Abstract
Supervised learning methods promise to improve integrated testing strategies (ITS), but must be adjusted to handle high dimensionality and dose–response data. ITS approaches are currently fueled by the increasing mechanistic understanding of adverse outcome pathways (AOP) and the development of tests reflecting these mechanisms. Simple approaches to combine skin sensitization data sets, such as weight of evidence, fail due to problems in information redundancy and high dimension-ality. The problem is further amplified when potency information (dose/response) of hazards would be estimated. Skin sensitization currently serves as the foster child for AOP and ITS development, as legislative pressures combined with a very good mechanistic understanding of contact dermatitis have led to test development and relatively large high-quality data sets. We curated such a data set and combined a recursive variable selection algorithm to evaluate the information available through in silico, in chemico and in vitro assays. Chemical similarity alone could not cluster chemicals’ potency, and in vitro models consistently ranked high in recursive feature elimination. This allows reducing the number of tests included in an ITS. Next, we analyzed with a hidden Markov model that takes advantage of an intrinsic inter-relationship among the local lymph node assay classes, i.e. the monotonous connection between local lymph node assay and dose. The dose-informed random forest/hidden Markov model was superior to the dose-naive random forest model on all data sets. Although balanced accuracy improvement may seem small, this obscures the actual improvement in misclassifications as the dose-informed hidden Markov model strongly reduced "false-negatives" (i.e. extreme sensitizers as non-sensitizer) on all data sets.
Keywords: LLNA, in vitro, skin sensitization, Integrated Testing Strategy, machine learning, Hidden Markov Model, QSAR, Feature Selection
Introduction
Skin sensitization, which clinically manifests in humans as allergic contact dermatitis, is an increasingly common concern for both regulators and the general population. Epidemiologic data indicate that an estimated 15–20% of the general population suffers from contact allergy (Thyssen et al., 2007a). Most common are allergies to preservatives and fragrances (Peiser et al., 2012). In the particular case of fragrance allergy, prevalence estimates range from 1.0 to 4.2% (Thyssen et al., 2007b). Occupational contact dermatitis is particularly prevalent in the personal services industry, with an estimated prevalence of 1.2% in the beauty/haircare industry (Warshaw et al., 2012), as well as the petrochemical, rubber, plastic, metal and automotive industries (McDonald et al., 2006). For several decades, animal testing has been used as predictive tool to identify and characterize skin sensitizers, with the guinea pig as the initial animal of choice, which over the last 15 years has increasingly been replaced by the mouse local lymph node assay (LLNA), which has also been validated as a stand-alone (OECD, 1992, 2010). The assay uses fewer animals (16 instead of 20), reduces time and suffering as it stops at the stage of lymph node swelling, and is thus considered a refinement alternative, and provides a sensitization potency estimate, in contrast to the guinea pig assay. However, during the last few decades, there has been a growing concern about using animals for product development and regulatory testing, particularly for cosmetic products and ingredients. The drive for this change resulted first in the implementation in Europe of Cosmetic Directive (76/768/EEC), now Cosmetics Regulation (European Union, 2009), which stipulates a progressive phasing out of animal tests for the purpose of assessing the safety of cosmetics and their ingredients, and ultimately, a complete testing ban, enforced with a marketing ban with deadline in 2013. The European chemicals legislation on the Registration, Evaluation, Authorization and Restriction of Chemicals (REACH) (regulation EC no. 1907/2006 requires that animal testing for hazard assessment should be conducted only as a last resort and authorize the usage of validated in vitro methods (Hartung, 2009b). Noteworthy, LLNA data or equivalent are requested for all REACH substances, i.e. 30 000 – 60 000 existing chemicals (Hartung and Rovida, 2009; Rovida and Hartung, 2009). In 2007 the US National Academy of Sciences released a report called "Toxicity Testing in the 21st Century: A Vision and a Strategy" outlining a strategy for toxicity testing that would be based on human rather than animal biology and suggests moving regulatory toxicology to a more mechanistic approach requiring substantially fewer or no animals (Hartung, 2009a; National Research Council, 2007). Furthermore, as knowledge of the molecular key steps of skin sensitization becomes more detailed, this presents both an opportunity and a challenge to improve the availability of alternative methods (Casati et al., 2005).
Newer alternative methods developed for skin sensitization are based on the specific, key mechanistic steps: the chemical’s ability to penetrate the skin (Basketter et al., 2007), its capacity to bind with proteins present in the skin, as well as the recognition of this protein complex by immune cells (Adler et al., 2011). The direct peptide reactivity assay (DPRA) is the first non-animal test method formally recommended by the European Centre for the Validation of Alternative Methods for skin sensitization (European Commission Joint Research Centre, 2013) and addresses the chemical’s reactivity to proteins by measuring depletion of synthetic peptides containing either cysteine or lysine (Gerberick et al., 2004, 2007). The accuracy of the DPRA for distinguishing sensitizers from non-sensitizers was 82% (sensitivity of 76%, specificity of 92%), excluding metal compounds for which the test is not applicable (Gerberick et al., 2007). More recently, the European Centre for the Validation of Alternative Methods also published a recommendation indicating the usefulness of the KeratinoSens™ assay (European Commission Joint Research Centre, 2014). The assay addresses the activation of the Keap1-Nrf2 ARE pathway in human keratinocytes (HaCaT), which is considered a major regulator of cytoprotective responses to electrophile and oxidative stress by controlling the expression of detoxification, antioxidant and stress response enzymes and proteins (Emter et al., 2010). The balanced accuracy was 77% based on testing of about 145 chemicals with 79% sensitivity and 72% specificity (Natsch et al., 2013). Until recently, none of these assays could be used as a stand-alone method and data should be considered in combination with other information. However, OECD Test guidelines 442C and 442D adopt DPRA and ARE-Nrf2 luciferase test methods (OECD, 2015a,b). A similar assay using the same cell system, including a combination of glutathione depletion and gene expression known to be activated by sensitizing agents (Keap 1/Nrf 2/ARE/EpRE, ARNT/AhR/XRE and Nrf1/MTF/MRE) shown an accuracy of 84%, with a sensitivity of 81% and specificity of 92% based on 102 chemicals (McKim et al., 2010). Other assays have shown promising results to test the activation of dendritic cells (DC), which includes cell line surrogates h-CLAT (THP-1 a human monocytic leukemia cell line) and U937 (a human histiocytic lymphoma cell line) with DC-like characteristics for phenotypic markers of activated DC (e.g. CD86 and CD54) (Ashikaga et al., 2006; Python et al., 2007; Sakaguchi et al., 2006). In addition, some commercially available in silico models such as TIMES (Dimitrov et al., 2005), DEREK (Sanderson & Earnshaw, 1991) and OECD Toolbox (http://www.qsartoolbox.org/) (Diderich, 2010) approaches are developed based on structure–activity relationships (SAR).
These data sources need to be considered in combination with other information, particularly as each assay typically attempts to distinguish only sensitizers from non-sensitizers. The emergence of in vitro tests that each addresses one aspect of skin sensitization necessitates the development of an integrated testing strategy (ITS). An ITS seeks neither to replace any specific animal test nor do away with all animal testing to-gether; instead, the aim is to develop a model that best exploits all data sources (epidemiological studies, animal data, in vitro, in chemico and in silico data such as quantitative SAR [QSAR]) and provides a more formal, systematic and quantitative approach to risk estimation (as distinct from a weight-of-evidence approach). Ideally, such an approach would leverage the extensive data available from non-animal sources to reduce animal testing and would be robust to incomplete data, allowing for the combination of data sets to improve predictability and extend the applicability domain. Additionally, efforts are needed to compile data from known mechanisms for toxicity into testing strategies (Hartung et al., 2013). Pipelines for integrated approaches to testing and assessment (Tollefsen et al., 2014) demonstrate an effort to compile relevant skin sensitization data from adverse outcome pathways (Patlewicz et al., 2014). Therefore, skin sensitization provides a strong domain for the use of modern machine learning techniques.
