Abstract
Muscular manifestation of myotonic dystrophy type 1 (DM1), a common inheritable degenerative multisystem disorder, is mainly caused by expression of RNA from a (CTG·CAG)n-expanded DM1 locus. Here, we report on comparative profiling of expression of normal and expanded endogenous or transgenic transcripts in skeletal muscle cells and biopsies from DM1 mouse models and patients in order to help us in understanding the role of this RNA-mediated toxicity. In tissue of HSALR mice, the most intensely used ‘muscle-only’ model in the DM1 field, RNA from the α-actin (CTG)250 transgene was at least 1000-fold more abundant than that from the Dmpk gene, or the DMPK gene in humans. Conversely, the DMPK transgene in another line, DM500/DMSXL mice, was expressed ∼10-fold lower than the endogenous gene. Temporal regulation of expanded RNA expression differed between models. Onset of expression occurred remarkably late in HSALR myoblasts during in vitro myogenesis whereas Dmpk or DMPK (trans)genes were expressed throughout proliferation and differentiation phases. Importantly, quantification of absolute transcript numbers revealed that normal and expanded Dmpk/DMPK transcripts in mouse models and DM1 patients are low-abundance RNA species. Northern blotting, reverse transcriptase–quantitative polymerase chain reaction, RNA-sequencing and fluorescent in situ hybridization analyses showed that they occur at an absolute number between one and a few dozen molecules per cell. Our findings refine the current RNA dominance theory for DM1 pathophysiology, as anomalous factor binding to expanded transcripts and formation of soluble or insoluble ribonucleoprotein aggregates must be nucleated by only few expanded DMPK transcripts and therefore be a small numbers game.
Introduction
Myotonic dystrophy type 1 (DM1, OMIM no. 160900) is an autosomal dominant repeat expansion disorder, affecting skeletal and smooth muscle as well as the heart, the endocrine system, the eye and the central nervous system (1). The multisystemic manifestation and progression of DM1 are caused by expansion of a (CTG·CAG)n repeat, located in the 3′-untranslated region (3′ UTR) of the dystrophia myotonica protein kinase (DMPK) gene (2) and in an overlapping antisense transcription unit in the DM1 locus (3). In DM1 families the expanded repeat is unstable, both somatically and intergenerationally, with a bias toward expansion, causing progression of disease symptoms during ageing and over successive generations (4).
Several mechanisms may contribute to the molecular pathogenesis of DM1 (5). Expanded DMPK transcripts are retained in the nucleus, where they form focal complexes in insoluble or diffuse-soluble state by abnormal association with transcription factors and RNA-binding proteins, like members of the muscleblind-like family (MBNL1–3), DEAD-box helicases and hnRNP proteins (1,6,7). In turn, abnormal phase transitions in RNP complexes lead to sequestering of factors needed for processing of other transcripts with in trans consequences for faithful alternative splicing and polyadenylation and expression of miRNAs (7,8). Production of proteins by ribosomes that decode the normally untranslated (CUG)n repeat tract in DMPK mRNA by a newly discovered process, coined repeat-associated non-ATG (RAN) translation, is also possible (9,10). Similar toxic events may occur with antisense transcripts originating from the complementary strand of the DM1 locus, overlapping the 3′ end of the DMPK gene. Abnormal RNAs are thus formed with an expanded (CAG)n repeat, potentially leading to the production of homopolymeric peptides by RAN translation of the (CAG)n repeat, which may evoke an imbalance in proteostasis (9,10). Finally, it cannot be excluded that problems with DNA replication across the repeat tract or abnormal epigenetic modification of the chromatin region containing the DM1 locus also contribute to pathology (3,11).
Together, alterations in the transcriptome, proteome and replisome may compromise the physiological integrity of cells and tissues in which the mutant DMPK and the DM1-antisense gene are expressed. Throughout development, growth and adulthood this imbalance may lead to the loss of function and ultimately to cell degeneration, causing the muscle wasting and CNS white matter loss in patients (4,12).
For study of biological mechanisms underlying DM1 pathology and for testing of possible therapeutic strategies in preclinical studies, several animal models are available, including Drosophila, zebra fish and mouse (13,14). Predominant focus is thereby oriented toward mechanisms involved in RNA-based disease etiology. Notably, DM1 animal models differ profoundly in nature, structural organization and chromatin context of their transgenic insert and in the length of the (CTG·CAG)n segment therein. Comparison of pathobiological findings between models and extrapolation to the situation in patients remain therefore difficult. Work of others has already demonstrated that the timing of DMPK expression, i.e. the onset of potential RNA toxicity, influences phenotypic severity (15). Expression of RNA with an abnormal repeat tract in satellite cells or neuronal progenitor cells may affect proper muscle and brain development (16–19) and have serious consequences for tissue regenerative capacity in adulthood. The absolute number of expanded RNAs and their structure at any given moment may also be crucial, as these ultimately will influence the extent of toxicity caused by abnormal RNP binding or abnormal properties of RAN translation products (20–22). The type of gene promoters, whether from endogenous or ectopic origin, that drive transcription during development and ageing, and the structure of the transcripts that entail the repeat segment are therefore critical parameters in animal models and patients.
Here, we report on comparison of expression and measurement of absolute numbers of (CUG)n-repeat containing RNAs in muscle cells and tissues of four commonly used mouse DM1 models and in cells and biopsies from patients. DM1 mouse models express transgenes with different promoters, different structural organization and different repeat lengths: DM500, DMSXL, Tg26 and HSALR (Table 1). DM500 and DMSXL mice are both descendants of the DM300-328 line, which was subject to intergenerational repeat expansion. These mice carry a complete human DM1 locus (23,24). The DMPK transgene in Tg26 mice carries a tandem insert of ∼25 copies of the complete human DMPK gene, with a normal-sized (CTG)11 repeat (25,26). In HSALR mice, the transgene is under control of the ACTA1 promoter and the repeat is embedded in the context of the ACTA1 gene (27). The rationale for quantification of repeat RNA expression in these models is that knowledge about toxic RNA concentration will provide us with more insight in pathophysiological cascades per se, especially as more and more anatomical, physiological and behavioral phenotype data become available, enabling relatively easy cross comparisons. Furthermore, some of the DM1 models have already been extensively used for preclinical translational studies in the past decade, but translation of findings in these models has been difficult.
Table 1.
Characteristics of DM1 mouse models used in this study.
| Mouse model | Transgene | Transgene copy number | Promoter | Expression | (CTG)n | Genetic background | References |
|---|---|---|---|---|---|---|---|
| DM500 (DM300-328 line) | Human DM1 locus (43 kb transgene) | 1 | Human DMPK (∼11.5 kb region upstream of main TSS) | All DM1-related tissues (e.g. skeletal muscle, heart and CNS) | 500–600 | >90% C57BL/6 | (23) |
| DMSXL (DM300-328 line) | Human DM1 locus (43 kb transgene) | 1 | Human DMPK (∼11.5 kb region upstream of main TSS) | All DM1-related tissues (e.g. skeletal muscle, heart and CNS) | ∼1300 | >90% C57BL/6 | (24) |
| Tg26 | Human DMPK gene (14 kb transgene) | ∼25 | Human DMPK (∼1.9 kb region upstream of main TSS) | All DM1-related tissues (e.g. skeletal muscle, heart and CNS) | 11 | FVB/n | (25) |
| HSALR (LR20b line) | Human α-actin gene; CTG repeat inserted in 3′ UTR (7.1 kb transgene) | 2 | Human α-actin (∼2.1 kb region upstream of TSS) | Skeletal muscle only | 220–250 | FVB/n | (27) |
| WT | No transgene | n.a. | n.a. | n.a. | n.a. | >90% C57BL/6 | n.a. |
TSS, transcription start site; n.a., not applicable.
We demonstrate that, in comparison with expression of normal and mutant DMPK transcripts in patient cells, considerable variation exists in level and developmental timing of transgene expression in DM1 cell and animal models. A remarkable low level of expression with absolute numbers of, at most, a few dozen RNA molecules per cell was observed for DMPK transcripts in human samples. Our findings highlight the hitherto unrecognized involvement of low-abundance RNA molecules in DM1 pathophysiology, altering our current view on the RNA gain-of-function theory, which explains the role of repeat RNA in DM1 manifestation. We discuss the possible implications of our findings for future interpretation of data from fundamental and translational studies in which these DM1 models and patient cells will be used.
Results
Derivation of myogenic cell lines from DM1 mouse models
Characteristics of mouse models included in this study are listed in Table 1. For profiling of transgene expression at the cellular level, we established conditionally immortalized myoblast populations from each model by pooling clones of individual cells derived from the calf muscle complex from double hemizygous mice carrying one transgenic DM1 allele and one H-2Kb-tsA58 allele (28). As there is strong evidence that satellite cells from different inbred mice behave intrinsically differently (29,30), it is important to note that crossings included different genetic backgrounds to generate the double hemizygous animals. The cell populations have therefore distinct mixed genetic backgrounds with contributions of C57BL6, FVB/n, CBA/Ca and C57BL/10. We do believe, however, that these differences have no major impact on transcriptome composition and therefore should not overtly confound our comparison.
An important feature of the immortalized myoblasts is that during prolonged passaging in culture under permissive conditions cells have the tendency to undergo polyploidization, as an effect of the presence of the temperature-sensitive SV40 large T-antigen (31). This leads to a mix of 2N and 4N cells in populations of the different lineages and to variation in the absolute abundance of individual transcripts per cell (Supplementary Material, Fig. S1A). Hence, all possible care was taken to compare myoblasts from similar passage numbers.
