Abstract
By means of two supramolecular systems - peptide amphiphiles engaged in hydrogen-bonded β-sheets, and chromophore amphiphiles driven to assemble by π-orbital overlaps - we show that the minima in the energy landscapes of supramolecular systems are defined by electrostatic repulsion and the ability of the dominant attractive forces to trap molecules in thermodynamically unfavourable configurations. These competing interactions can be selectively switched on and off, with the order of doing so determining the position of the final product in the energy landscape. Within the same energy landscape, the peptide-amphiphile system forms a thermodynamically favoured product characterized by long bundled fibres that promote biological cell adhesion and survival, and a metastable product characterized by short monodisperse fibres that interfere with adhesion and can lead to cell death. Our findings suggest that, in supramolecular systems, function and energy landscape are linked, superseding the more traditional connection between molecular design and function.
The design strategy for supramolecular systems1 has been to use building blocks programmed to assemble into a desired nanoscale structure when thermodynamic equilibrium is reached. Molecular design of the building blocks can potentially create functional systems, and this approach has led to the demonstration of catalytically active2, bioactive3, chemically4, light-5 or pH-6 responsive materials, among many others. For supramolecular polymers7, it has recently been shown that assemblies can remain trapped and the targeted thermodynamic minimum is often not reached. Several examples have been reported in which the building blocks assemble into a number of architectures that depend on the pathway selected for the preparation of the supramolecular system8,9,10,11,12. The dependence of structure on preparative pathway is then encoded in the design of the molecular building blocks. The structure should forecast what might constitute the dominant interactions between molecules, and most importantly predict how these interactions will compete with one another. For instance, the attractive π-orbital overlap interactions that drive conjugated molecules to stack could face competition from repulsive forces, either electrostatic or steric. The competition will define the structural energy landscape of the system and thereby the potential for function at different coordinates. This connection between landscape coordinates and function, as well as the strategy of rationally switching on and off specific dominant interactions to navigate within the landscape has not been well explored in supramolecular systems. In this work we offer a model for this strategy by focusing on peptides that are mainly held together by β-sheet hydrogen bonds but also repel one another as a result of electrostatic forces. We chose this particular system because these two competitive forces are ubiquitous in the folding and self-assembly of proteins and other biological molecules. To generalize our finding, we also studied a supramolecular system formed by an anionic perylene-monoimide amphiphile of potential importance in energy functions13, where self-assembly is dominantly driven by π-orbital overlap that can be outcompeted by repulsion among carboxylate groups. Finally, we study how the position in the energy landscape affects the properties of the peptide-based supramolecular materials.
Peptide amphiphiles (PAs) are molecules in which an aliphatic hydrophobic segment is covalently linked to an amino acid sequence. In a subset of these molecules developed in our laboratory the peptide sequence includes a β-sheet forming domain that leads to self-assembly into supramolecular nanofibres14. These molecules have been of interest because the supramolecular fibres can have high potency to signal cells and create artificial extracellular matrices for regenerative medicine3,15. The resulting structures can be rendered bioactive by the conjugation of specific peptide signals at the terminus of the molecules to promote cell proliferation16, differentiation3, and migration16. Furthermore, the efficacy of these bioactive filaments also depends on their physical properties, such as the internal supramolecular cohesion of fibres17 and their morphology18. In this work, we studied the energy landscapes of self-assembly of a PA with the sequence V3A3K3 conjugated to a 16-carbon alkyl chain at the N-terminus, which is dominantly controlled by β-sheet formation in the V3A3 domain and charge repulsion in the three consecutive lysine residues. We studied the energy landscapes above and below a critical ionic strength (Ic=6 mM) that determines if electrostatic forces outcompete β-sheet formation (Fig. 1).
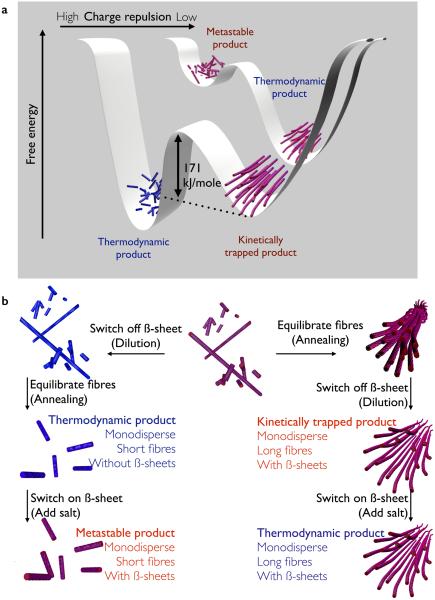
Figure 1. Energy landscapes of PA self-assembly and pathways to access each well.
a, Schematic representation of the free energy landscapes of PA assemblies under conditions with relatively high (front) and low (back) charge repulsion between PA molecules. At low intermolecular repulsion, long fibres with β-sheets are favoured and monodisperse short fibres represent a metastable state. At high repulsion, a kinetically trapped assembly and a thermodynamically favoured product separated by an energy barrier (Eb) of 171 kJ/mole were found. b, Freshly prepared PA solutions at 4.4 mM in water contain polydisperse fibrous assemblies with β-sheet internal structure. By first switching off the β-sheets (dilution) and then equilibrating the assemblies (annealing), the thermodynamically favoured product of monodisperse ~150 nm fibres without β-sheet is obtained. Addition of salt switches back on the β-sheet, giving rise to short metastable monodisperse fibres. In contrast, if the fibres are first equilibrated (annealing) and then diluted kinetically trapped fibres with β-sheets are obtained. Addition of salt to those solutions does not alter the morphology, as the β-sheets remain. Fibres with β-sheets are represented with red stripes; fibres with coil-like internal structure are blue.