As skin sensitization is a complex endpoint that needs more than one alternative assay to replace animal tests, the open question remains on how to integrate available information for predicting the skin sensitization hazard, and more specifically how to make the best use of the cumulative information in the most efficient way possible as well as guide for future testing in such a way that the information gain is maximized and accomplished with fewest possible tests (Jaworska et al., 2011). Recently, the use of an ITS of methods of in vitro tests combined with in silico models have been proposed for the replacement of the LLNA (Bauch et al., 2012; Hartung et al., 2013; Hirota et al., 2013; Jaworska et al., 2011, 2013; Maxwell et al., 2014; McKim et al., 2010, 2012; Nukada et al., 2013). ITS provides a more formal, systematic, and quantitative approach to risk estimation (as distinct from a weight-of-evidence approach) than a fixed battery of tests (Hartung et al., 2013, Rovida et al., 2015). As suggested earlier, ITS is "an algorithm to combine (different) test result(s) and, possibly, non-test information (existing data, in silico extrapolations from existing data or modeling) to give a combined test result. They often will have interim decision points at which further building blocks may be considered" (Hartung et al., 2013).
As the volume of data – in silico, in chemico and in vitro – to be considered increases at a rapid rate and is becoming more heterogeneous in nature, there is a keen need for new ways to combine them that offer both a robust and powerful approach to estimate hazard and support a risk decision. Most likely, this has to be done in a probabilistic way, where the different input parameters are combined to generate an overall probability of hazard and risk. Further-more, understanding the effects of test substances at different doses is an essential aspect of safety. We believe that an ITS based on a machine learning approach offers a robust means to combine data for estimating hazard. To this end, we combined a variable selection algorithm to evaluate the information available through both in silico, in chemico and in vitro assays with the hidden Markov model (HMM) that takes advantage of an intrinsic inter-relationship among the LLNA classes, i.e. the connection between LLNA and dose.
Materials and Methods
Data Set
The data set included a total of 145 distinct chemicals, including 145 chemicals with in vitro assays from Jaworska et al. (2013), which included the chemicals of the LLNA data set (Gerberick et al., 2005). In addition, we obtained for a subset of the original chemicals from Mckim et al. (2010) and Natsch et al. (2009) additional in vitro assays. The total number of descriptors: 7, 9 and 10 in vitro/in chemico for data sets 1, 2 and 3, respectively, and 1666 DRAGON descriptors (Tetko et al., 2005). For all distinct 145 chemicals, LLNA classifications were available as reference classification. SMILES strings were obtained via Pubchem (Bolton et al., 2010). DRAGON features were calculated with VCLABS E-DRAGON software (Tetko et al., 2005; Todeschini et al., 2009). An overview can be seen in Table 1.
Table 1.
Overview on data sets 1–3 as described the section data set
| Chemicals | Descriptors | Source | |
|---|---|---|---|
| Data Set 1 | 145 | TIMES, Dragon Descriptors, KeratinoSens KEC 1.5 and KEC 3.0, Cytotoxicic_ IC50, DPRACys, DPRALys, Cfree, CD86 |
Jaworska et al., 2013 |
| Data Set 2 | 83 | Dragon Descriptors, KeratinoSens KEC 1.5 and KEC 3.0, Cytotoxicic_ IC50, DPRACys, DPRALys, CDFree, CD86, ARE EC 1.5, Imax, ARE Cmax |
Jaworska et al., 2013
Natsch et al., 2009 |
| Data Set 3 | 64 | Subset of Data Set 1 with additional Glutathione depletion data available |
Jaworska et al., 2013
McKim et al., 2010 Natsch et al., 2009 |
ARE, antioxidant response element; CD86, concentration at 50% activation of cell surface marker CD86; Cfree, maximum free concentration in mid-epidermis; Cytotoxic_IC50, cytotoxicity value for KeratinoSens; DPRACys, direct peptide reactivity assay for cysteine; DPRALys, direct peptide reactivity assay for lysine; Dragon descriptors, chemical descriptors from VCCLab E-Dragon; KEC, KeratinoSens luciferase nrf2 reporter assay; TIMES, a QSAR for skin sensitization potential.
The initial data set (data set 1, 145 distinct chemicals) was based on the work of Jaworska et al. (2013), combined with Dragon descriptors. Data set 1 was subdivided into smaller data sets based on additional available in vitro results as follows: data set 2 included values for ARE EC 1.5, ARE Cmax, and Imax for 83 chemicals from Natsch et al. (2009), and data set 3 included glutathione depletion from McKim et al. (2010) for a subset of 64 chemicals. Data sets are available in Supplement 1.
Chemical Similarity Generation
A chemical similarity map was generated by the ChemViz plug-in and Cytoscape 2.8.3 (http://www.cgl.ucsf.edu/cytoscape/chemViz/). Tanimoto distances were calculated based on SMILES strings using the Klekota and Roth fingerprint algorithm (Klekota and Roth, 2008), and any chemical with a Tanimoto similarity of greater than 0.70 was considered a link.
Random Forest
We used the scikit-learn Random Forest (Pedregosa et al., 2011) version 0.14 implementation in these analyses. Random forest is an ensemble supervised learning model. Briefly, a random forest model (Breiman, 2003) is trained on a subset of all the data; during training we construct 100 random trees. Each tree is constructed via recursively splitting training data using a random selection of the available features with each permitted up to log_2 of the available features and splitting continued until the split data contains only one chemical (tree split criterion: entropy; min-samplesleaf: 1).
During each chemical prediction, the class is passed down each random tree (using the feature values for that test chemical). Each tree reports the class of the chemical in the leaf node most closely matching that of the test chemical. The random forest then makes a prediction by picking the class most voted for (so-called ensemble method).
Recursive Feature Elimination
Recursive feature elimination involves first evaluating feature importance and then eliminating low importance features. Feature importance was calculated with the scikit-learns implementation of the Breiman random forest variable importance algorithm (Breiman, 2003). This algorithm evaluates a given feature’s importance in a trained model by randomly permuting all available values and recording the subsequent loss in model accuracy and the permutation that results in the greatest loss is given greater accuracy. Variable importance was normalized by dividing each feature importance value by that of the maximally important feature, which was thereby assigned a value of 1.
Dose Transformation
To encode the data using LLNA classes we transformed the LLNA classification into a binary classification for each dose (Table 2).
Table 2.
Sensitization class to dose specific binary class transformation
| Class | Low dose | Medium dose | High dose |
|---|---|---|---|
| Non-sensitizer | Negative | Negative | Negative |
| Moderate sensitizer | Negative | Negative | Positive |
| Strong sensitizer | Negative | Positive | Positive |
| Extreme sensitizer | Positive | Positive | Positive |
This transformation allows us to train a dose-informed random forest that can classify chemicals combined with categories as toxic or non-toxic. Thus for a given chemical our new model can make three predictions (Table 3).
Table 3.