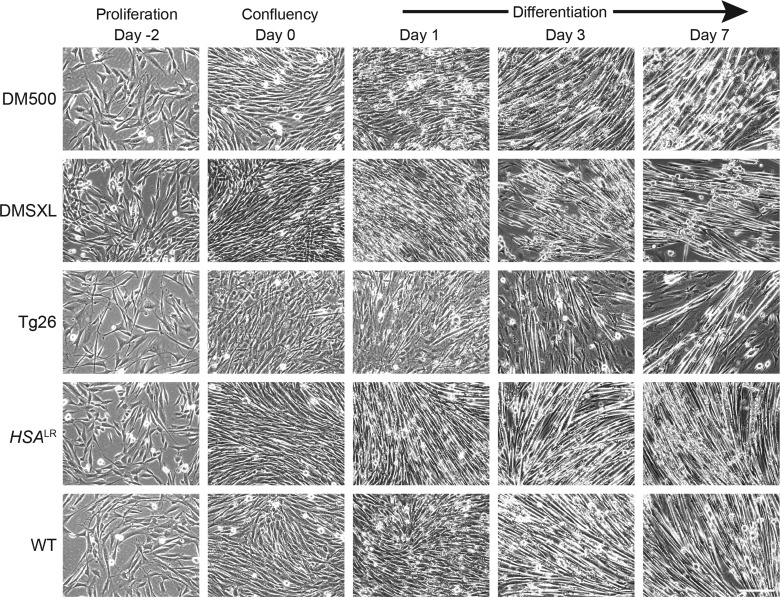
DM500, DMSXL, HSALR and WT myoblast populations had normal morphological appearance and showed apparently normal proliferative capacity and terminal differentiation features upon shift to myogenesis-promoting conditions (Fig. 1). Multinuclear contractile myotubes appeared after 3–5 days of differentiation. The Tg26 cell population had normal morphological appearance, but showed slightly disordered cell alignment and diminished fusion capacity upon induction of differentiation. This may be due to overproduction of certain DMPK protein isoforms (26,32,33).
Figure 1.
Morphology of myoblasts derived from DM1 and control mouse models. Conditionally immortalized myoblasts were derived from the GPS muscle complex of DM1 mouse models. Myoblasts differentiated into contractile myotubes under low-serum conditions. Representative images of cultures during proliferation (Day -2), confluency (Day 0) and differentiation (Days 1, 3 and 7) are shown. Bar 100 µm.
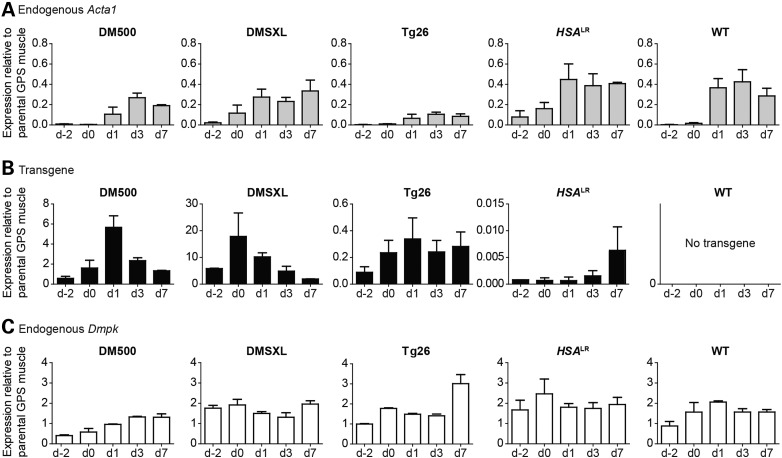
To quantify gene expression during proliferation and differentiation of the myoblast populations, RNA was isolated at various time points and analyzed by northern blotting and reverse transcriptase–quantitative polymerase chain reaction (RT-qPCR). Progression of myogenic differentiation in vitro was accompanied by a clear increase in skeletal muscle α-actin (Acta1) mRNA (Fig. 2A), a well-known differentiation marker encoding a major constituent of the contractile apparatus (34–36). The increase was less profound in Tg26 cultures, in accordance with their diminished fusion capacity. Whereas Acta1 was induced, β-actin (Actb) expression decreased during the 7-day differentiation period (Supplementary Material, Fig. S2) (37).
Figure 2.
Expression profile of proliferating and differentiating myoblasts derived from DM1 and control mouse models. Analysis of endogenous Acta1 (A), transgene (B) and endogenous Dmpk (C) mRNA levels by northern blotting. Each hybridization signal was normalized to that of 18S rRNA and then compared with the normalized value measured for the same (trans)gene in the GPS muscle from that particular DM1 mouse model. Data were obtained from two independent culture series per cell line, for which triplicate cultures per series were pooled and analyzed; bars represent mean + SEM. For profiling of endogenous Dmpk (C) in Tg26 samples RT-qPCR was used, because signals of transgenic DMPK and endogenous Dmpk overlapped on northern blot. Bars represent mean + SEM. Timing of differentiation on the x-axis refers to Figure 1.
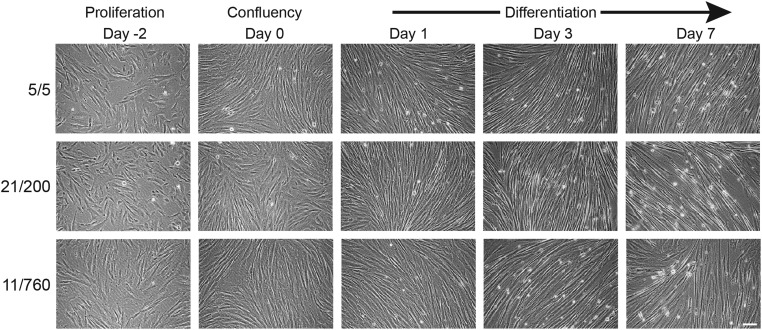
For reference, primary human muscle cells obtained from a healthy individual and two DM1 patients were also included in our study. Myoblasts with healthy (CTG)5/(CTG)5 or disease-specific (CTG)21/(CTG)200 or (CTG)11/(CTG)760 repeat combinations appeared morphologically diverse, which can be explained by their different origin and culture history (Fig. 3). Multinucleated myotubes were formed during differentiation, but spontaneous contractions were never observed (Fig. 3, data not shown). RNA expression analysis corroborated this observation by showing that ACTA1 expression was minimal at all time points measured and remained low in comparison with Acta1 levels in DM500 GPS tissue (Fig. 4A). We, therefore, conclude that human cultures did not attain the same endpoint of terminal differentiation as mouse myoblasts under our in vitro conditions.
Figure 3.
Morphology of primary human DM1 and healthy myogenic cells. Human primary myoblasts aligned and fused under low-serum conditions. Representative images at different time points during proliferation (Day -2), confluency (Day 0), and differentiation (Days 1, 3 and 7) are shown. The number of CTG triplets for the two DMPK alleles are indicated for the one healthy (5/5) and two DM1 (21/200 and 11/760) cultures. Bar 100 µm.
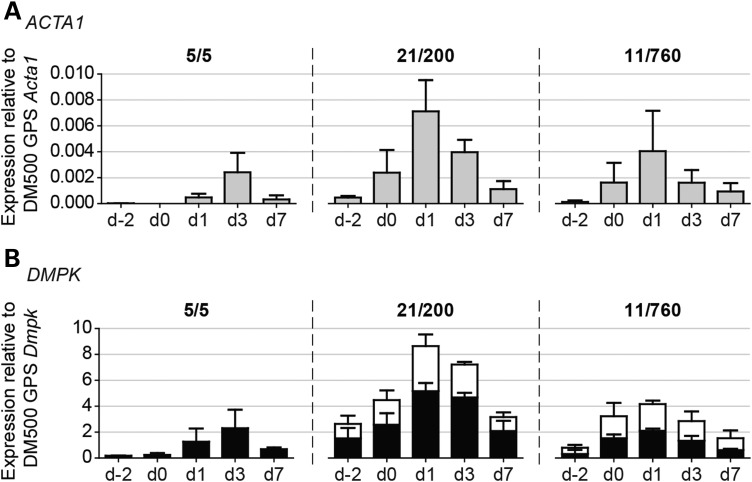
Figure 4.
Expression profile of proliferating and differentiating primary human DM1 myogenic cells. Assessment of ACTA1 (A) and DMPK (B) transcript levels by northern blot analysis. Signal strength in each sample was normalized to that of 18S rRNA, as outlined in the legend of Figure 2. To facilitate direct comparison with DM1 mouse model-derived myoblasts, ratios were related to Acta1 and Dmpk levels in DM500 GPS muscle. Data were obtained from two independent culture series per cell line, for which triplicate cultures per series were pooled and analyzed; bars represent mean + SEM. Stacked bar graphs show levels of normal-sized DMPK (black part) and expanded DMPK (white part) in patient-derived cultures. On average, the ratio expanded versus normal-sized DMPK transcripts was 0.63 for 21/200 and 0.84 for 11/760. Timing of differentiation on the x-axis refers to Figure 3.
Transgene expression differs between DM1 myoblast models
Transgene expression was assayed in the myoblast populations during proliferation and differentiation (Fig. 2B and Supplementary Material, Fig. S3). DMPK transgene expression in differentiating DM500 and DMSXL cells transiently increased 3- to 10-fold and then returned to basal levels in proliferating myoblasts. A modest increase in transgene expression was also observed in differentiating Tg26 myoblasts, but this change appeared to be more permanent. In contrast, transcript levels from the Dmpk gene hardly varied with differentiation state in any of the five myoblast populations (Fig. 2C). The observed differential regulation of endogenous/transgenic Dmpk/DMPK expression may be caused by (i) species differences between the structure and function of Dmpk/DMPK promoters, (ii) differences in the length of the region upstream of the TSS in the human transgenes used (Table 1), (iii) differences between chromosomal insert sites and the endogenous Dmpk locus or (iv) different epigenetic alterations across the transgenic loci. Influence of the (CUG)n-repeat length on RNA stability is unlikely, because temporal profiles of transgenic RNA expression in DM500 and DMSXL cells during myogenic differentiation levels were highly similar.