Energy landscape below the critical ionic strength
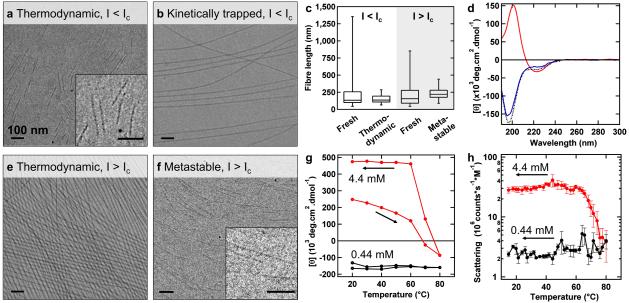
We first investigated the energy landscape of the PA system below an ionic strength of 6 mM by means of circular dichroism (CD) spectroscopy, cryogenic transmission electron microscopy (cryo-TEM) and dynamic light scattering (DLS). Freshly prepared aqueous solutions of PAs at 0.44 mM (I=2.6 mM) contained fibres with polydisperse lengths, 9 nm diameter, and a CD signal characteristic of peptides in random coil configuration (Supplementary Fig. 2). After annealing those solutions (heating to 80°C, gently cooling down), cryo-TEM revealed fibres with relatively monodisperse lengths of about 150 nm (Fig. 2a, c). Throughout the annealing step, the PA assemblies maintained their random coil secondary structure and DLS revealed a constant scattering intensity of 2 Mcps/M (Fig. 2g-h). Altering the cooling rate during annealing did not result in significant morphological differences. All the above observations point to the fact that the short monodisperse fibres with random coil secondary structure are the thermodynamically favoured product at a low ionic strength, and thus correspond to the deepest well in this energy landscape (Fig. 1).
Figure 2. Micrographs and spectroscopic assessment of morphologies of all products.
Cryo-TEM micrographs of (a) thermodynamic product and (b) kinetically trapped product under low ionic strength conditions (I<Ic). c, Fibre-lengths as measured by cryo-TEM. Box-and-whisker plots show mean and 5th-95th percentiles from data of n>180. d, CD-spectra of thermodynamic product at low ionic strength (blue), kinetically trapped product at low ionic strength before (red) and after (black) re-annealing. Cryo-TEM micrographs of (e) thermodynamic product and (f) metastable product under high ionic strength conditions (I>Ic). g, The extremes in CD intensity around 200 nm as a measure of β-sheet content (positive) or random coil content (negative) and (h) molar scattering intensity by DLS during the process of annealing at high and low ionic strengths. All scale bars are 100 nm.
In the low ionic strength energy landscape, a second energy minimum corresponding to a kinetically trapped state could be obtained in which long β-sheet fibres are stable. These long fibres can only be obtained by diluting long fibres that are thermodynamically favoured under high ionic strength conditions (vide infra) to an ionic strength below Ic, effectively transferring between the two energy landscapes investigated (Fig. 1). Cryo-TEM of the diluted PA solutions at 0.44 mM showed long fibres that exhibited a characteristic CD signal for a β-sheet (Fig. 2b, d). Moreover, the solutions were birefringent, and fluorescence microscopy revealed the presence of aligned domains (Supplementary Fig. 3). Cryo-TEM and DLS measurements did not reveal any morphological changes after ageing for up to 7 days. Only re-annealing the long fibres fully transitioned the system into the thermodynamically favoured monodisperse short random coil fibres (Fig. 2d and Supplementary Fig. 4). We further investigated the transition from long to short fibres by DLS in the temperature range of 50 to 70°C. By means of an Arrhenius plot, the energy barrier for this transition was calculated to be 171 kJ/mol (Supplementary Fig. 5). The height of this barrier implies a half-life of months at room temperature, and we can therefore conclude that the long fibres with β-sheets are kinetically trapped in this energy landscape. In order to investigate the mechanism behind this phenomenon, we incubated a sample of the kinetically trapped fibres at 50°C and plunged samples into liquid ethane at −188°C after 4, 15 or 30 minutes, effectively allowing us to take snapshots of the morphological transition by cryo-TEM (Supplementary Fig. 5). We found only long fibres, similar to the kinetically trapped ones after 4 minutes. Interestingly, at 15 minutes we found that a bimodal population of fibres emerges; short and monodisperse fibres next to long fibres. The long fibres could not be distinguished from the kinetically trapped product and the short fibres were identical to the thermodynamically favoured product. As expected, after 30 minutes, the population of supramolecular fibres became enriched in the short monodisperse fibres at the expense of long ones.
Energy landscape above the critical ionic strength
We then studied the energy landscape in an environment with an ionic strength above Ic (Fig. 1). Dissolving PAs at 4.4 mM (I=26.4 mM) resulted in fibres polydisperse in length, with a 9 nm diameter and with β-sheet secondary structure (Supplementary Fig. 2). Heating these PA solutions resulted in a gradual loss of β-sheet secondary structure with a clear recovery upon cooling. While the turbidity of the solutions did not change during the heating process, it abruptly increased from 2 to 30 Mcps/M below 74°C. Cryo-TEM of the annealed solutions revealed fibres with a 9 nm diameter and a length much greater than their unannealed counterparts (Fig. 2e, g-h). In fact, fibre ends were rarely found within the area of individual micrographs, suggesting that fibres are effectively of infinite length. Moreover, the long fibres appeared aligned into micrometre-wide domains with periodic inter-fibre spacing of 34±4 nm. When using fluorescently labelled molecules, these domains could be observed by fluorescence microscopy, and also between cross polarizers (Supplementary Fig. 3). This fibre alignment with periodic spacing has been previously attributed by us to electrostatic repulsion among PA assemblies19, and the birefringent, organized domains indicate liquid crystalline behaviour20. Taken together, the observations showed that the additional thermal energy provided by the annealing process under high ionic strength conditions enables the nucleation and rapid growth of fibres with β-sheet secondary structure, following a cooperative nucleation-elongation model21. Furthermore, we found that thermodynamically favoured long fibres can be obtained from PA solutions at concentrations between 100 μM and 10 mM, provided the total ionic strength is above Ic. In other words, if PAs were dissolved at lower concentrations, but NaCl was added to increase the ionic strength above 6 mM, long fibres became favoured, consistent with the assembling behaviour in the energy landscape under high ionic strength (Fig. 1 and Supplementary Fig. 6).
