Abstract
Complex diseases such as cancer are often associated with aberrant gene expression at both the transcriptional and post-transcriptional level. In the past several years, competing endogenous RNAs (ceRNAs) have emerged as an important class of post-transcriptional regulators that alter gene expression through a microRNA-mediated mechanism. Recent studies in both solid tumors and hematopoietic malignancies showed that ceRNAs play significant roles in cancer pathogenesis by altering the expression of key tumorigenic or tumor suppressive genes. Characterizing the identity, function and mechanism of the ceRNAs will not only further our fundamental understanding of RNA-mediated cancer pathogenesis, but also may shed light on developing new RNA-based therapeutic strategies for treating cancer.
Non-coding RNAs (ncRNAs) are prevalently transcribed in the human genome
The central dogma of molecular biology is that DNA is transcribed into messenger RNAs (mRNAs), which in turn serve as the template for protein synthesis. The conventional view that most transcribed RNAs are information carriers rather than functional molecules has been challenged with the discovery of regulatory RNAs that do not code for proteins [1]. Large-scale transcriptome profiling of human cells revealed that over 70% of the human genome is transcribed however, many of these transcripts are not translated into protein but function as ncRNAs [2]. Based on their transcript size, ncRNAs can be classified as small (≤200 base pairs) or (>200 base pairs) long non-coding RNA (lncRNA). Human ncRNAs include about 10,000 small ncRNA genes, around 14,500 pseudogenes and almost 16,000 lncRNA genes according to the GENCODE Release 24 annotation. The small ncRNAs include transfer RNAs, microRNAs (miRNAs), small-interfering RNAs, small nuclear RNAs, small nucleolar RNAs, PIWI-interacting RNAs, transcription initiation RNAs, promoter upstream transcripts and promoter-associated small RNAs. These small ncRNAs participate in gene regulation at the transcriptional or post-transcriptional level. The functions and molecular mechanisms of most small ncRNAs are relatively well characterized. In contrast to small ncRNAs, the function of most lncRNAs remains to be elucidated despite their functional importance in both development and complex diseases such as cancer [3, 4]. Recent studies showed that many lncRNAs encode small open reading frames (sORFs), some of which may be functional [5–7]. These findings underscore the complexity of lncRNA function. Given the significant role of ncRNA-mediated pathogenesis in cancer, we discuss the emerging function and mechanism of competing endogenous RNA (ceRNA), a class of RNAs that may play an important role in cancer pathogenesis, and suggest directions for future studies.
The microRNA sponge, ceRNA hypothesis and its generalization
miRNAs are a class of small (~22 nts), noncoding regulatory RNAs that are part of the RNA-induced silencing complex and bind to their target mRNAs to modulate target gene expression [8]. Although miRNAs are well conserved in both plants and animals, the mode of target recognition and regulation is different between the two kingdoms. Plant miRNAs usually have a near-perfect pairing with their targets, which induces the cleavage and subsequent degradation of the target transcripts [9]. In contrast, animal miRNAs predominantly recognize their targets using as few as 6–8 nucleotides (nts) in the 5' end of the miRNA. This region, usually nucleotides 2–7, forms Watson–Crick (WC) pairs with the target mRNA 3’ untranslated region (UTR) [8, 10]. miRNAs have also been found to target the 5’UTR and coding sequence of the mRNAs as well as pseudogenes and lncRNAs [11–14]. Animal miRNAs post-transcriptionally repress target gene expression via transcript degradation and/or translation inhibition [15, 16]. Given the relatively short miRNA recognition sequence in animals, the miRNA binding sites, also known as miRNA response elements (MREs), may occur in the 3’ UTRs of many mRNAs. Therefore miRNA regulation in animals is combinatorial in nature: an individual miRNA may target hundreds of different mRNAs, and each mRNA may be regulated by multiple miRNAs [17].
It is conceivable that when multiple mRNAs are targeted by the same miRNA or miRNA family, increasing the expression of one target transcript might decrease the repression of the other targets. The rationale of competition-based cross-regulation was first exploited to block the endogenous activity of a specific miRNA or miRNA family by using synthetic competitive inhibitors called miRNA sponges [18–20]. Synthetic miRNA sponges are usually engineered to carry multiple binding sites for a miRNA or miRNA family of interest and are ectopically expressed from strong promoters. The synthetic miRNA sponges have been shown to effectively inhibit miRNA activity and thus prevent the repression of miRNA targets both in vitro and in vivo [20–24].
The theoretical and practical development of the synthetic miRNA sponge laid an important foundation for the hypothesis of competing endogenous RNAs (ceRNAs) [25]. The ceRNA hypothesis postulated that all endogenous coding and non-coding RNAs sharing common MREs may crosstalk and indirectly regulate the expression of each other by competing for miRNA binding. [25–27]. The first naturally occurring ceRNA was discovered in plants [28]. The non-coding RNA IPS1 from Arabidopsis thaliana was found to alter the abundance of PHO2 mRNA, an important component of the phosphate-signaling pathway, and to modulate the shoot phosphate content by sequestering the phosphate starvation-induced miRNA, miR-399. Unlike most miRNA targets in plants that have almost perfect complementarity with miRNAs, the effective sequestration of miR-399 by IPS1 is due to a mismatched loop at the miRNA cleavage site that prevents IPS1 RNA from miRNA-targeting induced endonucleolytic cleavage [28]. Since this initial discovery, additional ceRNAs have been shown to function in viruses and animals in a variety of biological processes, including muscle differentiation, host pathogen interaction and cancer [29–33].