Example chemical 1-bromobutane - LLNA reference classification: non-sensitizer
| LLNA | Low dose | Medium dose | High dose |
|---|---|---|---|
| Transformed LLNA classification |
Negative | Negative | Negative |
| Possible problematic supervised model prediction |
Negative | Positive | Negative |
In this example, a non-sensitizer is transformed into a compound that is non-toxic at all dose levels. This transformation allows us to use the predictions made by the random forest to build a HMM. It should be noted that a supervised model trained with this dose transformation might predict/classify a chemical (Table 3).
This prediction series is concerning because our previous knowledge tells us that if a chemical is toxic at low dose it will remain toxic at higher doses, particularly in case of acute skin sensitization. This knowledge was factored in selecting the prediction sequence of highest probability given this constraint (see also the section on "Cross-validation").
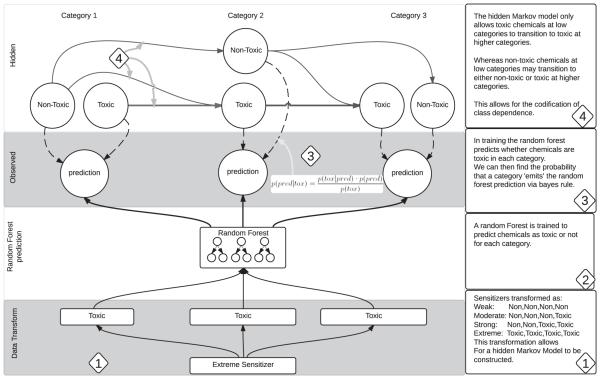
Hidden Markov Model
A HMM allows us to enforce proper prediction series by encoding our knowledge of allowable toxicity transformations. We assume in this model that a chemical that is toxic at low dose will be just as toxic at higher doses, and a chemical that is non-toxic at high dose will be non-toxic at lower doses; the relaxation of this assumption is addressed in the discussion.
The HMM contains several important properties:
Hidden states: These are states that cannot be directly observed. In our case, a given chemical contains six hidden states, one for toxic or non-toxic at each of the three dose categories.
Transition probabilities: Transition probabilities tell us the probability for transitioning from one hidden state to another. Transition probabilities allow us to encode our previous knowledge about toxicity changes. By disallowing a transformation from the hidden state corresponding to low dose and toxic to the hidden state corresponding to moderate dose and non-toxic, we can ensure that no prediction sequences will contain this transition.
Empirically speaking, transition probabilities can be obtained from the data by counting how often a chemical transitions from one hidden state to another. Thus, no special treatment is needed to encode our previous knowledge about chemical transformations as, for instance, the chemical data will contain no instances where a chemical transitions from toxic at low dose to non-toxic at higher dose.
Emission probabilities: In our case, emission probabilities inform about the probability that a given hidden state will emit the prediction given by our dose-informed supervised model. This emission probability can be obtained empirically by counting how often a given prediction aligns with the given hidden state divided by the number of predictions.
The HMM was built using the scikit-learn HMM-module (Pedregosa et al., 2011). Transmission probabilities were built by enumeration from data and emission probabilities by counting classifier outputs matched with actual toxicity class (10 iterations, 0.01 threshold). The trained Markov model chemical predictions were obtained using the Viterbi algorithm (Viterbi, 1967) scikit-learn implementation. For an introduction to HMM please see e.g. Baum and Petrie (1966).
Cross-validation
To ensure a training data set that closely resembled the testing data set, we used 100 iterations of train/test set splits created via scikit-learn’s stratified shuffle split-cross validation approach (Pedregosa et al., 2011). In testing both the dose-informed and dose-naive approaches to skin-sensitization classification we allowed training on 90% of the available data and testing on the remaining unseen 10% of the data separately for each data set, avoiding peeking by ensuring that no model was trained on data it would later be tested on except unavoidably in the case of the comparison of data set 1 with and without the TIMES predictions as TIMES was trained on a number of chemicals included in the data set.
Distance Weighted Error
To evaluate changes in model accuracy relative to distances between predictions and classes we used a distance weighted error approach. This metric allows us to penalize models that make large misclassification errors (e.g. classifying a non-sensitizer as an extreme sensitizer). The equation for the distance weighted error metric E is:
p is a vector of predictions for the given class. p_i is the predicted probability the given chemical was predicted to be class i. d(class, i) is a distance function for which d(class, i)=0 when i = class, d(class, i) = 1 when i is separated by one from class (i.e. none and moderate), or d(class, i)=2 when i is separated by two from class.
Results
Chemical Diversity of Data Set
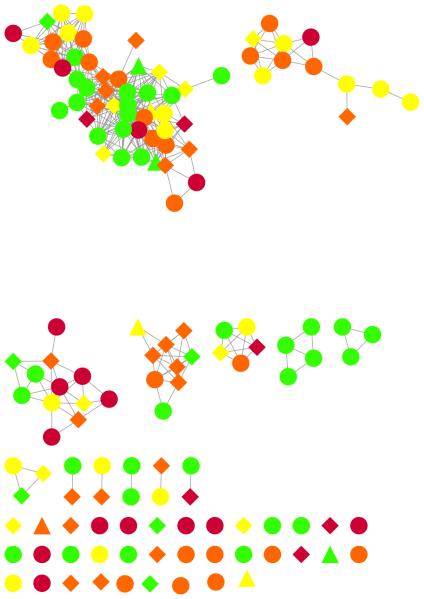
The chemical similarity map (Fig. 1) indicates that many of the chemicals were highly similar compounds, but that clusters of similar chemicals did not necessarily share LLNA status – skin sensitization is therefore difficult to predict using chemicals descriptors alone. Furthermore, the data set included several chemicals that were chemically dissimilar – meaning they had a Tanimoto similarity of less than 0.70 – from all other chemicals in the data set. Interestingly, the largest cluster of similar chemicals contained several instances of chemicals from all four classes, but had only one chemical with a class error greater than 1, indicating that the model performed well in differentiating LLNA class among structurally similar chemicals.
Figure 1.
Chemical similarity map. Chemicals are colored according to local lymph node assay status (red = extreme sensitizer; orange = strong; yellow = moderate; green = weak/none). Distance is proportional to Tanimoto similarity. The difference between predicted and actual class are denoted by shape: no difference between predicted and actual are indicated by circles; one class difference by squares; and two class difference by triangles. Chemical pairs with Tanimoto distance > 0.7 were linked and the resulting network visualized in Cytoscape.
Feature Selection and Variable Importance
As skin sensitization is difficult to predict from chemoinformatics methods/QSARs alone, it is desirable to combine in silico data with in vitro and in chemico assays. Feature selection methods typically improve predictive models by avoiding overfitting due to high dimensionality, shortening computational time and improving model comprehensibility. Recursive feature elimination can be used to trim a data set with a large number of features – in essence, a random forest is trained on the data set and the resulting features are ranked according to the Breiman feature importance test (Breiman, 2003). After ranking the data set, it is modified by the removal of the least valuable feature. The process is then repeated until the number of features in the data set is reduced to the 20 most informative variables that were subsequently selected for building the prediction model.