In the high ionic strength energy landscape, a second thermodynamic state was found containing short monodisperse nanofibres with β-sheets. These short fibres could only be obtained by transferring short fibres without β-sheets to the high ionic strength energy landscape through the addition of NaCl or phosphate buffered saline (PBS). For instance, addition of PBS to an ionic strength of 19 mM to a solution of the previously described short fibres at 0.44 mM resulted in the reformation of β-sheets (Supplementary Fig. 2). The monodispersity of these fibres was unaffected as evidenced by cryo-TEM (Fig. 2c, f). Over the course of days at room temperature, these fibres transformed into long ones and eventually could not be differentiated from those in the thermodynamically favoured product. This finding implies that the short fibres are metastable and the two minima in the energy landscape are separated from one another by a relatively low energy barrier (Fig. 1).
The origin of energy landscapes
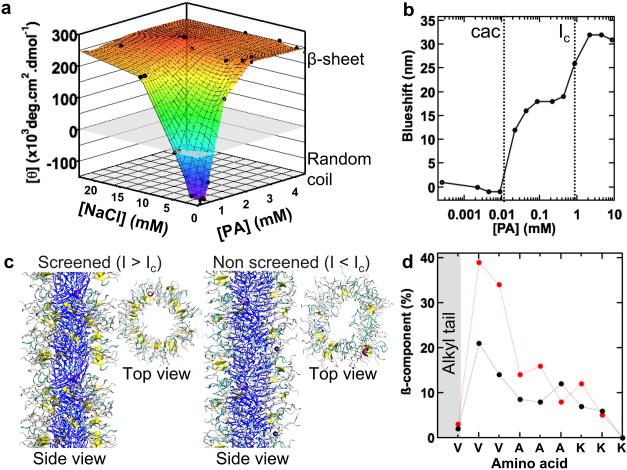
In order to understand the origin of the two energy landscapes and their dependence on ionic strength, we studied the response of the system to dilution. Fresh PA solutions at 4.4 mM showed a CD-signal typical of β-sheets. Upon dilution, the signal for β-sheet secondary structure weakened rapidly and transitioned to a random coil conformation signal. A plot of molar ellipticity at 202 nm as a function of concentration showed that the transition from a positive signal for β-sheets to a negative signal for random coils occurred around a concentration of PA equal to 1 mM, which corresponds to an ionic strength of 6 mM (Fig. 3a). The Nile Red fluorescence assay (NRF), which probes the internal hydrophobicity of assemblies, revealed a decrease in blueshift when diluting beyond 1 mM, suggesting that the transformation to random coil decreases the internal hydrophobicity of the fibres. Moreover, the NRF assay showed a critical aggregation concentration (cac) of 10 μM (Fig. 3b). We also analysed by CD a series of solutions of varying PA concentration kept at a fixed ionic strength by addition of NaCl and did not find a transition to random coil nanostructures. Moreover, when NaCl was added to PA solutions of 0.22 or 0.44 mM, a transition was observed from random coil to β-sheet structures once the total ionic strength reached values above 6 mM (Fig. 3a). Based on these experiments, we hypothesized that at total ionic strengths above 6 mM, the ions of cationic PA molecules are sufficiently screened to allow β-sheet formation. At lower ionic strengths and consequently higher Debye screening lengths, the cationic PAs are not sufficiently screened to favour β-sheet formation. To support this hypothesis, we carried out atomistic simulations22 on PA fibres in the presence or absence of charge screening. The simulations with or without counterions mimic the conditions above and below the critical ionic strength, respectively. From these simulations, we can calculate for each amino acid in the peptide sequence the distribution of their hydrogen bonds in an α-helix, β-sheet or random coil configuration. The simulation that mimics the screened conditions (I>Ic) yields a cylindrical supramolecular fibre with a relatively high population of β-sheet conformation for the β-sheet forming sequence V3A3 of the peptide (Fig. 3c-d). In contrast, the simulation that mimics non-screened conditions (I<Ic) reveals a supramolecular structure with non-circular cross-section and a drastically lower β-sheet population for the same peptide region. We also calculated the relative free energies of charged fibres in high and low ionic strength environments. Interestingly, we found a minimum in fibre length, Lo, at low ionic strengths, which is nearly independent of salt concentration due to the competition of attractive hydrophobic and repulsive electrostatic contributions (see Supplementary Information for details). This finding is consistent with the experimental observation that at low ionic strengths we observe the breaking of effectively infinite fibres into nearly monodisperse ones. The main expression derived applicable at low ionic strengths is given by,
| (1) |
where β’ is an effective gain in energy due to attractive interactions (hydrogen bonds between amino acids and hydrophobic collapse of alkyl tails) per aggregated PA molecule which depends on charge density of a PA taking into account the condensed counterions, 1/b is the PA bare linear charge density, fc is the fraction of condensed counterions which is determined by minimizing the total free energy of the system and lB is the Bjerrum length (the separation at which the electrostatic interaction between two elementary charges is comparable in magnitude to thermal energy). Interestingly, at a critical salt concentration, the minimum at a finite fibre length disappears and fibres transition sharply to acquire infinite length due to complete screening of electrostatics by counterions. In this regime, β-sheet formation is the dominant interaction, explaining the presence of long supramolecular fibres at the deepest well of the energy landscape at high salt concentration. Our calculation is also consistent with the presence of fairly monodisperse fibres in the deepest well of the energy landscape at low salt concentration when electrostatic repulsion competes with attractive forces (fibres of Lo dimension in the context of the calculation).
Figure 3. Assessment of morphological transitions of PA assemblies as a function of ionic strength.
a, CD signal at 202 nm spectra plotted as a function of PA and salt concentrations. Negative CD intensities denote a random coil structure; positive values correspond to a β-sheet signal. b, The blueshift of Nile Red fluorescence as an indicator of internal hydrophobicity of the assemblies as a function of PA concentration. c, Atomistic modelling at a relatively high and low ionic strength showing difference in β-sheet hydrogen bonding between the two conditions. Dark blue: alkyl tails, yellow: β-sheet hydrogen bonding, cyan: β-turn, grey: random coil. In the top views the alkyl tail is left out for clarity. d, Intensities and distribution of β-sheet hydrogen bonding within the peptide region of PA assemblies calculated from atomistic simulations that mimic conditions below (black) and above (red) Ic.