For DMPK expression in human myoblasts, we performed separate quantitative analysis of healthy and expanded transcripts, which migrate differentially on northern gels. Steady-state levels of normal and expanded DMPK RNA molecules were approximately similar for both the 21/200 and the 11/760 patient cell cultures (Fig. 4B). Comparison of total DMPK expression levels between healthy 5/5 and patient cell cultures showed variation. However, temporal profiles had a similar shape with a peak in expression during early differentiation, alike expression behavior of DMPK transcripts from the DMPK transgene in mouse myoblasts. This supports our idea that species-specific differences in the DMPK/Dmpk promoter explain the differential expression pattern of DMPK and Dmpk transcripts.
For better interpretation of in vitro and in vivo findings, we compared DMPK transgene expression in myogenic cultures to that in parental GPS muscle in the DM1 models. Transgene expression was ∼0.6- to 6-fold higher, 6- to 18-fold higher and 3- to 11-fold lower for DM500, DMSXL and Tg26 myoblast/myotube cultures, respectively (Fig. 2B). Of particular, interest was the expression profile of the expanded ACTA1 transgene in differentiating myoblasts from the HSALR model, showing that transcripts were expressed only late in differentiation (Fig. 2B). Even in fully differentiated hemizygous HSALR myotubes in vitro, expanded ACTA1 transgene expression remained ∼200-fold lower than in GPS muscle from homozygous HSALR mice.
Transgene expression in distinct DM1 mouse models differs over a 1000-fold range
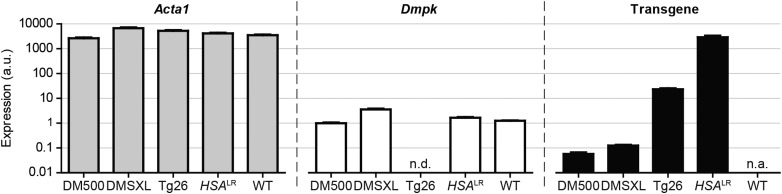
By extending our analysis to mouse muscle in vivo, using the GPS complex as RNA source and 18S rRNA for normalization, our comparison revealed that Acta1 expression was remarkably similar between models (Fig. 5 and Supplementary Material, Fig. S4A), despite differences in their genetic background (Table 1). Also Dmpk expression showed only minor fluctuations. Whereas Acta1 and Dmpk expression did not vary >2- to 4-fold between mouse models, the mean level of Acta1 RNA was ∼2,500-fold higher than that of Dmpk RNA.
Figure 5.
Transgene expression in DM1 mouse skeletal muscle. Expression of Acta1, Dmpk and transgenes was determined in GPS muscle of each DM1 mouse model. Transcripts were quantified by northern blotting using 18S rRNA for normalization (n ≥ 4). Endogenous Dmpk RNA levels in DM500 GPS muscle were used as reference (set to one). Mean + SEM are shown. a.u., arbitrary units; n.d., not determined (Dmpk and transgenic signals overlap); n.a., not applicable.
We next compared transgene expression in GPS muscles from DM500, DMSXL and Tg26 mice. Quantification was straightforward, since all models expressed differentially expanded, but otherwise similar and intact DMPK transgenes. Transgenic DMPK RNA accumulated in muscle from hemizygous DM500 and DMSXL mice to a 10-fold lower level than RNA from the endogenous Dmpk genes (Fig. 5). In contrast, hemizygous Tg26 GPS muscle expressed 20- to 25-fold more DMPK mRNA than Dmpk mRNA [assuming that Dmpk expression in Tg26 mice (FVB background) was similar to that in DM500 mice (C57BL/6 background)]. Determination of relative levels of transgenic RNA in HSALR muscle appeared more challenging. Use of a (CAG)9 probe to detect (CUG)n segments, the only sequence shared between transgenic products, proved unreliable to compare RNAs with different repeat lengths (data not shown). Therefore, a balanced mix of ACTA1 and DMPK probes, generated by random-primed labeling on cDNA templates of equal size was used instead (see the Materials and Methods’ section). With this approach we found ACTA1 (CUG)n RNA over 1000-fold higher expressed than Dmpk mRNA in homozygous HSALR mice (Fig. 5). This means that the concentration of expanded transcripts in HSALR muscle is extraordinarily high and similar to that of Acta1 mRNA. This finding is perhaps not too surprising as both promoter and backbone of the human transgene and the Acta1 gene share strong homology.
Analysis of DMPK RNA levels in human muscle allows direct interpretation of transgene dosage
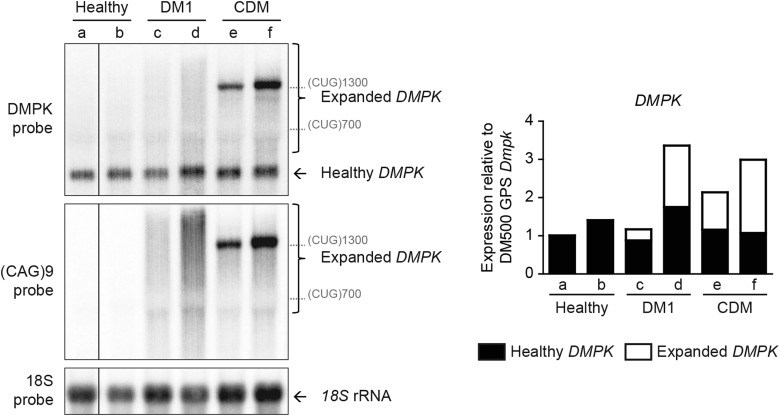
Skeletal muscle samples from healthy humans and DM1 patients were included in our study to extend our comparisons. DMPK mRNA expression in the samples varied, but was not >3-fold higher than the level of Dmpk mRNA in mouse GPS tissue (Fig. 6). Expression of normal-sized and of expanded DMPK alleles were about equal in each of the patient samples. Since DMPK levels in human muscles and Dmpk levels in mouse muscle show high similarity, observations about transgene expression dosage effects in DM1 mice may be directly translated to relevance for DM1 patients. Any difference in DMPK mRNA content between human samples probably represents variation in fiber type and muscle origin or must be caused by differentiation or disease state. Note that, effects of ageing on repeat length heterogeneity were clearly visible on northern blot: in samples from adult DM1 patients a smear was observed for RNA from the expanded allele, whereas in congenital DM (CDM) patient material a defined signal was apparent (Fig. 6) (38).
Figure 6.
DMPK expression in human control and DM1 patient tissue. Northern blot (left) of human control and DM1 muscle tissue using a DMPK and a (CAG)9 repeat probe. Healthy DMPK mRNA appeared as a defined band in all tissues. Expanded DMPK mRNA appeared as a defined band in congenital DM1 tissue (CDM-e and -f), but as a smear (representing somatic mosaicism of repeat size) in adult DM1 tissue (DM1-c and -d). As size markers, we indicated the location of DMPK (CUG)700 and (CUG)1300 transcripts. DMPK transcript levels, normalized to those of 18S rRNA, were plotted relative to Dmpk transcript levels in DM500 GPS muscle to allow for comparison with DM1 mouse models (right). Stacked bar graphs show levels of normal-sized DMPK (black part) and expanded DMPK (white part) in patient-derived samples. The ratio expanded versus normal-sized DMPK transcripts was 0.33 in DM1-c; 0.92 in DM1-d; 0.84 in CDM-e and 1.79 in CDM-f.
Multi-pronged analysis reveals low copy number of expanded DMPK transcripts
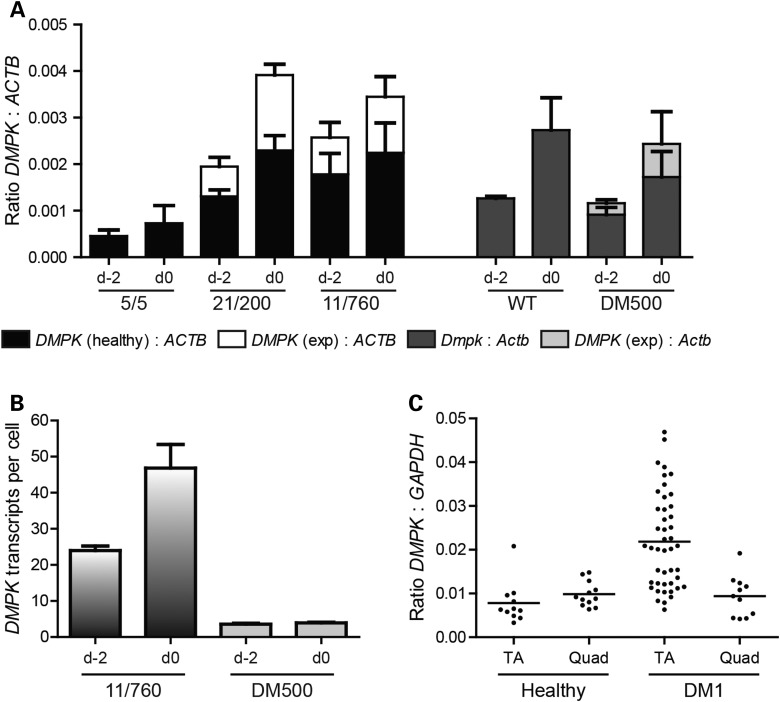
For further stoichiometric and pathomechanistic considerations, we decided to determine the absolute number of DMPK mRNA copies per cell. Quantification on northern blot, after normalization for signal strength and correction for probe length, revealed that DMPK/Dmpk expression was 400- to 2000-fold lower than that of ACTB/Actb in proliferating human/mouse myoblasts (Fig. 7A and Supplementary Material, Fig. S4B). Based on known values for the copy number of ACTB/Actb mRNA, which ranges from 350 to 8000 per cell (39–42) (Supplementary Material, Table S1), we inferred that the absolute number of DMPK transcripts must be in the range of 1–20 per cell. In patient myoblasts, this population consists of approximately equal numbers of normal-sized and expanded DMPK transcript molecules (Figs. 4 and 7A).
