Abstract
Protein ubiquitination, the covalent attachment of ubiquitin to target proteins, has emerged as one of the most prevalent posttranslational modifications (PTMs), regulating nearly every cellular pathway. The diversity of signaling associated with this particular PTM stems from the myriad ways in which a target protein can be modified by ubiquitin, e.g., monoubiquitin, multi-monoubiquitin, and polyubiquitin linkages. In this Review, we focus on developments in both enzymatic and chemical methods that engender ubiquitin with new chemical and physical properties. Moreover, we highlight how these methods have enabled studies directed toward (i) characterizing enzymes responsible for reversing the ubiquitin modification, (ii) understanding the influence of ubiquitin on protein function and crosstalk with other PTMs, and (iii) uncovering the impact of polyubiquitin chain linkage and length on downstream signaling events.
Keywords: Ubiquitin, Polyubiquitin chains, Ubiquitin E1 activating enzyme, Ubiquitin E2 conjugating enzyme, Deubiquitinase (DUB), Ubiquitin-binding domain (UBD), Native chemical ligation, Unnatural amino acid incorporation, Directed disulfide bond formation
Graphical Abstract
Covalent attachment of ubiquitin (Ub) and ubiquitin-like proteins (Ubls) to the ε-amino group of lysine residues in a target protein, a process termed ubiquitination or ubiquitylation, is one of the most prevalent mechanisms for regulating protein function and stability in eukaryotes.1,2 Indeed, sequence annotations suggest nearly 5% of the human genome is dedicated to the coupling and removal of Ub/Ubls to and from proteins. Given the central role of the Ub network in cellular physiology, misregulation is often associated with numerous human diseases, including cancer, immune disorders, neurodegenerative diseases, and congestive heart failure.3–6
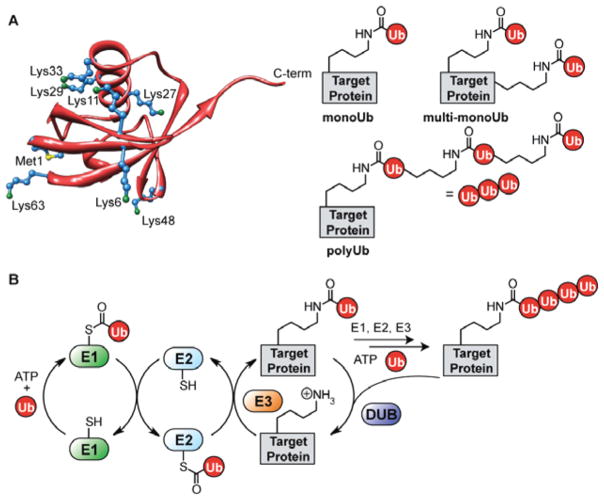
Ubiquitination is unique among the ensemble of posttranslational modifications (PTMs), specifically from the standpoint of signal diversity.7–9 For example, in contrast to other prevalent PTMs such as phosphorylation, proteins can be modified with Ub on a single lysine residue (monoUb), multiple lysines (multi-monoUb), or a single lysine with a polymeric chain of Ub (polyUb). With regards to polyUb chain formation, Ub possesses seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) and an N-terminal methionine residue (M1), each of which can form isopeptide or peptide linkages with the carboxy terminus of another Ub molecule (Figure 1A). This feature adds significant complexity to intracellular Ub signaling networks as it permits the assembly of chains with many different types of linkages and lengths with the potential to control distinct biological processes.10
Figure 1.
The many facets of protein ubiquitination. (A) The structure of Ub (PDB code 1UBQ118) showing all seven lysines (blue, with green nitrogen atom) and the N-terminal methionine (blue, with yellow sulfur atom). Also, each of the different types of Ub modifications are shown: monoUb (where a single lysine of the target is modified), multi-monoUb (where multiple lysines in the target are modified), and polyUb (where one of the eight amino groups in Ub serves as the point of attachment for a growing polymer chain). (B) The enzymatic cascade leading to Ub conjugation and removal: E1 activating, E2 conjugating, E3 ligase, and deubiquitinating (DUB) enzymes.
New experimental approaches have been central to unraveling how Ub signals are formed, recognized, and transduced into various biological responses. In this Review, we describe a combination of enzymatic and chemical methods used to develop molecular probes. We provide a comprehensive overview of enzymatic syntheses of monoUb and polyUb derivatives and compare these to chemoselective reactions used to construct Ub variants unobtainable by other means. Moreover, we highlight how each method enables the interrogation of Ub signaling, such as the specificity of deubiquitinating enzymes (DUBs), recognition of polyUb chains, and function of ubiquitinated target proteins. Finally, we discuss the utility of each approach in future applications to dissect the importance of polyUb linkage type and chain length.
SYNTHESIS OF UBIQUITIN DERIVATIVES: ENZYMATIC CONTROL OVER REGIOSPECIFICITY
Enzymatic transformations provide an efficient and selective means to engender Ub with new chemical and physical properties. In particular, two types of enzymes are used to alter Ub: those that selectively target the C-terminus and others that modify lysines to afford specific homotypic polyUb chains. These approaches have significantly advanced the Ub field by identifying DUBs and discovering tetraUb, with each Ub monomer linked through K48, as the minimal signal for proteasomal substrate targeting.
C-TERMINAL UB MODIFICATIONS
Use of Ub-Conjugating E2 Enzymes
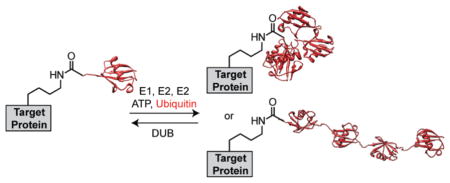
An intricate network of enzymes couple Ub to target proteins (Figure 1B). Specifically, three types, i.e., E1, E2, and E3, attach Ub to targets through a sequential enzymatic cascade.11 E1 initiates the cascade by first catalyzing the adenylation of Ub at the C-terminal glycine residue (Gly76).12 Following adenylation, a catalytic cysteine of E1 attacks the adenylate affording an E1-S-Ub thioester intermediate. The ubiquityl moiety is then shuttled from E1 to an active site cysteine of an E2 through a transthioesterification event affording a Ub-charged E2 (E2-S-Ub).13 To complete the enzymatic sequence, an E3 associates with a substrate and E2-S-Ub to mediate the delivery of Ub to an ε-amino group of a substrate lysine residue.2,14
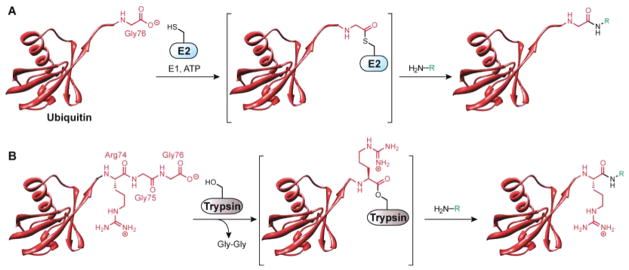
Early biochemical studies of the Ub pathway indicated the ubiquityl moiety could be directly transferred from E2-S-Ub to amine-based nucleophiles in the absence of an E3.15 This observation led to the supposition that the reactivity of E2s could be exploited to modify the C-terminus of Ub (Figure 2A). Using this approach, a series of Ub C-terminal adducts bearing either small residues (single amino acids or small amines) or large molecules (proteins such as cytochrome c and lysozyme) were synthesized. More recently, several studies have suggested that the molecular basis for the transfer of Ub from E2-S-Ub to amines relies on cooperativity between E2 and the donor Ub moiety in organizing an active site for nucleophilic activation of an amine.16–19
Figure 2.
Enzymatic modification of the Ub C-terminus. (A) Use of Ub conjugation machinery to selectively modify the C-terminus of Ub Gly76. (B) Trypsin-mediated transpeptidation reactions as a means to modify Ub Arg74.