Navigating through landscapes by switching interactions on and off
We have identified three distinct assemblies of the supramolecular system, each associated with a well in two different energy landscapes: monodisperse short fibres with or without internal β-sheets, and periodic bundles of infinitely long fibres with β-sheets (Fig. 1). In analogy to the folding of proteins, the pathway selected to form the system determines the position in the energy landscape, and thus the final supramolecular structure. The first of the two crucial steps are changes of ionic strength in the aqueous environment to control repulsive interactions among charged PA molecules and thus either switch “off” or “on” formation of hydrogen bonded β-sheets. The second step is the use of thermal energy that effectively equilibrates the assemblies into the favoured size (the length of the one-dimensional structure in this case). One surprising finding was the ability of β-sheets in fibres of effectively infinite length to kinetically lock molecules in configurations that are thermodynamically unfavourable. As a result, changing the order of these two crucial steps in the preparation of supramolecular systems, dilution and annealing, drastically affects structural outcomes. To investigate the generality of the concept, we set out to explore whether the order of these two steps also influenced the position in the landscape of another supramolecular system completely different chemically from PAs. We chose a chromophore amphiphile from our library of self-assembling molecules (CA, see Supplementary Fig. 7) that forms a supramolecular system of particular interest as an effective light-harvesting structure for the production of solar fuels13.
Similar to PAs, CAs self-assemble as a result of the balance between a strong attractive interaction (π-π orbital stacking as opposed to hydrogen bonds in the case of PAs) in competition with repulsive electrostatic forces. In water with ionic strengths above 30 mM (I>Ic), CAs assemble into supramolecular thin ribbons that showed a UV/Vis absorption spectrum corresponding to highly ordered crystalline arrangements of molecules13 (Supplementary Fig. 7). Annealing these solutions resulted in ribbons extending from nanometres to microns in width and tens of microns in length, which is analogous to the formation of nearly infinite supramolecular assemblies in PAs when electrostatic repulsions are switched off. These wide crystalline ribbons kinetically lock the CAs so that upon dilution below Ic the same crystalline structures were maintained as observed by cryo-TEM and UV/Vis. This is also analogous to PA assemblies, which become kinetically trapped even under conditions where repulsive electrostatic forces are switched on again. Moreover, re-annealing the kinetically trapped crystalline wide ribbons gave rise to the thermodynamically favoured thin, non-crystalline ribbons. At the same time, when non-annealed crystalline thin ribbons were first diluted below Ic to turn on electrostatic forces, their UV/Vis absorption spectra revealed an immediate loss of crystallinity driven by these repulsive interactions. Annealing these assemblies did not change the supramolecular system, which remains in a non-crystalline, thin ribbon architecture, thus indicating it is in its thermodynamically favoured state (Supplementary Fig. 7). These results on chromophore amphiphiles, with covalent structures that differ drastically from peptides and involving different attractive forces, demonstrate that the presence of the competing interactions enable us to generalize the nature of their supramolecular energy landscapes. We hypothesize that this competition with repulsive electrostatic forces may involve attractive forces such as hydrogen bonding, π-orbital overlaps, and also other interactions. Having the ability to enhance the electrostatic repulsion, and consequently switch off a dominant attractive force, will transition the supramolecular system into a new energy landscape and generate kinetically trapped states provided the assemblies are of large enough dimensions. An important aspect of these findings is that functions of supramolecular systems are likely to be dictated by positions in their various energy landscapes, which effectively allows one to study the influence of the nanoscale structure on properties of materials without altering the molecular design. In this work, we have therefore explored the functional consequences of the peptide supramolecular systems positioned in different coordinates of their landscapes.
Materials properties vs energy landscape position
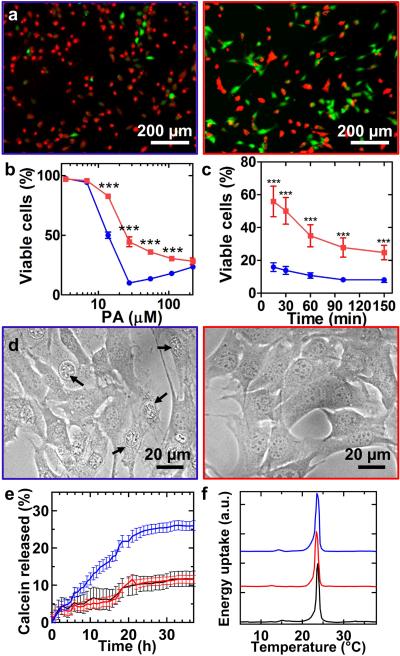
Given the great interest on bioactive functions of peptide supramolecular systems23, we proceeded to examine their ability to interact with cells depending on their position in a given energy landscape. The monodisperse short fibres and long fibres both with β-sheets at 0.44 mM were prepared as shown in Fig. 1. On the time scale of the assays in this study, the fibres remained unchanged, therefore allowing us to compare bioactivities between nanofibres of short or infinite length. Since cationic assemblies are known to be cytotoxic, the first material property we investigated was the viability of cells after exposure to the solutions containing short or long PA fibres. After three hours and at concentrations between 10 and 110 μM, the solutions containing short fibres led to a significantly higher cell death compared to those containing long fibres (Fig. 4a-b). The cell death by short fibres occurred rapidly since the amount of viable cells decreased to 16% within 15 minutes. Moreover, live-cell imaging also revealed that the majority of cells treated with short fibres, even though appeared to have a spread morphology similar to that of living cells, were not moving and exhibited a bright nucleus by that time point (Fig. 4c-d). These observations suggest that cell death is likely mediated though PA interactions with cell membrane. In order to probe the PA-phospholipid membrane interactions, we prepared cell-sized liposomes of egg phosphatidylcholine immobilized on a surface and subsequently exposed them to either short or long fibres. Time-lapse microscopy showed that over the course of an hour very few liposomes disappeared when treated with long fibres. In contrast, roughly 10% of the liposomes treated with short fibres had disappeared over the same timeframe (Supplementary Fig. 8). A closer examination showed that liposomes tended to deflate over the course of minutes when their membrane was compromised by the short fibres. To quantify membrane leakage, we monitored the fluorescence recovery of calcein loaded in liposomes at self-quenching concentrations24. In the presence of long fibres, the calcein release profile was similar to untreated liposomes and roughly 10% leaked out over the course of 36 hours. In contrast, short fibres showed a faster calcein release with a cumulative amount of 25% over the same time (Fig. 4e). Next, we examined whether the disruption was caused by incorporation of PA monomers into their lipid membrane. Differential scanning calorimetry (DSC) can monitor phase changes of lipids and has been used previously to evaluate the ability of amphiphiles to co-assemble with liposomes17. The DSC thermograms for 100 or 400 nm DMPC liposomes in the presence of short or long fibres showed no changes compared to an untreated control, indicating that no significant amount of PA monomers had incorporated into the membrane (Fig. 4f). Based on the above assays, we hypothesized that the cell death occurred through disruption of cell membrane by intact fibres, as opposed to single PA monomers. Finally, we studied by cryo-TEM egg-PC liposomes of 100 nm in diameter subjected to PA fibres for 15 minutes. The micrographs showed that some of the liposomes exposed to short fibres had discontinuous membranes, pronounced membrane distortions or contained smaller (roughly 20 nm) liposomes budding out from their bilayers (Supplementary Fig. 9). As further evidence of this last observation, we also found many small (20-30 nm) liposomes separated from the original 100 nm liposomes. Although such small liposomes were also found inside larger ones in control samples, free small liposomes were rarely found in the presence of long fibres or untreated controls. Interestingly, the defective liposomes described above were effectively not observed in the presence of long fibres.