Parallel studies in prokaryotes have demonstrated that RNA–RNA crosstalk can be mediated by small regulatory RNAs (sRNAs) other than miRNAs [34]. Unlike miRNAs, the mechanisms by which these bacterial sRNAs exert their regulatory effects are diverse. They can bind near the translation initiation site and promote or inhibit mRNA stability and/or translation [35]. In rare cases, the sRNA can also bind to the 3’ end of its target RNA, and stabilize its target [36]. Moreover, there are many cases in eukaryotes of RNA–RNA crosstalk being mediated by RNA-binding proteins (RBPs) [37], some of which had been discovered before the proposal of the ceRNA hypothesis. RBP-mediated RNA-RNA crosstalk has been implicated in a class of neurological or muscular diseases such as myotonic dystrophy (dystrophia myotonica, DM) and fragile X tremor ataxia syndrome (FXTAS), the so-called RNA dominant diseases [38]. A common feature of these diseases is that the mutations in non-protein-coding regions, often in the form of an expansion of repetitive sequences, give rise to a deleterious gain-of-function disease phenotype. For example, the DM type 1 (DM1) is caused by an expansion of CUG repeats in the 3′-UTR of the DM protein kinase transcript. The CUG repeats bind to the Muscleblind-like 1 (MBNL1) protein, a splicing regulator, and sequester it from binding to its target precursor mRNAs (pre-mRNAs). This leads to abnormal alternative splicing [38]. Therefore the ceRNA concept can be generalized for any RNA–RNA crosstalk that acts through competitive binding to common regulators, including, but not limited to sRNAs, miRNAs and RNA-binding proteins.
Different types of ceRNAs
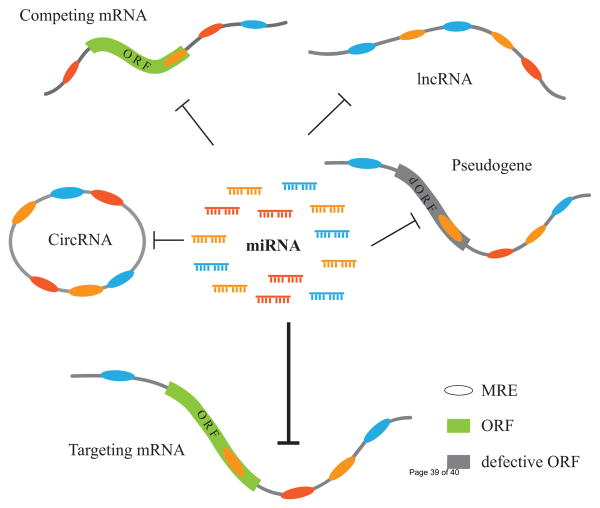
In theory, any RNA molecule can potentially serve as an active ceRNA if it shares miRNA binding with other RNAs. Thus it is not completely unexpected that RNAs that serve as ceRNAs come in a variety of forms, including pseudogenes, protein-coding transcripts, lncRNAs and circular RNAs (circRNAs)(Figure 1, Key Figure). One of the first examples of pseudogenes as ceRNAs is PTENP1, a pseudogene of the tumor suppressor PTEN (phosphatase and tensin homolog) [30–32, 39]. The knockdown of endogenous PTENP1 reduces PTEN expression and promotes cancer cell proliferation. Moreover, PTENP1 is deleted in various human malignancies, suggesting a tumor-suppressing function of PTENP1 [30].
Figure 1 (Key Figure).
Different types of ceRNAs including circRNA, lncRNA, pseudogene and protein-coding mRNA may regulate a target mRNA expression by competing for miRNAs binding.
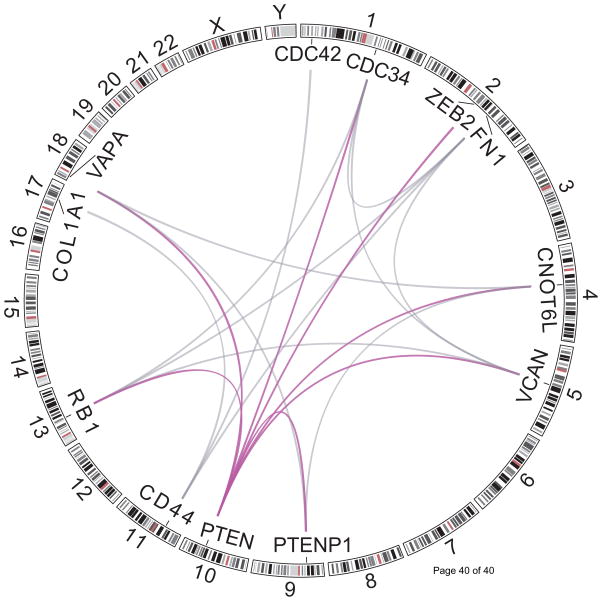
Since the discovery of PTENP1, the list of PTEN ceRNAs has been largely expanded (Figure 2), with the addition of many protein-coding transcripts [40]. Through an integrative analysis of gene expression and matched miRNA profiles, Sumazin et al. computationally predicted and experimentally validated 13 protein-coding ceRNAs of PTEN in glioblastoma multiforme (GBM), for which the locus deletions are predictive of reduced PTEN expression [39]. The down-regulation of these ceRNAs in GBM cell lines decreased PTEN in a 3’ UTR-dependent fashion and enhanced cancer cell growth. Two parallel studies identified two protein-coding genes, CNOT6L and VAPA, as PTEN ceRNAs in prostate cancer [31], and ZEB2 as a PTEN ceRNA in melanoma [32]. These ceRNAs were found to regulate PTEN protein expression in a miRNA-dependent and protein coding independent manner, counteracting downstream PI(3)K signaling and exerting a tumor-suppressing function.
Figure 2.
Computationally predicted and experimentally validated competing activities between members in the PTEN ceRNA network are shown. The ceRNAs directly competing with PTEN are shown in purple.
With the discovery of nearly 16,000 lncRNAs in the human genome, many lncRNAs were found to function as ceRNAs [33, 41–45]. When compared to protein-coding genes, lncRNAs show more tissue specific expression, but have lower abundance [46]. Although many lncRNAs are expressed at low levels that may not be high enough to effectively function as ceRNAs, individual lncRNAs may be potent ceRNAs in specific developmental stages or disease states in which their expression is upregulated. HULC, one of the first examples of a lnc-ceRNA, was indeed identified as being among the most significantly upregulated transcripts in hepatocellular carcinoma (HCC) [42]. In addition to their function in cancer, lnc-ceRNAs are important regulators of developmental processes, such as muscle differentiation [33] and the self-renewal and differentiation of embryonic stem cells [44].