The utility of E2-mediated carboxy-terminal modifications is particularly evident when considering the history of DUBs, a class of enzymes whose function is to oppose that of E3 ligases.20 By 1980 it was clear that the cellular pool of Ub conjugates was dynamic; however, the factors responsible for this activity had not been identified.21 A likely culprit was an abundant protein in rabbit reticulocytes, now known as Ub C-terminal hydrolase (UCH-L3).22 Initially, UCH-L3 had been shown to promote the hydrolysis of Ub carboxy-terminal thioesters but its actual function remained unclear.23 To examine the reactivity and specificity of UCH-L3, a series of E2-dervived Ub variants were employed to monitor the rate at which UCH-L3 catalyzed the release of free Ub. The results of these experiments revealed that Ub harboring small C-terminal adducts displayed release rates close to 3 orders of magnitude higher than Ub-protein conjugates, indicating UCH-L3 has a strong preference for the former class of substrates.15 Importantly, these investigations were the first to suggest UCHs play a role in the cotranslational processing of Ub gene products and in recycling Ub from either small molecular weight adducts or Ub-protein conjugates.20 Furthermore, these experiments established a foundation for the identification and characterization of other DUBs, specifically in the UCH subfamily.24
Use of Limited Proteolysis
Protease-mediated trans-peptidations are another method for modifying the C-terminus of Ub since the latter is relatively intransigent to proteolytic cleavage by trypsin.25 In fact, the only site that is efficiently cleaved by trypsin is R74, which leads to the release of the C-terminal GG dipeptide (Figure 2B). Accordingly, proteolysis can be performed in the presence of excess amine-based nucleophile to effectively equip Ub with unique functional groups at the C-terminal position of R74.26 This approach was pioneered with the Ub C-terminal ethyl ester27 and subsequently exploited to synthesize an array of Ub-like reagents, including Ub-aldehyde,28 Ub-nitrile,29 the fluorogenic Ub-7-amido-4-methyl coumarin (UbAMC) probe,30 Ub vinyl sulfone (UbVS) derivatives,31 and nonhydrolyzable Ub-isopeptide isosteres.32 All of these probes have been utilized to illuminate different aspects of DUBs (some of which have been reviewed in refs 33 and 34). For example, UbAMC has been instrumental in characterizing the biochemical activity of newly discovered DUBs. That is, by examining a putative DUB against a panel of UblAMC derivatives, the specificity of that particular enzyme can be determined. Another area in which C-terminal probes have been valuable is in the identification of DUBs that are differentially expressed between normal, virus-infected, and tumor-derived human cells.35
ENZYMATIC SYNTHESIS OF POLYUBIQUITIN CHAINS
Over the past 10 years, a remarkable degree of complexity has been uncovered in the range of Ub signals governing distinct cellular processes.36 All seven lysine residues of Ub and the amino (N) terminus are used to form chains in vivo.37,38 Currently, the purpose of many of these chain types remains elusive, with the exception of canonical K48- and K63-linked homopolymeric chains. K48-linked chains mediate degradation of a conjugated target protein by shuttling the latter to the proteasome.39 By contrast, K63-linked chains have non-proteolytic functions, e.g., regulating the target’s function during DNA repair and inflammatory signaling pathways.40 Insight into the physiological roles of these two chain types comes largely from in vitro methods that allow synthesis of well-defined polyUb chains. Here, we discuss how these chains are constructed enzymatically and used to characterize their distinct functions. Moreover, we highlight how “atypical” polyUb chains, such as K11-linked homopolymers, can be synthesized in a similar manner using dedicated enzymes.
Building polyUb Chains
Due to the recalcitrant nature of isopeptide bonds, enzymatic construction remains one of the most effective methods currently available to synthesize well-defined polyUb chains. However, specific requirements must be met in order to obtain chains of specific linkages in quantities suitable for functional studies: (1) the enzyme must exhibit linkage specificity and (2) the enzyme must be active with free Ub. Currently, three E2 conjugating enzymes satisfy these criteria: K48-specific E2–25K (also known as UbcH1 or UBE2K),41 K63-specific Ubc13/Mms2,42,43 and K11-specific UBE2S.44
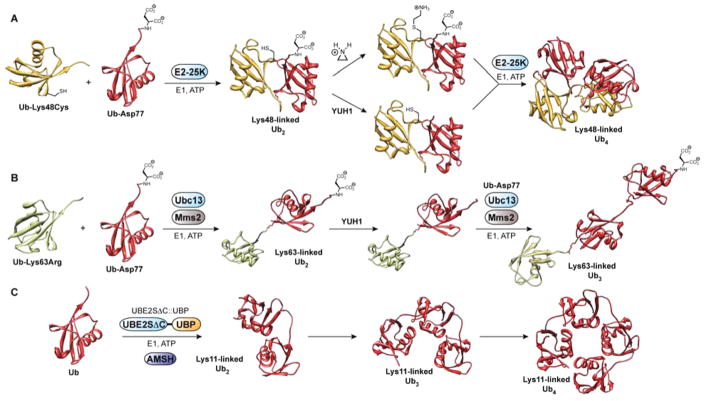
To control K48-linked chain synthesis using E2–25K, two Ub variants were required: one bearing a K48C mutation and another harboring a C-terminal aspartate cap (Ub-D77).45,46 Conjugation of these two monomers is performed with E2–25K, E1, and ATP (Figure 3A). It is important to note that if the desired end product is a K48-linked diUb, Ub K48C is replaced with Ub K48R to prevent elongation at the distal end. However, if longer chains are required, Ub K48C serves as the distal monomer and additional steps must be conducted to extend the chain. Namely, the C-terminal D77 cap is displaced using a UCH enzyme, often yeast Ub hydrolase 1 (YUH1), and the C48 residue is site-specifically alkylated with ethyleneimine affording an S-aminoethylcysteine lysine surrogate. Subsequently, ligation of the deprotected dimers with E2–25K furnishes K48-linked Ub tetramers. Importantly, structural analysis shows that Ub dimers linked through S-amino-ethylcysteine resemble native dimers.47 At this time, the molecular basis for K48 selectivity is unclear, but the C-terminal Ub-associated (UBA) domain of E2–25K, which binds free Ub, is thought to play a role.48
Figure 3.
Controlled synthesis of Lys48-, Lys63-, and Lys11-linked polyUb chains. (A) Enzymatic synthesis of Lys48-linked polyUb chains using Lys48-linkage specific E2 conjugating enzyme E2–25K. In the final step leading to Lys48-linked Ub4, two different Lys48-linked Ub dimers are required: one with a C-terminal aspartate cap and thiolysine residue at Lys48, and another with a free C-terminal Gly76 and Lys48 blocked with cysteine. The dimers are coupled through the thiolysine and free C-terminus. The proximal Ub (i.e., the only monomer not tethered to another Ub molecule) has an Asp77 cap, while the distal Ub (i.e., the last Ub in the tetramer) contains a Lys48Cys mutation. YUH1: yeast Ub hydrolase 1. PDB code: 2O6V.47 (B) Enzymatic synthesis of Lys63-linked polyUb chains using heterodimeric Lys63-linkage specific E2 conjugating system Ubc13/ Mms2. The proximal Ub has an Asp77 cap, while the distal Ub (shown in green) contains a Lys63Arg mutation. PDB code: 3HM3.119 (C) Enzymatic synthesis of Lys11-linked polyUb chains using an engineered Lys11-linkage specific E2 conjugating enzyme UBE2SΔC::UBP. Using UBE2SΔC::UBP alone leads to a mixture of Lys11- and Lys63-linked chains, thus AMSH is added to cleave all Lys63-linkages and obtain a homogeneous product. Dimers, trimers, and tetramers are produced in the same reaction. AMSH: associated molecule with the SH3 domain of STAM. PDB code: 2XEW.55
An analogous approach has been used to assemble K63-linked polyUb chains, with a few critical differences (Figure 3B). For instance, E2–25K was replaced with a heterodimeric complex consisting of Ubc13 and Mms2, an E2 enzyme and an E2 variant (UEV) lacking the active site cysteine normally present in an E2. Notably, structural details have suggested Mms2 plays a role in properly arranging K63 of the acceptor Ub in the active site of Ubc13.49 The inefficiency with which Ubc13/Mms2 utilizes tagged Ub substrates, including Ub containing the β-thiolysine at position 63, necessitates a synthetic strategy using native K63 residues as the nucleophile in chain extension.50 Thus, instead of the K48-linked chain-building strategy, K63-linked chains are generated by successive addition of Ub monomers.
