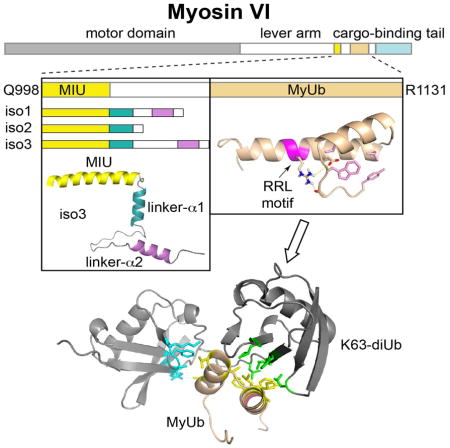
SUMMARY
Myosin VI is critical for cargo trafficking and sorting during early endocytosis and autophagosome maturation, with abnormalities linked to cancers, neurodegeneration, deafness, and hypertropic cardiomyopathy. Herein, we identify a structured domain in myosin VI, MyUb (Myosin VI Ubiquitin-binding domain), that binds to ubiquitin chains, especially those linked via K63, K11, and K29. We solve the solution structure of MyUb, and of MyUb:K63-linked diubiquitin. MyUb folds as a compact, helix-turn-helix-like motif and nestles between the ubiquitins of K63-linked diubiquitin, interacting with distinct surfaces of each. A nine amino acid extension at the C-terminal helix (Helix2) of MyUb is required for myosin VI interaction with endocytic and autophagic adaptors. Structure-guided mutations revealed that a functional MyUb is necessary for optineurin interaction. In addition, we found that an isoform-specific helix restricts MyUb binding to ubiquitin chains. This work provides fundamental insights into myosin VI interaction with ubiquitinated cargo and functional adaptors.
Graphical Abstract
INTRODUCTION
Myosins are a superfamily of quintessential molecular motors that power movements on actin filaments by converting ATP hydrolysis to mechanical energy and force. A highly conserved N-terminal motor domain undergoes conformational changes during the ATPase cycle that modulate actin affinity, as reviewed in (Geeves and Holmes, 1999). These motions are amplified into the myosin powerstroke by a variable calmodulin-binding lever arm causing nanometer-scale movement, reviewed in (Spudich and Sivaramakrishnan, 2010). With the exception of myosin VI, movement is toward the barbed (plus) end of actin filaments (Wells et al., 1999). Myosin VI contains an additional calmodulin-binding insertion that redirects the effective lever arm toward the pointed (minus) end of actin filaments (Menetrey et al., 2005). The C-terminal tail region is divergent among myosins and confers specificity for cargo and distinct interactions that define subcellular localization and specialized functions.
Humans express ~40 known or predicted myosins (Berg et al., 2001) that participate in diverse activities, including conventional skeletal myosin IIs for muscle contraction and unconventional myosins that function in intracellular trafficking, cell division and motility, actin cytoskeletal organization, and cell signaling, as reviewed in (Sellers, 2000). Myosin malfunction has been implicated in hypertrophic cardiomyopathy (Mohiddin et al., 2004), Usher syndrome (Hasson et al., 1995; Weil et al., 1995), deafness (Avraham et al., 1995; Gibson et al., 1995), Griscelli syndrome (Kumar et al., 2001; Takagishi and Murata, 2006), and cancer (Dunn et al., 2006; Yoshida et al., 2004); thus prompting the development of small molecule myosin inhibitors, as reviewed in (Bond et al., 2013).
The myosin VI cargo-binding tail (Figure 1A) interacts with multiple adaptor proteins, including regulators of clathrin-mediated endocytosis and autophagy, as reviewed in (Tumbarello et al., 2013). Some of these ligands require a myosin VI Arg-Arg-Leu (RRL) motif (Figure 1C), including nuclear dot protein 52 (NDP52) (Morriswood et al., 2007), Traf6-binding protein (T6BP) (Morriswood et al., 2007), optineurin (Sahlender et al., 2005), and GAIP-interacting protein C-terminus (GIPC) (Bunn et al., 1999; Spudich et al., 2007). Others engage a Trp-Trp-Tyr (WWY) triplet present in the cargo-binding domain (CBD, Figure 1A), including Tom1/Tom1L2 (Finan et al., 2011; Tumbarello et al., 2012), Dab2 (Inoue et al., 2002; Morris et al., 2002; Spudich et al., 2007), and lemur tyrosine kinase-2 (LMK2) (Chibalina et al., 2007).
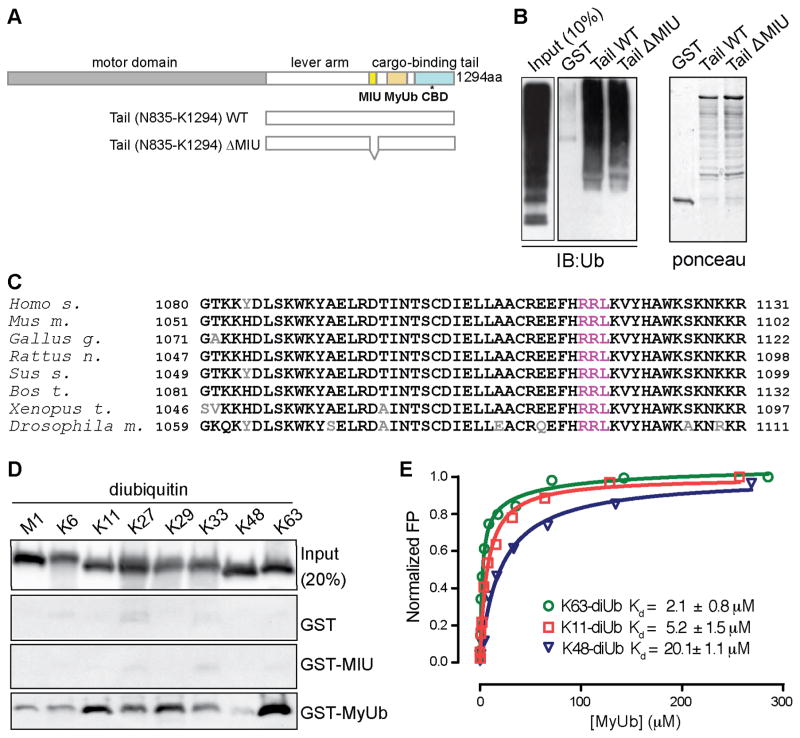
Figure 1. Identification and characterization of the MyUb domain.
(A) Domain architecture of myosin VI highlighting the motor domain (grey), lever arm (white), MIU (yellow), MyUb (beige), and cargo-binding domain (CBD, light blue). The WWY motif is in the CBD and noted with an asterisk. aa, amino acid. Depicted below are the tail constructs used for the experiment described in (B).
(B) GST-tagged myosin VI tail (N835-K1294) wild type and mutant carrying an internal deletion of the MIU domain (S997-I1024) were used to pull-down polyubiquitinated proteins from HEK293T cellular lysates. Immunoblotting (IB) as indicated. Right panel, ponceau showing equal loading of the GST proteins.
(C) Sequence alignment of MyUb from Homo sapiens, Mus musculus, Gallus gallus, Rattus vorvegicus, Sus scrofa, Bos Taurus, Xenopus tropicalis, and Drosophila melanogaster with conserved and non-conserved amino acids in black and grey, respectively. The RRL motif is highlighted in magenta.
(D) GST pull-down assay with indicated myosin VI constructs or GST as a control. GST-fusion proteins were incubated with purchased diubiquitins linked by M1, K6, K11, K27, K29, K33, K48, or K63 (Boston Biochem, Inc.) and analyzed by immunoblotting with anti-ubiquitin antibody.