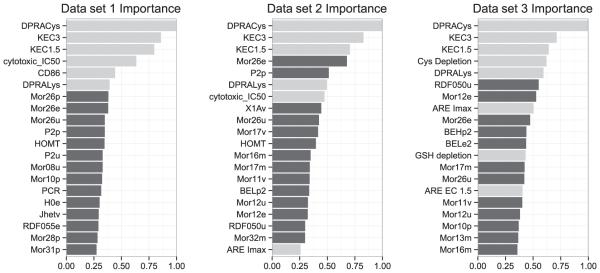
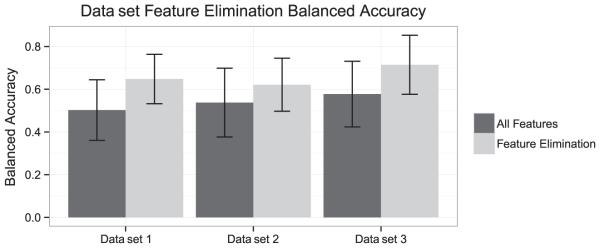
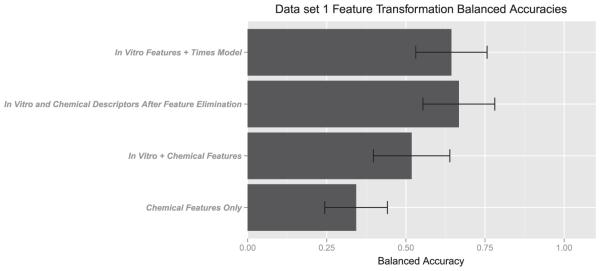
Recursive feature selection indicated that the available in vitro tests were providing substantial information relative to chemical descriptors. In vitro tests consistently ranked within the top 20 descriptors (see Fig. 2). As data accumulate, recursive feature elimination will likely allow for a more informed ranking of in vitro assays and a better choice in terms of what test to perform next when presented with a chemical with limited available in vitro data, or in cases where a QSAR has predicted the potential for skin sensitization either on the basis of skin permeability or electrophilicity (which is predictive of protein binding). Figure 3 shows that the use of feature selection (20 most informative features) consistently improved balanced accuracy. Chemical descriptors alone show very poor overall accuracy for prediction. Combining in vitro assays with the chemical descriptors selected by the recursive feature elimination algorithm performed seemingly as well as the in vitro models with TIMES (shown for data set 1 in Fig. 4). Furthermore, TIMES performance is probably overstated in this case by "peeking," i.e. this data set includes chemicals that were part of the TIMES training set.
Figure 2.
Variable importance: 20 most informative features were selected by the recursive feature elimination algorithm. In vitro/in chemico assays are shown in gray and DRAGON descriptors are shown in black. The unit free x-axis is relative importance normalized to the most important variable. In vitro assays consistently ranked among the top features selected. For more details on the DRAGON descriptors, see Supplement 1.
Figure 3.
Balanced accuracy (four-class prediction model) of data with feature (20 most informative features) and without feature selection consistently improved the balanced accuracy compared to a non-balanced accuracy. Error bars indicate standard deviations of balanced accuracy estimates calculated from cross-validation.
Figure 4.
Balanced accuracies for different feature subsets of data set 1.
While the balanced accuracy of these methods performed at the 60–70% level, they demonstrate the value of feature selection in combining SAR data with in vitro data. Given the utilized four class discretization, 60–70% accuracy is well above chance. The use of cross-validation outside of the feature selection loop helps to reduce the possibility for overfitting, and the avoidance of QSAR algorithms prevents problems with peeking, i.e. the common problem that chemicals were part of the training set to build the (Q)SAR. Our results for feature selection establish that models integrating SAR and in vitro data can perform as well or better than those containing QSAR models as a variable and do not suffer from the same issues with testing.
Hidden Markov Model Generation and Validation
HMMs are a formal method for making probabilistic models of sequential data problems; a Markov system typically has N discrete states and T discrete time steps; in this case, however, instead of time the Markov chain is based on dose. This required transforming our data from pairs of chemicals/LLNA class into chemical–dose pairs. In other words, each chemical was classified as toxic/non-toxic at a low, medium or high dose (see Fig. 5) with thresholds defined by LLNA category, which meant that the dose-informed model in essence predicts a binary question, i.e. whether the chemical was toxic or non-toxic at a given dose increment, instead of trying to predict a four-class problem. In principle, a model that uses this extra information – a "dose-informed" hidden Markov/random forest approach – should perform better than a "dose-naïve" random forest. Figure 5 shows how HMM has been implemented to build the dose-informed HMM/random forest approach.
Figure 5.
Visual description of dose transformation use in a Hidden Markov Model.
Average Class Error
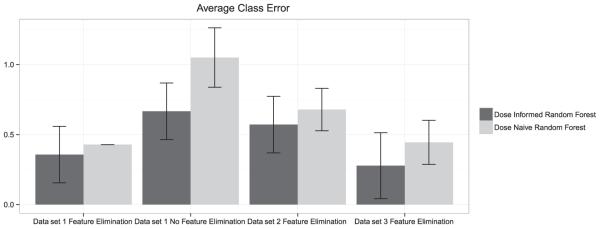
With the addition of the DRAGON descriptors, the total number of features available for data sets was quite large. In this case, a comparison of both dose-informed and dose-naïve models by average class error using all available chemical descriptors from DRAGON and all in vitro assays performed seemingly worse than using only the 20 most informative features selected by recursive feature selection (see Fig. 6). Significantly, average class error for dose-informed random forest/HMMs outperformed the dose-naive random forest models on all data sets. Although from the stand-point of balanced accuracy the improvement is not very impressive, this obscures the actual improvement in misclassifications.
Figure 6.
Average class error and standard deviation from cross validation: For all comparisons the dose-informed model gave smaller average class errors compared to the dose-naive model. Y-axis is a measure of the average distance between predicted class and actual chemical class (non-sensitizer, moderate, strong, extreme). Furthermore, feature selection improved the results – using all chemical descriptors significantly worsened the performance of the random forest.
The best performing dose-informed models (data sets 1 and 2) had no misclassifications greater than 2 classes, i.e. no extreme sensitizers classified as non-sensitizers and no non-sensitizers classified as extreme sensitizers, indicating overall a very small rate of extreme false-negatives (an extreme sensitizer classified as a non-sensitizer) and no extreme false-positives (non-sensitizers classified as extreme sensitizers) in any data set (see Table 2). Interestingly, the extreme sensitizer misclassified (phthalic anhydride [CAS 85-44-9] in data Set 3) hydrolyzes in water at pH 6.8–7.24 with half-lives of 0.5–1 min at 25 °C, forming phthalic acid and is therefore not within the applicability domain of in vitro assays (OECD SIDS Initial Assessment Report, 2005). Phthalic acid (CAS 88-99-3) is classified as a non-sensitizer by a modification of the Maguire method and the LLNA (ECHA database on registered substances, searched on July 25, 2014), which corresponds with the classification as a non-sensitizer by our approach.
The dose-informed HMM had 95.8%, 92.6% and 92.1% accuracy in predicting the LLNA class ±1 one class, versus just 90.4%, 88.6% and 90.6% balanced accuracy for the dose-naive models for data set 1, 2 and 3, respectively. While these accuracies are high, they are not indicative of overall model accuracy. These accuracies demonstrate improvements on the off-by-one accuracy of dose-informed approaches versus dose-naive.
While our average class error differences elude confidence measures, the consistent drop in average class error combined with theoretical considerations demonstrate the HMM dose-informed approach as an improvement over dose-naïve supervised learning approaches (Table 4).
Table 4.