To avoid complications in chain assembly caused by unnatural isopeptide linkages, a hybrid approach was recently developed using the genetically encoded lysine analogue Nε(tert-butyloxycarbonyl)-L-lysine, also referred to as H-Lys-(Boc)-OH.51,52 In this case, a pyrrolysyl (Pyl)-tRNA-synthetase (PylRS)/tRNAPyl pair from the methanogen Methanosarcina barkeri facilitates incorporation of non-proteinogenic pyrrolysine analogue H-Lys(Boc)-OH into the genetic code of E. coli. Ub is then produced with a single Lys(Boc), site-specifically encoded by a TAG stop codon through a process referred to as amber suppression (for a review of this topic, see ref 53). Thus, in conjunction with E2–25K or Ubc13/Mms2, controlled syntheses of well-defined polyUb chains with natural linkages can be achieved following chain extension of Boc-deprotected Ub dimers.
Once considered an atypical linkage, K11-linked polyUb chains have recently made a mark as an abundant form of polyUb in vivo, accounting for approximately 30% of all linkages in yeast and 2% in mammalian cells.54 To understand the function of these chains, the E2 UBE2S, which is selective for K11, was engineered to synthesize free chains.55 Critical to the success of this approach was the replacement of the lysine-rich C-terminal tail of UBE2S, a target for in cis autoubiquitination, with the Ub-binding zinc finger UBP domain of USP5/IsoT. This domain binds Ub with nanomolar affinity and, importantly, does not block the K11 side chain from serving as the nucleophile during chain extension. Using the UBE2S variant (UBE2SΔC::UBP) in combination with K63-specific DUB AMSH to prevent the formation of mixed linkage chains, K11-linked polymers of defined length can be assembled with high efficiency (Figure 3C).
In contrast to methods designed to produce chains of controlled length through multistep processes, linkage-specific E2 conjugating enzymes have also been used to produce multimilligram quantities of pure wild type polymers after a single elongation step.56 Given the simplicity of this approach and the ability to readily incorporate fluorescent probes, this strategy may prove the most effective for studying the molecular details of these polymers.
Understanding the Function of polyUb Chains
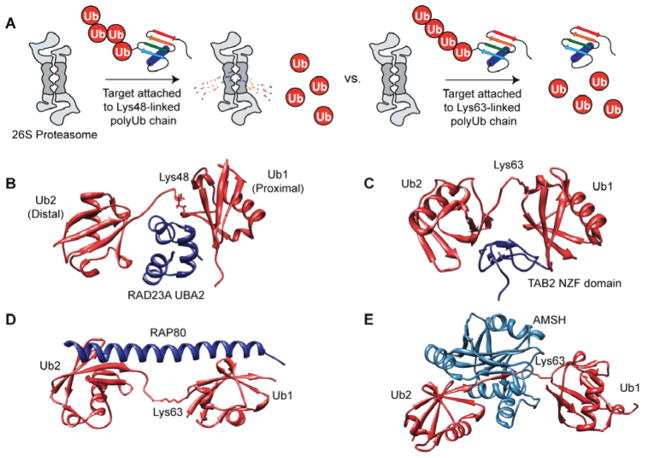
Enzymatic access to K48- and K63-linked polyUb chains has proven vital to understanding how the 26S proteasome processes substrates bearing polyUb chains. Using a model substrate for the 26S proteasome (dihydrofolate reductase (DHFR) fused to structurally defined K48-linked polyUb chains), several key aspects of proteasomal targeting have been unveiled.57 For instance, tetraUb was identified as the minimal signal for proteasomal targeting, with markedly increased degradation rates observed for substrates carrying longer polyUb chains. The molecular basis of this phenomenon is that by increasing chain length the affinity to the proteasome increases, which confers a greater commitment to degradation with substrates carrying long polyUb chains (>4 monomers). Subsequent to this report, K48- and K63-linked chains of the same length were shown to possess similar affinities to the proteasome despite significant differences in the structures of the two chains:58 K48-linked chains are compact with significant contacts between monomeric units,47,59 whereas K63-linked chains form more extended structures with minimal intersubunit contacts.60,61 Why, then, are substrates tethered to K48-linked polyUb chains degraded while those fused to K63-linked chains are not? The answer to this question may rest in the kinetic partitioning between deubiquitination and unfolding of the proteasome-bound substrate.62 In the case of K63-linked polyUb chains, the rate of deubiquitination by proteasome-associated DUBs is faster than the rate of unfolding, resulting in release of the intact substrate from the proteasome prior to degradation. By contrast, the residence time for a substrate bearing a K48-linked polyUb chain is sufficient for both unfolding and proteolytic cleavage (Figure 4A). As a result, substrates bearing K48-linked chains are preferentially degraded by the 26S proteasome despite both types of chains displaying the ability to interact with the proteasome.
Figure 4.
PolyUb signal processing and recognition. (A) Differences between protein degradation through the 26S proteasome when a target is tethered to a Lys48-linked polyUb chain versus a Lys63-linked chain. A target tethered to a Lys48-linked chain is translocated into the proteolytic core of the proteasome and rapidly degraded. By contrast, a target conjugated to a Lys63-linked chain binds to the proteasome, but the rate at which deubiquitination occurs exceeds the rate of translocation, ultimately leading to release of the intact target protein and formation of free Ub. (B) Structure of the Ub-associated 2 (UBA2) domain of RAD23A (dark blue) bound to a Lys48-linked Ub dimer (red). PDB code: 1ZO6.66 UBA2 forms extensive contacts with the linker region and the residues surrounding the isopeptide bond, providing a structural basis for the Lys48-linkage specificity of UBA domains. (C) Structure of the NZF (Npl4 Zinc Finger) domain of TAB2 (dark blue), a subunit of the TAK1 kinase, bound to a Lys63-linked Ub dimer (red). PDB code: 2WWZ.68 This structure reveals how the linker region is no longer involved in binding and how this particular UBD (Ub binding domain) interacts equally with both monomers. (D) Structure of receptor associated protein 80 (RAP80) tandem Ub interaction motif (tUIM) (dark blue) bound to Lys63-linked Ub dimer (red). PDB code: 3A1Q.70 (E) Structure of the DUB AMSH (associated molecule with the SH3 domain of STAM) (light blue) bound to Lys63-linked Ub dimer (red). PDB code: 2ZNV.74
Access to well-defined polyUb chains has also been instrumental in understanding how binding partners other than the proteasome recognize Ub signals. Current estimates predict the human genome encodes approximately 200 Ub binding domain (UBD)-containing proteins.63 UBDs are effector molecules that transduce specific Ub signals to downstream cellular events. A particular family of UBDs that has received a significant amount of attention is the class of Ub-associated (UBA) domain-containing proteins, whose function is usually ascribed to proteosomal targeting and regulation. In a set of groundbreaking studies, UBAs were found to preferentially bind K48-linked chains over K63-linked chains, providing the first sign that UBDs exhibit linkage selective recognition.64 However, when UBAs work in concert (i.e., upon homodimerization or heterodimerization) their intrinsic linkage selectivity can be overcome. This results in the ability of other linkages, e.g., K63, to effectively compete with K48-linkages for binding.65 From a structural standpoint, monomeric UBAs favor K48-polyUb because the latter presents an ideal compact epitope to which a contiguous surface can bind.66 That is, UBAs have a tendency to directly interact with the isopeptide linkage as well as distinct surfaces on each monomer (Figure 4B).