(E) Fluorescence polarization (FP) assays to determine MyUb binding affinities for K63-, K48- and K11-linked diubiquitin. Results are representative of at least three independent experiments. Dissociation constants with their respective SD are reported.
See also Figure S1.
We previously reported the existence of a Motif Interacting with Ubiquitin(MIU) domain C-terminal to the myosin VI coiled-coil region (Penengo et al., 2006) (Figure 1A, in yellow). In this manuscript, we identify a second ubiquitin-binding domain in myosin VI - which we name MyUb (Myosin VI Ubiquitin-binding domain) - that contains the RRL motif. We use NMR spectroscopy to find that MyUb adopts a compact protein fold that is required for ubiquitin-binding and disrupted by amino acid substitutions in the RRL motif. We evaluate MyUb in the context of myosin VI binding to autophagy adaptor optineurin and the distinct myosin VI isoforms expressed in humans.
RESULTS
Identification of a ubiquitin-binding domain in myosin VI: the MyUb
In Rabex-5, the MIU domain binds to ubiquitinated EGFR and promotes coupled monoubiquitination (Penengo et al., 2006). To obtain insight into the function of the MIU (Q998-S1025) in the context of myosin VI (Figure 1A), we analyzed the myosin VI tail spanning N835-K1294, which recapitulates myosin VI interaction and localization (Buss et al., 2001). Surprisingly, deletion of the MIU domain (S997-I1024) did not abrogate myosin VI tail binding to ubiquitinated species from cellular lysates (Figure 1B). Deletion analysis led to the identification of a second ubiquitin-binding domain (UBD) C-terminal to the identified MIU (Figure 1A), which we narrowed down to a 43-amino acid fragment (G1080-H1122 and Figures S1.A–S1C). This region is highly conserved in myosin VI from various species (Figure 1C), and appears to be unique to myosin VI, as bioinformatics analysis was unable to detect its presence in other genes (Kay Hoffman, personal communication). Since this region lacks sequence similarity with any previously described UBD, we henceforth refer to it as MyUb.
We used diubiquitin molecules made with the eight possible linkages (M1, K6, K11, K27, K29, K33, K48, K63) to test whether MyUb or MIU exhibit preference for a specific chain type. GST-MIU binding to diubiquitin was barely detectable by this method (Figure 1D); on the contrary, GST-MyUb bound robustly to ubiquitin chains, with preference for K63-, K11-, and K29-linked diubiquitin (Figure 1D). Weak interaction was detected with K48-linked diubiquitin (Figure 1D), a linkage type associated with proteasomal degradation (Ehlinger and Walters, 2013). This difference in affinity was confirmed by fluorescence polarization (FP) assays. MyUb bound to K11-, K48-, and K63-linked diubiquitin with low micromolar affinity, with the K63- and K11-linked diubiquitin exhibiting 10- and 4-fold greater affinity compared to K48-linked diubiquitin, respectively (Figure 1E).
Solution structure of MyUb reveals a compact fold that is critical for binding to ubiquitin and adaptors for endocytosis and autophagy
We used NMR techniques (described in Experimental Procedures) to solve the MyUb (G1080-H1122) structure. The twenty lowest energy structures calculated from 100 extended starting ones converged to fit recorded NMR data (Table S1), with a backbone root mean square deviation (RMSD) of 0.20 Å (Figure S2A). From these data we concluded that MyUb adopts a compact helix-turn-helix-like fold with two helices spanning Y1091 to T1100 (Helix1) and I1104 to K1119 (Helix2) (Figure 2A). Importantly, the structural fold is stabilized by numerous interactions with an N-terminal region (Figure 2B). Y1084 and L1086 from the N-terminal region pack against hydrophobic amino acids (L1094 and L1106) from Helix 2 while W1089 is partially buried by L1094 and L1086 (Figure 2B, pink). Alanine substitution of L1086 or L1106 causes misfolding of the MyUb domain (Figures S2B and S2C) and significantly decreased binding to K63-linked ubiquitin chains (Figure 2C). A W1089A mutation was instead tolerated, as it did not fully disrupt the MyUb structure (Figure S2D) and did not prevent MyUb binding to K63-linked diubiquitin (Figures 2C and S2E). Changing the temperature and salt conditions did not alter MyUb structure (Figures S2F–S2H) or interaction with K63-linked diubiquitin (Figure S2I).
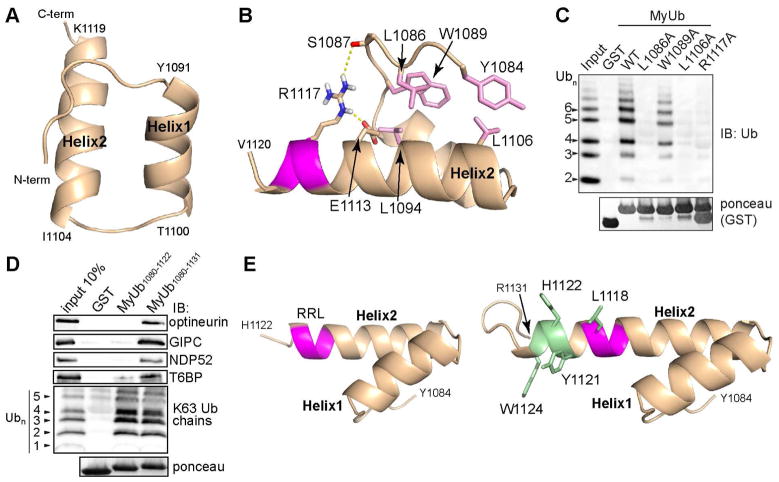
Figure 2. Myosin VI contains a compact ubiquitin-binding domain.
(A, B) Ribbon representation of MyUb (G1080-H1122) highlighting the helix boundaries and the interactions that stabilize the fold, including hydrogen bonds from R1117 to S1087 and E1113 (dashed yellow lines) and van der Waals interactions engaging Y1084, L1086, W1089, L1094, and L1106 (pink).
(C) GST pull-down assay performed with GST-MyUb (G1080-R1131) wild type (WT) or amino acid substituted-protein as indicated. GST fusion proteins were incubated for 2 hours at 4°C with K63-linked polyUb1-7 and analyzed by immunoblotting (IB) with anti-ubiquitin antibody. Ponceau staining shows comparable loading of GST-tagged proteins (bottom panel). GST was used as a negative control.
(D) GST-MyUb (G1080-H1122), GST-MyUb (G1080-R1131), or GST (as a negative control) were incubated with synthetic K63-linked polyubiquitin1-7 or cellular lysates from HEK293T cells transfected with GFP-optineurin, FLAG-T6BP, His-GIPC, or FLAG-NDP52. Immunoblotting (IB) was performed using the specific anti-TAG antibodies. Ponceau staining shows comparable loading of GST-tagged proteins (bottom panel).
(E) Ribbon representations of MyUb (G1080-H1122) (left) and MyUb (G1080-R1131) with extended hydrophobic surface highlighted in green formed by addition of A1123-R1131 (right) were shown. The RRL motif was colored magenta.
See also Figure S2.