Figure 4. Implications of the thermodynamic state of PA assemblies on their cytotoxicity.
Images or graphs in blue represent short fibre conditions and those in red represent long fibre conditions. a, Live (green) and dead (red) staining of C2C12 pre-myoblasts 3 h after treated with media containing short or long cationic PA assemblies at 27.5 μM. b, Quantification of viable cells after 3 h. c, Quantification of viable cells over time at 110 μM. Error bars depict ± standard error of the mean for n ≥ 3, and a two-way ANOVA with a Bonferroni post-test is used: *** p<0.001, compared among assemblies at each concentration or time. d, Phase micrographs showing morphology of cells 15 min after exposure to media containing short or long PA fibres at 110 μM. Arrows indicate dead cells. e, Release profiles of calcein from liposomes after exposure to media alone (black) or media containing short or long fibres. f, Representative DSC thermograms for DMPC liposomes in media alone (black) or media containing short or long fibres.
In the context of our mechanistic experiments for rapid cell death by short fibres, a similar observation was reported for short fibres of amyloidogenic peptides25. A subsequent study using liposomes attributed this phenomenon to the ability of fibre endcaps to damage lipid bilayers26. In our opinion this is a reasonable proposal, which by default may explain why cells are more inclined to survive in the presence of long fibres and liposomes appear to coexist undamaged in their presence. Additional mechanisms which may operate when cells (or liposomes) are surrounded by long versus short fibres could be associated with differences in their relative ability to remove mass from membranes following electrostatic attachment. It is interesting to consider that our results indicate clearly that synthetic supramolecular systems designed to interact with cells can have drastically different biological consequences determined by their positions within an energy landscape.
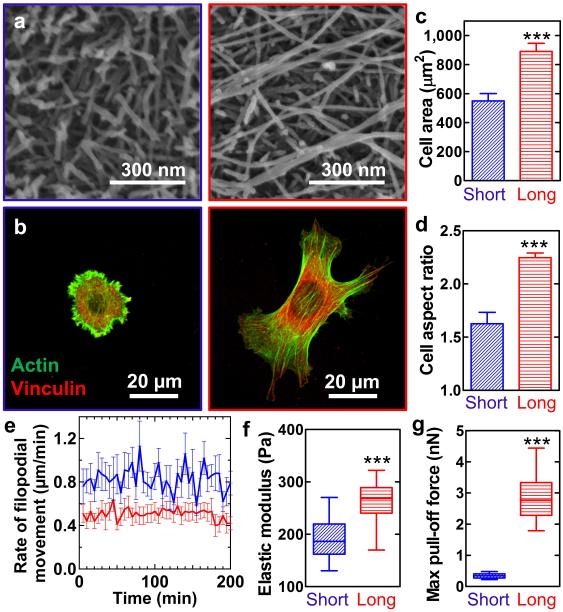
As a second function dependent on position in the energy landscape, we studied the efficacy of PA fibres at forming scaffolds that promote biological cell adhesion. For this purpose, PAs were functionalized at the peptide terminus with the fibronectin-derived RGDS sequence27 via a triglycine spacer (C16-V3A3K3G3RGDS, RGDS PA). The short and long PA fibres were prepared following Fig. 1, and subsequently immobilized on an alginate-coated glass coverslip through electrostatic attraction since alginate is negatively charged. Scanning electron microscopy (SEM) confirmed a homogenous coverage of short fibres or long fibres (Fig. 5a and Supplementary Fig. 10). Binding of cellular transmembrane integrin receptors to RGDS displayed on a scaffold is known to trigger the formation of focal adhesion complexes and improves cell adhesion, survival, and proliferation. Using C2C12 myoblast progenitor cells, we observed attachment and survival of cells on both short and long fibre substrates. Interestingly, spreading and proliferation of cells were significantly greater on the surfaces prepared with long fibres compared to short fibres (Fig. 5b-c and Supplementary Fig. 10). Moreover, cells on the long fibre substrate showed more apparent clustering of the focal adhesion protein vinculin, more evident formation of actin stress fibres, as well as a higher projected surface area28 compared to cells on the short fibres. The most striking difference was observed in the cells’ ability to acquire an elongated morphology. C2C12 cells on short fibres remained relatively round with an aspect ratio of 1.5, where the aspect ratio is defined as the ratio of the major to the minor axis of a fitted ellipse and 1 corresponds to a perfect circle. In contrast, on the long fibres substrate, the aspect ratio of cells was drastically higher at 2.2 (Fig. 5d). Such cell elongation has been linked to the stability of focal adhesion sites, following a mechanism where cell polarization can only occur upon forming static adhesion sites on substrates29. In order to investigate the stability of cell adhesions on PA substrates, we measured the focal adhesion dynamics using live cell imaging and tracking software. Filopodia on short fibres contracted or expanded with an average rate of 0.5 μm/min, whereas filopodia on long fibres moved with an average rate of 0.8 μm/min, which indicates more stable focal adhesion sites (Fig. 5e and Supplementary Fig. 10).