Figure 7.
DMPK RNA copy number in human and mouse myoblasts. The absolute number of DMPK transcripts per cell was calculated based on experimental data from northern blotting (A), RT-qPCR (B) and RNA-sequencing (C). (A) Stacked bar graph showing DMPK/Dmpk:ACTB/Actb transcript ratios determined from signal strengths on northern blots for human myoblast lines 5/5, 21/200 and 11/760, and mouse myoblasts lines WT and DM500 (Supplementary Material, Fig. S4B). (B) DMPK transcript copy number in 11/760 and DM500 myoblasts was determined on the basis of known amounts of synthetic DMPK RNA fragments in RT-qPCR (Supplementary Material, Fig. S5). Note that, normal-sized and expanded DMPK transcripts could not be measured independently. (C) DMPK:GAPDH ratio based on RNA-sequencing signal of healthy and DM1 TA and quadriceps (Quad) tissue (www.dmseq.org). Each data point represents a single tissue. Mean + SEM (A and B), or mean only (C) are shown. Descriptions d-2 and d0 refer 938 to Figures 1 and 3.
To validate this rough estimate, DMPK RNA copy number was quantified using two in vitro transcribed DMPK RNAs as standard references in RT-qPCR (Supplementary Material, Fig. S5). We found 20–25 DMPK transcripts per proliferating 11/760 myoblast and 45–50 DMPK molecules in myoblasts just prior to the onset of myogenic differentiation (Fig. 7B). This number includes both healthy and expanded DMPK transcripts. On average about four expanded DMPK transcripts per cell were detected in DM500 myoblasts.
Thirdly, we used RNA-sequencing data of healthy and DM1 skeletal muscle (available via www.dmseq.org) to estimate DMPK transcript copy number. We calculated the DMPK:GAPDH ratio based on RNA-sequencing signal and found that DMPK mRNA molecules were 60- to 160-fold less abundant than GAPDH mRNA (Fig. 7C). Based on current estimates for the prevalence of GAPDH RNA, which is in the order of 250–2900 molecules per cell (39,42–45) (Supplementary Material, Table S2), DMPK transcript copy number must be between 2 and 50 per cell.
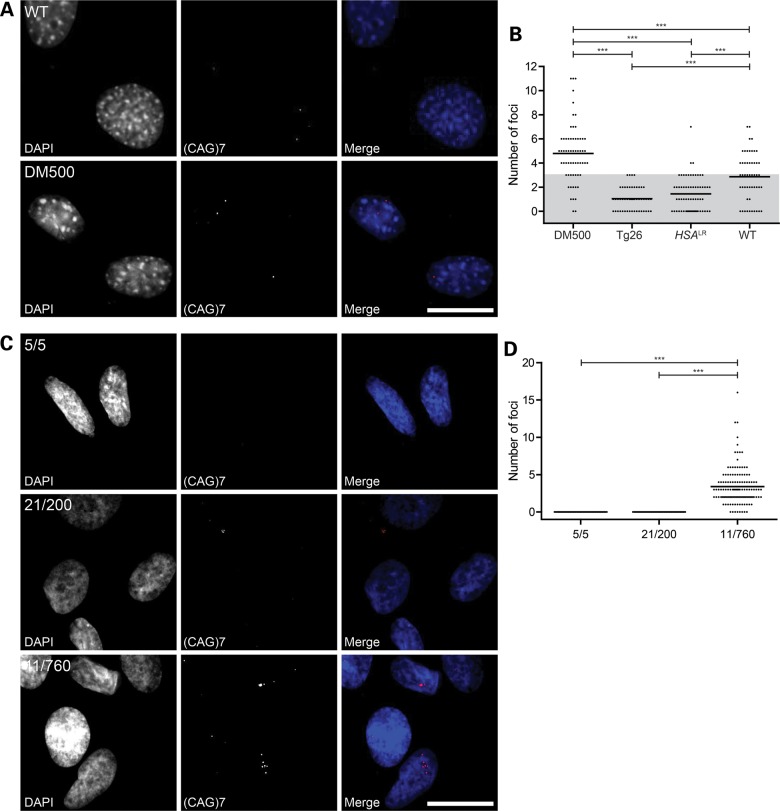
Finally, we determined the number of fluorescent in situ hybridization (FISH)-detectable RNP complexes in mouse and human myoblasts using a Cy3-labeled (CAG)7 probe. Initial analyses demonstrated that the number of FISH signals slightly varied between myoblast pools and experiments and was influenced by culture conditions, such as cell density, feeding regime or changes in the culture medium (data not shown). This variation in foci number appeared suppressible by maintaining a very strict scheme for cell culture. On average, 5–9 foci per cell were detected in DM500 myoblasts (Fig. 8A and B). The number of foci differed between diploid and tetraploid DM500 myoblasts, with averages of ∼5 and ∼9 foci per cell, respectively (Supplementary Material, Fig. S1B). For reasons unknown, we also consistently observed signals in WT mouse myoblasts (Fig. 8A and B). These signals could not be distinguished from expanded DMPK (CUG)n mRNA signals and may also be present in other cell types from mouse. In cells from human origin, we did not observe overt background signals, so the problem may be species specific. To deal with this issue, we decided to apply a correction by defining a background interval in Figure 8. After this correction, an average of two transgenic expanded (CUG)n-specific foci per cell remained in DM500 myoblasts. Foci counts in Tg26 and HSALR myoblasts were within the background range, as expected, based on the idea that the (CTG)11 repeat in Tg26 RNA cannot yield sufficient signal and our finding that the HSALR transgene is barely expressed in proliferating myoblasts. In 11/760 patient myoblasts 3–4 foci per nucleus were detected, whereas no signal was observed in 21/200 and 5/5 myoblasts (Fig. 8C and D), obviously caused by lack of FISH-signal strengths by the limited length of the repeat target.
Figure 8.
(CUG)n foci number in DM1 mouse and human myoblasts. Representative RNA FISH images using a (CAG)7 oligo probe (A and C) and quantification (B and D) of (CUG)n foci in DM1 mouse and human myoblasts. (A and B) RNA FISH signal in WT and DM500 myoblasts. In WT mouse myoblasts foci were observed which cannot be specific for the transgenic expanded (CUG)n transcripts. As these signals may also be present in cell lines derived from other mouse lineages, including the transgenic models, we defined a level of uncertainty, plotted as a shaded area in (B). Thus, an average of two expanded (CUG)n-specific foci per cell remained in DM500 myoblasts and no specific foci were identified in Tg26 and HSALR myoblasts. (C and D) RNA FISH on human healthy (5/5) and DM1 (21/200 and 11/760) myoblasts. Foci were only detected in 11/760 cells. Each data point represents foci number in one nucleus; mean is plotted in graph. Bar 20 µm. ***P < 0.001.
All data combined, we conclude that mouse and human myoblasts each contain up to half a dozen foci. We have to keep in mind that FISH protocols may detect transcripts with an efficiency of only 30–50% (41), for example, due to inaccessibility of the transcripts to the probe or loss of RNA molecules from the fixed cells during washing. Thus, the actual number of RNP complexes that appear as foci may be 2- to 3-fold higher. Given our estimates for the absolute number of expanded transcripts, our data suggest that every RNP complex that forms a FISH-visible aggregate is nucleated by one or only few expanded (CUG)n transcripts.
Discussion
DM1 is considered a prototypical RNA-dominant disorder, because its neurodegenerative and myopathic manifestation is thought to be based on processes wherein repeat-containing RNAs play a crucial role. Much of our current knowledge on the presumed toxic role of RNA in DM1 pathophysiology originates from direct comparison of findings in mutation carriers with findings in transgenic animals or genome edited cells. This is often done with simple bypassing of the fact that models may differ profoundly in nature, structural organization and chromatin context of the transgene and in length of the contained (CTG·CAG)n repeat (13,14). A major unsolved aspect of DM1 etiology is how repeat length and dose and nature of abnormally expanded RNA transcripts affect onset and complexity of disease manifestation and its rate of progression and severity. Quantitative studies of transcript production could thus help to explain differential experimental findings with distinct DM1 models and add to conceptual progress. Here, we used gene expression profiling to compare relative abundance and absolute copy number of expanded repeat RNAs between muscle cells from patients and mouse models that are among the most commonly used in the DM1 field.
Our analyses unveiled differences in timing of production during myogenic differentiation as well as in the abundance of (CUG)n transcripts between DM1 mouse model and patient muscle cells. Expression of DMPK transgenes in DM500, DMSXL and Tg26 mice is at a basal level in proliferating myoblasts and peaks early in differentiation in a manner similar to the profile of endogenous DMPK expression in human myoblasts. Increase in DMPK expression at the start of myogenic differentiation was earlier reported for human cells (46) and also seen for C2C12 mouse myoblasts (47). Our group (25) and others (32) have demonstrated that the actual onset of DMPK expression is already seen in somites in the developing embryo, well before the actual commitment to specific muscle cell fate and the onset of myogenesis.
DM500/DMSXL- and Tg26-derived myoblasts carry transgenes of different size that may lack regulatory elements upstream of the DMPK TSS, but the proximal promoter sequence and the enhancer sequence in Intron 1 (elements responsive to MyoD via conserved E-boxes (48)), are present. Thus, regulatory sequences from the human locus may drive ‘near-natural’ behavior of these transgenes and control the peak-shaped up-regulation with transient increase in transcriptional activity during early differentiation. The ‘human-like’ regulation of the DMPK transgene renders these mouse myoblasts useful models for study of toxic effects of normal or expanded DMPK RNA in early development. Others have demonstrated that early-onset expression of repeat-containing RNA may influence phenotypic severity by affecting proper tissue development in animals in vivo (16–19).