Confusion matrix of predicted chemical’s sensitizing potency versus LLNA reference classification for data sets 1–3, including balanced accuracy and balanced error. The data for this table can be found in Supplement 2
| Dose Informed |
Dose Naïve |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LLNA Reference Classification |
LLNA Reference Classification |
|||||||||||
| Predicted | Non | Moderate | Strong | Extreme | Sum of Predictions |
Non | Moderate | Strong | Extreme | Sum of Predictions |
||
| Data Set I | ||||||||||||
| Non | 35 | 5 | 3 | 0 | 43 | Balanced | 36 | 6 | 5 | 3 | 50 | Balanced |
| Moderate | 6 | 18 | 5 | 2 | 31 | Accuracy | 2 | 18 | 5 | 1 | 26 | Accuracy |
| Strong | 1 | 10 | 25 | 10 | 46 | 0.65 | 4 | 8 | 22 | 9 | 43 | 0.63 |
| Extreme | 0 | 0 | 7 | 18 | 25 | 0 | 1 | 8 | 17 | 26 | ||
| Occurrences | 42 | 33 | 40 | 30 | 145 | Balanced | 42 | 33 | 40 | 30 | 145 | Balanced |
| Accuracy | 0.83 | 0.55 | 0.63 | 0.60 | Error | 0.86 | 0.55 | 0.55 | 0.57 | Error | ||
| Distance | 0.19 | 0.45 | 0.45 | 0.47 | 0.39 | 0.24 | 0.48 | 0.58 | 0.67 | 0.49 | ||
| Weighted | ||||||||||||
| Error | ||||||||||||
| Data Set II | ||||||||||||
| Non | 13 | 3 | 4 | 0 | 20 | Balanced | 14 | 1 | 5 | 2 | 22 | Balanced |
| Moderate | 5 | 10 | 2 | 1 | 18 | Accuracy | 5 | 11 | 2 | 2 | 20 | Accuracy |
| Strong | 2 | 4 | 19 | 6 | 31 | 0.62 | 1 | 5 | 20 | 5 | 31 | 0.63 |
| Extreme | 0 | 0 | 4 | 10 | 14 | 0 | 0 | 2 | 8 | 10 | ||
| Occurrences | 20 | 17 | 29 | 17 | 83 | Balanced | 20 | 17 | 29 | 17 | 83 | Balanced |
| Accuracy | 0.65 | 0.59 | 0.66 | 0.59 | Error | 0.70 | 0.65 | 0.69 | 0.47 | Error | ||
| Distance | 0.45 | 0.41 | 0.48 | 0.47 | 0.45 | 0.35 | 0.35 | 0.48 | 0.88 | 0.52 | ||
| Weighted | ||||||||||||
| Error | ||||||||||||
| Data Set III | ||||||||||||
| Non | 11 | 1 | 2 | 1 | 15 | Balanced | 12 | 1 | 2 | 1 | 16 | Balanced |
| Moderate | 4 | 9 | 3 | 1 | 17 | Accuracy | 1 | 9 | 2 | 1 | 13 | Accuracy |
| Strong | 0 | 2 | 15 | 4 | 21 | 0.65 | 2 | 3 | 16 | 5 | 26 | 0.66 |
| Extreme | 0 | 1 | 3 | 7 | 11 | 0 | 0 | 3 | 6 | 9 | ||
| Occurrences | 15 | 13 | 23 | 13 | 64 | Balanced | 15 | 13 | 23 | 13 | 64 | Balanced |
| Accuracy | 0.73 | 0.69 | 0.65 | 0.54 | Error | 0.80 | 0.69 | 0.70 | 0.46 | Error | ||
| Distance | 0.27 | 0.38 | 0.43 | 0.69 | 0.44 | 0.33 | 0.31 | 0.39 | 0.77 | 0.45 | ||
| Weighted | ||||||||||||
| Error | ||||||||||||
LLNA, local lymph node assay.
Discussion
Although toxicology has a handful of (OECD-accepted) in vitro test methods that are well established (e.g. phototoxicity, skin irritation, assay’s growth in importance and corrosion), such approaches could be improved via combination of modern high-throughput in vitro data sets and chemical feature data. In vitro assay’s growth in importance and availability necessitates objective means of evaluation. Merely using weight-of-evidence for large numbers of validated in vitro assays will result in an accumulation of false-positives and ultimately inaccurate risk assessment and distrust in in vitro methods, as for example increasingly observed for mutagenicity assays.
As toxicological machine learning models grow in popularity, new problems will arise in the use of existing QSAR models. QSAR models are built on chemical training data sets; when these models are incorporated into new models they violate principles of supervised learning as they are testing models on chemicals on which they have been trained. Our results demonstrate that feature elimination algorithms show promise for abrogating the necessity of QSAR models for improving in vitro supervised learning approaches. Using physical chemical properties directly in concert with in vitro assays and applying feature elimination results in stronger accuracies than using preformed QSAR models (at least here in the case of skin sensitization). It will be interesting to see whether this holds for other hazards and data sets.
Feature elimination can be useful for determining the mechanistic pathways behind toxicity. In Fig. 2 we saw DPRAcys, KEC3 and KEC1.5 in the top three features in all three data sets, indicating the KeratinoSens and direct peptide reactivity assay as strong models for skin sensitization. The chemical descriptors most heavily represented in variable importance included many descriptors of electrophilicity, molecular weight and descriptors related to the ability to penetrate skin. These descriptors make sense in the absence of skin permeability information provided from in vitro assays.
While skin sensitization provides a strong domain for the use of modern machine learning techniques, it also presents some challenges: as the models will be applied for regulatory purposes, we need hazard estimation models that are easily understood and visualized. However, the existing data sets have several traits that make impractical the more straightforward approaches, such as decision trees. The curse of dimensionality presents a strong stumbling block to conventional supervised learning approaches. Toxicological data sets frequently present a "high-p/low-n" problem; i.e. the number of samples is typically very low relative to the number of parameters. Here, we show that in vitro tests contribute substantial predictive information compared to the chemical descriptors alone. Furthermore, we show that given the expansion of chemical descriptors, in vitro tests and in chemico tests machine learning techniques probably require pruning the information used in a model; something that will become even more important as ToxCast and other high-throughput data become available. It is always a temptation to assume that using all available data will improve accuracy; however, the reality is that more descriptors may simply be adding more noise and not offering additional information. In toxicology, we have the prominent example of the accumulation of false-positives that have made the battery of tests for mutagenicity cumbersome (Kirkland et al., 2005).
Some probabilistic models already exist for skin sensitization, including the Bayesian ITS (Jaworska et al., 2013). The Bayesian ITS shows a remarkable balanced accuracy (94% on a small external test set). While this and some other approaches show high accuracies on test sets, some fail to use cross-validation. Thus, the possibility remains that accuracy on test sets is not representative of the method. The Bayesian ITS method, while demonstrating strong accuracy and a valuable approach (Bayesian networks), also used the TIMES QSAR, which may cause problems due to peeking (as mentioned above). Our approach improves over existing models by incorporating SAR data without using QSARs and by proposing a method to incorporate the range of dose–response data rather than summary statistics alone. Combining these approaches with new data sets will be interesting for future research.