Figure 5. Implications of the thermodynamic state of PA assemblies on their bioactivity as a scaffold.
Images or graphs in blue represent short fibre conditions and those in red represent long fibre conditions. a, SEM micrographs of surfaces coated with short or long fibres assembled using RGDS PA. b, Immunofluorescent staining revealing morphology of C2C12 pre-myoblasts cultured on the RGDS PA coated substrates. Analysis of cell morphology by (c) projected surface area and (d) aspect ratio. Graphs plot mean ± standard error of the mean of n≥4 from two individual experiments with a total of more than 200 cells per substrate. e, Filopodial movement rate against time. Graphs plot mean ± standard error of the mean of n≥4. f, Elastic modulus and (g) adhesion force of the substrates obtained by AFM force spectroscopy. Adhesion force is defined by a maximum pull-off force measured from an unloading curve. Box-and-whisker plots show mean and 5th-95th percentiles from data of n>150 from two separated experiments. All statistical analysis was performed using unpaired two-tailed Student's t-test; *** p<0.001).
All the above observations point to an established cellular mechanism of mechanosensing where cells probe the surface rigidity by extending filopodia. Materials that cannot elastically sustain the stresses exerted by cells inhibit maturation of focal adhesion, resulting in faster filopodia retraction. On the other hand, materials that do sustain such stresses enable focal adhesion maturation, thereby promoting stable focal adhesion sites and cell elongation29. To identify differences in mechanical properties between short and long fibres substrates, we probed them with atomic force microscopy (AFM). The AFM indentation measurements showed that both materials were relatively soft with a Young's modulus in the range of 0.1-0.3 kPa (Fig. 5f and Supplementary Fig. 10). We also measured the adhesive forces between a negatively charged AFM probe that mimics the expected net charge on the cell membrane and the positively charged PA substrates. Interestingly, we found an average adhesion force of 0.3 nN for short fibres and a ten-fold greater force of 3.0 nN for the long ones (Fig. 5g). Based on these findings we hypothesize that the fibres of effectively infinite length are able to sustain the forces exerted by the cells to a greater extent than the short fibres and thereby allow cells to form more static focal contacts and consequently spread and proliferate more.
Outlook
Our work has shown that self-assembling molecules that form supramolecular systems access different thermodynamic wells in their energy landscapes depending on their environment and thermal history. The notion of an energy landscape with multiple energy minima, useful in the context of protein folding, is not commonly contemplated for self-assembling supramolecular systems, especially in its opportunities for variable structures that have different functional values. In this context, we have suggested an approach to navigate the landscape experimentally through an initial judgement of dominant forces involved in the formation of the supramolecular assembly, which is then explored by the possibility of selectively switching “on” or “off” these forces. Most importantly we have also shown that assemblies at different positions in their energy landscape can have drastically different functional attributes. To further develop our ability to design supramolecular systems formed through self-assembly and unravel the connections between their structures and their potential functions, will require great advances in computation with atomistic detail. On the experimental side, it will require exploring such systems efficiently in a rich variety of environments and thermal trajectories.
Methods
Materials
All reagents were purchased from Sigma-Aldrich and used without any further purification unless otherwise indicated. C2C12 mouse premyoblasts and Dulbecco's modified Eagle's medium (DMEM) were obtained from ATCC. Foetal bovine serum (FBS), penicillin/streptomycin (P/S), phosphate buffered saline (PBS), Modified Eagle's Medium (MEM), Hank's Balanced Salt Solution (HBSS) were ordered from Gibco® (Life Technologies). Normal goat serum, modified nucleotide 5-ethynyl-2′-deoxyuridine (EdU), 4',6-diamidino-2-phenylindole HCl (DAPI) and all Alexa Fluor® conjugated products for immunofluorescence staining were ordered from Life Technologies. Sodium alginate was purchased from FMC Biopolymer.
Peptide synthesis and purification
All peptides were synthesized according to previously reported protocols28. Their purity was determined by analytical HPLC as well as electrospray ionization mass spectrometry in positive mode (Supplementary Fig. 1).
General sample preparation
All samples were prepared by dissolving PAs in Milli-Q water and sonicating the solution for 20 min to induce dissolution. All solutions were in the pH range between 6.2 and 6.4 which remained unchanged upon dilution or annealing. Annealing was performed in a thermal cycler (Mastercycler® pro, Eppendorf) set to incubate at 80°C for 30 min before cooling the samples to 20°C over the course of 1 h (1°C/min). This cooling rate was maintained as much as possible for experiments where cooling was done in situ (circular dichroism, fluorescence spectroscopy, and dynamic light scattering).
Circular Dichroism
The secondary structure of the PAs was probed using circular dichroism (CD). Samples loaded in a quartz cuvette were measured as soon as possible after preparation using a JASCO J-715 CD spectrophotometer equipped with a Peltier temperature controller. Spectra were collected at 20°C over a wavelength range of 190 - 300 nm with a step size of 1 nm. A variable-temperature CD experiment was done between 20 and 80°C with the rate of heating or cooling at 1°C/min and a spectrum collected every 10°C.
Nile Red Assay
The self-assembly of the PAs was assessed by incorporation of the hydrophobic solvatochromic probe Nile Red (9-diethylamino-5-benzo[α]phenoxazinone) according to literature procedure28,30.
Cryogenic-Transmission Electron Microscopy
Cryogenic transmission electron microscopy and sample preparation was performed following previously reported methods17. The length of fibres was measured from cryo-TEM micrographs using ImageJ software (NIH). At least six areas each with >180 fibres were analysed.
Dynamic Light Scattering
The scattering rates of the samples were determined by dynamic light scattering measurement on a Malvern Zetasizer Nano ZSP following a previously reported method31. As the objects in solutions are anisotropic and the models used by Malvern software are fitting for spherical objects, we chose not to use the diameters given by the software, but only use the scattering rates as a measure for assembly size. This rate was normalized by the concentration of the solution to yield the molar scattering rate (in Mcounts / s / M).
Arrhenius plot using DLS data
The DLS data was used to calculate a rate constant for the transformation from long to short fibres following ref 32.
Molecular Dynamics Simulations
MD simulations were carried out similarly to experiments in ref 17 and 22. See the Supplementary Information for more details.