Several myosin VI adaptor proteins, namely optineurin, GIPC, T6BP and NDP52, were shown to require an intact 1116RRL1118 motif for interaction with myosin VI (Bunn et al., 1999; Morriswood et al., 2007; Sahlender et al., 2005; Spudich et al., 2007). In particular, amino acid substitution of this motif with the alanine triple AAA was reported to abolish myosin VI interaction with these autophagy adaptor proteins (Morriswood et al., 2007). The RRL motif resides in Helix2 where R1117 forms hydrogen bonds to S1087 and E1113 (Figure 2B), suggesting that it is critical for MyUb (G1080-H1122) structural integrity. As expected, replacement of R1117 with alanine resulted in misfolding, as measured by a 2D NMR experiment (Figure S2J, red compared to black) and abolished interaction with ubiquitin (Figure 2C).
This result prompted us to re-examine the interaction of myosin VI with previously characterized autophagy adaptor proteins using various RRL-containing fragments. The minimal binding region was found to be a MyUb construct spanning G1080-R1131 (Figure 2D). The shorter construct spanning G1080-H1122 was unable to bind optineurin, GIPC, T6BP and NDP52, even though it was competent for binding to ubiquitin (Figures 2D and S2K) and adopted a stable protein fold (Figure 2A). Thus, the interaction surface between myosin VI and its known partners extends beyond the previously identified 1116RRL1118 motif towards the C-terminal part of Helix2 of the MyUb domain. We next used NMR to solve the structure of this longer construct (Figure S2L; Table S1). The backbone RMSD of the extended MyUb (G1080-R1131) to MyUb (G1080-H1122) was 0.46 Å for the overlapping region. Helix2 was extended however by six amino acids forming an additional 1.5 helical turns, with an expanded hydrophobic surface contributed by Y1121, H1122 and W1124 (Figure 2E, right). We speculate that this extended hydrophobic surface is involved in myosin VI binding to autophagy adaptor proteins.
MyUb nestles between ubiquitins of K63-linked diubiquitin
The MyUb bound well to K11-, K29, and K63-linked ubiquitin chains (Figure 1D). We therefore used K63-linked diubiquitin as a model to dissect the molecular mechanism of MyUb binding to ubiquitin chains. Initially, we determined the binding stoichiometry of the MyUb:K63-linked ubiquitin chain complex by using size exclusion chromatography. 2- or 4-fold molar excess MyUb incubated with 100 μM diubiquitin or triubiquitin was loaded on a size exclusion column (Figures 3A and 3B). At 2-fold molar excess, MyUb and K63-linked diubiquitin co-eluted at the expected molecular weight for a 1:1 complex with the excess MyUb eluting separately (Figure 3A, blue). By contrast, very little MyUb eluted separately from K63-linked triubiquitin (Figure 3B, blue) and the presence of free MyUb in the mixture with triubiquitin increased significantly when MyUb was at 4-fold molar excess (Figure 3B, purple). Altogether, these data indicate a 1:1 stoichiometry for the MyUb:diubiquitin complex and a 2:1 stoichiometry for the MyUb:triubiquitin complex.
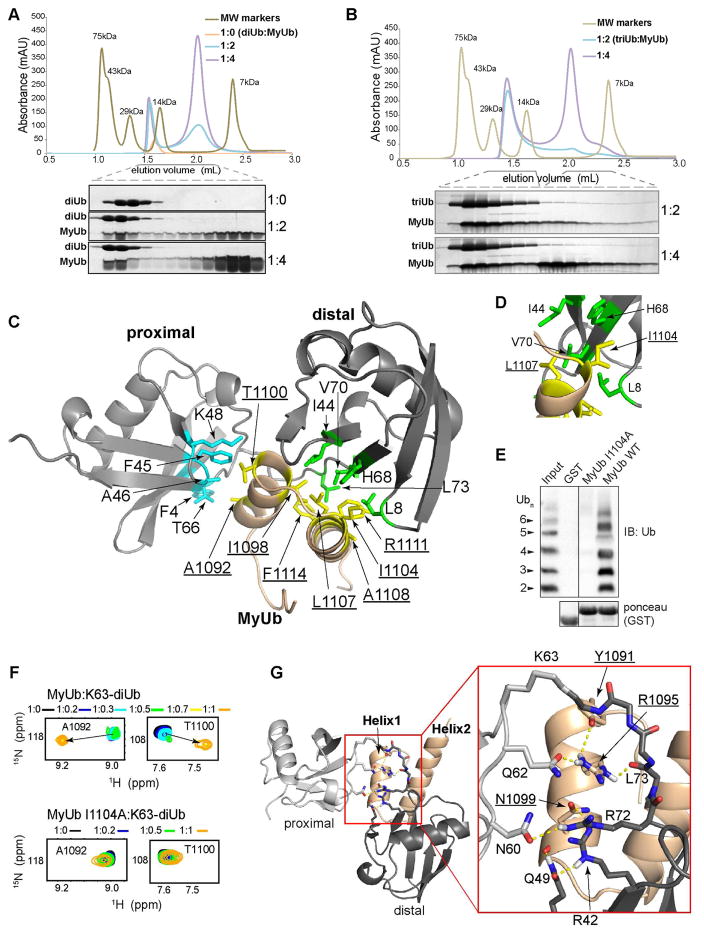
Figure 3. MyUb nestles between the moieties of K63-linked diubiquitin.
(A, B) Stoichiometry of the complex formed by MyUb and K63-linked (A) diubiquitin or (B) triubiquitin, as indicated. 100 μM of purified K63-linked diubiquitin or K63-linked triubiquitin were incubated (1 hr, 20°C) with either 200 or 400 μM MyUb, followed by fractionation on a Superdex75 column. Selected fractions were separated by SDS-PAGE gel and stained with Coomassie.
(C) Ribbon diagram of MyUb (G1080-H1122):K63-linked diubiquitin with MyUb (beige) nestled between the proximal ubiquitin (light grey) and distal ubiquitin (dark grey) with interactions to a I44-centered surface on distal ubiquitin (green) and a F45-centered surface of proximal ubiquitin (blue). Interacting MyUb amino acids are displayed in yellow. Labels for MyUb amino acids are underlined.
(D) Expanded view of (C) showing critical interactions of I1104 from MyUb with L8 and H68 of the distal K63-linked diubiquitin.
(E) GST pull-down assay performed with GST-MyUb (G1080-R1131) wild type (WT) or I1104A. GST fusion proteins were incubated for 2 hours at 4°C with K63-linked polyUb1-7 and analyzed by immunoblotting (IB) with anti-ubiquitin antibody. Ponceau staining shows comparable loading of GST-tagged proteins (bottom panel). GST was used as a negative control.
(F) Selected regions that include A1092 and T1100 signals from 1H-15N HSQC spectra acquired on 15N-labeled MyUb wild type (top) or MyUb I1104A (bottom) with K63-linked diubiquitin at the indicated molar ratio. Arrows point to new peak positions caused by addition of diubiquitin. All spectra were recorded at 850 MHz on MyUb sample concentrations of 0.1 mM.
(G) Ribbon diagrams of the MyUb:K63-linked diubiquitin complex showing the opposite surface compared to (C), rotated 145 degrees along the x-axis, to highlight hydrogen bond (yellow dashed line) interactions involving MyUb and K63-linked diubiquitin. An expanded region (boxed in red) is displayed to the right.
See also Figure S3.
