achilli F., Bros-Facer V., Williams H.P., Banks G.T., AlQatari M., Chia R., Tucci V., Groves M., Nickols C.D., Seburn K.L. et al. (2009) An ENU-induced mutation in mouse glycyl-tRNA synthetase (GARS) causes peripheral sensory and motor phenotypes creating a model of Charcot-Marie-Tooth type 2D peripheral neuropathy. Dis. Model Mech., 7, 359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chihara T., Luginbuhl D., Luo L. (2007) Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nat. Neurosci., 7, 828–837. [DOI] [PubMed] [Google Scholar]
- 21.Beis D., Bartman T., Jin S.W., Scott I., D'Amico L., Ober E., Verkade H., Frantsve J., Field H., Wehman A. et al. (2005) Genetic and cellular analyses of zebrafish atrio-ventricular cushion and valve development. Development, 132, 4193–4204. [DOI] [PubMed] [Google Scholar]
- 22.Turner R.J., Lovato M., Schimmel P. (2000) One of two genes encoding glycyl-tRNA synthetase in Saccharomyces cerevisiae provides mitochondrial and cytoplasmic functions . J. Biol. Chem., 275, 27681–27688. [DOI] [PubMed] [Google Scholar]
- 23.Panzer J.A., Gibbs S.M., Dosch R., Wagner D., Mullins M.C., Granato M., Balice-Gordon RJ (2005) Neuromuscular synaptogenesis in wild-type and mutant zebrafish. Dev. Biol., 285, 340–357. [DOI] [PubMed] [Google Scholar]
- 24.Westerfield M., Eisen J.S. (1988) Neuromuscular specificity: pathfinding by identified motor growth cones in a vertebrate embryo. Trends Neurosci., 11, 18–22. [DOI] [PubMed] [Google Scholar]
- 25.Postel R., Vakeel P., Topczewski J., Knöll R., Bakkers J. (2008) Zebrafish integrin-linked kinase is required in skeletal muscles for strengthening the integrin-ECM adhesion complex. Dev. Biol., 318, 92–101. [DOI] [PubMed] [Google Scholar]
- 26.Smith L.L., Beggs H.A., Gupta A.V. (2013) Analysis of skeletal muscle defects in larval zebrafish by birefringence and touch-evoke escape response assays. J. Vis. Exp., 82, 50925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sehnert A.J., Huq A., Weinstein B.M., Walker C., Fishman M., Stainier D.Y. (2002) Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet., 31, 106–110. [DOI] [PubMed] [Google Scholar]
- 28.Christodoulou K., Kyriakides T., Hristova A.H., Georgiou D.M., Kalaydjieva L., Yshpekova B., Ivanova T., Weber J.L., Middleton L.T. (1995) Mapping of a distal form of spinal muscular atrophy with upper limb predominance to chromosome 7p. Hum. Mol. Genet., 4, 1629–1632. [DOI] [PubMed] [Google Scholar]
- 29.Sambuughin N., de Bantel A., McWilliams S., Sivakumar K (2003) Deafness and CMT disease associated with a novel four amino acid deletion in the PMP22 gene. Neurology, 60, 506–508. [DOI] [PubMed] [Google Scholar]
- 30.Antonellis A., Ellsworth R.E., Sambuughin N., Puls I., Abel A., Lee-Lin S.Q., Jordanova A., Kremensky I., Christodoulou K., Middleton L.T. et al. (2003) Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet., 72, 1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivakumar K., Kyriakides T., Puls I., Nicholson G.A., Funalot B., Antonellis A., Sambuughin N., Christodoulou K., Beggs J.L., Zamba-Papanicolaou E. et al. (2005) Phenotypic spectrum of disorders associated with glycyl-tRNA synthetase mutations. Brain, 128, 2304–2314. [DOI] [PubMed] [Google Scholar]
- 32.He W., Bai G., Zhou H., Wei N., White N.M., Lauer J., Liu H., Shi Y., Dumitru C.D., Lettieri K. et al. (2015) CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature, 526, 710–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakasugi K., Slike B.M., Hood J., Otani A., Ewalt K.L., Friedlander M., Cheresh D.A., Schimmel P. (2002) A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl Acad. Sci. USA, 99, 173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herzog W., Müller K., Huisken J., Stainier D.Y. (2009) Genetic evidence for a noncanonical function of seryl-tRNA synthetase in vascular development. Circ. Res., 104, 1260–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin S.W., Beis D., Mitchell T., Chen J.N., Stainier DY (2005) Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development, 132, 5199–5209. [DOI] [PubMed] [Google Scholar]
- 36.Moro E., Ozhan-Kizil G., Mongera A., Beis D., Wierzbicki C., Young R.M., Bournele D., Domenichini A., Valdivia L.E., Lum L. et al. (2012) In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev. Biol., 366, 327–340. [DOI] [PubMed] [Google Scholar]
- 37.Salazar-Grueso E.F., Kim S., Kim HE (1991) mbryonic mouse spinal cord motor neuron hybrid cells. Neuroreport, 2, 505–508. [DOI] [PubMed] [Google Scholar]
- 38.Müller J.S., Jepson C.D., Laval S.H., Bushby K., Straub V., Lochmüller H. (2010) Dok-7 promotes slow muscle integrity as well as neuromuscular junction formation in a zebrafish model of congenital myasthenic syndromes. Hum. Mol. Genet., 19, 1726–1740. [DOI] [PubMed] [Google Scholar]
- 39.Ramesh T., Lyon A.N., Pineda R.H., Wang C., Janssen P.M., Canan B.D., Burghes A.H., Beattie C.E. (2010) A genetic model of amyotrophic lateral sclerosis in zebrafish displays phenotypic hallmarks of motoneuron disease. Dis. Model. Mech., 3, 652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monte Westerfield. (1995) The Zebrafish Book: A Guide for the Laboratory use of Zebrafish (Danio Rerio). University of Oregon Press, Eugene. [Google Scholar]
- 41.Kalogirou S., Malissovas N., Moro E., Argenton F., Stainier D.Y., Beis D. (2014) Intracardiac flow dynamics regulate atrioventricular valve morphogenesis. Cardiovasc. Res., 104, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thisse B., Thisse C. (2014) In situ hybridization on whole-mount zebrafish embryos and young larvae. Methods Mol. Biol., 1211, 53–67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.