Another emerging type of ceRNA is circRNA. A circRNA is an RNA that forms a circle by covalently–joining 3’ and 5’ ends of exons in a non-linear order (e.g through “back-splicing” [47–51]). The development of RNase R-based enrichment methods and novel bioinformatics methods for identifying junction reads between non-linear exons, together with the high-throughput sequencing of ribosomal RNA (rRNA) and/or polyA-depleted libraries, have enabled the identification of over 7,000 circRNAs in human and over 600 circRNAs in mouse, respectively [52]. The ceRNA hypothesis states that ceRNAs crosstalk by competing for miRNA binding. A circRNA has several unique features that are favorable for being ceRNAs, including a lack of free ends (resistance to exonucleases and conferring high stability), predominant cytoplasm localization (right subcellular localization for effective miRNA-targeting) and a largely noncoding nature (effective miRNA targeting without ribosome interference). However, so far there have been only a handful of circRNAs found to serve as ceRNAs [53–56]. In addition, Guo et al. found that, among many putative human and mouse circRNAs, only two circRNAs harbor more MREs than would be expected by chance [52]. Therefore, despite the unique features of circRNAs as potentially potent ceRNAs, it remains unclear how prevalent the contribution from circRNAs may be to the functional ceRNA crosstalk.
Identifying ceRNA crosstalk and decoding its mechanism
Computational methods have been instrumental in the large-scale prediction of ceRNA interactions (Box 1). Since ceRNA crosstalk occurs through a competition for miRNA binding between RNAs, the prediction of ceRNA interactions depends on the identification of MREs within the transcripts of interest. Different miRNA-target prediction algorithms, such as TargetScan [17], PicTar [57], miRanda [58], rna22 [53] and PITA [59] have greatly facilitated the discovery of ceRNA crosstalk. However, miRNA-target prediction methods that are solely based on sequence information may have many false positives in target prediction. False positive predictions of the miRNA target could lead to spurious predictions of ceRNA interactions. As alternatives to computational methods, high-throughput biochemical approaches (Box 2) have enabled genome-wide identification of miRNA–RNA target interactions. These unbiased methods have uncovered many non-canonical MREs [60, 61] as well as MREs outside 3’UTR regions [12–14, 61] that have not been well captured by current computational methods. Given the complementary information provided by the high-throughput biochemical methods and computational predictions about miRNA–target interactions, it is important to combine both approaches in order to identify bona fide miRNA-target interactions. The prediction of miRNA-target interactions is often the first step toward identifying active ceRNA crosstalk. The integration of additional genomic datasets, such as a compendium of transcriptome profiles across related conditions (e.g. across many tumors of the same cancer type), is often useful for predicting ceRNA interactions [31, 39, 62]. In particular, the miRNA-dosage-dependent change in the correlation or co-variation of expression between ceRNAs is an important feature that can be utilized to predict ceRNA interactions [31, 39, 62].
Box 1. Computational methods and resources for studying ceRNAs.
For mRNAs, usually the 3’ UTR regions are used for target searching, although MREs can also be found in the coding and 5’ UTR regions in some cases. The popular target prediction methods include TargetScan [17], PicTar [57], miRanda [58], PITA [59] and RNA22 [113]. These methods differ by how they score the sequence complementary between miRNA and RNA in the seed region as well as the features outside the seed region, how the evolutionary conservation is incorporated into the modeling and whether they use biophysical parameters, such as the free energy of miRNA:RNA duplex or miRNA binding sites accessibility in prediction.
Hermes is an information theory-based multivariate analysis method to systematically infer ceRNA regulations by estimating Mutual Information (MI) and Conditional Mutual Information (CMI) from gene and miRNA expression profiles [39].
starBase 2.0 applies hypergeometric tests to each of the ceRNA pairs that are supported by both computational methods and Ago CLIP experiments. All p values are false discovery rate (FDR) corrected to infer the potential ceRNA regulations [114].
Cupid is a framework for context-specific miRNA target inference by combining sequence-based evidence and expression data. Cupid first applies a support vector machine (SVM) classifier to predict candidate miRNA-target interactions, and then utilizes expression-based evidence to further infer ceRNA activities [62].
Box 2. Experimental technologies for studying ceRNAs.
Affinity-based enrichment techniques
CLIP-seq, also known as HITS-CLIP (High-throughput sequencing of RNA isolated by CLIP) uses crosslinking and immunoprecipitation of a RBP of interest to pull down the RBP-interacting RNAs, followed by high-throughput sequencing [115].
PAR-CLIP (Photoactivatable-Ribonucleoside-Enhanced CLIP) improves the crosslink efficiency by incorporating photoreactive ribonucleoside analogs into nascent RNA transcripts before crosslinking [12].
iCLIP (Individual-nucleotide resolution CLIP) enables PCR amplification of the cDNAs that are truncated at protein-RNA crosslinking sites and thus allows for identifying protein-RNA direct interaction site at single-nucleotide resolution [116].
FAST-iCLIP (Fully Automated and Standardized iCLIP) simplifies the standard iCLIP procedure with improved biochemistry and reduces experiment time by half [117].
TAP-Tar (Tandem affinity purification of miRNA target mRNAs) sequentially pulls down mRNA/miRNA complex, first the Argonaute and then the miRNA, through tandem affinity purification [118].
LAMP (Labeled microRNA pull-down) uses biotin labeled miRNA mimics [119] or digoxigenin (DIG) labeled pre-miRNAs [120] to pull down the miRNA targeting mRNAs.
MS2-TRAP (MS2-tagged RNA affinity purification) labels miRNAs with tandem MS2 RNA hairpin tags, and then co-purifies their targeting RNAs [121]
miTRAP (miRNA trapping by RNA in vitro affinity purification) is reverse to MS2-TRAP, which labels the RNA of interest with tandem MS2 RNA hairpin tags by in vitro transcription and co-purifies the bound miRNAs [122].
RNA pull down assay with biotinylated antisense oligonucleotides uses biotin labeled antisense DNA oligonucleotides to affinity pull down the RNA of interest and its interacting RNAs [123].
CLASH(crosslinking, ligation and sequencing of hybrids) can systematically identify RNA-RNA interactions by UV cross-linking, affinity-purification of RNPs, ligation to record the spatial arrangement of RNA–RNA hybrids and sequencing to obtain the chimeric reads [124].
Single-cell related technologies
Single-cell gene expression analysis includes the SINCE-PCR (single-cell PCR) method, which allows for measuring the expression of a subset of transcripts in individual cells [91], and the scRNA-seq (single-cell RNA-sequencing) method, which allows for measuring the whole transcriptome expression at single-cell level [92].