To explore the molecular basis of MyUb binding to ubiquitin, we used NMR to solve the structure of MyUb (G1080-H1122) in complex with K63-linked diubiquitin. The MyUb structure was unchanged upon ubiquitin binding, as demonstrated by an almost identical interaction network for free and K63-linked diubiquitin-bound MyUb (Figure S3A). We isotopically labeled either the proximal (defined by a free G76 that can be in principle conjugated to a substrate protein) or the distal ubiquitin of K63-linked diubiquitin and used 13C half-filtered NOESY experiments to detect interactions between MyUb and each ubiquitin (Figures S3B–S3D). We were able to assign 59 intermolecular interactions between MyUb and the proximal ubiquitin and 47 intermolecular interactions between MyUb and the distal ubiquitin; these were used to solve the structure of the MyUb:K63-linked diubiquitin complex (Figures S3E and S3F; Table S1). We found that MyUb nestles between the two ubiquitins, making extensive contacts to both ubiquitin moieties (Figure 3C).
On the distal ubiquitin, an exposed MyUb hydrophobic surface formed by the two helices contacts the classic hydrophobic patch centered on I44 (Liu and Walters, 2010) (Figure 3C, green). This contact surface includes I1098 from Helix1 and I1104, L1107, A1108, R1111 and F1114 from Helix2 (Figure 3C, yellow). In particular, I1104 forms critical interactions with L8 and H68 of distal ubiquitin (Figure 3D). Its replacement with alanine does not affect the overall MyUb fold (Figure S3G), but abolishes binding to K63-linked diubiquitin, as shown in a pull-down experiment (Figure 3E) and by NMR titration experiments (Figure 3F). In the slow exchange regime on the NMR time scale (Walters et al., 2001), amino acids A1092 and T1100 shift to a new position upon K63-linked diubiquitin addition (Figure 3F, top panels). This effect is lost in the MyUb I1104A mutant (Figure 3F, bottom panels). These data were confirmed by FP analysis, which demonstrated a Kd > 400 μM for the I1104A mutant (Figure S3H).
On the proximal ubiquitin, aforementioned A1092 and T1100 from Helix1 of MyUb contact a surface formed by F4, F45, A46, K48, and T66 (Figure 3C, blue). Likely, the involvement of K48 at this location explains the relatively poor affinity of the MyUb for this chain type (Figures 1D and 1E). Some NOEs were assigned to the L8-I44-V70 hydrophobic patch of the proximal ubiquitin, indicating that MyUb can form a lower affinity interaction with this region (Figures S3C, S3F orange, and S3I). However, binding to this second, low affinity site was not retained in the size exclusion chromatography experiment (Figure 3A), indicating that this interaction is weak and possibly not present in the context of the full-length protein.
MyUb interaction with K63-linked diubiquitin is stabilized by electrostatic contacts that surround the ubiquitin isopeptide region (Figure 3G). MyUb R1095 forms hydrogen bonds with distal ubiquitin L73 and proximal ubiquitin Q62. Distal ubiquitin R42 and Q49 are spatially close to MyUb N1099 and form hydrogen bonds with its side chain and backbone, respectively.
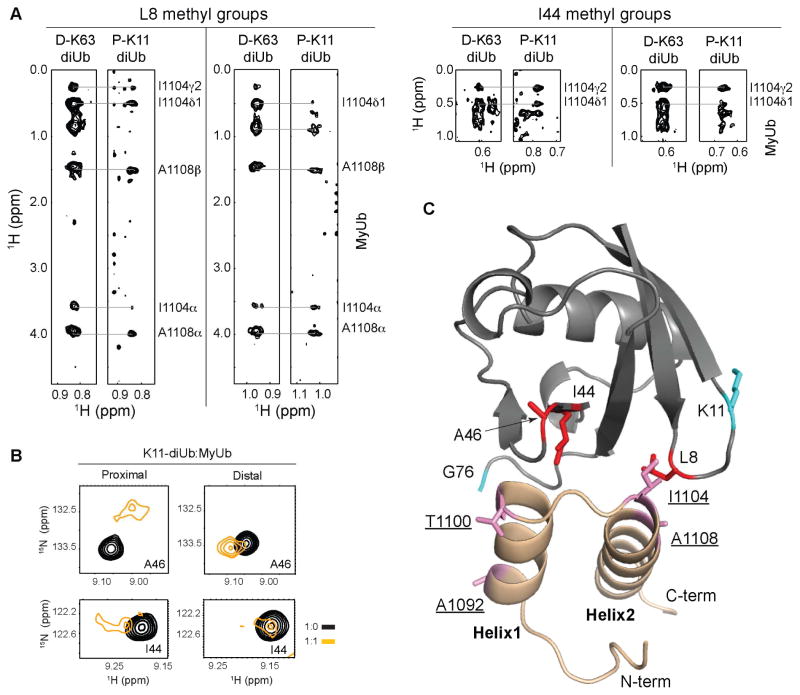
We also explored how MyUb binds to K11-linked diubiquitin taking advantage of K11-linked diubiquitin in which either the proximal or distal ubiquitin was 13C and 15N labeled. We found in a 13C half-filtered NOESY experiment that L8 and I44 from the proximal K11-linked ubiquitin formed similar contacts to MyUb as was observed for the distal K63-linked ubiquitin component (Figure 4A). For example, L8 and I44 from the proximal ubiquitin of K11-linked diubiquitin were similarly shown to directly interact with I1104 from MyUb (Figure 4A). No such intermolecular NOE interactions were observed for L8 or I44 from the distal ubiquitin of K11-linked diubiquitin (data not shown). Furthermore, the I44 and A46 amide signals from the proximal, but not distal ubiquitin, demonstrated significant shifting following addition of equimolar MyUb (Figure 4B, orange versus black). Taken together, our data suggest that MyUb prefers the hydrophobic patch of proximal ubiquitin to distal ubiquitin in the context of K11-linked diubiquitin, and that the proximal ubiquitin hydrophobic patch of K11-linked diubiquitin binds to MyUb in a similar mode as that observed for distal ubiquitin of K63-linked diubiquitin (Figures 4C and 3C). Further studies are needed to solve the structure of this MyUb complex, but we concluded that MyUb binds to K11-linked chains in a distinct manner that involves contacts between MyUb I1104 and the L8 and I44 methyl groups (Figures 4A and 4C).
Figure 4. MyUb binds to K11-linked diubiquitin through the hydrophobic patch of the proximal ubiquitin moiety.
(A) Selected regions showing methyl groups from L8 and I44 from a 3D 13C half-filtered NOESY experiment acquired on ~0.5 mM unlabeled MyUb (G1080-H1122) mixed with equimolar K63- (left) or K11-linked (right) diubiquitin with the proximal or distal ubiquitin moiety 15N and 13C labeled, respectively. NOEs assigned uniquely to MyUb atoms are labeled. D-K63, K63-linked diubiquitin with the distal moiety 15N and 13C labeled; P-K11, K11-linked diubiquitin with the proximal moiety 15N and 13C labeled.
(B) Selected regions that include A46 and I44 amide signals from 1H-15N HSQC spectra acquired on 13C, 15N-labeled proximal (left) or distal (right) K11-linked diubiquitin (black) and with equimolar unlabeled MyUb (orange).
(C) Model structure of MyUb bound to a signal ubiquitin based on Figure 3C, illustrating the amino acids involved in complex formation as revealed in (A–B).
The ubiquitin-binding ability of MyUb contributes to optineurin interaction
Optineurin is a well-characterized myosin VI interactor. Previous work has suggested a direct interaction mediated by the RRL motif in myosin VI and the UBDs of optineurin (Sahlender et al., 2005; Shen et al., 2015). Our results suggest that ubiquitin may be part of the myosin VI interaction with this adaptor protein.