UBDs selective for K63- and M1-linkages also interact with multiple Ub moieties simultaneously, but the actual isopeptide linker plays less of a role in comparison to UBAs. For instance, the Npl4 zinc-finger (NZF) domains in TAB2 and TAB3, both of which activate the NF-κB pathway, selectively recognize K63-linked polyUb chains by serving as a molecular ruler that measures the length between the two hydrophobic patches in a Ub dimer and interacts with both monomers equally (Figure 4C).67,68 This strategy is also observed for the NF-κB essential modulator (NEMO), which interacts with linear chains through the UBAN domain (Ub binding in A20-binding inhibitor of NF-κB proteins), and the K63 specific tandem UIMs (Ub interaction motifs) in receptor associated protein 80 (RAP80) (Figure 4D), a critical responder to DNA damage.69–71 In each of these examples, the isopeptide linkage does not participate in the interaction and, instead, likely facilitates proper orientation of the two Ub surfaces.
DUBs are another class of UBDs that exhibit linkage selectivity. For instance, both USP14, the DUB associated with the 26S proteasome, and OTUB1 show specificity for K48-linked chains.72,73 By contrast, several DUBs selectively cleave K63-linked chains, including CYLD, TRABID, OTUD5, AMSH, CSN5, BRCC36, POH1, and MYSM1.20 Structural analyses suggest that linkage specificity is achieved by interactions that encompass a specific region of Ub surrounding the Lys residue forming the isopeptide bond. This is best illustrated by the structure obtained for a catalytically inactive variant of AMSH bound to K63-linked Ub dimer (Figure 4E). Here, AMSH binds the surface surrounding K63 of the proximal Ub (i.e., the Ub with a free C-terminus), providing a molecular basis of K63-linkage specificity.74
Collectively, these studies have dramatically expanded our knowledge of how different Ub signals are recognized and processed. To further illuminate the dynamics of the Ub network, future work must focus on atyptical polyUb chains of defined length. Although significant advances have been made along these lines, e.g., in identifying the K11 linkage-specific DUB Cezanne and developing K11 linkage-specific antibodies used to dissect the role of these chains in mitosis,55,75 further developments must rely on the arduous task of identifying enzymes that synthesize chains of specific linkage. Thus, chemical approaches to synthesizing polyUb chains of defined linkage and length will likely have an immediate impact on uncovering salient features of atypical polyUb chains.
SYNTHESIS OF UB DERIVATIVES: CHEMICAL CONTROL OVER REGIOSPECIFICITY
Recent advances in bioconjugate chemistry have led to the discovery of numerous routes to regiospecifically conjugate Ub to target proteins. In this section, we highlight these breakthroughs while showcasing how each method has been employed to illuminate details associated with Ub biology (for other general reviews, see refs 76 and 77).
NATIVE CHEMICAL LIGATION (NCL)
Semisynthetic Approaches to Ub-Protein Conjugates
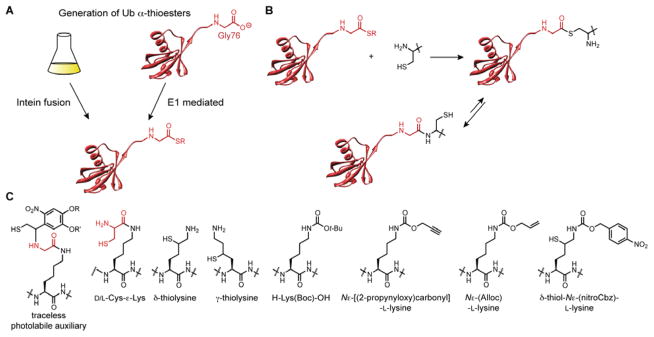
Expressed protein ligation (EPL) is the most common method to construct Ub-protein conjugates (Figure 5A). In general, EPL involves a chemoselective native chemical ligation (NCL) between a recombinant protein harboring a C-terminal thioester (α-thioester) and a synthetic peptide with a free thiol adjacent to a primary or secondary amine.78 α-Thioesters are typically generated upon thiolysis of an intein fusion protein, while the peptide component bearing a non-proteinogenic amino acid specifically is obtained through solid-phase peptide synthesis (SPPS). With these building blocks, EPL commences with transthioesterification between the thiol of a modified amino acid and α-thioester, followed by a spontaneous intramolecular S-to-N acyl transfer resulting in a native amide bond (Figure 5B).79
Figure 5.
Components of EPL. (A) Two different methods are available for generating recombinant Ub α-thioesters. The first method is based on the production of Ub-intein fusion proteins as thiolysis of the fusion protein leads to the desired α-thioester. The second method exploits the reactivity of E1 Ub-activating enzymes. This reaction proceeds through an E1-S-Ub acyl enzyme intermediate that is captured by a small molecule bearing a free thiol to afford Ub(1–76) α-thioester. (B) The mechanism of EPL with 1,2-aminothiols. The reaction commences with a transthioesterification followed by a rapid S-to-N acyl shift to afford a native amide bond. (C) Structures of various non-proteinogenic amino acids used to construct isopeptide linkages with Ub α-thioesters. Both H-Lys(Boc)-OH and Nε-[(2-propynyloxy)carbonyl]-L-lysine have not been used to forge an isopeptide linkage with Ub α-thioester; rather, other Ub C-terminal variants are required.102,104
As an alternative to intein-derived Ub α-thioesters, the intrinsic reactivity of the E1 Ub-activating enzyme can be exploited to generate similar products, namely Ub(1–76) α-thioester (Figure 5A).80 Recall that E1 activates Ub as an adenylate, which is then attacked by a cysteine residue to afford an E1-S-Ub thioester intermediate. Since E1 does not possess the basic residues necessary for deprotonating amine-based nucleophiles, the E1-S-Ub intermediate readily reacts with nucleophilic thiols to afford a Ub(1–76) α-thioester.81 This method has proven extremely effective, as yields approaching 80% have been observed for the 2-mercaptoethanesulfonate (MESNa) thioester.82 An additional benefit to using the E1 approach is the ability to label a single monomer in a polyUb chain with a chemical tag.
With regards to the peptide component of EPL, several strategies have been developed to promote ligations with Ub α-thioesters. A common approach is to use the canonical cysteine-mediate ligation,80,83,84 but several recent studies have focused on the use of non-proteinogenic amino acids (Figure 5C). The distinction between these two is that the cysteine-mediated ligation affords a peptide linkage while non-proteinogenic amino acids promote isopeptide bond formation. Due to limitations in space our discussion is centered on the latter.
A Nε-(Gly)-L-lysine variant decorated with a photolabile auxiliary group was the first non-proteinogenic amino acid used to forge an isopeptide tether between Ub and the synthetic peptide.85 After difficulties were encountered in scaling up this protocol, an alternative method was pursued involving the classic cysteine-mediated ligation strategy with Nε-(L-Cys)-L-Lys.86 Unlike the strategy based on photolabile auxiliary groups, the cysteine-mediated approach, albeit more effective in terms of yield, is no longer traceless as subsequent desulfurization reactions are required to remove the residual thiols. Moreover, the use of Nε-(L-Cys)-L-Lys ultimately results in the replacement of Ub Gly76 with alanine, which could be problematic if one is to use this method to study the activity of DUBs; Ala76-ε-Lys is more resistant to cleavage than the native Gly76-ε-Lys. To overcome some of these limitations, the Brik and Liu laboratories independently reported on the utility of thiolysines.87–89 Both γ-thiolysine and δ-thiolysine benefit from the same principles that promote NCL with cysteine residues, although δ-thiolysine is thought to stimulate faster ligations relative to γ-thiolysine due to differences in rates associated with five-membered versus six-membered ring formation.