In HSALR myoblasts, expression of repeat-containing RNA was hardly detectable until late in differentiation in vitro. Conspicuously, the Acta1 gene, whose human counterpart served as the recipient body for the transgene with (CTG)250 repeat insertion, showed an earlier onset of expression in differentiating mouse cells. The explanation for this differential behavior may thus be that the proximal promoter is present in the transgene, but that a cis-regulatory module in a region that activates transcription in differentiating myoblasts, >20 kb downstream of the basal promoter (49), is lacking.
We found pronounced differences in the abundance of (CUG)n-repeat RNAs from transgenes and RNA products from the endogenous Dmpk gene and between expanded RNAs from cell lineages and the transgenic mouse tissues from which they originate. For proliferating cells, comparison was only meaningful for Tg26, DM500 and DMSXL myoblasts, as HSALR myoblasts essentially lacked expression of expanded (CUG)n RNA. As explained by the presence of multiple copies of the DMPK gene in the transgenic insert in Tg26 cells, DMPK levels were relatively high. Transcripts from the DMPK transgene in DM500 and DMSXL cells were ∼10-fold lower than transcripts from the Dmpk gene in these myoblasts. Even if we take into account that these observations reflect the production rate from one DMPK transgene versus two Dmpk genes in the DM500 cell model, transgenic RNA production is still ∼5-fold lower. Intrinsic differences in regulatory capacity of DMPK and Dmpk promoters may partly explain this observation, but we cannot exclude involvement of progressive epigenetic alterations, like DNA methylation or heterochromatization of the transgene or the DMPK promoter. Brouwer et al. (50) observed decreased transgene expression with age in the DM300-328 lineage, the ancestral mouse model from which DM500 and DMSXL mice and thus DM500 and DMSXL cells originate. Cis effects of repeat presence may have contributed to methylation state alterations in the DMPK transgene (50). Also transient propagation of mice in the homozygous state may have triggered partial gene silencing even before the myoblasts were derived in vitro (51–53). Finally, we cannot exclude effects of experimental handling, for example, by the use of dimethylsulfoxide during repeated cell freezing, which can affect the epigenetic state and is a known modulator of differentiation programming (54). Although we did not systematically study expression levels over time, we have the impression that DMPK expression in DM500 cells decreases with increasing passage number [compare data shown in (55)]. This could point to the influence of epigenetic silencing, explaining also variability in observations of different research groups in the DM1 field.
Also in muscle tissue obtained from hemizygous DM500 and DMSXL mice DMPK transcripts were 10-fold less abundant than Dmpk RNAs, confirming the observation by Huguet et al. (56). The overall low level of transgene expression may explain why disease manifestation in these models is relatively mild, even in homozygous animals. Conversely, however, one can also argue that the presence of these few expanded RNAs is already sufficient to cause mild myotonia, slow progressive muscle weakness, nuclear (CUG)n foci and splicing abnormalities (1,14,57), for which compelling evidence was provided in various studies.
Our most conspicuous observation was that ACTA1 (CUG)n expression in GPS muscle of homozygous HSALR mice was over 1000-fold higher than that of Dmpk, and remarkably similar to Acta1 expression. The high (CUG)n RNA expression may explain why the HSALR mouse lineage has by far the most severe DM1-like phenotype of all models, including myotonia, extensive splice abnormalities and a conspicuous abnormal density of intranuclear foci, all limited to skeletal muscle (1,14,27).
The use of expression ratios between transgenic and endogenous transcripts (e.g. Dmpk, Acta1, Actb and 18S RNA) helped us to compare repeat RNA levels between muscle cells and mouse and human tissues, but we reasoned that the knowledge of absolute transcript copy numbers in muscle would be mechanistically more informative. Absolute numbers of molecules can only be meaningfully assessed if studied on a per cell basis. Analysis of DMPK expression in the three human myoblast lineages with no, intermediate or long repeat expansions confirmed that transcripts from the wild-type and mutant allele are about equally abundant. We found no evidence for an inverse correlation between repeat the length and RNA expression level (46,58). Because endogenous DMPK/Dmpk transcripts appeared equally abundant in human and mouse myoblasts, quantitative comparison between patients and DM1 models was straightforward. Estimates for RNA expression ratios of ACTB and GAPDH, data from northern blot and RNA-sequencing analysis and absolute RT-qPCR experiments all point to the presence of at most 50 DMPK transcripts per cell. We thus propose that in patient cells <25 are expanded (CUG)n transcripts. In myoblasts from the DM500 and DMSXL mouse models around four RNA molecules per cell originate from the DMPK transgene.
Estimates for DMPK transcript numbers correspond remarkably well with the observed foci number in DM500 cell nuclei. Also in patient myoblasts the number of foci per nucleus and the number of DMPK transcripts per cell are in the same order of magnitude. Other research groups, studying different cell types from DM1 patients, have reported similar amounts of foci from their FISH experiments (59–62). Different and variable amounts of foci were observed for MyoD-transduced DM1 fibroblasts, which may be best explained by the distinct MyoD induction of DMPK expression, caused by differential vector systems or MyoD promoters (60,63). We, therefore, predict that RNP complexes that assemble in (CUG)n foci contain, on average, one to maximally six expanded transcripts. Along the same line of arguments, we propose that RNP aggregate formation is nucleated by one or very few expanded RNA molecules, thereby acting as individual entities. Phase transition of RNP complexes to insoluble aggregates (which appear as foci after FISH or MBNL antibody staining) may therefore not require further fusion with additional naked or RNP-decorated RNAs, but could be merely a protein-based event.
Many mechanistic avenues are now awaiting further exploration as our findings have important implications for the RNA-gain-of-function hypothesis that is currently in use for explaining DM1 features. How low numbers of only 1–25 abnormally folded RNA scaffolds can have a negative impact on ribostasis regulation—via titration of protein molecules from the total available pools of RNP proteins—is only one of the mechanistic questions that must be answered. The extent of loss of function of proteins may determine the gain of function toxicity of mutant RNA—and ultimately control the extent of cell stress caused by the DM1 mutation. Whether there is a direct relationship with the magnitude of temporal or permanent sequestration from the cellular pool of these RNP proteins is still difficult to answer. Stochastic events could be involved, as formation of an abnormal type of—microscopically visible or invisible diffusive—RNP aggregate on a repeat-containing transcript (6,7) might cause temporal or permanent perturbation of a specific nuclear pathway for mRNA processing or transport. Dominant effects of low-abundance triplet repeat RNA on RNA splicing and polyadenylation may be expected if effects on inhomogeneity and compartmentalization of protein factor pools (e.g. MBNL isoforms) in different nuclear trajectories for mRNA RNP processing play a decisive role. Finally, amplification of toxic effects by rare triplet repeat RNAs may occur otherwise: various studies have demonstrated that activation of stress-signaling cascades that involve GSK3β (64), PKC (65), double-stranded RNA-dependent protein kinase PKR (66) or sensors of foreign RNA that normally initiate immune responses (67) is involved in disease manifestation in DM1. Whatever the exact course of cellular events in these scenarios, even with low-expressed, abnormally expanded repeat RNAs the probability of exceeding the toxic threshold increases over time for any permanently expressing cell in the tissue population. Ultimately, this may result in accumulation of stress in an increasing number of cells, and progressive cell loss and loss of function of tissue during ageing.
In a parallel study, we have recently found that expression of another possible player, a DM1 antisense transcript carrying a (CAG)n repeat, is even 10-fold lower than levels observed for DMPK (Gudde et al. manuscript in preparation). Thus also here, the question remains how such an extremely rare transcript could contribute to disease manifestation. One unifying answer may be that the rare DMPK sense RNA and the even rarer antisense transcript both serve as templates for RAN translation (9,10). Homopolymeric protein products thus formed could possibly contribute to disease manifestation via initiation of aggregation of metastable proteins, initiating a prion-like cascade of events (68–70). Again, further analysis of steady-state levels of DMPK (CUG)n and antisense transcripts in cycling or resting cells and study of the frequency of use of RAN translation on these rare RNAs is necessary to better understand this possible distinct aspect of DM1 etiology.
Our findings highlight that careful choice of cellular and animal model systems that take structural properties of transgenes and transgenic products into account in combination with quantitative modeling is imperative for such studies. For this work not only knowledge of the ‘per cell’ presence of individual endogenous and mutant RNA molecules is important, but also stoichiometric considerations on the binding of MBNL1–3, Staufen, DDX, HnRNP or other RNP proteins (7) by one (CUG)n repeat tract are needed to better understand nucleation and phase transition events in abnormal aggregates that could form around expanded DMPK mRNAs. Whatever the molecular mechanism involved, based on our findings, we propose that DM1 is caused by anomalous behavior of only very few mutant RNA molecules per muscle cell.
Materials and Methods
Human material
Human skeletal muscle samples were obtained from pre-consented post-mortem donors for research purposes in accordance with local guidelines in The Netherlands more than 18 years ago. No additional approval by an ethics committee was required at that time. Muscle autopsies were obtained from patients with a confirmed clinical and DNA diagnosis of adult-onset DM1 [male DM1-c, 55 years, psoas muscle (CTG)5/(CTG)exp; female DM1-d, 65 years, quadriceps muscle (CTG)16/(CTG)exp] and congenital DM1 [male CDM-e and -f, 14 days, sternocleidomastoideus and gastrocnemius muscle (CTG)12/(CTG)1300]. All tissues were snap frozen immediately after collection and stored at −135 to 80°C until further use. As control, skeletal muscle samples from healthy anonymous donors [healthy-a and -b (CTG)11/(CTG)12 and (CTG)5/(CTG)11, respectively] from our own repository were included in our study. These samples were collected long before the current guidelines for written consent were enforced and no detailed information could be traced.