Single-cell fluorescence reporter assays use synthetic mRNAs that carry engineered MREs and encode fluorescent proteins to quantitatively measure the corresponding miRNA activity in response to altering miRNA or ceRNA expression level in vivo [66].
Mass-spectrometry-related technologies
Mass-spectrometry based quantitative proteomics approachescan be classified into two categories. Label-free quantification quantifies the relative abundance of proteins in more than one sample based on precursor signal intensity or on spectral counting [125]. In contrast, the stable isotope labeling quantifies the relative abundance of proteins in different samples using stable isotopes that allow the mass spectrometer to distinguish between identical proteins in these samples, owing to a known mass shift of the labeled protein or peptide in the mass spectrum [126–129].
RNA interactome capture captures all the proteins bound to mRNAs by using UV crosslinking or Photoactivatable-Ribonucleoside-Enhanced Crosslinking coupled with mass spectrometry [87, 130].
In addition to their utility in predicting ceRNA regulation, the computational methods, especially biophysical and mathematical modeling approaches have provided significant insight into the mechanism of ceRNA crosstalk [34, 63, 64]. Levine et al. utilized mass-action equations to quantitatively model ceRNA-like sRNA-mediated RNA–RNA crosstalk in E. coli [34]. The quantitative modeling showed that the sRNA-mediated crosstalk effect is nonsymmetrical, but hierarchical: the expression of a weakly sRNA-interacting target is highly influenced by another more strongly interacting target, but not vice versa. In the same vein as the study in bacteria, Ala et al. utilized simple kinetic models of miRNA-mediated ceRNA crosstalk to evaluate the requirement of different factors for ceRNA cross-regulation [64]. In contrast to the non-catalytic nature of sRNA regulation in bacteria, the interaction between miRNA and its target can be purely catalytic, purely non-catalytic (i.e. stoichiometric) or a mixture of both. The kinetic modeling revealed that, at the steady state, no ceRNA crosstalk can occur if the miRNA-target interaction is purely catalytic. However, even in a purely catalytic scenario, ceRNA crosstalk can occur in the out-of-equilibrium regime as long as the timescale is much shorter than the time required to reach equilibrium. A variety of factors such as the relative abundance of ceRNAs and miRNAs, and the number and affinity of miRNA binding sites, contribute to the effectiveness of ceRNA crosstalk. Moreover, it was uncovered by theoretical modeling that even if two RNAs are not targeted by common miRNAs, they may form ceRNA crosstalk given that they have MREs in common with a third RNA [64]. These findings suggest a complex and highly context-dependent ceRNA regulatory network.
Despite the insight from biophysical and mathematical modeling studies into the ceRNA crosstalk mechanism, several studies have challenged the physiological relevance of the original ceRNA hypothesis, i.e. whether the competition for miRNAs is the major cause of the observed ceRNA effect under physiological conditions. Denzler et al. quantitatively assessed the stoichiometry between miR-122, a highly expressed miRNA in primary hepatocytes, and its target MRE sites [65]. By overexpressing the miR-122 target, aldolase mRNA, they found that only when the number of added aldolase MREs exceeded physiological levels, did significant de-repression of endogenous miR-122 targets occur. Additionally, using a simple biophysical model to analyze transcriptome data of monocytes for miR-20a target genes, Jen et al., reached a similar conclusion, that the substantial ceRNA crosstalk solely based on competition for miR-20a binding is very unlikely for its endogenous targets [37]. A recent study by Bosson et al. utilized an integrated approach to assess miRNA target competition in the mouse embryonic stem cell (mESC) [66]. Through an analysis of Ago2 iCLIP-seq, mRNA-seq and small RNA-seq data, together with mathematical modeling and a single-cell reporter assay, the authors showed that the difference in the relative abundance of miRNA and the target pool as well as the target affinity quantitatively differentiate not only in vivo Ago binding profiles, but also miRNA susceptibility to target competition [66]. Consistent with the results from the other two studies [37, 65], they found that the targets of highly expressed miRNA families such as miR-294 and let-7 in the mESC are unlikely to be susceptible to ceRNA crosstalk. In contrast, the targets of active miRNA families for which the ratio of the miRNA versus the target is low, such as miR-92/25, might be susceptible to ceRNA crosstalk under physiological conditions. The findings from these studies suggest that the current view of ceRNA crosstalk by competition for miRNAs between RNAs is likely to be simplistic and may require significant revisions to account for the experimentally observed ceRNA crosstalk.
The function of ceRNAs in cancer
Ever since the discovery of the PTEN pseudogene and protein-coding ceRNAs in melanoma, prostate cancer and GBM, there has been a rapid expansion in the discovery of ceRNA functions in both solid tumors and hematopoietic malignancies.
Solid tumors
Gastrointestinal cancer
In addition to the lnc-ceRNA HULC, several ceRNAs have been found to impact different aspects of hepatocellular tumorigenesis. LncRNA-ATB is a lncRNA that is upregulated by TGF-β signaling in metastatic HCC and is associated with a poor prognosis [67]. It promotes celluar invasion and metastasis partly by serving as a miR200 family-mediated ceRNA of two important epithelial–mesenchymal transition (EMT) activators ZEB1 and ZEB2. These findings suggest that lncRNA-ATB may serve as a potential antimetastatic target [67]. INTS6P1 is a pseudogene that exerts a tumor-suppressing function and upregulates its cognate gene INTS6 through competitive binding of oncogenic miR-17-5p in HCC [68].
Cir-ITCH is a circular RNA that originates from an E3 ubiquitin protein ligase named ITCH. It is downregulated in esophageal squamous cell carcinoma (ESCC) compared with matched normal tissues. It serves as a sponge of miR-7, miR-17, and miR-214, upregulates ITCH expression, and inhibits the Wnt/beta-catenin pathway and tumor growth in xenograft mouse models [55]. A parallel study showed that cir-ITCH is also downregulated and exerts a tumor-supressing function in colorectal cancer (CRC) through the same mechanism demonstrated in ESCC [56]. The miR-139-5p is an intronic miRNA encoded within the second intron of phosphodiesterase 2A (PDE2A) and exerts tumor-suppressing function in CRC, HCC and gastric cancer. In CRC, miR-139-5p was found to mediate ceRNA interactions between its targets, including IGF1R, ROCK2 and RAP1B. The overexpression of its target genes increases both tumor cell growth and motility, suggesting an oncogenic function of these ceRNAs in CRC [69]. GAPLINC is a lncRNA that is highly expressed in gastric cancer cells, and its upregulation is associated with shorter survival times in patients with gastric cancer. The knockdown of GAPLINC impaired the proliferation and invasion of gastric cancer cells, and GAPLINC exerted its tumor-promoting function by serving as a ceRNA of CD44 and competing for miR-211-3p [70].