Functional Studies of Ub Using Semisynthesis
Chromatin biology was the first area targeted by Ub semisynthesis (for more detailed reviews of this topic, see refs 90 and 91). Ub plays a critical role in orchestrating gene transcription, replication, DNA repair and recombination by spatially and temporally regulating interactions at the interface of chromatin.92 Covalent attachment of Ub to the core components of chromatin, histones H2A and H2B, is one of the most common modifications, often regulating additional modifications such as histone methylation. This level of interplay between ubiquitylation and methylation is referred to as “crosstalk”. Until recently, the molecular details associated with crosstalk were unclear because of the difficulty associated with enzymatically generating well-defined histone modifications. To address this issue, Muir and co-workers elegantly employed protein semisynthesis to construct Ub-modified H2B (uH2B).86,93 Semisynthetic uH2B was then incorporated into nucleosomes, the fundamental unit of chromatin, to obtain chemically defined probes for biochemical studies with the methyltransferases responsible for H3 K4 and K79 methylation (hSet1L and hDot1L). The results of these experiments revealed that (i) methylation was dependent on the presence of uH2B and (ii) a noncanonical surface of Ub (i.e., a surface not composed of the hydrophobic patch consisting of L8, I44, and V70) associates with the methyltransferases. One of the most surprising findings from these studies, however, was that Ub does not function by enhancing binding of the methyltransferases to nucleosomes but rather by stimulating a catalytically competent conformation of the enzyme through allosteric regulation.
Thiolysine-based semisynthetic approaches have also proven effective in understanding how Ub alters the function of a protein. In particular, δ-thiolysine was used to investigate the ubiquitination of α-synuclein K6 (Ub-α-synuclein).94 α-Synuclein is a presynaptic protein implicated in the etiology of several neurodegenerative diseases including Parkinson’s disease (PD) and is modified in myriad ways, e.g., through phosphorylation, glycosylation, truncation, and/or ubiquitination. Each of these modifications is associated with the pathogenesis of PD and commonly found in α-synuclein aggregates (also referred to as fibrils). Consequently, there is significant interest in understanding how fibrils are formed. In this context, recent biophysical studies using semisynthetic Ub-α-synuclein demonstrated that, contrary to some studies,95 ubiquitination of K6 does not promote aggregation relative to the unmodified form. These results suggest Ub stabilizes the monomeric form of α-synuclein, which would be consistent with previous observations indicating the majority of ubiquitinated species are found in soluble form and not in fibrils.96 Future work with defined mixtures of unmodified and modified α-synuclein (both monoUb and multi-monoUb) may shed more light on how Ub alters the morphology of fibrils.
Efforts to understand how K33-linked polyUb chains serve in a non-proteolytic capacity have also relied on the use of thiolysine-based semisynthesis.97 In this example, 15N-isotopic labeling of the distal Ub unit (obtained as a recombinant protein and activated by E1 as α-thioester) enabled spectroscopic interrogation of the conformation and ligand binding properties of K33-linked Ub dimer; δ-thiolysine was installed into the proximal Ub monomer through total synthesis. These studies revealed two critical differences between K33- and K48-linkages, the canonical signal for protein degradation. First, unlike K48-linked Ub dimers where both subunits form strong interdomain contacts through the hydrophobic patch of Ub (L8, I44, V70), the interface between Ub monomers in the K33-linked dimer is transient due to the engagement of only one hydrophobic patch. Second, K33-linked Ub dimer interactions with a proteasomal chaperone (the human homologue of protein Rad23, hHR23a) indicated that binding affinity is reduced by an order of magnitude relative to K48-linked Ub dimers. This result is consistent with studies on the ubiquitination of T-cell receptors,98 which suggest K33-linkages do not promote protein degradation through the 26S proteasome.
Total Synthesis of Ub-Protein Conjugates
Total chemical synthesis of Ub derivatives has emerged as a powerful strategy to generate Ub-target conjugates as evidenced by the recent synthesis of the 304-residue K48-linked tetraUb.99 Building on previous efforts in synthesizing all seven isopeptide-linked Ub dimers,100 the Brik group reported a linear synthesis of tetraUb in which each monomer is incorporated consecutively. Key to success of their approach was the development and implementation of a chemical synthesis of Ub α-thioester using a C-terminal N-methylcysteine residue.101 This allows for the incorporation of δ-thiolysine into the same Ub sequence without self-ligating, thus enabling chain elongation to proceed. Three distinct monomers, each constructed from two peptide fragments using NCL, were prepared, two of which contained both the thioester and orthogonally protected δ-thiolysine. After each round of ligation a desulfurization step led to the removal of the thiols and formation of native isopeptide bonds. Ultimately, K48-linked tetraUb was produced in a 5% overall yield starting from a single Ub monomer. As mentioned by the authors, further improvements could be made by exploiting recent developments in SPPS.82 Upon implementing these approaches, this methodology may lend itself to the construction other types of homopolymers (e.g., K6, K27, K29, and K33) as well as linear heteropolymers (monomers linked through different lysine residues), both of which are unattainable by enzymatic means in a well-defined manner.
Incorporation of Unnatural Amino Acids
Undoubtedly, the most streamlined method to synthesizing Ub-protein conjugates is the regiospecific ligation of recombinant Ub α-thioester to another recombinant protein. This strategy avoids the laborious synthetic steps required for semisynthesis and total synthesis and takes advantage of the ease with which recombinant proteins can often be obtained. To this end, the PylRS/tRNAPyl pair from M. barkeri has been used to insert pyrrolysine analogues into specific sites of Ub and target proteins. Several different pyrrolysine analogues have successfully been incorporated into recombinant Ub, namely, H-Lys(Boc)-OH,102 D/L-Cys-ε-Lys,103 Nε-[(2-propynyloxy)-carbonyl]-L-lysine,104 Nε-(Alloc)-L-lysine,105 and a latent form of δ-thiolysine referred to as δ-thiol-Nε-(p-nitrocarbobenzyloxy)-L-lysine or δ-thiol-Nε-(nitroCbz)-L-lysine106 (Figure 5C). Of these, δ-thiol-Nε-(nitroCbz)-L-lysine may prove the most versatile as it allows for a traceless synthesis of Ub dimers as well as Ub-Ubl heterodimers with user-defined control over the linkage site. Another interesting feature of δ-thiol-Nε-(nitroCbz)-L-lysine is that the nitroCbz is removed in situ, enabling direct isolation of Ub bearing δ-thiolysine.
Functional Studies Based on Unnatural Amino Acid Incorporation
Methods utilizing PylRS/tRNAPyl pairs to obtain native Ub dimers have exposed important details related to atypical linkages.102 For instance, structural analysis of K6-linked diUb showed a compact conformation distinct from previously characterized K11-, K48-, and K63-linked diUb derivatives. This observation suggests K6-linked polyUb chains may bind novel UBDs and mediate distinct biological processes, which would be congruent with the prevalence of this linkage in DNA repair pathways.107 Having dimers linked through atypical lysine residues also provides an opportunity to profile the selectivity and specificity of DUBs. For example, biochemical studies with 10% of the known human DUBs led to the identification of TRABID, an ovarian tumor (OTU)-domain containing DUB, which cleaves K29-linked diUb 40-fold more efficiently than K63-linked diUb. Importantly, these synthetic strategies provide a means of investigating the biological effects of distinct Ub and Ubl chain topologies in vitro, as well as developing other biochemical tools for in vivo studies, e.g., linkage-specific antibodies.