Mice
Mice were housed and procedures performed with approval of the Animal Ethics Committee of Radboud University Nijmegen (Permit number: RU-DEC 2014-099). Characteristics of the mouse models are summarized in Table 1. DM500 and DMSXL mice both originated from the DM300-328 lineage (23). Due to intergenerational triplet instability the repeat in the DMPK gene grew to 500–600 and ∼1300 CTG triplets in DM500 and DMSXL mice, respectively (24). For isolation of immortalized myoblasts, each of the DM1 mouse models was crossed with hemizygous H-2Kb-tsA58 transgenic mice (ImmortoMouse®, Charles River Laboratories) (71), harboring the gene for thermolabile TAg from SV40. Nine-day old pups carrying one DMPK or ACTA1 transgene copy and one H-2Kb-tsA58 transgene copy were selected and used for myoblast generation.
Cell culture
Conditionally immortalized myoblasts from the gastrocnemius–plantaris–soleus (GPS) muscle of DMSXL, Tg26 and HSALR mice after crossing with H-2Kb-tsA58 mice were derived as described (28,55,71). Individual myoblast lineages were obtained by ring cloning and selected for myotube formation ability. Myoblast populations used in this study were established by forming equal mixtures of five cell clones for DMSXL, seven cell clones for Tg26 and four cell clones for HSALR. DM500 and WT myoblasts were derived previously (55).
Myoblasts were grown on 0.1% (w/v) gelatin-coated culture dishes in the proliferation medium containing Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 20% (v/v) fetal bovine serum (PAA Laboratories), 4 mm l-glutamine (Gibco), 1 mm pyruvate (Sigma), 50 µg/ml gentamicin (Gibco), 20 units/ml γ-interferon (BD Biosciences) and 2% (v/v) chicken embryo extract (Sera Laboratories International) at 7.5% CO2 and 33°C. Differentiation to myotubes was induced by placing myoblasts, grown to confluency on Matrigel (BD Biosciences), in the differentiation medium containing DMEM supplemented with 5% (v/v) horse serum and 50 µg/ml gentamicin at 7.5% CO2 and 37°C. Differentiation conditions were maintained for a maximum of 7 days. Spontaneous contractions started to appear around Day 3.
Human myoblast cultures isolated from skeletal muscle of fetuses, one healthy control (5/5) and two DM1 affected lines [21/200 with (CCG·CGG)n interruptions in the 3′ end of the expanded (CTG·CAG)n repeat; data not shown] and 11/760 were a gift of Dr Furling et al. (46). Myoblasts were maintained on 0.1% (w/v) gelatin-coated culture dishes in proliferation medium containing Ham's F10 medium (Gibco) supplemented with GlutaMAX, 20% bovine growth serum (Thermo Scientific) and 25 µg/ml gentamicin at 7.5% CO2 and 37°C. Differentiation to myotubes was induced by placing confluent myoblast cultures in the differentiation medium containing DMEM supplemented with 4 mm l-glutamine, 1 mm pyruvate, 10 μg/ml insulin (Sigma), 100 μg/ml apo-transferrin (Sigma) and 25 μg/ml gentamicin.
Phase contrast images of cell cultures were taken with a Zeiss Axiovert 35 M light microscope, 10×/0.30 objective.
Fluorescence-activated cell sorting
Immortalized WT mouse myoblasts were detached from the culture surface using trypsin, washed in phosphate-buffered saline (PBS) and fixed in 70% (v/v) ethanol in PBS overnight at −20°C. After two times PBS wash, cells were stained in 20 µg/ml propidium iodide, 0.2 mg/ml RNase A, 0.1% Triton X-100 in PBS for 15 min at 37°C. Fluorescence-activated cell sorting analysis was performed on the Beckman Coulter Epics Altra cell sorter. Immortalized DM500 myoblasts were cultured in proliferation medium at 37°C for 24 h, detached and incubated with 3 µg/ml Hoechst 33342 at 37°C for 30 min and then sorted. Diploid and tetraploid DM500 cells were seeded on gelatin-coated glass cover slips, cultured for another 24 h at 37°C and analyzed by FISH.
RNA isolation
RNA from cultured cells was isolated using the Aurum Total RNA Mini Kit (Bio-Rad), according to the manufacturer's protocol. RNA from muscle tissue was isolated using TRIzol reagent (Invitrogen), according to the manufacturer's protocol.
Northern blotting
Northern blotting was performed according to standard procedures. RNA was subjected to electrophoresis in a 1.2% agarose gel under denaturing conditions. Depending on sample availability, 5–12 µg RNA was loaded per lane. RNA was transferred to Hybond-XL nylon membrane (Amersham Pharmacia Biotech) by capillary transfer in 10× SSC and hybridized with random-primed 32P-labeled cDNA-based probes or 32P-end-labeled oligonucleotides: a 2.6 kb DMPK cDNA (covering the entire ORF and 3′ UTR) (55) and the complete 1.9 kb human 18S rRNA cDNA were used as templates in random priming reactions. A (CAG)9 oligo (55), a Acta1 oligo (5′-ACCCTGCAACCACAGCACGATTGTCGATTG-3′) (72) and a mixture of three oligos complementary to both Acta1 and ACTA1(5′-GCGGTGGTCTCGTCTTCGTCGCACAT-3′, 5′-TGGCATACAGGTCCTTCCTGATGTCGATGTC-3′, 5′-GCCTCGTCGTACTCCTGCTTGGTGATCC-3′) were 5′-end labeled. The Acta1/ACTA1 oligo mix and the DMPK cDNA probe were used to quantify ACTA1 and DMPK mRNA levels, respectively, in human samples. In murine samples, the single Acta1 oligo, DMPK cDNA and (CAG)9 oligo probes were used to quantify Acta1, Dmpk and transgenic DMPK (DM500 and DMSXL) and transgene (Tg26 and HSALR) mRNA levels, respectively. Blots were washed and exposed to X-ray film (Kodak, X-OMAT AR). Quantification of signals was performed by phosphor-imager analysis (Molecular Imager FX, Bio-Rad) and analyzed with Quantity One (Bio-Rad) and FIJI (73) software. 18S rRNA levels were used for normalization.
To be able to compare transgene expression in the DM1 mouse models, carrying different transgenes (i.e. DMPK- or ACTA1-type), a mix of ACTA1 cDNA probe (1.3 kb, Exon 2–7, 27% G-content) and DMPK cDNA probe (1.3 kb, Exon 3–12, 34% G-content) was used. Both genes are ∼90% identical between mouse and human, so simultaneous detection of endogenous and transgenic transcripts was possible. [α-32P]-dCTP label was diluted with non-radioactive dCTP for labeling of the ACTA1 probe to obtain comparable signal strengths for simultaneous phosphor-imager detection of Acta1/ACTA1 and Dmpk/DMPK on the same blot. Signals were corrected for label dilution and G-content in the two probes. Since ACTA1 and Actb are 84% identical, the ACTA1 probe was also used for detection of Actb. A similar method was used to compare DMPK/Dmpk and ACTB/Actb expression. A 1.3 kb ACTB cDNA probe was used (Exon 1–6, 27% G-content), which has a 90% identical sequence with the mouse variant.
In vitro transcription
DNA templates corresponding to DMPK regions Exon 1–6 (0.6 kb) and Exon 11–15 (1.0 kb) were generated by PCR using primers 5′-GAATTTAATACGACTCACTATAGGGAGAGGCGGCTCCAGCAGC-3′ and 5′-CCGCAGCTTGA-GGCAAGAG-3′, and 5′-GAATTTAATACGACTCACTATAGGGAGATGGAGGCCATCCG-3′ and 5′-GTCCTGTAGCCTGTCAGCGA-3′, respectively (T7 promoter underlined). DNA purity and sequence identity were confirmed by sequencing. For in vitro transcription, 200 ng of the templates was used in the MEGAscript T7 Transcription Kit (Ambion) following manufacturer's instructions. RNA products were purified using the Aurum Total RNA Mini Kit (Bio-Rad). Transcript length, purity and concentration were verified by gel electrophoresis and absorbance at 260/280 nm (NanoVUE spectrophotometer, GE Healthcare Life Sciences). Known amounts of RNA fragments DMPK Exon 1–6 and Exon 11–15 were mixed with WT mouse total RNA (as carrier RNA) and used as standards in RT-qPCR.
Reverse transcriptase–quantitative polymerase chain reaction
Using 500 ng RNA template per reaction and random hexamers as primers, cDNA synthesis was performed with SuperScript™ II Reverse Transcriptase (Invitrogen) or iScript™ cDNA Synthesis Kit (Bio-Rad). For RT-qPCR, 3 µl of 10–100-fold diluted cDNA preparation was mixed in a final volume of 10 µl containing 5 µl 2× Sybr Green mix (Roche Applied Science) or 5 µl iQ™ SYBR® Green Supermix (Bio-Rad) and 4 pmol of each primer: DMPK Exon 1–2: 5′-ACTGGCCCAGGACAAGTACG-3′ and 5′-CCTCCTTAAGCCTCACCACG-3′; DMPK Exon 15(5′): 5′-AGAACTGTCTTCGACTCCGGG-3′ and 5′-TCGGAGCGGTTGTGAACTG-3′; ACTA1: 5′-CACCTCCAGCACGCGACTT-3′ and 5′-CGATGGCAGCAACGGAAGTTGT-3′; Dmpk: 5′-TTTTGAAGGTGATCGGGCGTG-3′ and 5′-CCTCTCTTCAGCATGTCCCACTTA-3′; 18S rRNA: 5′-GTAACCCGTTGAACCCCATT-3′ and 5′-CCATCCAATCGGTAGTAGCG-3′. Samples were analyzed using the CFX96 Real-time System (Bio-Rad). A melting curve was obtained for each sample in order to confirm single product amplification. cDNA samples from no template control and no reverse transcriptase control (RT-) were included as negative controls. Transgene (DMPK and ACTA1) and endogenous Dmpk mRNA levels were normalized to 18S rRNA expression.