Free energy calculation
See the Supplementary Information for details.
UV/Vis
UV/Vis spectra were obtained following a procedure previously published13.
Cell culture
C2C12 mouse premyoblasts were maintained in DMEM. The growth media was supplemented with 10% FBS and 1% P/S. Cells were grown in 75-mm2 flasks at 37°C, 5% CO2 and passaged every three days.
Cytotoxicity assays
The in vitro cytotoxicity of PAs was measured using the LIVE/DEAD® Viability/Cytotoxicity Assay Kit and the CyQUANT® Direct Cell Proliferation Assay Kit (Molecular Probes™) following the vendor's protocol. C2C12 cells in the growth media were seeded into 96-well cell-culture plate at 7-10 × 103 per well. After 24 h incubation, cell media were replaced with a fresh media containing PAs. PAs solutions were prepared following the general sample preparation to a final concentration of 440 μM in water. The solutions of PAs were further diluted to the desired concentrations in MEM media. Cells were either imaged immediately by time-lapse microscopy or incubated for 2-3 h prior to live-dead quantifications. Fluorescent imaging and colorimetric measurements were performed on a cell imaging multi-mode microplate reader (Cytation™ 3, BioTek). Cell viability was measured as a ratio of calcein positive cells over total number of cells. In the CyQUANT® assay, fluorescent intensities which linearly correlate with amount of viable cells were normalized by the intensity of a control without any PA treatment. All assays were done in at least triplicate.
Liposome preparation and their interactions with PA fibres
Liposomes were prepared and the interactions between PA fibres and lipid membrane were probed by calcein leakage experiment and differential scanning calorimetry (using a MicroCal VP-DSC) following a previously published protocol by our group17.
Preparation of PA-coated substrates
PA substrates were prepared using a layer-by-layer coating method following a published protocol from our laboratory34.
Scanning Electron Microscopy
Critical point dried samples were prepared following a published protocol34 and imaged using a Hitachi SU8030 FE-SEM secondary electron detector with an accelerating voltage of 2 keV.
Atomic Force Microscopy
AFM measurements were carried out similarly to experiments in ref 33. See the supplementary information for more details.
Cell adhesion experiments
Cells were seeded at a low density of 26 cells/mm2 to minimize cell-cell contact and either immediately used for live-cell imaging or incubated for 5 h in a growth medium prior to fixation and staining.
Cell proliferation experiments
The proliferation of cells was identified using incorporation of EdU and detection by click chemistry based fluorescence conjugation. Cells were seeded at 130 cells/mm2 in a growth medium and incubated for 5-6 h prior to addition of EdU at a final concentration 4 μM. Cells were further incubated for 16 h after which cells were fixed and stained.
Cell fixing and staining
Cells were fixed using a 4% paraformaldehyde. For immunostaining, fixed samples were stained for vinculin, actin, and nuclei following a published protocol34. For EdU staining, fixed samples were first permeabilised with 0.4% triton X-100 (10 min) and then EdU was detected using a detection solution consisting of 2 M Tris (pH 8.5), 50 mM CuSO4, 0.5 M ascorbic acid, and 1 μg/mL Alexa Fluor® azide. The samples were also stained for nuclei using DAPI for quantification of total cell counts.
Cell imaging and analysis
Live cell imaging was performed using BioStation IM-Q (Nikon). Filopodial dynamics were tracked using an ImageJ plugin MTrackJ, only for cells that did not divide during the time35. Fluorescence images were obtained using an inverted confocal laser scanning microscope (Nikon A1R) or TissueGnostics cell imaging and analysis system mounted to an upright microscope (Zeiss). Quantification of the cell morphology and proliferation was done on phalloidin and EdU stained fluorescent images which are analysed using routines incorporating a build-in ‘particle analysis’ function in ImageJ software28. Quantification was performed on images obtained from two independent experiments with at least 200 randomly selected cells per substrate.
Statistical Analysis
Cell cytotoxicity data was analysed using a two-way ANOVA with a Bonferroni post-test. Other comparisons were analysed using unpaired two-tailed Student's t-test. Graphs plot mean ± standard error of the mean, unless noted otherwise. ** p < 0.01; *** p < 0.001.
Supplementary Material
Acknowledgements
Synthesis of PAs, their morphological assessments and MD simulations were supported by the Center for Bio-Inspired Energy Sciences (CBES), an Energy Frontiers Research Center (EFRC) funded by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under award number DE-SC0000989. The biological studies were supported by National Institutes of Health NIDCR grant (R01DE015920). J.B, F.T. and J.L. are grateful for support by a Rubicon Fellowship of the Netherlands Organisation for Scientific Research (NWO), the Royal Thai Government scholarship and the Northwestern University Bioscientist program, respectively. We acknowledge the following core facilities at Northwestern University: the Peptide Synthesis Core at the Simpson Querrey Institute for BioNanotechnology, the Biological Imaging Facility (supported by the Northwestern University Office for Research), the Center for Advanced Microscopy (NCI CCSG P30 CA060553), Keck Biophysics Facility, the EPIC, SPID facility (NUANCE Center- NSF DMR-1121262 and NSF EEC-0647560). The authors thank Mark Seniw for help with graphics.
Footnotes
Author contributions F.T. and J.B. designed and performed experiments, analysed data, and wrote the manuscript. X.W., R.K., G.S., E.X.Z., R.Z., C.J.N, J.H.O., J.L. and L.C.P. performed experiments, analysed data and provided discussion. T.Y., G.C.S. and M.O.d.l.C. developed and performed theoretical calculations and provided discussion. S.I.S. wrote the manuscript and supervised the research.
Additional information Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at www.nature.com/reprints.
Competing financial interests The authors declare no competing financial interests.