ALTERNATIVES TO NCL
Thioether Ligations
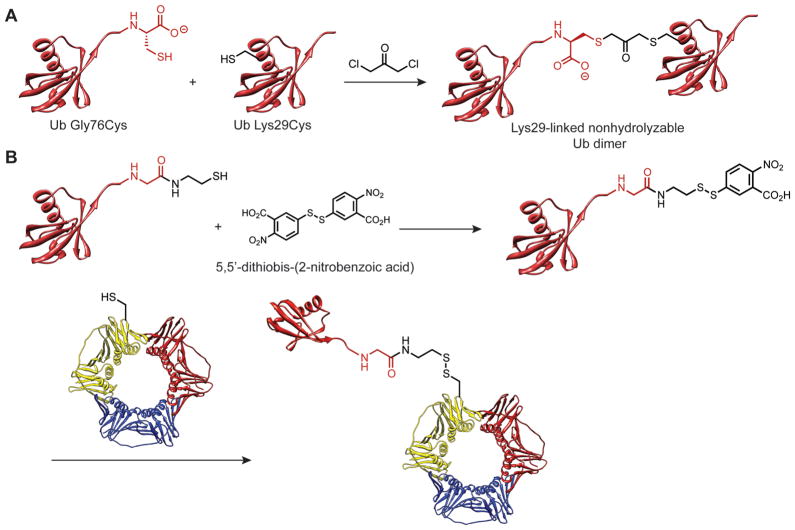
As highlighted above, the objective behind synthesizing all possible isopeptide Ub linkages is to enable studies designed to identify and characterize linkage-selective binding partners. The source of these binding partners is often an extract from a lysed population of cells. However, cellular extracts present a difficult environment in which to study polyUb linkage-selective interactions due to the vast number of DUBs that reduce the half-life of the specific polyUb chain under investigation. To overcome this problem, Wilkinson and co-workers pioneered the development of nonhydrolyzable linkages between Ub molecules based on thioether bonds.108 Their strategy was to fashion a thioether tether between recombinant Ub monomers by exploiting the absence of native cysteines. Two different Ub monomers are required for this site-specific ligation: one where G76 is mutated to cysteine and another where a specified lysine is mutated to cysteine (Figure 6A). Subsequent fusion of these two monomers through a thioether bridge results in a linkage that differs slightly from that of the native isopeptide bond in length (one carbon longer) and polarity (there is an acetone spacer along with a free carboxylate). Nevertheless, the ability of these dimers to serve as inhibitors of DUBs and Ub conjugation machinery is testament of their ability to target enzymes that process polyUb chains. Further extension of this methodology toward the synthesis of nonhydrolzable tetraUb chains immobilized on resin led to the development of novel tools for identifying binding partners in cell lysates.109 Indeed, a key finding from these experiments was the identification of Ufd3 (an important player in sorting membrane proteins into multivesicular bodies) as a novel interacting partner of what is ostensibly referred to as a nonhydrolyzable variant of K29-linked tetraUb.
Figure 6.
Synthesis of Ub-protein conjugates using methods other than NCL. (A) Synthesis of nonhydrolyzable Ub dimers by constructing a thioether bridge between monomers. (B) Conjugation of Ub to target protein using directed disulfide exchange. The target protein shown here is proliferating cell nuclear antigen (PCNA).111 PCNA PDB code: 1VYM.120
Directed Disulfide Bond Formation
Isopeptide-like linkages between recombinant proteins can also be constructed through a selective exchange of disulfide bonds.110,111 In this approach, the addition of cysteamine to a Ub-intein fusion affords Ub with a C-terminal aminoethanethiol linker. Combining this Ub derivative with 5,5′-dithiobis(2-nitrobenzoic acid) and a target containing a single cysteine residue results in the formation of a site-specific disulfide tether between the two proteins (Figure 6B). It is important to note that despite the disulfide linkage being ~2.4 Å longer than the native isopeptide bond, the function of the Ub-protein conjugate is not altered. The advantages of this approach relative to other methods are that (i) chemical synthesis is no longer required and (ii) the conjugation site can be genetically encoded with standard techniques of recombinant DNA similar to the thioether strategy.
Functional Studies with Directed Disulfide Bond Formation
The utility of the disulfide-directed methodology is highlighted by the ability to rapidly construct uH2Bss variants and glean information on the impact Ub has on nucleosome structure and function.110,112 For instance, by applying a positional scanning approach, whereby the Ub attachment is varied on the nucleosome surface, two key observations were made. First, the Ub attachment point is not important for stimulating hDot1L activity since methylation of H3 K79 occurred when Ub was attached to other positions on H2B as well as on H2A. Second, mononucleosomes harboring regioisomers of uH2Bss exhibited different structural characteristics, thus presaging a role for Ub in regulating chromatin structure. Indeed, experiments with nucleosome arrays led to the proposal that uH2B prevents folding of the entire chromatin fiber, thereby promoting the formation of a structure more conducive to active transcription.
Another example where disulfide bond formation has illuminated biochemical details is with proliferating cell nuclear antigen (PCNA).111 PCNA is a homotrimeric protein involved in processive DNA synthesis. When PCNA is ubiquitinated following DNA damage, the DNA polymerase Polη is recruited to the site of damage to promote translesion DNA synthesis (TLS). To investigate how PCNA-Ub specifically stimulates TLS, a chemical approach using the disulfide-directed methodology was employed. In this case, several PCNA mutants (K107C, K127C, and K164C) were coupled to Ub and another Ubl, SUMO (small ubiquitin-like modifier), both harboring a C-terminal aminoethanethiol linker to afford PCNA-Ubl conjugates at different positions. Using these probes, it was revealed that SUMO has little effect on the degree to which Polη suppresses Polδ-PCNA-catalyzed DNA synthesis. By contrast, ubiquitination of PCNA rapidly promotes the exchange between Polδ and Polη. An additional finding indicated that the position of Ub attachment on PCNA does not influence the inhibition of DNA synthesis through Polη-PCNA-Ub complex formation, suggesting a great deal of conformational flexibility in the latter ternary complex. These results, along with the uH2B studies described above, illustrate the power of the disulfide-directed approach in establishing structure–activity relationships.
SUMMARY AND OUTLOOK
Although Ub-based probes have played a central role in elucidating the basic principles of Ub signaling, there remains much to be discovered with regards to the function of diverse Ub signals in specific biological processes. In this Review, we have discussed methods, both enzymatic and non-enzymatic, that lead to the generation of specific Ub modifications. The resulting Ub-based probes have been used to (i) identify and characterize DUB activity, (ii) understand the influence of Ub on protein function and crosstalk with other PTMs, and (iii) help comprehend the impact of polyUb chain linkage and length on downstream signaling events. Despite these achievements, we are currently left with a vast number of uncharacterized polyUb signals, DUBs and UBDs, many of which play important roles in human disease.36
Recent developments in synthesizing specific isopeptide bonds between Ub molecules and other Ubls offer tremendous potential in unraveling many aspects of Ub signaling. The DNA damage response (DDR) is one pathway in particular where access to a diverse array of tools could help dissect the role of different polyUb chains (reviewed in ref 113). For example, upon induction of DNA double-strand breaks, chromatin is modified with K63-linked polyUb chains, which recruit additional E3 ligases including BRCA1. Since BRCA1 has been shown to assemble atypical K6-linked polyUb chains,107 the question is: What role(s) might these chains serve during DDR? Is it possible they act as an additional platform on which other proteins are mobilized to the response, as a switch to facilitate the exchange between repair mechanisms, or both? Another area of interest is in understanding how different linkage types promote degradation through the 26S proteasome. Quantitative proteomics experiments with yeast suggest the kinetics of chain disassembly via the proteasome correlate with the extent to which different chains accumulate during proteasomal inhibition.37 While this is consistent with K48 linkages functioning as the major degradative signal due to their overall abundance, the identification of linkage-selective chaperones responsible for delivering substrates to the proteasome implies there are additional factors that could regulate the rates of protein degradation.114,115 Finally, the emergence of mixed Ub-Ubl species, particularly Ub-SUMO and Ub-NEDD8, as distinct cellular signals presents an opportunity for non-enzymatic approaches to the construction of these molecules to play an important role in understanding their function.116,117
Acknowledgments
We apologize to those investigators whose important contributions were not cited in this Review due to limitations in space. We thank S. Shekhawat for critical reading of the manuscript. D.A.K. is supported by a Graduate Research Fellowship from the National Science Foundation (2011101911). Research in the E.R.S. laboratory is supported by the Susan G. Komen Foundation, the Greater Milwaukee Foundation Shaw Scientist Program. and the University of Wisconsin-Madison.