Fluorescent in situ hybridization
Human myoblasts (5/5, 21/200 and 11/760) and mouse myoblasts (DM500, Tg26, HSALR and WT) grown on gelatin-coated glass cover slips were washed once with PBS and fixed in 4% (w/v) formaldehyde, 2 mm MgCl2 in PBS for 10 min at room temperature. Cells were stored under 70% ethanol at 4°C. After two times PBS wash, cells were pre-hybridized in 40% (v/v) deionized formamide in 2× SSC for 20 min at room temperature, followed by overnight hybridization with a 0.1 ng/μl Cy3-(CAG)7 probe (2′-O-methyl phosphorothioate-modified) in 40% deionized formamide, 10% (w/v) dextran sulfate, 0.1% Triton X-100, 1 mg/ml herring sperm DNA, 100 μg/ml yeast tRNA, 0.2% (w/v) BSA, 2 mm VRC, 2× SSC in a humidified chamber at 37°C. After two times PBS wash, cells were counterstained with 0.33 μg/ml DAPI in PBS for 10 min at room temperature, followed by a PBS wash, dehydration in methanol and mounting in Mowiol. Images were acquired using the Zeiss Axiophot2 Fluorescence microscope or Olympus IX-71 wide field fluorescence microscope.
RNA-sequencing
RNA-sequencing data from healthy and DM1-affected human tissue samples [tibialis anterior (TA), quadriceps and heart] were obtained from www.dmseq.org. RNA-sequencing signals of full-length DMPK and GAPDH were used as an independent measure for transcript abundance.
Statistical analysis
Endogenous and transgenic mRNA levels were compared between time points and models, using a one-way analysis of variance (ANOVA) test, followed by a post-test for linear trend or Tukey's multiple comparison test. All values in graphs are presented as mean±SEM. Foci counts were compared between different myoblast lines with a one-way ANOVA test and Tukey's multiple comparisons post-test. Data are visualized as scatter plot, each dot representing one observation, and the mean is shown. Statistical analyses were performed with GraphPad Prism version 5.01 for Windows.
Supplementary Material
Funding
This work was supported by the Prinses Beatrix Spierfonds (grant number W.OR10-04; with contribution from the Stichting Spieren voor Spieren) and by ZonMw (TOP grant NL91212009). Funding to pay Open Access charges was provided by both organizations and Radboudumc.
Supplementary Material
Acknowledgements
We thank Geneviève Gourdon (Paris, France) for the DM500 and DMSXL mouse lines, Charles Thornton (Rochester, NY, USA) for the HSALR line and Denis Furling (Paris, France) for human myoblasts. We thank the founders and contributors of the Myotonic Dystrophy Deep Sequencing Data Repository available viawww.dmseq.org. We thank members of the Department of Cell Biology for discussions.
Conflict of Interest statement. None declared.
References
- 1.Udd B., Krahe R. (2012) The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol., 11, 891–905. [DOI] [PubMed] [Google Scholar]
- 2.Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barceló J., O'Hoy K. (1992) Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science, 255, 1253–1255. [DOI] [PubMed] [Google Scholar]
- 3.Cho D.H., Thienes C.P., Mahoney S.E., Analau E., Filippova G.N., Tapscott S.J. (2005) Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell, 20, 483–489. [DOI] [PubMed] [Google Scholar]
- 4.Harper P.S. (2001) Myotonic Dystrophy. 3rd edn London, Saunders W. B. [Google Scholar]
- 5.Sicot G., Gourdon G., Gomes-Pereira M. (2011) Myotonic dystrophy, when simple repeats reveal complex pathogenic entities: new findings and future challenges. Hum. Mol. Genet., 20, R116–R123. [DOI] [PubMed] [Google Scholar]
- 6.Wojciechowska M., Krzyzosiak W.J. (2011) Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet., 20, 3811–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettersson O.J., Aagaard L., Jensen T.G., Damgaard C.K. (2015) Molecular mechanisms in DM1—a focus on foci. Nucleic Acids Res., 43, 2433–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batra R., Charizanis K., Manchanda M., Mohan A., Li M., Finn D.J., Goodwin M., Zhang C., Sobczak K., Thornton C.A. et al. (2014) Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell, 56, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A.C. et al. (2011) Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl Acad. Sci. USA, 108, 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleary J.D., Ranum L.P.W. (2013) Repeat-associated non-ATG (RAN) translation in neurological disease. Hum. Mol. Genet., 22, R45–R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu G., Chen X., Gao Y., Lewis T., Barthelemy J., Leffak M. (2012) Altered replication in human cells promotes DMPK (CTG)(n) · (CAG)(n) repeat instability. Mol. Cell. Biol., 32, 1618–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minnerop M., Weber B., Schoene-Bake J.C., Roeske S., Mirbach S., Anspach C., Schneider-Gold C., Betz R.C., Helmstaedter C., Tittgemeyer M. et al. (2011) The brain in myotonic dystrophy 1 and 2: evidence for a predominant white matter disease. Brain, 134, 3527–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wansink D.G., Wieringa B. (2003) Transgenic mouse models for myotonic dystrophy type 1 (DM1). Cytogenet. Genome Res., 100, 230–242. [DOI] [PubMed] [Google Scholar]
- 14.Gomes-Pereira M., Cooper T.A., Gourdon G. (2011) Myotonic dystrophy mouse models: towards rational therapy development. Trends Mol. Med., 17, 506–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gladman J.T., Mandal M., Srinivasan V., Mahadevan M.S. (2013) Age of onset of RNA toxicity influences phenotypic severity: evidence from an inducible mouse model of myotonic dystrophy (DM1). PLoS One, 8, e72907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furling D., Lam L.T., Agbulut O., Butler-Browne G.S., Morris G.E. (2003) Changes in myotonic dystrophy protein kinase levels and muscle development in congenital myotonic dystrophy. Am. J. Pathol., 162, 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thornell L.-E., Lindstöm M., Renault V., Klein A., Mouly V., Ansved T., Butler-Browne G., Furling D. (2009) Satellite cell dysfunction contributes to the progressive muscle atrophy in myotonic dystrophy type 1. Neuropathol. Appl. Neurobiol., 35, 603–613. [DOI] [PubMed] [Google Scholar]
- 18.Todd P.K., Ackall F.Y., Hur J., Sharma K., Paulson H.L., Dowling J.J. (2014) Transcriptional changes and developmental abnormalities in a zebrafish model of myotonic dystrophy type 1. Dis. Model. Mech., 7, 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marteyn A., Maury Y., Gauthier M.M., Lecuyer C., Vernet R., Denis J.A., Pietu G., Peschanski M., Martinat C. (2011) Mutant human embryonic stem cells reveal neurite and synapse formation defects in type 1 myotonic dystrophy. Cell Stem Cell, 8, 434–444. [DOI] [PubMed] [Google Scholar]
- 20.Mahadevan M.S., Yadava R.S., Yu Q., Balijepalli S., Frenzel-McCardell C.D., Bourne T.D., Phillips L.H. (2006) Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet., 38, 1066–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krzyzosiak W.J., Sobczak K., Wojciechowska M., Fiszer A., Mykowska A., Kozlowski P. (2012) Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic Acids Res., 40, 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortimer S.A., Kidwell M.A., Doudna J.A. (2014) Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet., 15, 469–479. [DOI] [PubMed] [Google Scholar]
- 23.Seznec H., Lia-Baldini A.S., Duros C., Fouquet C., Lacroix C., Hofmann-Radvanyi H., Junien C., Gourdon G. (2000) Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum. Mol. Genet., 9, 1185–1194. [DOI] [PubMed] [Google Scholar]
- 24.Gomes-Pereira M., Foiry L., Nicole A., Huguet A., Junien C., Munnich A., Gourdon G. (2007) CTG trinucleotide repeat ‘big jumps’: large expansions, small mice. PLoS Genet., 3, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jansen G., Groenen P.J., Bächner D., Jap P.H., Coerwinkel M., Oerlemans F., van den Broek W., Gohlsch B., Pette D., Plomp J.J. et al. (1996) Abnormal myotonic dystrophy protein kinase levels produce only mild myopathy in mice. Nat. Genet., 13, 316–324. [DOI] [PubMed] [Google Scholar]
- 26.O'Cochlain D.F., Perez-Terzic C., Reyes S., Kane G.C., Behfar A., Hodgson D.M., Strommen J.A., Liu X.-K., van den Broek W., Wansink D.G. et al. (2004) Transgenic overexpression of human DMPK accumulates into hypertrophic cardiomyopathy, myotonic myopathy and hypotension traits of myotonic dystrophy. Hum. Mol. Genet., 13, 2505–2518. [DOI] [PubMed] [Google Scholar]
- 27.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. (2000) Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science, 289, 1769–1773. [DOI] [PubMed] [Google Scholar]
- 28.Morgan J.E., Beauchamp J.R., Pagel C.N., Peckham M., Ataliotis P., Jat P.S., Noble M.D., Farmer K., Partridge T.A. (1994) Myogenic cell lines derived from transgenic mice carrying a thermolabile T antigen: a model system for the derivation of tissue-specific and mutation-specific cell lines. Dev. Biol., 162, 486–498. [DOI] [PubMed] [Google Scholar]
- 29.Grounds M.D., McGeachie J.K. (1989) A comparison of muscle precursor replication in crush-injured skeletal muscle of Swiss and BALBc mice. Cell Tissue Res., 255, 385–391. [DOI] [PubMed] [Google Scholar]
- 30.Fukada S., Morikawa D., Yamamoto Y., Yoshida T., Sumie N., Yamaguchi M., Ito T., Miyagoe-Suzuki Y., Takeda S., Tsujikawa K. et al. (2010) Genetic background affects properties of satellite cells and mdx phenotypes. Am. J. Pathol., 176, 2414–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weaver B.A.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. (2007) Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell, 11, 25–36. [DOI] [PubMed] [Google Scholar]