Furthermore, it has become increasingly evident that characterizing the dose–response relationship in in vitro assays is key to using them effectively in machine learning techniques. Typically, LLNA prediction is a four-class problem (non-sensitizer, moderate, strong and extreme) in line with the Globally Harmonized System of Classification and Labeling of Chemicals. While some approaches try to solve this problem by predicting sensitizer versus non-sensitizer only, this model seeks to exploit the fact that LLNA follows a monotonous dose–response curve; i.e. if a chemical is a sensitizer at a low dose, it will also be a sensitizer at a high dose. By redefining the problem as predicting whether a chemical is a skin sensitizer at a given dose increment, the prediction becomes a binary problem. From a theoretical perspective, it is clear that a HMM will lessen extreme misclassifications; this is borne out in our data sets by the fairly small average class distance between predicted versus actual for the dose-informed HMM approach versus the dose-naïve one. From a practical standpoint, this can give users of the model some confidence that a predicted non-sensitizer is unlikely to be an extreme sensitizer and vice versa. Our dose-informed HMM generally outperformed the dose-naïve four-class random forest prediction models and minimized missclassifications of more than a two-class distance. Furthermore, a dose-informed HMM could potentially be extended when used with attributes that show a dose–response curve as opposed to the single value assays used here (notably, only the classifications resulting from the in vitro models were available, not their concentration–response curves).
This dose-informed approach already shows promising improvements in off-by-one accuracy and average class error. It can be improved by using descriptors with dose–response data and by accounting for dermal penetration data. Dermal penetration has the potential to change the proposed dose-informed model, by adjusting the "effective" dose of a given toxicant to account for penetration features (Basketter et al., 2007). Increased availability of dose–response data from sources such as ToxCast provides the potential to increase observed accuracy differences between dose-naive and dose-informed models. The proposed doseinformed approach could also be improved by accounting for chemicals with non-monotonic LLNA relationships. This correction could be realized by allowing transitions from toxic at low dose to non-toxic at a higher dose.
The proposed HMM transforms dose-naive machine learning models into dose-informed models. This approach appears to reduce the rate of extreme misclassifications and can be applied to all existing models for skin sensitization, thus allowing for an incremental improvement of any dose-naive approach to classifying toxicity.
The available data set has three further weaknesses. First, about 20 substances show in vitro effects close to cytotoxicity and it actually appears that these tend to be the ones wrongly predicted by the random forest approach. Cytotoxicity presents a problem for deriving predictions directly from assays. When in vitro behavior is observed near cytotoxic concentrations the behavior may be a confounder for the in vitro activity. In fact we see in data set 1 and 2 that KeratinoSens cytotoxicity is one of the more highly ranked features (fourth and seventh respectively). Second, the data set included five anhydrates with a hydrolyzing half-life of about a minute in water, i.e. they are clearly outside the applicability domain of in vitro assays and can cause wrong predictions if the hydrolyzed product has a different LLNA classification as discussed for the case of phthalic anhydride. Anhydrates pose a problem for making predictions from in vitro assays and are typically discarded as not being part of the applicability domain of the assay. We could improve the predictive ability by including structure information for the hydrolysis products as well as the original compound. Such pruning, for which criteria need to be defined, might further improve predictions but reduces the power and spread of the data set. Third, data for the different assays were not available as concentration–responses but as resulting classifications, which considerably reduces the information content.
Recently, Urbisch et al. (2015) published a large skin sensitization data set on 213 substances with LLNA and humans data. It would be informative to apply these approaches to that data set, and to data sets incorporating more dose–response specific data.
Taken together, this study shows for the example of skin sensitization, a key health effect for the evaluation of chemicals, that a computational combination of in silico, in chemico and in vitro information approximates with about 90% accuracy the in vivo classification. Whether the determination of (arbitrarily chosen) 20 parameters, among them six to seven in vitro tests, is feasible and justifies the resources might be discussed, though the ToxCast program shows that even hundreds of automated assays can be conducted on a given substance at affordable prices. Compared to earlier approaches predicting skin sensitization potency (Jaworska et al., 2013), we apparently avoided extreme misclassifications by introducing the HMM approach to dose–response, replaced commonly used QSAR by feature elimination and achieved robustness of results for the first time shown by cross-validation. Given the fact that none of these assays was developed to serve as a complementing component of an ITS but as a stand-alone test, there is room for improvement for "lighter" ITS. The proof-of-principle remains that computational approaches can integrate and thereby optimize in vitro and in chemico information for skin sensitization potency at levels of certainty normally accepted in validation studies for alternative methods replacing animal tests.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Joanna Jaworska (P&G, Brussels), Dr. Andreas Natsch from Givaudan (Dübendorf, Switzerland) for the fruitful discussions and sharing unpublished experimental data. This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq (no. 238194/2012-4). CNPq had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Financial support to Alexandra Maertens by an NIEHS Research Supplement to Promote Diversity in Health-Related Research (PA-08-190) and to Thomas Luechtefeld by an NIEHS training grant (T32 ES007141) is greatfully appreciated.
Footnotes
Conflict of interest
The authors did not report any conflict of interest.
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Adler S, Basketter D, Creton S, Pelkonen O, Van Benthem J, Zuang V, Andersen KE, Angers Loustau A, Aptula A, Bal-Price A, Benfenati E, Bernauer U, Bessems J, Bois FY, Boobis A, Brandon E, Bremer S, Broschard T, Casati S, Coecke S, Corvi R, Cronin M, Daston G, Dekant W, Felter S, Grignard E, Gundert-Remy U, Heinonen T, Kimber I, Kleinjans J, Komulainen H, Kreiling R, Kreysa J, Leite SB, Loizou G, Maxwell G, Mazzatorta P, Munn S, Pfuhler S, Phrakonkham P, Piersma A, Poth A, Prieto P, Repetto G, Rogiers V, Schoeters G, Schwarz M, Serafimova R, Tahti H, Testai E, Van Delft J, Van Loveren H, Vinken M, Worth A, Zaldivar JM. Alternative (non-animal) methods for cosmetics testing: current status and future prospects – 2010. Arch. Toxicol. 2011;85:367–485. doi: 10.1007/s00204-011-0693-2. [DOI] [PubMed] [Google Scholar]