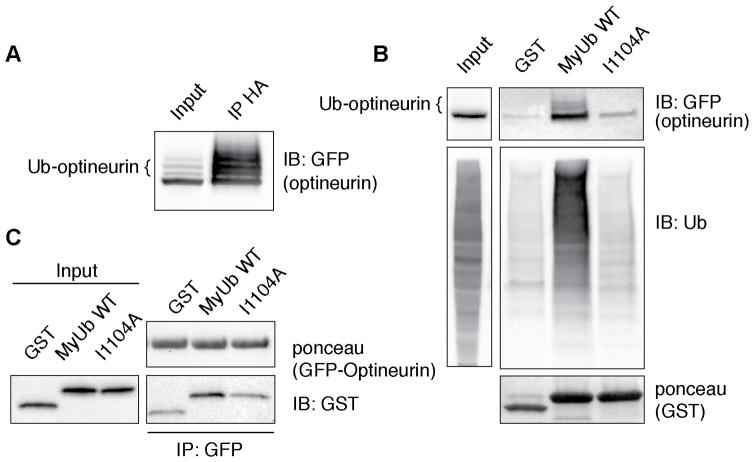
To investigate this issue, we first analyzed whether optineurin undergoes ubiquitination. HEK293T cells were transfected with GFP-optineurin and HA-ubiquitin and the cell lysate subjected to immunoprecipitation using anti-HA antibody. As visible in Figure 5A, optineurin is poly- or multi-ubiquitinated under these conditions. The lysate was next used for a pull-down assay in which we compared MyUb wild-type (WT) with the ubiquitin binding-impaired I1104A mutant (Figure 3E). Wild-type MyUb bound strongly to the ubiquitinated form of optineurin while the I1104A mutant retained only a basal interaction with this autophagic adaptor (Figure 5B). Notably, the I1104A mutant also showed a clear impairment for ubiquitin binding in vivo where ubiquitinated proteins were robustly immunoprecipitated with the WT protein (Figure 5B). Similar results were obtained incubating immunoprecipitated GFP-optineurin with GST proteins eluted from the beads (Figure 5C). Altogether, these results indicate that the ubiquitin-binding surface of the MyUb participates in binding to this RRL interactor, either by direct binding or by binding to ubiquitin that is conjugated to optineurin.
Figure 5. Ubiquitin binding by MyUb contributes to myosin VI;optineurin interaction.
(A) Lysate from HEK293T cells expressing GFP-optineurin and HA-ubiquitin was immunoprecipitated and immunoblotted as indicated.
(B) 2 μM of the indicated GST-fusion proteins were incubated with the same cellular lysate (2 mg) shown in (A). Immunoblotting was performed using anti-GFP or anti-ubiquitin antibodies. Ponceau staining shows comparable loading of GST-tagged proteins (bottom panel).
(C) GFP-Trap beads were used to purify GFP-optineurin from cellular lysates of HEK293T transfected cells. Washed beads were incubated with the indicated eluted GST proteins and immunoblotting was performed using anti-GST. Ponceau staining shows comparable loading of GFP-optineurin (upper panel).
MIU exists within a long α-helix that kinks into a shorter linker helix
We have previously identified a MIU domain that is C-terminal to the myosin VI coiled-coil region (Penengo et al., 2006) (Figure 6A). We tested whether we could detect binding between a peptide that encompasses the myosin VI MIU (spanning Q998-S1025) and ubiquitin by NMR. Unlabeled ubiquitin was added to 15N labeled MIU and the effects recorded (Figure 6B). Shifting and disappearance of MIU signals was observed with ubiquitin addition (Figures 6B and S4A), including signals from A1013 and L1014, which were predicted to be at the center of the MIU ubiquitin-binding surface (Penengo et al., 2006). By contrast, unlabeled MyUb did not affect NMR spectra recorded on 15N MIU (Figure S4B), indicating that these two structural elements do not interact. In support of this finding, the MIU is not significantly different in spectra recorded on the MIU by itself (Q998-S1025) compared to in the context of the MIU-MyUb region (Q998-R1131, Figure S4C), similarly demonstrating that it does not interact with these other regions of the protein.
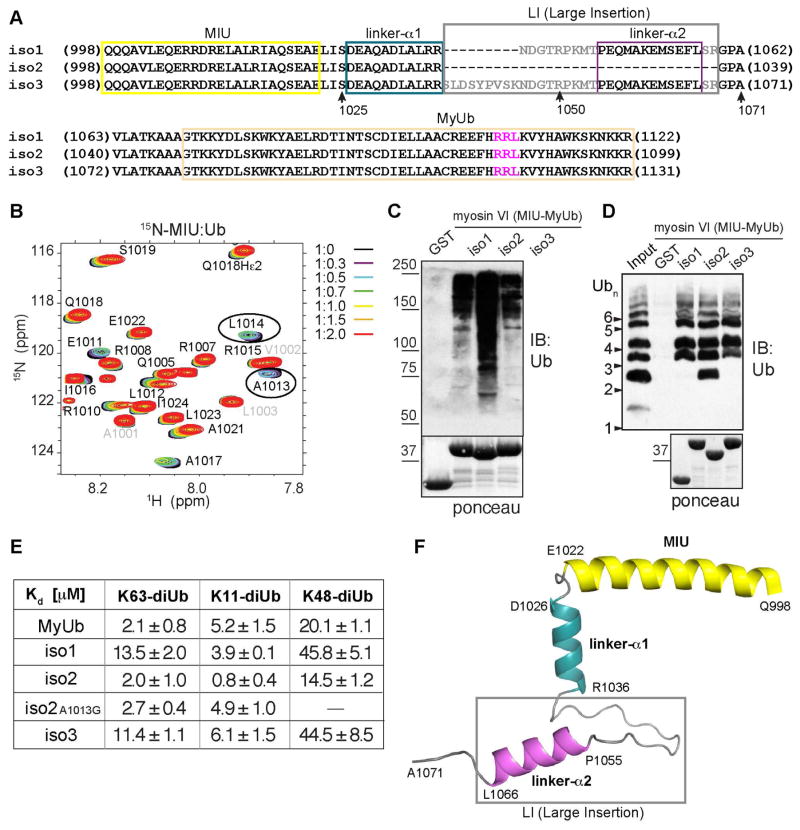
Figure 6. The presence of an isoform-specific helix in the MIU-MyUb region of myosin VI affects its affinity for ubiquitin chains.
(A) Sequence alignment of myosin VI isoforms spanning MIU (yellow box) to MyUb (beige box). Linker-α1 (boxed in blue) and linker-α2 (boxed in purple) are indicated. The RRL motif is highlighted in magenta. Amino acids from the LI region are boxed in gray.
(B) Expanded region of 1H-15N HSQC spectra recorded on 0.1 mM 15N MIU (Q998-S1025) with unlabeled monoubiquitin added at the indicated molar ratio. This dataset was acquired at 700 MHz.
(C) GST-tagged myosin VI MIU-MyUb constructs from the different isoforms were used to pull-down polyubiquitinated proteins from HEK293T cellular lysates (1 μg). GST was used as a control, immunoblotting (IB) was done with anti-ubiquitin antibody, and ponceau staining is included below to indicate loading of GST-tagged proteins.
(D) GST pull-down assay for 2 μM indicated myosin VI GST-MIU-MyUb or GST (as a control) with 2 hour incubation with K63-linked ubiquitin chains at 4°C. Analysis was done by immunoblotting (IB) with anti-ubiquitin antibody. All six isoforms bind longer chains, but only iso2 MIU-MyUb retains binding to K63-linked diubiquitin. Bottom panel, ponceau staining showing equal loading of the GST proteins.
(E) Binding affinities (± SD) determined by FP between K63-, K11-, and K48-linked diubiquitin and the indicated myosin VI constructs.