Breast and endometrial cancer
TUSC2P, a pseudogene of TUSC2, was found to exert a tumor-suppressing function in mouse and human breast cancer cells by serving as a ceRNA of TUSC2, TIMP2 and TIMP3 [71]. Linc-ROR was shown to regulate the self-renewal of human embryonic stem cells and induced pluripotent stem cells by serving as a ceRNA and regulating Oct4, Nanog, and Sox2 [44]. Interestingly, the linc-ROR was also found to play an important role in maintaining the pluripotent state of endometrial cancer by operating as a miR-145 sponge [72] . Furthermore, the linc-ROR was found to be upregulated in breast cancers and to promote EMT transition and metastasis by upregulating some mir-205 target genes, including the EMT inducer ZEB2 [67]. This finding suggests that the linc-ROR is a potential therapeutic target for aggressive and metastatic breast cancer.
Thyroid cancer, clear cell renal carcinoma and lung cancer
In contrast to the lnc-ceRNA HULC that is upregulated in cancer cells, the ceRNA PTCSC3 was found to be dramatically downregulated in thyroid cancers and to exert a tumor-suppressing effect. The overexpression of PTCSC3 in thyroid cancer cells can induce cancer cell growth inhibition, cell cycle arrest and apoptosis by acting as a sponge of miR-574-5p [43] . PTENP1, a pseudogene of PTEN that exerts a tumor-suppressing function in prostate cancer was also found to serve as a PTEN ceRNA in clear cell renal cell carcinoma (CCRCC), suggesting that some ceRNAs may influence tumorigenesis in multiple cancers [73]. A protein-coding gene AEG-1 was found to promote EMT in human non-small cell lung cancer (NSCLC) by regulating miR-30a activity and operating as ceRNAs of Snail and Vimentin, the EMT inducer and marker [74].
Hematopoietic malignancies
Lymphoma
To establish a causal role of putative pseudogene ceRNAs in cancer development in vivo, Karreth et al. engineered mice to overexpress a full-length murine B-Raf pseudogene, Braf-rs1. They found that the engineered mice developed an aggressive malignancy that resembles human diffuse large B cell lymphoma. Braf-rs1 and its human ortholog, BRAFP1, exert their oncogenic function, at least in part, by serving as ceRNAs to elevate BRAF expression and enhance MAPK signaling in vitro and in vivo. Notably, BRAFP1 harbors frequent transcriptional or genomic aberrations in B cell lymphoma as well as in other human cancers [75].
Leukemia
In a study to identify the lncRNAs that regulate Bcr-Abl-mediated cellular transformation, Guo et al. found that the overexpression of lncRNA-BGL3 in mice alone was sufficient to impair Bcr-Abl-mediated primary bone marrow transformation. LncRNA-BGL3 exerted a tumor-suppressing function by serving as a PTEN ceRNA and its expression was repressed by Bcr-Abl through c-Myc-dependent DNA methylation [76].
Concluding Remarks
Significant progress has been made in discovering ceRNAs and in understanding their function and mechanism in cancers. However, one of the fundamental questions that remains to be addressed is how effective ceRNA crosstalk occurs physiologically in the cell (See Outstanding Questions box) [65]. Both experiments and quantitative biophysical modeling [37, 65] have suggested that it may be atypical for endogenous changes in mRNA levels to significantly relieve miRNA repression on other mRNAs by sequestration alone. The current ceRNA hypothesis based on competition alone is likely to be oversimplified in many cases and cannot fully account for the experimentally observed functional crosstalk. Therefore, the detailed mechanisms of functional ceRNA crosstalk remains to be elucidated and will be facilitated by a combination of mathematical modeling and genome-wide and/or quantitative approaches.
Outstanding questions.
How does effective ceRNA crosstalk occur within the physiologically accessible concentrations in the cell?
What are the mechanisms by which ceRNA crosstalk is regulated and exploited by cancer cells?
How does ceRNA crosstalk differ from cell to cell in a genetically identical or a genetically heterogeneous population?
How does a ceRNA regulatory network rewire during tumor evolution and contribute to the therapeutic resistance mechanism of cancer cell?
Further improvement of computational methods for predicting miRNA-target interactions that occur through non-canonical MREs [60, 61] or MREs outside 3’UTR regions [12–14, 61, 77] will facilitate the discovery of new pathways regulated by ceRNA crosstalk in cancer. Moreover, the development of computational methods that are capable of quantitatively integrating heterogeneous datasets such as those generated from small RNA-seq, mRNA-seq and HITS-CLIP/iCLIP/PAR-CLIP-seq experiments will allow for a quantitative prediction of ceRNA regulatory networks and will be critical for developing ceRNA-based cancer therapeutic strategies.
Aside from the importance of the relative abundance of miRNA and RNAs in modulating the strength of ceRNA crosstalk, the other mechanisms, whereby ceRNA crosstalk is regulated and exploited by cancer cells remain poorly understood. The 3’UTR length of protein-coding transcripts is known to be highly regulated during cancer development and progression [78, 79]. Because the 3’UTR length influences the targeting of miRNAs to individual mRNA, it is conceivable that ceRNA crosstalk could be altered or rewired when the 3’UTR length of involved ceRNAs changes.
Another post-transcriptional mechanism that could impact ceRNA crosstalk is RNA editing. RNA editing could influence miRNA target interactions and thus ceRNA crosstalk by either altering miRNA sequence and its target spectrum [80] or modifying miRNA-binding sites in RNA transcripts [81, 82].
The subcellular localization change [45] and/or the binding of RBPs to MREs of those RNAs that are involved in ceRNA crosstalk could affect their ceRNA interactions by altering the physical accessibility to miRNA or Ago proteins [26, 40]. Technologies such as HITS-CLIP/iCLIP/PAR-CLIP, multiplexed subcellular RNA sequencing [83] or imaging [84, 85], and the mass-spectrometry based quantitative proteomics approaches for capturing RNA-interactome [86, 87] or measuring RBP abundance [88, 89] will be useful tools to assess the effect of subcellular localization change and/or competitive binding of RBP to MREs on ceRNA crosstalk.