- Ashikaga T, Yoshida Y, Hirota M, Yoneyama K, Itagaki H, Sakaguchi H, Miyazawa M, Ito Y, Suzuki H, Toyoda H. Development of an in vitro skin sensitization test using human cell lines: the human Cell Line Activation Test (h-CLAT). I. Optimization of the h-CLAT protocol. Toxicol. In Vitro. 2006;20:767–773. doi: 10.1016/j.tiv.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Basketter D, Pease C, Kasting G, Kimber I, Casati S, Cronin M, Diembeck W, Gerberick F, Hadgraft J, Hartung T, Marty JP, Nikolaidis E, Patlewicz G, Roberts D, Roggen E, Rovida C, Van De Sandt J. Skin sensitisation and epidermal disposition: the relevance of epidermal disposition for sensitisation hazard identification and risk assessment. The report and recommendations of ECVAM workshop 59. Altern. Lab. Anim. 2007;35:137–154. doi: 10.1177/026119290703500124. [DOI] [PubMed] [Google Scholar]
- Bauch C, Kolle SN, Ramirez T, Eltze T, Fabian E, Mehling A, Teubner W, Van Ravenzwaay B, Landsiedel R. Putting the parts together: combining in vitro methods to test for skin sensitizing potentials. Regul. Toxicol. Pharmacol. 2012;63:489–504. doi: 10.1016/j.yrtph.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Baum LE, Petrie T. Statistical inference for probabilistic functions of finite state Markov chains. Ann. Math. Statist. 1966;37:1554–1563. [Google Scholar]
- Bolton EE, Wang YL, Thiessen PA, Bryant SH. PubChem: Integrated Platform of Small Molecules and Biological Activities. Annu. Rep. Comput. Chem. 2010;4:217–241. [Google Scholar]
- Breiman L. Random Forests. Machine Learning. 2003;45:5–32. [Google Scholar]
- Casati S, Aeby P, Basketter DA, Cavani A, Gennari A, Gerberick GF, Griem P, Hartung T, Kimber I, Lepoittevin JP, Meade BJ, Pallardy M, Rougier N, Rousset F, Rubinstenn G, Sallusto F, Verheyen GR, Zuang V. Dendritic cells as a tool for the predictive identification of skin sensitisation hazard. Report and Recommendations of ECVAM Workshop 51. Altern. Lab. Anim. 2005;33:47–62. doi: 10.1177/026119290503300108. [DOI] [PubMed] [Google Scholar]
- Diderich B. Tools for category formation and read-across: overview of the OECD (Q) SAR Application Toolbox. In: Cronin MTD, Madden JC, editors. In Silico Toxicology: Principles and Applications. RSC Publishing; Cambridge: 2010. pp. 385–407. [Google Scholar]
- Dimitrov SD, Low LK, Patlewicz GY, Kern PS, Dimitrova GD, Comber MH, Phillips RD, Niemela J, Bailey PT, Mekenyan OG. Skin sensitization: modeling based on skin metabolism simulation and formation of protein conjugates. Int. J. Toxicol. 2005;24:189–204. doi: 10.1080/10915810591000631. [DOI] [PubMed] [Google Scholar]
- Emter R, Ellis G, Natsch A. Performance of a novel keratinocyte-based reporter cell line to screen skin sensitizers in vitro. Toxicol. Appl. Pharmacol. 2010;245:281–290. doi: 10.1016/j.taap.2010.03.009. [DOI] [PubMed] [Google Scholar]
- European Commission Joint Research Centre . EUR 26383 – EURL ECVAM Recommendation on the Direct Peptide Reactivity Assay (DPRA) for Skin Sensitisation Testing. Publications Office of the European. Luxembourg; Union: 2013. [Google Scholar]
- European Commission Joint Research Centre . EUR 26427 – EURL ECVAM Recommendation on the KeratinoSensTM Assay for Skin Sensitisation Testing. Publications Office of the European. Luxembourg; Union: 2014. [Google Scholar]
- European Union Regulation (EC) No 1223/2009 of the European Parliament and of the Council of 30 November 2009 on Cosmetic Products. Off. J. Eur. Union. 2009;L342:59–209. [Google Scholar]
- Gerberick GF, Ryan CA, Kern PS, Dearman RJ, Kimber I, Patlewicz GY, Basketter DA. A chemical dataset for evaluation of alternative approaches to skin-sensitization testing. Contact Dermatitis. 2004;50:274–288. doi: 10.1111/j.0105-1873.2004.00290.x. [DOI] [PubMed] [Google Scholar]
- Gerberick GF, Ryan CA, Kern PS, Schlatter H, Dearman RJ, Kimber I, Patlewicz GY, Basketter DA. Compilation of historical local lymph node data for evaluation of skin sensitization alternative methods. Dermatitis. 2005;16:157–202. [PubMed] [Google Scholar]
- Gerberick GF, Vassallo JD, Foertsch LM, Price BB, Chaney JG, Lepoittevin JP. Quantification of chemical peptide reactivity for screening contact allergens: a classification tree model approach. Toxicol. Sci. 2007;97:417–427. doi: 10.1093/toxsci/kfm064. [DOI] [PubMed] [Google Scholar]
- Hartung T. A toxicology for the 21st century – mapping the road ahead. Toxicol. Sci. 2009a;109:18–23. doi: 10.1093/toxsci/kfp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T. Toxicology for the twenty-first century. Nature. 2009b;460:208–212. doi: 10.1038/460208a. [DOI] [PubMed] [Google Scholar]
- Hartung T, Luechtefeld T, Maertens A, Kleensang A. Integrated testing strategies for safety assessments. ALTEX. 2013;30:3–18. doi: 10.14573/altex.2013.1.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T, Rovida C. Chemical regulators have overreached. Nature. 2009;460:1080–1081. doi: 10.1038/4601080a. [DOI] [PubMed] [Google Scholar]
- Hirota M, Kouzuki H, Ashikaga T, Sono S, Tsujita K, Sasa H, Aiba S. Artificial neural network analysis of data from multiple in vitro assays for prediction of skin sensitization potency of chemicals. Toxicol. In Vitro. 2013;27:1233–1246. doi: 10.1016/j.tiv.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Jaworska J, Dancik Y, Kern P, Gerberick F, Natsch A. Bayesian integrated testing strategy to assess skin sensitization potency: from theory to practice. J. Appl. Toxicol. 2013;33(11):1353–1364. doi: 10.1002/jat.2869. [DOI] [PubMed] [Google Scholar]
- Jaworska J, Harol A, Kern PS, Gerberick GF. Integrating non-animal test information into an adaptive testing strategy – skin sensitization proof of concept case. ALTEX. 2011;28:211–225. doi: 10.14573/altex.2011.3.211. [DOI] [PubMed] [Google Scholar]
- Kirkland DJ, Henderson L, Marzin D, Muller L, Parry JM, Speit G, Tweats DJ, Williams GM. Testing strategies in mutagenicity and genetic toxicology: an appraisal of the guidelines of the European Scientific Committee for Cosmetics and Non-Food Products for the evaluation of hair dyes. Mutat. Res. 2005;588:88–105. doi: 10.1016/j.mrgentox.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Klekota J, Roth FP. Chemical substructures that enrich for biological activity. Bioinformatics. 2008;24:2518–2525. doi: 10.1093/bioinformatics/btn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell G, Mackay C, Cubberley R, Davies M, Gellatly N, Glavin S, Gouin T, Jacquoilleot S, Moore C, Pendlington R, Saib O, Sheffield D, Stark R, Summerfield V. Applying the skin sensitisation adverse outcome pathway (AOP) to quantitative risk assessment. Toxicol In Vitro. 2014;28:8–12. doi: 10.1016/j.tiv.2013.10.013. [DOI] [PubMed] [Google Scholar]