(F) Snapshot ribbon representation of the solution structure of iso3 Q998-A1071. MIU is part of a 6.5-turn helix (yellow), which is followed by a 3-residue kink and 2.5-turn helix (linker-α1, blue). A long, flexible linker region separates linker-α1 from isoform-specific linker-α2 (pink) and the orientation of these two helices is not defined. Amino acids from the LI region are boxed in gray.
See also Figure S4.
Myosin VI is generated by alternatively spliced isoforms, which are differentially expressed in tissues and cell lines, and associated with specific subcellular compartments (Au et al., 2007; Buss et al., 2001; Tomatis et al., 2013). These isoforms differ by insertions in the cargo-binding tail. Notably, the large insert (LI) (Buss et al., 2001) separates the two myosin VI UBDs, the MIU (Penengo et al., 2006) and the MyUb (Figure 6A). We first tested whether the presence of the LI variable exons (as expressed in iso1 and iso3) has an impact on MyUb binding to ubiquitin by using pull-down assays. Compared to iso2 MIU-MyUb, iso1/3 MIU-MyUb exhibited reduced overall binding to ubiquitinated proteins from cellular lysates (Figure 6C). Moreover, the iso2 MIU-MyUb fragment that lacks the LI bound short K63-linked ubiquitin chains more robustly (Figure 6D). FP analysis with K63-linked diubiquitin corroborated this data, indicating iso1 and iso3 MIU-MyUb to have a 4- to 5-fold reduction in ubiquitin-binding strength compared to iso2 MIU-MyUb (Figures 6E and S4D–S4F). Thus, the presence of the LI led to reduced affinity of the myosin VI ubiquitin-binding region for K63-linked ubiquitin chains.
We also tested whether the MIU-MyUb region from iso1/3 and iso2 exhibit differences in affinity for K11- and K48-linked diubiquitin by using FP. We similarly found higher affinity for iso2 compared to iso1/3, with ~5- and ~3-fold greater affinity for K11- and K48-linked chains, respectively (Figure 6E). Interestingly, binding to K63- and K48-linked diubiquitin is similar between MyUb and iso2, whereas K11-linked diubiquitin binds with a greater affinity to iso2 compared to MyUb (Figures 6E and S4E), indicating a possible contribution of the MIU domain in myosin VI for K11-linked ubiquitin chains. To test the contribution of the MIU region further, we analyzed by FP the behavior of a myosin VI iso2 construct in which the alanine at position 1013 was mutated to glycine to abrogate MIU binding (Penengo et al., 2006). The results validated the hypothesis, as the MIU mutation did not change the iso2 affinity for K63-linked diubiquitin, but reduced affinity towards K11-linked diubiquitin 6-fold (Figures 6E and S4G).
In an effort to understand why longer isoforms (iso1/3) exhibit reduced affinity for ubiquitin chains compared to the isoform with no insert (iso2), we produced a fragment that extends from the MIU region to just before the MyUb, spanning Q998-A1071 of the longer myosin VI isoform (iso3, Figure 6A). This region includes the LI, which is deleted in the short myosin VI isoform (iso2, Figure 6A). By using NMR techniques, we solved the 3-dimensional structure of the iso3 Q998-A1071 fragment, which contains the MIU domain and the LI (a snapshot is provided in Figure 6F). This analysis revealed the presence of three helices, including the anticipated MIU (Q998-E1022), a common linker helix (linker-α1, D1026-R1036) and an isoform-specific helix (linker-α2, P1055-L1066). A 3-residue kink follows the MIU while an 18-amino acid flexible region separates linker-α1 and linker-α2 (Figure 6F). Long-range NOE interactions between the three helices were not observed (data not shown). The relative configuration of MIU and linker-α1 is constrained by the short intervening region (Figures S4H and S4I), whereas linker-α1 and linker-α2 are widely distributed due to their long, flexible loop sequence (Figures S4I and S4J).
Based on these results, we hypothesized that linker-α2 is sufficient to limit MyUb:ubiquitin interaction. To test this idea, we evaluated binding to K63-linked diubiquitin of various MyUb constructs. As previously shown by a GST pull-down assay (Figure 2D), G1080-R1131 and G1080-H1122 constructs demonstrated an almost identical affinity for K63 diubiquitin (Figure S2K). By contrast, myosin VI 1050-1131, which includes linker-α2, exhibited a 4-fold reduction in binding affinity compared to the MyUb domain alone (Figure S2K). Altogether, these data led us to conclude that the isoform-specific helix linker-α2 restricts MyUb binding to ubiquitin
DISCUSSION
Myosin VI as a ubiquitin receptor
We describe here a previously unanticipated structural domain in myosin VI with a helix-turn-helix-like configuration that is stabilized by a folded back N-terminal loop. This small domain, which we name MyUb, binds to ubiquitin chains, with preference for K63, K11, and K29 linkages (Figure 1D). We find that MyUb nestles between K63-linked ubiquitins and that K48 is part of a unique binding surface on the proximal ubiquitin (Figure 3C). This recognition mode provides a rationale for MyUb binding to K48-linked diubiquitin with 10-fold lower affinity compared to K63-linked diubiquitin (Figure 1E). In binding to K11-linked diubiquitin, MyUb engages the classic hydrophobic patch on the proximal ubiquitin moiety in a manner akin to its interaction with the distal ubiquitin of K63-linked chains (Figure 4). Future experiments are needed to determine how MyUb recognizes K11- and K29-linked ubiquitin chains; however, we provide evidence that the MIU plays a role in binding to K11-linked ubiquitin chains that is not recapitulated for K63-linked ubiquitin chains (Figures 6E, S4E, and S4G).
The demonstrated binding with K63-linked ubiquitin chains is consistent with the well-established role played by myosin VI in intracellular transport of endocytic vesicles (Buss et al., 2004; Tumbarello et al., 2013). Indeed, K63-linked ubiquitin chains function in cell signaling and membrane trafficking events (Acconcia et al., 2009). K63- and K11-linked ubiquitin chains may also co-exist in ubiquitinated substrates, such as in the case of the major histocompatibility complex class I (MHC I) where these signals have been found to be required for efficient epsin1-dependent internalization (Boname et al., 2010; Goto et al., 2010). In this context, ubiquitin binding could contribute to the association of myosin VI with known interactors, adding an additional interaction layer of regulation. As a proof of principle, we tested optineurin and our experiment appears to support this idea (Figure 5).
As a K11-linked ubiquitin receptor, myosin VI remains to be explored. During cell cycle progression, the anaphase-promoting complex/cyclosome (APC/C) modifies substrates with ubiquitin chains linked by multiple lysines (heterotypic chains) that contain stretches of K11 linkages, allowing for their expedient degradation by the proteasome (Meyer and Rape, 2014). A recent study suggests that the need for heterotypic chains may be derived from poor binding to mammalian proteasome of ubiquitin chains that are linked exclusively by K11 (Grice et al., 2015). Notably, a few reports suggest a nuclear function for myosin VI (Vreugde et al., 2006; Zorca et al., 2015). In this subcellular compartment, myosin VI may interact with proteins modified by K11-linked chains to regulate transcription and viral infection.