Not only do cancer cells between tumors show genetic and phenotypical distinctions (i.e. inter-tumor heterogeneity), but cancer cells within the same tumor (i.e. intra-tumor heterogeneity) also do. Intra-tumor heterogeneity is considered an important contributor to neoplasia, cancer progression, and therapeutic resistance [90]. So far most studies of ceRNA regulation have been performed at a cell-population level. It remains to be elucidated how ceRNA crosstalk differs from cell to cell in a genetically identical or a genetically heterogeneous population. It also remains unknown how ceRNA regulatory networks rewire during tumor evolution and contributes to the therapeutic resistance mechanism of a cancer cell. By combining the powerful single-cell techniques [91, 92], bioinformatics and mathematical modeling approaches, we may be able to shed light on how ceRNA crosstalk contributes to tumor heterogeneity, which can have far-reaching clinical implications.
Taken together, different contributing factors that may be utilized by cancer cells to modulate ceRNA interactions await further characterization. With a better understanding of both genetic and non-genetic factors that are important for modulating ceRNA interactions in different cancer subtypes and ultimately individual tumors from the same cancer, we may be able to exploit the context-dependency of ceRNA crosstalk for precise/personalized RNA-based therapeutics. The development of new computational methods and modeling approaches in combination with the state-of-art omic techniques and single-cell methods are key to understanding the quantitative nature of ceRNA crosstalk and unveiling their contribution to cancer development, progression and therapeutic resistance.
Table 1.
Cancer-related ceRNAs.
| ceRNA | Shared miRNA | Competing target | Cancer type | Reference |
|---|---|---|---|---|
|
mRNA
| ||||
| AEG-1 3'UTR | miR-30a | Vimentin, Snail | NSCLC | [74] |
| CNOT6L/VAPA/ SERINC1 | miR-17, miR-19a, miR-20a/b, miR-106a/b, miR-93 | PTEN | Prostate cancer | [31] |
| ZEB2 | miR-25, miR-92a, miR-181a, miR-200b | PTEN | Melanoma | [32] |
| CYP4Z2P-3'UTR | miR-211, miR-197, miR-204, miR-125a-3p, miR-1226 | CYP4Z1 | Breast cancer | [93] |
| EPO/EPOR | miR-125b | ERBB2/HER2 | Breast cancer | [94] |
| FOXO1 3'UTR | miR-9 | E-cadherin | Breast cancer | [95] |
| OCT4B | miR-145, miR-20a/b, miR-106a/b, miR-335 | OCT4A | CRC Prostate cancer |
[96] |
| IGF1R/ROCK2/ RAP1B 3'UTR | miR-139-5p | IGF1R/ROCK2/ RAP1B | CRC | [69] |
| VERSICAN 3'UTR | miR-136, miR-199a-3p, miR-144 | Rb1, PTEN | Breast cancer | [97] |
| miR-133a, miR-199a-3p, miR-144,miR-431 | CD34, Fibronectin | HCC | [98] | |
|
| ||||
|
lncRNA
| ||||
| BARD1 9'L | miR-203, miR-101 | BARD1 | Multiple cancers | [99] |
| CCAT1 | miR-218-5p | BMI1 | Gallbladder cancer | [100] |
| let-7 | HMGA2, c-Myc | HCC | [101] | |
| DANCR | miR-214, miR-320a, miR-199a | CTNNB1 | HCC | [102] |
| GAPLINC | miR-211-3p | CD44 | Gastric cancer | [70] |
| GAS5 | miR-21 | N/A | Breast cancer | [103] |
| HOTTIP | miR-125b | N/A | HCC | [104] |
| HOTAIR | miR-331-3p | HER2 | Gastric cancer | [105] |
| HULC | miR-372 | PRKACB | HCC | [42] |
| LINC00974 | miR-642 | KRT19 | HCC | [106] |
| LncRNA-BGL3 | miR-20a/b, miR-106a/b, miR-17, miR-93 | PTEN | Leukemia | [76] |
| LncRNA-FER1L4 | miR-106a-5p | RB1 | Gastric cancer | [107] |
| LncRNA-ATB | miR-200 family | ZEB, ZEB2 | HCC | [67] |
| Linc-ROR | miR-145 | N/A | Endometrial cancer | [72] |
| miR-205 | ZEB2 | Breast cancer | [67] | |
| miR-145 | ARF6 | Breast cancer | [108] | |
| PCAT1 | miR-3667-3p | c-Myc | Prostate cancer | [109] |
| PTCSC3 | miR-574-5p | N/A | Thyroid cancer | [43] |
| UCA1 | miR-216b | FGFR1 | HCC | [110] |
| XIST | miR-152 | N/A | GBM | [111] |
|
| ||||
|
Pseudogene
| ||||
| BRAFP1 | miR-30a, miR-876, miR-182 | BRAF | Lymphoma | [75] |
| INTS6P1 | miR-17-5p | INTS6 | HCC | [68] |
| KRAS1P | N/A | K-RAS | Prostate cancer | [30] |
| PTENP1 | miR-17, miR-19b, miR-20a | PTEN, PHLPP | HCC | [112] |
| miR-17, miR-19, miR-21, miR-26, miR-214 | PTEN | Prostate cancer | [30] | |
| miR-21 | PTEN | CCRCC | [73, 112] | |
|
| ||||
|
circRNA
| ||||
| Circ-ITCH | miR-7,miR-17,miR-214 | ITCH | CRC ESCC |
[55, 56] |
Abbreviations:
CRC is colorectal cancer; GBM is glioblastoma multiforme; HCC is hepatocellular cancer; NSCLC is nonsmallcell lung cancer; CCRCC is clear cell renal cell carcinoma; ESCC is esophageal squamous cell carcinoma;N/A is Not Available
Trends.
In recent years, competing endogenous RNAs (ceRNAs) have emerged as an important class of post-transcriptional regulators that alter gene expression through a microRNA-mediated mechanism.
Studies in both solid tumors and hematopoietic malignancies showed that ceRNAs played important roles in different aspects of cancer etiology, suggesting that ceRNAs have great potential as therapeutic targets.
A combination of computational and experimental approaches have been instrumental to the characterization of identity, function and mechanism of the ceRNAs.