References
- 1.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 2.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 3.Kirkin V, Dikic I. Ubiquitin networks in cancer. Curr Opin Genet Dev. 2011;21:21–28. doi: 10.1016/j.gde.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Chiu YH, Zhao M, Chen ZJ. Ubiquitin in NF-kappaB signaling. Chem Rev. 2009;109:1549–1560. doi: 10.1021/cr800554j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 6.Patterson C, Ike C, Willis PWt, Stouffer GA, Willis MS. The bitter end: the ubiquitin-proteasome system and cardiac dysfunction. Circulation. 2007;115:1456–1463. doi: 10.1161/CIRCULATIONAHA.106.649863. [DOI] [PubMed] [Google Scholar]
- 7.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda F, Crosetto N, Dikic I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143:677–681. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 15.Pickart CM, Rose IA. Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides. J Biol Chem. 1985;260:7903–7910. [PubMed] [Google Scholar]
- 16.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 17.Rodrigo-Brenni MC, Foster SA, Morgan DO. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol Cell. 2010;39:548–559. doi: 10.1016/j.molcel.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 21.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 22.Mayer AN, Wilkinson KD. Detection, resolution, and nomenclature of multiple ubiquitin carboxyl-terminal esterases from bovine calf thymus. Biochemistry. 1989;28:166–172. doi: 10.1021/bi00427a024. [DOI] [PubMed] [Google Scholar]
- 23.Rose IA, Warms JV. An enzyme with ubiquitin carboxy-terminal esterase activity from reticulocytes. Biochemistry. 1983;22:4234–4237. doi: 10.1021/bi00287a012. [DOI] [PubMed] [Google Scholar]
- 24.Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- 25.Wilkinson KD, Audhya TK. Stimulation of ATP-dependent proteolysis requires ubiquitin with the COOH-terminal sequence Arg-Gly-Gly. J Biol Chem. 1981;256:9235–9241. [PubMed] [Google Scholar]
- 26.Homandberg GA, Laskowski M., Jr Enzymatic resynthesis of the hydrolyzed peptide bond(s) in ribonuclease S. Biochemistry. 1979;18:586–592. doi: 10.1021/bi00571a006. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson KD, Cox MJ, Mayer AN, Frey T. Synthesis and characterization of ubiquitin ethyl ester, a new substrate for ubiquitin carboxyl-terminal hydrolase. Biochemistry. 1986;25:6644–6649. doi: 10.1021/bi00369a047. [DOI] [PubMed] [Google Scholar]
- 28.Dunten RL, Cohen RE. Recognition of modified forms of ribonuclease A by the ubiquitin system. J Biol Chem. 1989;264:16739–16747. [PubMed] [Google Scholar]
- 29.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 30.Dang LC, Melandri FD, Stein RL. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 31.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shanmugham A, Fish A, Luna-Vargas MP, Faesen AC, El Oualid F, Sixma TK, Ovaa H. Nonhydrolyzable ubiquitin-isopeptide isosteres as deubiquitinating enzyme probes. J Am Chem Soc. 2010;132:8834–8835. doi: 10.1021/ja101803s. [DOI] [PubMed] [Google Scholar]
- 33.Ovaa H. Active-site directed probes to report enzymatic action in the ubiquitin proteasome system. Nat Rev Cancer. 2007;7:613–620. doi: 10.1038/nrc2128. [DOI] [PubMed] [Google Scholar]
- 34.Love KR, Pandya RK, Spooner E, Ploegh HL. Ubiquitin C-terminal electrophiles are activity-based probes for identification and mechanistic study of ubiquitin conjugating machinery. ACS Chem Biol. 2009;4:275–287. doi: 10.1021/cb9000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ovaa H, Kessler BM, Rolen U, Galardy PJ, Ploegh HL, Masucci MG. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc Natl Acad Sci USA. 2004;101:2253–2258. doi: 10.1073/pnas.0308411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, Rush J, Peng J. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hershko A, Ciechanover A. The ubiquitin system. Ann Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 40.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Chen Z, Pickart CM. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J Biol Chem. 1990;265:21835–21842. [PubMed] [Google Scholar]
- 42.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 43.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 44.Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci USA. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piotrowski J, Beal R, Hoffman L, Wilkinson KD, Cohen RE, Pickart CM. Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. J Biol Chem. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- 46.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 47.Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol. 2007;367:204–211. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 48.Wilson RC, Edmondson SP, Flatt JW, Helms K, Twigg PD. The E2-25K ubiquitin-associated (UBA) domain aids in polyubiquitin chain synthesis and linkage specificity. Biochem Biophys Res Commun. 2011;405:662–666. doi: 10.1016/j.bbrc.2011.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 50.Hofmann RM, Pickart CM. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem. 2001;276:27936–27943. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- 51.Castaneda CA, Liu J, Kashyap TR, Singh RK, Fushman D, Cropp TA. Controlled enzymatic synthesis of natural-linkage, defined-length polyubiquitin chains using lysines with removable protecting groups. Chem Commun. 2011;47:2026–2028. doi: 10.1039/c0cc04868b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J, Castaneda CA, Wilkins BJ, Fushman D, Cropp TA. Condensed E. coli cultures for highly efficient production of proteins containing unnatural amino acids. Bioorg Med Chem Lett. 2010;20:5613–5616. doi: 10.1016/j.bmcl.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Ann Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 54.Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- 55.Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong KC, Helgason E, Yu C, Phu L, Arnott DP, Bosanac I, Compaan DM, Huang OW, Fedorova AV, Kirkpatrick DS, Hymowitz SG, Dueber EC. Preparation of distinct ubiquitin chain reagents of high purity and yield. Structure. 2011;19:1053–1063. doi: 10.1016/j.str.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 57.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peth A, Uchiki T, Goldberg AL. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol Cell. 2010;40:671–681. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 60.Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 62.Jacobson AD, Zhang NY, Xu P, Han KJ, Noone S, Peng J, Liu CW. The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome. J Biol Chem. 2009;284:35485–35494. doi: 10.1074/jbc.M109.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 65.Sims JJ, Haririnia A, Dickinson BC, Fushman D, Cohen RE. Avid interactions underlie the Lys63-linked polyubiquitin binding specificities observed for UBA domains. Nat Struct Mol Biol. 2009;16:883–889. doi: 10.1038/nsmb.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 67.Sato Y, Yoshikawa A, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J. 2009;28:3903–3909. doi: 10.1038/emboj.2009.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol. 2009;16:1328–1330. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- 69.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 70.Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 2009;28:2461–2468. doi: 10.1038/emboj.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edelmann MJ, Iphofer A, Akutsu M, Altun M, di Gleria K, Kramer HB, Fiebiger E, Dhe-Paganon S, Kessler BM. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J. 2009;418:379–390. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- 74.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 75.Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, Kelley RF, Dixit VM. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 76.Fekner T, Li X, Chan MK. Nonenzymatic ubiquitylation. ChemBioChem. 2011;12:21–33. doi: 10.1002/cbic.201000625. [DOI] [PubMed] [Google Scholar]