- 32.Harmon E.B., Harmon M.L., Larsen T.D., Paulson A.F., Perryman M.B. (2008) Myotonic dystrophy protein kinase is expressed in embryonic myocytes and is required for myotube formation. Dev. Dyn., 237, 2353–2366. [DOI] [PubMed] [Google Scholar]
- 33.Oude Ophuis R.J.A., Mulders S.A.M., Van Herpen R.E.M.A., van de Vorstenbosch R., Wieringa B., Wansink D.G. (2009) DMPK protein isoforms are differentially expressed in myogenic and neural cell lineages. Muscle Nerve, 40, 545–555. [DOI] [PubMed] [Google Scholar]
- 34.Smith C.K., Janney M.J., Allen R.E. (1994) Temporal expression of myogenic regulatory genes during activation, proliferation, and differentiation of rat skeletal muscle satellite cells. J. Cell. Physiol., 159, 379–385. [DOI] [PubMed] [Google Scholar]
- 35.Laing N.G., Dye D.E., Wallgren-Pettersson C., Richard G., Monnier N., Lillis S., Winder T.L., Lochmüller H., Graziano C., Mitrani-Rosenbaum S. et al. (2009) Mutations and polymorphisms of the skeletal muscle alpha-actin gene (ACTA1). Hum. Mutat.,30, 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stern-Straeter J., Bonaterra G.A., Kassner S.S., Faber A., Sauter A., Schulz J.D., Hörmann K., Kinscherf R., Goessler U.R. (2011) Impact of static magnetic fields on human myoblast cell cultures. Int. J. Mol. Med., 28, 907–917. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz R.J., Rothblum K.N. (1981) Gene switching in myogenesis: differential expression of the chicken actin multigene family. Biochemistry, 20, 4122–4129. [DOI] [PubMed] [Google Scholar]
- 38.Wong L.J., Ashizawa T., Monckton D.G., Caskey C.T., Richards C.S. (1995) Somatic heterogeneity of the CTG repeat in myotonic dystrophy is age and size dependent. Am. J. Hum. Genet.,56, 114–122. [PMC free article] [PubMed] [Google Scholar]
- 39.Battich N., Stoeger T., Pelkmans L. (2013) Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods, 10, 1127–1133. [DOI] [PubMed] [Google Scholar]
- 40.Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S. (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods, 5, 877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsson C., Grundberg I., Söderberg O., Nilsson M. (2010) In situ detection and genotyping of individual mRNA molecules. Nat. Methods, 7, 395–397. [DOI] [PubMed] [Google Scholar]
- 42.Wills Q.F., Livak K.J., Tipping A.J., Enver T., Goldson A.J., Sexton D.W., Holmes C. (2013) Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nat. Biotechnol., 31, 748–752. [DOI] [PubMed] [Google Scholar]
- 43.Weiss Sachdev S., Sunde R.A. (2001) Selenium regulation of transcript abundance and translational efficiency of glutathione peroxidase-1 and -4 in rat liver. Biochem. J., 357, 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White A.K., Vaninsberghe M., Petriv O.I., Hamidi M., Sikorski D., Marra M.A., Piret J., Aparicio S., Hansen C.L. (2011) High-throughput microfluidic single-cell RT-qPCR. Proc. Natl Acad. Sci. USA, 108, 13999–14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcus J.S., Anderson W.F., Quake S.R. (2006) Microfluidic single-cell mRNA isolation and analysis. Anal. Chem., 78, 3084–3089. [DOI] [PubMed] [Google Scholar]
- 46.Furling D., Lemieux D., Taneja K., Puymirat J. (2001) Decreased levels of myotonic dystrophy protein kinase (DMPK) and delayed differentiation in human myotonic dystrophy myoblasts. Neuromuscul. Disord.,11, 728–735. [DOI] [PubMed] [Google Scholar]
- 47.Carrasco M., Canicio J., Palacín M., Zorzano A., Kaliman P. (2002) Identification of intracellular signaling pathways that induce myotonic dystrophy protein kinase expression during myogenesis. Endocrinology, 143, 3017–3025. [DOI] [PubMed] [Google Scholar]
- 48.Storbeck C.J., Sabourin L.A., Waring J.D., Korneluk R.G. (1998) Definition of regulatory sequence elements in the promoter region and the first intron of the myotonic dystrophy protein kinase gene. J. Biol. Chem., 273, 9139–9147. [DOI] [PubMed] [Google Scholar]
- 49.McCord R.P., Zhou V.W., Yuh T., Bulyk M.L. (2011) Distant cis-regulatory elements in human skeletal muscle differentiation. Genomics, 98, 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brouwer J.R., Huguet A., Nicole A., Munnich A., Gourdon G. (2013) Transcriptionally repressive chromatin remodelling and CpG methylation in the presence of expanded CTG-repeats at the DM1 Locus. J. Nucleic Acids, 2013, 567435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer P. (2013) Transgenes and their contributions to epigenetic research. Int. J. Dev. Biol., 57, 509–515. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Y., Zhang T., Zhang Q.K., Jiang Y., Xu D.G., Zhang M., Shen W., Pan Q.J. (2014) Unstable expression of transgene is associated with the methylation of CAG promoter in the offspring from the same litter of homozygous transgenic mice. Mol. Biol. Rep., 41, 5177–5186. [DOI] [PubMed] [Google Scholar]
- 53.Brummelkamp T.R., van Steensel B. (2015) A HUSH for transgene expression. Science, 348, 1433–1434. [DOI] [PubMed] [Google Scholar]
- 54.Chetty S., Pagliuca F.W., Honore C., Kweudjeu A., Rezania A., Melton D.A. (2013) A simple tool to improve pluripotent stem cell differentiation. Nat. Methods, 10, 553–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mulders S.A.M., van den Broek W.J.A.A., Wheeler T.M., Croes H.J.E., van Kuik-Romeijn P., de Kimpe S.J., Furling D., Platenburg G.J., Gourdon G., Thornton C.A. et al. (2009) Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl Acad. Sci. USA, 106, 13915–13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huguet A., Medja F., Nicole A., Vignaud A., Guiraud-Dogan C., Ferry A., Decostre V., Hogrel J.Y., Metzger F., Hoeflich A. et al. (2012) Molecular, physiological, and motor performance defects in DMSXL mice carrying >1,000 CTG repeats from the human DM1 locus. PLoS Genet., 8, e1003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seznec H., Agbulut O., Sergeant N., Savouret C., Ghestem A., Tabti N., Willer J.C., Ourth L., Duros C., Brisson E. et al. (2001) Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum. Mol. Genet., 10, 2717–2726. [DOI] [PubMed] [Google Scholar]
- 58.Krahe R., Ashizawa T., Abbruzzese C., Roeder E., Carango P., Giacanelli M., Funanage V.L., Siciliano M.J. (1995) Effect of myotonic dystrophy trinucleotide repeat expansion on DMPK transcription and processing. Genomics, 28, 1–14. [DOI] [PubMed] [Google Scholar]
- 59.Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H. (1995) Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol., 128, 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larsen J., Pettersson O.J., Jakobsen M., Thomsen R., Pedersen C.B., Hertz J.M., Gregersen N., Corydon T.J., Jensen T.G. (2011) Myoblasts generated by lentiviral mediated MyoD transduction of myotonic dystrophy type 1 (DM1) fibroblasts can be used for assays of therapeutic molecules. BMC Res. Notes, 4, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xia G., Ashizawa T. (2015) Dynamic changes of nuclear RNA foci in proliferating DM1 cells. Histochem. Cell Biol., 143, 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michel L., Huguet-Lachon A., Gourdon G. (2015) Sense and Antisense DMPK RNA foci accumulate in DM1 tissues during development. PLoS One, 10, e0137620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davis B.M., McCurrach M.E., Taneja K.L., Singer R.H., Housman D.E. (1997) Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl Acad. Sci. USA, 94, 7388–7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones K., Wei C., Iakova P., Bugiardini E., Schneider-Gold C., Meola G., Woodgett J., Killian J., Timchenko N.A., Timchenko L.T. (2012) GSK3β mediates muscle pathology in myotonic dystrophy. J. Clin. Invest.,122, 4461–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuyumcu-Martinez N.M., Wang G.-S., Cooper T.A. (2007) Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell, 28, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian B., White R.J., Xia T., Welle S., Turner D.H., Mathews M.B., Thornton C.A. (2000) Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA, 6, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olejniczak M., Urbanek M.O., Krzyzosiak W.J. (2015) The role of the immune system in triplet repeat expansion diseases. Mediators Inflamm.,2015, 873860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jucker M., Walker L.C. (2013) Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature, 501, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y.-J., Jansen-West K., Xu Y.-F., Gendron T.F., Bieniek K.F., Lin W.-L., Sasaguri H., Caulfield T., Hubbard J., Daughrity L. et al. (2014) Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol.,128, 505–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wen X., Tan W., Westergard T., Krishnamurthy K., Markandaiah S.S., Shi Y., Lin S., Shneider N.A., Monaghan J., Pandey U.B. et al. (2014) Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron, 84, 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jat P.S., Noble M.D., Ataliotis P., Tanaka Y., Yannoutsos N., Larsen L., Kioussis D. (1991) Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proc. Natl Acad. Sci. USA, 88, 5096–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wheeler T.M., Sobczak K., Lueck J.D., Osborne R.J., Lin X., Dirksen R.T., Thornton C.A. (2009) Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science, 325, 336–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.