Acknowledgments
We sincerely apologize to all investigators whose contribution was not cited due to space limitation. This work was partially funded by NIH R00CA175290, University of Texas Rising STARs award, and Texas CPRIT grant RR140071 to Y.C, and by the UT Southwestern Endowed Scholar Program, the Cancer Prevention and Research Institute of Texas (CPRIT R1103), the Welch Foundation (I-1800), National Institutes of Health (NIH) (GM114160), a career development award associated with University of Texas Specialized Program of Research Excellent in Lung Cancer (NIH CA70907), American Cancer Society (Research Scholar Grant, RSG-15-062-01-TBE and Institutional Research Grant, IRG-02-196-07) to Y. Y. It was also supported in part by U.S. National Cancer Institute (NCI; MD Anderson TCGA Genome Data Analysis Center) grant number CA143883, the Cancer Prevention Research Institute of Texas (CPRIT) grant number RP130397, the Mary K. Chapman Foundation, the Michael & Susan Dell Foundation (honoring Lorraine Dell), and MD Anderson Cancer Center Support Grant P30 CA016672 (the Bioinformatics Shared Resource). Y. Y. is a Virginia Murchison Linthicum Scholar in Medical Research and a CPRIT Scholar in Cancer Research.
Footnotes
Author Contributions Statement
Y.W., J.H. and Y.C. wrote most of the manuscript. P.Z. contributed to the writing of different types of ceRNAs section. Y.W. generated the figures. D.H. surveyed the existing computational and experimental methods for prediction and validation of ceRNAs (Box 1,2). M.S. collected literature evidence for cancer related ceRNAs and made Table 1 (Cancer-related ceRNAs). Y.Y. contributed to the writing of proteomics methods. Y.C. drafted the outline, supervised personnel and revised the manuscript. All authors reviewed the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nature structural & molecular biology. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 4.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer discovery. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chew GL, et al. Ribosome profiling reveals resemblance between long non-coding RNAs and 5' leaders of coding RNAs. Development. 2013;140:2828–2834. doi: 10.1242/dev.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magny EG, et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- 7.Anderson DM, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhoades MW, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 10.Lewis BP, et al. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Clark PM, et al. Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types. Scientific reports. 2014;4:5947. doi: 10.1038/srep05947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hausser J, et al. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013;23:604–615. doi: 10.1101/gr.139758.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helwak A, et al. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yekta S, et al. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 16.Davis E, et al. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Current biology : CB. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- 17.Friedman RC, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nature biotechnology. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 19.Care A, et al. MicroRNA-133 controls cardiac hypertrophy. Nature medicine. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 20.Ebert MS, et al. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentner B, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nature methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 22.Valastyan S, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Luna JM, et al. Hepatitis C Virus RNA Functionally Sequesters miR-122. Cell. 2015;160:1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. Rna. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salmena L, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Current biology : CB. 2010;20:R858–861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seitz H. Redefining microRNA targets. Current biology : CB. 2009;19:870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 28.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature genetics. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 29.Cazalla D, et al. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tay Y, et al. Coding-Independent Regulation of the Tumor Suppressor PTEN by Competing Endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karreth FA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine E, et al. Quantitative characteristics of gene regulation by small RNA. PLoS biology. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irnov I, et al. Identification of regulatory RNAs in Bacillus subtilis. Nucleic acids research. 2010;38:6637–6651. doi: 10.1093/nar/gkq454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Opdyke JA, et al. GadY, a small-RNA regulator of acid response genes in Escherichia coli. Journal of bacteriology. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nature reviews. Genetics. 2015;16:113–126. doi: 10.1038/nrg3853. [DOI] [PubMed] [Google Scholar]
- 38.Osborne RJ, Thornton CA. RNA-dominant diseases. Human molecular genetics. 2006;15(Spec No 2):R162–169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 39.Sumazin P, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Giorgio A, et al. Emerging roles of competing endogenous RNAs in cancer: insights from the regulation of PTEN. Molecular and cellular biology. 2013;33:3976–3982. doi: 10.1128/MCB.00683-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon JH, et al. LincRNA-p21 Suppresses Target mRNA Translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic acids research. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan M, et al. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Experimental and therapeutic medicine. 2013;5:1143–1146. doi: 10.3892/etm.2013.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Du, et al. Integrative analyses reveal a long non-coding-RNA mediated sponge regulatory network in prostate cancer. Nature communications. 2016 doi: 10.1038/ncomms10982. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang DM, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Gene Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna-a Publication of the Rna Society. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen TB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salzman J, et al. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. Plos One. 2012;7 doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang XO, et al. Complementary Sequence-Mediated Exon Circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Guo JU, et al. Expanded identification and characterization of mammalian circular RNAs. Genome biology. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 54.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 55.Li F, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang G, et al. cir-ITCH Plays an Inhibitory Role in Colorectal Cancer by Regulating the Wnt/beta-Catenin Pathway. Plos One. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krek A, et al. Combinatorial microRNA target predictions. Nature genetics. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 58.John B, et al. Human MicroRNA targets. PLoS biology. 2004;2:1862–1879. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kertesz M, et al. The role of site accessibility in microRNA target recognition. Nature genetics. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 60.Chi SW, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loeb GB, et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiu HS, et al. Cupid: simultaneous reconstruction of microRNA-target and ceRNA networks. Genome Res. 2015;25:257–267. doi: 10.1101/gr.178194.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan Y, et al. Model-guided quantitative analysis of microRNA-mediated regulation on competing endogenous RNAs using a synthetic gene circuit. P Natl Acad Sci USA. 2015;112:3158–3163. doi: 10.1073/pnas.1413896112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ala U, et al. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci U S A. 2013;110:7154–7159. doi: 10.1073/pnas.1222509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Denzler R, et al. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bosson AD, et al. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuan JH, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 68.Peng H, et al. Pseudogene INTS6P1 regulates its cognate gene INTS6 through competitive binding of miR-17-5p in hepatocellular carcinoma. Oncotarget. 2015;6:5666–5677. doi: 10.18632/oncotarget.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen K, et al. Post-transcriptional regulation of the tumor suppressor miR-139-5p and a network of miR-139-5p-mediated mRNA interactions in colorectal cancer. The FEBS journal. 2014;281:3609–3624. doi: 10.1111/febs.12880. [DOI] [PubMed] [Google Scholar]