References
- 1.Stupp SI, et al. Supramolecular materials: Self-organized nanostructures. Science. 1997;276:384–389. doi: 10.1126/science.276.5311.384. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann PA, Luisi PL, Lang J. Autocatalytic self-replicating micelles as models for prebiotic structures. Nature. 1992;357:57–59. [Google Scholar]
- 3.Silva GA, et al. Selective differentiation of neural progenitor cells by high-epitope density nanofibres. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 4.Boekhoven J, Hendriksen WE, Koper GJ, Eelkema R, van Esch JH. Transient assembly of active materials fueled by a chemical reaction. Science. 2015;349:1075–1079. doi: 10.1126/science.aac6103. [DOI] [PubMed] [Google Scholar]
- 5.Aggeli A, et al. Responsive gels formed by the spontaneous self-assembly of peptides into polymeric β-sheet tapes. Nature. 1997;386:259–262. doi: 10.1038/386259a0. [DOI] [PubMed] [Google Scholar]
- 6.de Jong JJ, Lucas LN, Kellogg RM, van Esch JH, Feringa BL. Reversible optical transcription of supramolecular chirality into molecular chirality. Science. 2004;304:278–281. doi: 10.1126/science.1095353. [DOI] [PubMed] [Google Scholar]
- 7.Aida T, Meijer EW, Stupp SI. Functional supramolecular polymers. Science. 2012;335:813–817. doi: 10.1126/science.1205962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boekhoven J, et al. Catalytic control over supramolecular gel formation. Nature Chem. 2013;5:433–437. doi: 10.1038/nchem.1617. [DOI] [PubMed] [Google Scholar]
- 9.Hirst AR, et al. Biocatalytic induction of supramolecular order. Nature Chem. 2010;2:1089–1094. doi: 10.1038/nchem.861. [DOI] [PubMed] [Google Scholar]
- 10.Korevaar PA, et al. Pathway complexity in supramolecular polymerization. Nature. 2012;481:492–496. doi: 10.1038/nature10720. [DOI] [PubMed] [Google Scholar]
- 11.Korevaar PA, Newcomb CJ, Meijer EW, Stupp SI. Pathway Selection in Peptide Amphiphile Assembly. J. Am. Chem. Soc. 2014;136:8540–8543. doi: 10.1021/ja503882s. [DOI] [PubMed] [Google Scholar]
- 12.Gröschel AH, et al. Precise hierarchical self-assembly of multicompartment micelles. Nature Commun. 2012;3:710. doi: 10.1038/ncomms1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weingarten AS, et al. Self-assembling hydrogel scaffolds for photocatalytic hydrogen production. Nature Chem. 2014;6:964–970. doi: 10.1038/nchem.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartgerink JD, Beniash E, Stupp SI. Self-assembly and mineralization of peptide-amphiphile nanofibres. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 15.Tysseling-Mattiace VM, et al. Self-assembling nanofibers inhibit glial scar formation and promote axon elongation after spinal cord injury. J Neurosci. 2008;28:3814–3823. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webber MJ, et al. Supramolecular nanostructures that mimic VEGF as a strategy for ischemic tissue repair. Proc. Natl. Acad. Sci. U S A. 108:13438–13443. doi: 10.1073/pnas.1016546108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newcomb CJ, et al. Cell death versus cell survival instructed by supramolecular cohesion of nanostructures. Nature Commun. 2014;5:3321. doi: 10.1038/ncomms4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldberger JE, Berns EJ, Bitton R, Newcomb CJ, Stupp SI. Electrostatic control of bioactivity. Angew. Chem. Int. Ed. 2011;50:6292–6295. doi: 10.1002/anie.201100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui H, et al. Spontaneous and x-ray-triggered crystallization at long range in self-assembling filament networks. Science. 2010;327:555–559. doi: 10.1126/science.1182340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, et al. A self-assembly pathway to aligned monodomain gels. Nature Mater. 2010;9:594–601. doi: 10.1038/nmat2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smulders MM, et al. How to distinguish isodesmic from cooperative supramolecular polymerisation. Chemistry. 2010;16:362–367. doi: 10.1002/chem.200902415. [DOI] [PubMed] [Google Scholar]
- 22.Lee OS, Stupp SI, Schatz GC. Atomistic molecular dynamics simulations of peptide amphiphile self-assembly into cylindrical nanofibers. J Am Chem Soc. 2011;133:3677–3683. doi: 10.1021/ja110966y. [DOI] [PubMed] [Google Scholar]
- 23.Boekhoven J, Stupp SI. Supramolecular Materials for Regenerative Medicine. Adv. Mater. 2014;26:1642–1659. doi: 10.1002/adma.201304606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen TM, Cleland LG. Serum-induced leakage of liposome contents. Biochim. Biophys. Acta (BBA)-Biomembranes. 1980;597:418–426. doi: 10.1016/0005-2736(80)90118-2. [DOI] [PubMed] [Google Scholar]
- 25.Xue WF, et al. Fibril fragmentation enhances amyloid cytotoxicity. J. Biol. Chem. 2009;284:34272–34282. doi: 10.1074/jbc.M109.049809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milanesi L, et al. Direct three-dimensional visualization of membrane disruption by amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20455–20460. doi: 10.1073/pnas.1206325109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 28.Boekhoven J, Rubert Pérez CM, Sur S, Worthy A, Stupp SI. Dynamic Display of Bioactivity through Host-Guest Chemistry. Angew. Chem. Int. Ed. 2013;46:12077–12080. doi: 10.1002/anie.201306278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prager-Khoutorsky M, et al. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nature Cell. Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- 30.Boekhoven J, et al. Alginate–peptide amphiphile core–shell microparticles as a targeted drug delivery system. RSC Adv. 2015;5:8753–8756. doi: 10.1039/C4RA16593D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boekhoven J, et al. Dissipative Self-Assembly of a Molecular Gelator by Using a Chemical Fuel. Angew. Chem. Int. Ed. 2010;49:4825. doi: 10.1002/anie.201001511. [DOI] [PubMed] [Google Scholar]
- 32.Greenfield NJ. Analysis of the kinetics of folding of proteins and peptides using circular dichroism. Nat Protoc. 2006;1:2891–2899. doi: 10.1038/nprot.2006.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, et al. Nano-Biomechanical Study of Spatio-Temporal Cytoskeleton Rearrangements that Determine Subcellular Mechanical Properties and Endothelial Permeability. Scientific reports. 2015;5 doi: 10.1038/srep11097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sur S, Matson JB, Webber MJ, Newcomb CJ, Stupp SI. Photodynamic Control of Bioactivity in a Nanofiber Matrix. ACS Nano. 2012;6:10776–10785. doi: 10.1021/nn304101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.