- Mcdonald JC, Beck MH, Chen Y, Cherry NM. Incidence by occupation and industry of work-related skin diseases in the United Kingdom, 1996–2001. Occup. Med. (Lond.) 2006;56:398–405. doi: 10.1093/occmed/kql039. [DOI] [PubMed] [Google Scholar]
- Mckim JM, Jr, Keller DJ, 3rd, Gorski JR. A new in vitro method for identifying chemical sensitizers combining peptide binding with ARE/EpRE-mediated gene expression in human skin cells. Cutan. Ocul. Toxicol. 2010;29:171–192. doi: 10.3109/15569527.2010.483869. [DOI] [PubMed] [Google Scholar]
- Mckim JM, Jr, Keller DJ, 3rd, Gorski JR. An in vitro method for detecting chemical sensitization using human reconstructed skin models and its applicability to cosmetic, pharmaceutical, and medical device safety testing. Cutan. Ocul. Toxicol. 2012;31:292–305. doi: 10.3109/15569527.2012.667031. [DOI] [PubMed] [Google Scholar]
- National Research Council . Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC; National Academies Press: 2007. [Google Scholar]
- Natsch A, Emter R, Ellis G. Filling the concept with data: integrating data from different in vitro and in silico assays on skin sensitizers to explore the battery approach for animal-free skin sensitization testing. Toxicol. Sci. 2009;107:106–121. doi: 10.1093/toxsci/kfn204. [DOI] [PubMed] [Google Scholar]
- Natsch A, Ryan CA, Foertsch L, Emter R, Jaworska J, Gerberick F, Kern P. A dataset on 145 chemicals tested in alternative assays for skin sensitization undergoing prevalidation. J. Appl. Toxicol. 2013;33(11):1337–1352. doi: 10.1002/jat.2868. [DOI] [PubMed] [Google Scholar]
- Nukada Y, Miyazawa M, Kazutoshi S, Sakaguchi H, Nishiyama N. Data integration of non-animal tests for the development of a test battery to predict the skin sensitizing potential and potency of chemicals. Toxicol. In Vitro. 2013;27:609–618. doi: 10.1016/j.tiv.2012.11.006. [DOI] [PubMed] [Google Scholar]
- OECD . Test No. 406: Skin Sensitisation. Paris, France; OECD Publishing: 1992. [Google Scholar]
- OECD . Test No. 429: Skin Sensitisation. Paris, France; OECD Publishing: 2010. [Google Scholar]
- OECD . Test No. 442C. Skin Sensitisation. Paris, France; OECD Publishing: 2015a. In Chemico. [Google Scholar]
- OECD . Test No. 442D. Skin Sensitisation. Paris, France; OECD Publishing: 2015b. In Vitro. [Google Scholar]
- Patlewicz G, Kuseva C, Kesova A, Popova I, Zhechev T, Pavlov T, Roberts DW, Mekenyan O. Towards AOP application – implementation of an integrated approach to testing and assessment (IATA) into a pipeline tool for skin sensitization. Regul. Toxicol. Pharmacol. 2014;69:529–545. doi: 10.1016/j.yrtph.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine Learning in Python. J. Machine Learning Res. 2011;12:2825–2830. [Google Scholar]
- Peiser M, Tralau T, Heidler J, Api AM, Arts JH, Basketter DA, English J, Diepgen TL, Fuhlbrigge RC, Gaspari AA, Johansen JD, Karlberg AT, Kimber I, Lepoittevin JP, Liebsch M, Maibach HI, Martin SF, Merk HF, Platzek T, Rustemeyer T, Schnuch A, Vandebriel RJ, White IR, Luch A. Allergic contact dermatitis: epidemiology, molecular mechanisms, in vitro methods and regulatory aspects. Current knowledge assembled at an international workshop at BfR, Germany. Cell Mol. Life Sci. 2012;69:763–781. doi: 10.1007/s00018-011-0846-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Python F, Goebel C, Aeby P. Assessment of the U937 cell line for the detection of contact allergens. Toxicol. Appl. Pharmacol. 2007;220:113–124. doi: 10.1016/j.taap.2006.12.026. [DOI] [PubMed] [Google Scholar]
- Rovida C, Alepee N, Api AM, Basketter DA, Bois FY, Caloni F, Corsini E, Daneshian M, Eskes C, Ezendam J, Fuchs H, Hayden P, Hegele-Hartung C, Hoffmann S, Hubesch B, Jacobs MN, Jaworska J, Kleensang A, Kleinstreuer N, Lalko J, Landsiedel R, Lebreux F, Luechtefeld T, Locatelli M, Mehling A, Natsch A, Pitchford JW, Prater D, Prieto P, Schepky A, Schuurmann G, Smirnova L, Toole C, Van Vliet E, Weisensee D, Hartung T. Integrated Testing Strategies (ITS) for safety assessment. ALTEX. 2015;32:25–40. doi: 10.14573/altex.1411011. [DOI] [PubMed] [Google Scholar]
- Rovida C, Hartung T. Re-evaluation of animal numbers and costs for in vivo tests to accomplish REACH legislation requirements for chemicals - a report by the transatlantic think tank for toxicology (T4) ALTEX. 2009;26:187–208. [PubMed] [Google Scholar]
- Sakaguchi H, Ashikaga T, Miyazawa M, Yoshida Y, Ito Y, Yoneyama K, Hirota M, Itagaki H, Toyoda H, Suzuki H. Development of an in vitro skin sensitization test using human cell lines; human Cell Line Activation Test (h-CLAT). II. An inter-laboratory study of the h-CLAT. Toxicol. In Vitro. 2006;20:774–784. doi: 10.1016/j.tiv.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Sanderson DM, Earnshaw CG. Computer prediction of possible toxic action from chemical structure; the DEREK system. Hum. Exp. Toxicol. 1991;10:261–273. doi: 10.1177/096032719101000405. [DOI] [PubMed] [Google Scholar]
- SIDS, OECD . PHTHALIC ANHYDRIDE CAS N: 85-44-9. Paris, France; UNEP Publications: 2005. [Google Scholar]
- Tetko IV, Gasteiger J, Todeschini R, Mauri A, Livingstone D, Ertl P, Palyulin VA, Radchenko EV, Zefirov NS, Makarenko AS, Tanchuk VY, Prokopenko VV. Virtual computational chemistry laboratory – design and description. J. Comput. Aided Mol. Des. 2005;19:453–463. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- Thyssen JP, Johansen JD, Menne T. Contact allergy epidemics and their controls. Contact Dermatitis. 2007a;56:185–195. doi: 10.1111/j.1600-0536.2006.01058.x. [DOI] [PubMed] [Google Scholar]
- Thyssen JP, Linneberg A, Menne T, Johansen JD. The epidemiology of contact allergy in the general population – prevalence and main findings. Contact Dermatitis. 2007b;57:287–299. doi: 10.1111/j.1600-0536.2007.01220.x. [DOI] [PubMed] [Google Scholar]
- Todeschini R, Consonni V, Todeschini R. Molecular Descriptors for Chemoinformatics. Weinheim; Wiley-VCH: 2009. [Google Scholar]
- Tollefsen KE, Scholz S, Cronin MT, Edwards SW, De Knecht J, Crofton K, Garcia-Reyero N, Hartung T, Worth A, Patlewicz G. Applying Adverse Outcome Pathways (AOPs) to support Integrated Approaches to Testing and Assessment (IATA) Regul. Toxicol. Pharmacol. 2014;70:629–640. doi: 10.1016/j.yrtph.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Urbisch D, Mehling A, Guth K, Ramirez T, Honarvar N, Kolle S, Landsiedel R, Jaworska J, Kern PS, Gerberick F, Natsch A, Emter R, Ashikaga T, Miyazawa M, Sakaguchi H. Assessing skin sensitization hazard in mice and men using non-animal test methods. Regul. Toxicol. Pharmacol. 2015;71:337–351. doi: 10.1016/j.yrtph.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Viterbi AJ. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans. Inform. Theor. 1967;13:260–269. [Google Scholar]
- Warshaw EM, Wang MZ, Mathias CG, Maibach HI, Belsito DV, Zug KA, Taylor JS, Zirwas MJ, Fransway AF, Deleo VA, Marks JG, Jr, Pratt MD, Storrs FJ, Rietschel RL, Fowler JF, Jr, Sasseville D. Occupational contact dermatitis in hairdressers/cosmetologists: retrospective analysis of North American contact dermatitis group data, 1994 to 2010. Dermatitis. 2012;23:258–268. doi: 10.1097/DER.0b013e318273a3b8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.