- 70.Hu Y, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer research. 2014;74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 71.Rutnam ZJ, et al. The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nature communications. 2014;5:2914. doi: 10.1038/ncomms3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou X, et al. Linc-RNA-RoR acts as a "sponge" against mediation of the differentiation of endometrial cancer stem cells by microRNA-145. Gynecologic oncology. 2014;133:333–339. doi: 10.1016/j.ygyno.2014.02.033. [DOI] [PubMed] [Google Scholar]
- 73.Yu G, et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Molecular cancer therapeutics. 2014;13:3086–3097. doi: 10.1158/1535-7163.MCT-14-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu K, et al. AEG-1 3'-untranslated region functions as a ceRNA in inducing epithelial-mesenchymal transition of human non-small cell lung cancer by regulating miR-30a activity. European journal of cell biology. 2015;94:22–31. doi: 10.1016/j.ejcb.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 75.Karreth FA, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319–332. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo G, et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene. 2015;34:1768–1779. doi: 10.1038/onc.2014.131. [DOI] [PubMed] [Google Scholar]
- 77.Tay Y, et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–U1112. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 78.Mayr C, Bartel DP. Widespread Shortening of 3 ' UTRs by Alternative Cleavage and Polyadenylation Activates Oncogenes in Cancer Cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xia Z, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3 '-UTR landscape across seven tumour types. Nature communications. 2014;5 doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawahara Y, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nature biotechnology. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 82.Borchert GM, et al. Adenosine deamination in human transcripts generates novel microRNA binding sites. Human molecular genetics. 2009;18:4801–4807. doi: 10.1093/hmg/ddp443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee JH, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen KH, et al. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cabili MN, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome biology. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Castello A, et al. System-wide identification of RNA-binding proteins by interactome capture. Nature protocols. 2013;8:491–500. doi: 10.1038/nprot.2013.020. [DOI] [PubMed] [Google Scholar]
- 87.Baltz AG, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 88.Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annual review of biochemistry. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- 89.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nature reviews. Genetics. 2009;10:617–627. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 90.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dalerba P, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nature biotechnology. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramskold D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nature biotechnology. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng L, et al. The 3'UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in breast cancer by acting as a ceRNA for CYP4Z1. Breast cancer research and treatment. 2015;150:105–118. doi: 10.1007/s10549-015-3298-2. [DOI] [PubMed] [Google Scholar]
- 94.Ferracin M, et al. miR-125b targets erythropoietin and its receptor and their expression correlates with metastatic potential and ERBB2/HER2 expression. Molecular cancer. 2013;12:130. doi: 10.1186/1476-4598-12-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang J, et al. FOXO1 3'UTR functions as a ceRNA in repressing the metastases of breast cancer cells via regulating miRNA activity. FEBS letters. 2014;588:3218–3224. doi: 10.1016/j.febslet.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 96.Li D, et al. OCT4B modulates OCT4A expression as ceRNA in tumor cells. Oncology reports. 2015;33:2622–2630. doi: 10.3892/or.2015.3862. [DOI] [PubMed] [Google Scholar]
- 97.Lee DY, et al. Expression of versican 3'-untranslated region modulates endogenous microRNA functions. Plos One. 2010;5:e13599. doi: 10.1371/journal.pone.0013599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fang L, et al. Versican 3'-untranslated region (3'-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:907–919. doi: 10.1096/fj.12-220905. [DOI] [PubMed] [Google Scholar]
- 99.Pilyugin M, Irminger-Finger I. Long non-coding RNA and microRNAs might act in regulating the expression of BARD1 mRNAs. The international journal of biochemistry & cell biology. 2014;54:356–367. doi: 10.1016/j.biocel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 100.Ma MZ, et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell death & disease. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Deng L, et al. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. Journal of experimental & clinical cancer research : CR. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yuan SX, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2015 doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 103.Zhang Z, et al. Negative regulation of lncRNA GAS5 by miR-21. Cell death and differentiation. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tsang FH, et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver international : official journal of the International Association for the Study of the Liver. 2015;35:1597–1606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 105.Liu XH, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Molecular cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tang J, et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell death & disease. 2014;5:e1549. doi: 10.1038/cddis.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xia T, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Scientific reports. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eades G, et al. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Molecular cancer research : MCR. 2015;13:330–338. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prensner JR, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–908. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang F, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yao Y, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer letters. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 112.Chen CL, et al. Suppression of hepatocellular carcinoma by baculovirus- mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials. 2015;44:71–81. doi: 10.1016/j.biomaterials.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 113.Miranda KC, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 114.Li JH, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic acids research. 2013;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Flynn RA, et al. Dissecting noncoding and pathogen RNA-protein interactomes. Rna-a Publication of the Rna Society. 2015;21:135–143. doi: 10.1261/rna.047803.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nonne N, et al. Tandem affinity purification of miRNA target mRNAs (TAP-Tar) Nucleic acids research. 2010;38:e20. doi: 10.1093/nar/gkp1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Orom UA, Lund AH. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods. 2007;43:162–165. doi: 10.1016/j.ymeth.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 120.Hsu RJ, et al. Labeled microRNA pull-down assay system: an experimental approach for high-throughput identification of microRNA-target mRNAs. Nucleic acids research. 2009;37 doi: 10.1093/nar/gkp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yoon JH, et al. MS2-TRAP (MS2-tagged RNA affinity purification): tagging RNA to identify associated miRNAs. Methods. 2012;58:81–87. doi: 10.1016/j.ymeth.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Braun J, et al. Rapid identification of regulatory microRNAs by miTRAP (miRNA trapping by RNA in vitro affinity purification) Nucleic acids research. 2014;42:e66. doi: 10.1093/nar/gku127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hassan T, et al. Isolation and identification of cell-specific microRNAs targeting a messenger RNA using a biotinylated anti-sense oligonucleotide capture affinity technique. Nucleic acids research. 2013;41:e71. doi: 10.1093/nar/gks1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kudla G, et al. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci U S A. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bantscheff M, et al. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Analytical and bioanalytical chemistry. 2012;404:939–965. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
- 126.Gygi SP, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature biotechnology. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 127.Thompson A, et al. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 128.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 129.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 130.Castello A, et al. Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]