MyUb binding to K63-linked ubiquitin chains
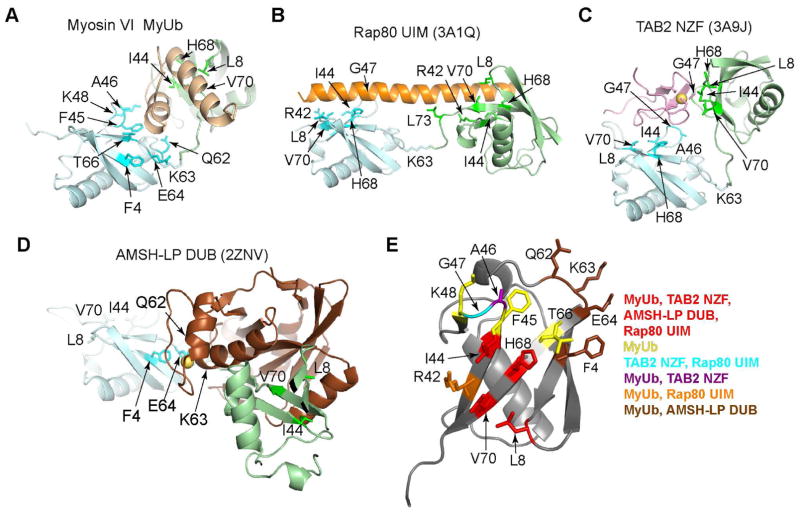
Our structural results indicate that the MyUb nestles between the neighboring moieties of a K63-linked diubiquitin, making extensive contacts around the ubiquitin ligation site (Figures 3C, 3G and 7A). We compared our MyUb:K63-linked diubiquitin structure to other solved structures of ubiquitin receptors in complex with K63-linked ubiquitin chains. All of them make direct interaction with ubiquitin at position L8, I44, H68, and V70 (Figures 7B–7D). Rap80 uses two UIMs that are registered in a contiguous helix to bind neighboring ubiquitins of K63-linked ubiquitin chains (Sims and Cohen, 2009) (Figure 7B). A small compact TAB2 NZF domain uses two distinct surfaces to bind the I44-centered surface of both ubiquitins in K63-linked ubiquitin chains (Kulathu et al., 2009; Sato et al., 2009) (Figure 7C) in a binding mode similar to hHR23A UBA2 domain binding to K48-linked ubiquitin chains (Varadan et al., 2005). Deubiquitinase AMSH-LP forms direct interactions with the K63 linker region to achieve linkage specificity engaging K63 and surrounding Q62, E64 and F4 (Sato et al., 2008) (Figure 7D). In contrast to the above-mentioned domains, MyUb binds to K63-linked ubiquitin chains by engaging the canonical hydrophobic surface on distal ubiquitin and a unique surface on proximal ubiquitin that includes F45, K48, N60, Q62, and T66 (Figures 7A and 7D).
Figure 7. MyUb interaction with K63-linked ubiquitin chains is unique.
(A–D) Structures of (A) myosin VI MyUb: K63-linked diubiquitin, (B) Rap80 UIM: K63-linked diubiquitin (3A1Q), (C) TAB2 NZF: K63-linked diubiquitin (3A9J), and (D) AMSH-LP DUB: K63-linked diubiquitin (2ZNV) displayed with their proximal ubiquitin in the same orientation. The proximal ubiquitin is depicted in light blue and the distal ubiquitin in light green. Amino acids in proximal and distal ubiquitin at the contact surface are displayed in blue and green, respectively, and labeled.
(E) Structure of monomeric ubiquitin highlighting the amino acids involved in binding to K63-linked chain specific receptors, as assessed by coordinates deposited in the Protein Data Bank (3A1Q, 3A9J, 2ZNV) and our structure (A). A legend is included to the right illustrating the color coding with corresponding amino acids on monoubiquitin for the listed binding domains. Amino acids in yellow are unique to MyUb binding.
Finally, it is important to appraise the MyUb in the context of the myosin VI protein. Our study established that the binding of MyUb to ubiquitin is regulated by two other nearby structural elements. The first one is the MIU, which shows limited ubiquitin-binding ability by itself (Figures 1E and 6B), and does not appear to contribute to the overall binding of myosin VI to K63-linked diubiquitin, but does participate in its binding to K11-linked diubiquitin (Figure 6E). These data suggest that myosin VI domains evolved in order to recognize and accommodate specific diubiquitin signals. The second element of regulation is the isoform-specific helix linker-α2, which is capable of modulating myosin VI interaction with ubiquitin, as iso1 and iso3 showed reduced binding compared to iso2 for all diubiquitin chain types tested (Figure 6E). This region could potentially regulate isoform-specific myosin VI functions. For a thorough understanding of these elements and their functions, additional structural and functional studies are needed.
Implications for RRL interactors
Embedded in the MyUb is the RRL motif previously identified as critical for interaction with endocytic and autophagic adaptors (Tumbarello et al., 2013). We demonstrate here that R1117 from this motif is crucial for MyUb structure integrity. Our findings underline the requirement of an extended hydrophobic surface on Helix2 of the MyUb (H1122-R1131) for binding to the functional adaptors optineurin, GIPC, T6BP and NDP52 (Figures 2D and 2E). In addition, we showed that optineurin may undergo ubiquitination and that I1104 on MyUb is critical for myosin VI binding to this autophagic adaptor (Figure 5). Although we cannot formally exclude the possibility that I1104 is part of the direct binding site, our data suggest that MyUb binding to ubiquitin on optineurin may regulate the interaction between myosin VI and optineurin in space and time, a hypothesis that is consistent with previous literature (Liu et al., 2014) This additional layer of regulation can be particularly relevant in the context of selective autophagy where both proteins were recently implicated (Tumbarello et al., 2015; Tumbarello et al., 2012).
Altogether, the fundamental insights we have generated within this study provide the basis for future studies that interrogate how myosin VI interacts with functional adaptors for downstream signaling.
EXPERIMENTAL PROCEDURES
Reagents, antibodies, protein expression and purification, and cell lines
Information regarding reagents, including the production of protein samples, antibodies, and cell culturing can be found in Supplementary Materials and Experimental Procedures. GFP-optineurin, FLAG-T6BP, His-GIPC, and FLAG-NDP52 were generously provided by Dr. Alain Israel, Dr. Folma Buss, Dr. Guido Serini, and Dr. Felix Randow, respectively. HA-ubiquitin was previously described (Polo et al., 2002).
Size exclusion chromatography
100 μM K63-linked diubiquitin or triubiquitin was incubated at different molar ratios with MyUb (G1080-R1131) for 1 hour at 20°C and subjected to size exclusion chromatography on a Superdex75 (5/150) column using the ÄKTA microsystem (GE Healthcare). Fractions containing complex or single proteins were analyzed by SDS-PAGE and Coomassie staining.
Fluorescence polarization assay
The thiol-reactive fluorescent probe BODIPY® TMR C5-maleimide (Invitrogen) was conjugated to diubiquitin G76C linked via K11, K48 or K63 (100 μM, in 20 mM Tris-HCl pH 7.6, 200 mM NaCl, 5% glycerol, 1 mM TCEP) following the manufacturer’s instructions. Briefly, a 20 mM stock solution of lyophilized dye suspended in DMSO was diluted into the protein solution at 10-fold molar excess and the conjugation reaction allowed to proceed for two hours, in the dark at 4°C. The reaction mixture was dialyzed against 20 mM Tris-HCl pH 7.6, 200 mM NaCl, 5% glycerol to remove excess dye, and then loaded on a Superdex75 10/300 column (GE Healthcare) to isolate labeled proteins, monitored by absorbance at 544 nm. Fluorescence polarization assays were performed at 22°C with λexcitation at 535 nm and λemission at 580 nm on a 384 well plate by using an Infinite 200 instrument (Tecan). Concentrations in the nanomolar range (20–50 nM) of labeled proteins were titrated with MyUb (280–300 μM). Polarization readings from three independent replicates were averaged and fitted as previously described (Eletr et al., 2005). Experiments were repeated at least three times and the reported Kd values represent the mean ± standard deviation (SD).