- 77.Martin LJ, Raines RT. Carpe diubiquitin. Angew Chem. 2010;49:9042–9044. doi: 10.1002/anie.201005946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 79.Johnson EC, Kent SB. Insights into the mechanism and catalysis of the native chemical ligation reaction. J Am Chem Soc. 2006;128:6640–6646. doi: 10.1021/ja058344i. [DOI] [PubMed] [Google Scholar]
- 80.Burchak ON, Jaquinod M, Cottin C, Mugherli L, Iwai K, Chatelain F, Balakirev MY. Chemoenzymatic ubiquitination of artificial substrates. ChemBioChem. 2006;7:1667–1669. doi: 10.1002/cbic.200600283. [DOI] [PubMed] [Google Scholar]
- 81.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 82.El Oualid F, Merkx R, Ekkebus R, Hameed DS, Smit JJ, de Jong A, Hilkmann H, Sixma TK, Ovaa H. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew Chem, Int Ed. 2010;49:10149–10153. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu X, Olsen SK, Capili AD, Cisar JS, Lima CD, Tan DS. Designed semisynthetic protein inhibitors of Ub/ Ubl E1 activating enzymes. J Am Chem Soc. 2010;132:1748–1749. doi: 10.1021/ja9088549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chatterjee C, McGinty RK, Pellois JP, Muir TW. Auxiliary-mediated site-specific peptide ubiquitylation. Angew Chem. 2007;46:2814–2818. doi: 10.1002/anie.200605155. [DOI] [PubMed] [Google Scholar]
- 86.McGinty RK, Kohn M, Chatterjee C, Chiang KP, Pratt MR, Muir TW. Structure-activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem Biol. 2009;4:958–968. doi: 10.1021/cb9002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ajish Kumar KS, Haj-Yahya M, Olschewski D, Lashuel HA, Brik A. Highly efficient and chemoselective peptide ubiquitylation. Angew Chem. 2009;48:8090–8094. doi: 10.1002/anie.200902936. [DOI] [PubMed] [Google Scholar]
- 88.Yang R, Pasunooti KK, Li F, Liu XW, Liu CF. Synthesis of K48-linked diubiquitin using dual native chemical ligation at lysine. Chem Commun. 2010;46:7199–7201. doi: 10.1039/c0cc01382j. [DOI] [PubMed] [Google Scholar]
- 89.Castaneda CA, Spasser L, Bavikar SN, Brik A, Fushman D. Segmental isotopic labeling of ubiquitin chains to unravel monomer-specific molecular behavior. Angew Chem. 2011;50:11210–11214. doi: 10.1002/anie.201104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chatterjee C, Muir TW. Chemical approaches for studying histone modifications. J Biol Chem. 2010;285:11045–11050. doi: 10.1074/jbc.R109.080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dhall A, Chatterjee C. Chemical approaches to understand the language of histone modifications. ACS Chem Biol. 2011;6:987–999. doi: 10.1021/cb200142c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 93.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hejjaoui M, Haj-Yahya M, Kumar KS, Brik A, Lashuel HA. Towards elucidation of the role of ubiquitination in the pathogenesis of Parkinson’s disease with semisynthetic ubiquitinated alpha-synuclein. Angew Chem. 2011;50:405–409. doi: 10.1002/anie.201005546. [DOI] [PubMed] [Google Scholar]
- 95.Lee JT, Wheeler TC, Li L, Chin LS. Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Hum Mol Genet. 2008;17:906–917. doi: 10.1093/hmg/ddm363. [DOI] [PubMed] [Google Scholar]
- 96.Nonaka T, Iwatsubo T, Hasegawa M. Ubiquitination of alpha-synuclein. Biochemistry. 2005;44:361–368. doi: 10.1021/bi0485528. [DOI] [PubMed] [Google Scholar]
- 97.Castaneda CA, Spasser L, Bavikar SN, Brik A, Fushman D. Segmental isotopic labeling of ubiquitin chains to unravel monomer-specific molecular behavior. Angew Chem, Int Ed. 2011;50:11210–11214. doi: 10.1002/anie.201104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang H, Jeon MS, Liao L, Yang C, Elly C, Yates JR, 3rd, Liu YC. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity. 2010;33:60–70. doi: 10.1016/j.immuni.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kumar KS, Bavikar SN, Spasser L, Moyal T, Ohayon S, Brik A. Total chemical synthesis of a 304 amino Acid k48-linked tetraubiquitin protein. Angew Chem. 2011;50:6137–6141. doi: 10.1002/anie.201101920. [DOI] [PubMed] [Google Scholar]
- 100.Kumar KS, Spasser L, Erlich LA, Bavikar SN, Brik A. Total chemical synthesis of di-ubiquitin chains. Angew Chem. 2010;49:9126–9131. doi: 10.1002/anie.201003763. [DOI] [PubMed] [Google Scholar]
- 101.Erlich LA, Kumar KS, Haj-Yahya M, Dawson PE, Brik A. N-methylcysteine-mediated total chemical synthesis of ubiquitin thioester. Org Biomol Chem. 2010;8:2392–2396. doi: 10.1039/c000332h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat Chem Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 103.Li X, Fekner T, Ottesen JJ, Chan MK. A pyrrolysine analogue for site-specific protein ubiquitination. Angew Chem. 2009;48:9184–9187. doi: 10.1002/anie.200904472. [DOI] [PubMed] [Google Scholar]
- 104.Eger S, Scheffner M, Marx A, Rubini M. Synthesis of defined ubiquitin dimers. J Am Chem Soc. 2010;132:16337–16339. doi: 10.1021/ja1072838. [DOI] [PubMed] [Google Scholar]
- 105.Castaneda C, Liu J, Chaturvedi A, Nowicka U, Cropp TA, Fushman D. Nonenzymatic assembly of natural polyubiquitin chains of any linkage composition and isotopic labeling scheme. J Am Chem Soc. 2011;133:17855–17868. doi: 10.1021/ja207220g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Virdee S, Kapadnis PB, Elliott T, Lang K, Madrzak J, Nguyen DP, Riechmann L, Chin JW. Traceless and site-specific ubiquitination of recombinant proteins. J Am Chem Soc. 2011;133:10708–10711. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu-Baer F, Lagrazon K, Yuan W, Baer R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem. 2003;278:34743–34746. doi: 10.1074/jbc.C300249200. [DOI] [PubMed] [Google Scholar]
- 108.Yin L, Krantz B, Russell NS, Deshpande S, Wilkinson KD. Nonhydrolyzable diubiquitin analogues are inhibitors of ubiquitin conjugation and deconjugation. Biochemistry. 2000;39:10001–10010. doi: 10.1021/bi0007019. [DOI] [PubMed] [Google Scholar]
- 109.Russell NS, Wilkinson KD. Identification of a novel 29-linked polyubiquitin binding protein, Ufd3, using polyubiquitin chain analogues. Biochemistry. 2004;43:4844–4854. doi: 10.1021/bi035626r. [DOI] [PubMed] [Google Scholar]
- 110.Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 111.Chen J, Ai Y, Wang J, Haracska L, Zhuang Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat Chem Biol. 2010;6:270–272. doi: 10.1038/nchembio.316. [DOI] [PubMed] [Google Scholar]
- 112.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 114.Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu-Baer F, Ludwig T, Baer R. The UBXN1 protein associates with autoubiquitinated forms of the BRCA1 tumor suppressor and inhibits its enzymatic function. Mol Cell Biol. 2010;30:2787–2798. doi: 10.1128/MCB.01056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 118.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 119.Datta AB, Hura GL, Wolberger C. The structure and conformation of Lys63-linked tetraubiquitin. J Mol Biol. 2009;392:1117–1124. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kontopidis G, Wu SY, Zheleva DI, Taylor P, McInnes C, Lane DP, Fischer PM, Walkinshaw MD. Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc Natl Acad Sci USA. 2005;102:1871–1876. doi: 10.1073/pnas.0406540102. [DOI] [PMC free article] [PubMed] [Google Scholar]