GST pull-down experiments
Detailed procedures used for GST pull-down experiments can be found in Supplementary Materials and Experimental Procedures.
NMR spectroscopy and structure determination
Detailed procedures for collecting and analyzing NMR data as well as for calculating, evaluating, and presenting the protein structures can be found in Supplementary Materials and Experimental Procedures.
Supplementary Material
Highlights.
Myosin VI contains a compact structural motif that binds ubiquitin chains
MyUb nestles between ubiquitins of K63-linked chains
Optineurin interaction requires an expanded MyUb and capacity to bind ubiquitin
An isoform-specific helix restricts MyUb binding to ubiquitin chains
Acknowledgments
This research was funded by a grant from the National Institutes of Health (CA136472 to KJW) and by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research to K.J.W and by the Association for International Cancer Research (AICR, grant 11-0051) and from Italian Ministry of Education, Universities and Research (PRIN 20108MXN2J) to S.P. We are grateful to Dr. Janusz Koscielniak for his technical assistance with the NMR facility.
Footnotes
Accession numbers
The structural coordinates and chemical shift data have been deposited in the Protein Data Bank (PDB) and Biological Magnetic Resonance Data Bank (BMRB) with respective accession codes 2N0Z and 25541 for MyUb (G1080-H1122), 2N10 and 25542 for MyUb (G1080-R1131), 2N11 and 25543 for iso3 Q998-A1071, 2N13 and 25545 for MyUb (G1080-H1122):K63-linked diubiquitin.
AUTHOR CONTRIBUTIONS
F.H. performed, analyzed, and interpreted all NMR experiments with the exception of backbone assignments for MyUb (G1080-R1131), which was done with assistance from A.E.; H-P.W. biochemically characterized the MyUb domain and its selective interaction with Ub and adaptor proteins with the help of M.B.; E.V. carried out all FP analysis; U.N. helped F.H. with NMR titration experiments and generating ubiquitin species for FP experiments; F.A. initially identified the MyUb domain; E.M. performed the experiment shown in Figure 5; S.P. and K.J.W. conceived of and directed the project; all authors contributed to the writing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Experimental cell research. 2009;315:1610–1618. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Au JS, Puri C, Ihrke G, Kendrick-Jones J, Buss F. Myosin VI is required for sorting of AP-1B-dependent cargo to the basolateral domain in polarized MDCK cells. The Journal of cell biology. 2007;177:103–114. doi: 10.1083/jcb.200608126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham KB, Hasson T, Steel KP, Kingsley DM, Russell LB, Mooseker MS, Copeland NG, Jenkins NA. The mouse Snell’s waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nature genetics. 1995;11:369–375. doi: 10.1038/ng1295-369. [DOI] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. A millennial myosin census. Molecular biology of the cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boname JM, Thomas M, Stagg HR, Xu P, Peng J, Lehner PJ. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic. 2010;11:210–220. doi: 10.1111/j.1600-0854.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond LM, Tumbarello DA, Kendrick-Jones J, Buss F. Small-molecule inhibitors of myosin proteins. Future medicinal chemistry. 2013;5:41–52. doi: 10.4155/fmc.12.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremm A, Komander D. Emerging roles for Lys11-linked polyubiquitin in cellular regulation. Trends in biochemical sciences. 2011;36:355–363. doi: 10.1016/j.tibs.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Bunn RC, Jensen MA, Reed BC. Protein interactions with the glucose transporter binding protein GLUT1CBP that provide a link between GLUT1 and the cytoskeleton. Molecular biology of the cell. 1999;10:819–832. doi: 10.1091/mbc.10.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F, Arden SD, Lindsay M, Luzio JP, Kendrick-Jones J. Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis. The EMBO journal. 2001;20:3676–3684. doi: 10.1093/emboj/20.14.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F, Spudich G, Kendrick-Jones J. Myosin VI: cellular functions and motor properties. Annual review of cell and developmental biology. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- Chibalina MV, Seaman MN, Miller CC, Kendrick-Jones J, Buss F. Myosin VI and its interacting protein LMTK2 regulate tubule formation and transport to the endocytic recycling compartment. Journal of cell science. 2007;120:4278–4288. doi: 10.1242/jcs.014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn TA, Chen S, Faith DA, Hicks JL, Platz EA, Chen Y, Ewing CM, Sauvageot J, Isaacs WB, De Marzo AM, et al. A novel role of myosin VI in human prostate cancer. The American journal of pathology. 2006;169:1843–1854. doi: 10.2353/ajpath.2006.060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlinger A, Walters KJ. Structural insights into proteasome activation by the 19S regulatory particle. Biochemistry. 2013;52:3618–3628. doi: 10.1021/bi400417a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nature structural & molecular biology. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- Finan D, Hartman MA, Spudich JA. Proteomics approach to study the functions of Drosophila myosin VI through identification of multiple cargo-binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5566–5571. doi: 10.1073/pnas.1101415108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves MA, Holmes KC. Structural mechanism of muscle contraction. Annual review of biochemistry. 1999;68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- Gibson F, Walsh J, Mburu P, Varela A, Brown KA, Antonio M, Beisel KW, Steel KP, Brown SD. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature. 1995;374:62–64. doi: 10.1038/374062a0. [DOI] [PubMed] [Google Scholar]
- Goto E, Yamanaka Y, Ishikawa A, Aoki-Kawasumi M, Mito-Yoshida M, Ohmura-Hoshino M, Matsuki Y, Kajikawa M, Hirano H, Ishido S. Contribution of lysine 11-linked ubiquitination to MIR2-mediated major histocompatibility complex class I internalization. The Journal of biological chemistry. 2010;285:35311–35319. doi: 10.1074/jbc.M110.112763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice GL, Lobb IT, Weekes MP, Gygi SP, Antrobus R, Nathan JA. The Proteasome Distinguishes between Heterotypic and Homotypic Lysine-11-Linked Polyubiquitin Chains. Cell reports. 2015;12:545–553. doi: 10.1016/j.celrep.2015.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson T, Heintzelman MB, Santos-Sacchi J, Corey DP, Mooseker MS. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:9815–9819. doi: 10.1073/pnas.92.21.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Sato O, Homma K, Ikebe M. DOC-2/DAB2 is the binding partner of myosin VI. Biochemical and biophysical research communications. 2002;292:300–307. doi: 10.1006/bbrc.2002.6636. [DOI] [PubMed] [Google Scholar]
- Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nature structural & molecular biology. 2009;16:1328–1330. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- Kumar M, Sackey K, Schmalstieg F, Trizna Z, Elghetany MT, Alter BP. Griscelli syndrome: rare neonatal syndrome of recurrent hemophagocytosis. Journal of pediatric hematology/oncology. 2001;23:464–468. doi: 10.1097/00043426-200110000-00015. [DOI] [PubMed] [Google Scholar]
- Liu F, Walters KJ. Multitasking with ubiquitin through multivalent interactions. Trends in biochemical sciences. 2010;35:352–360. doi: 10.1016/j.tibs.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng H, Xu X, Wang H, Yang M, Liu X, et al. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer cell. 2014;26:106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetrey J, Bahloul A, Wells AL, Yengo CM, Morris CA, Sweeney HL, Houdusse A. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohiddin SA, Ahmed ZM, Griffith AJ, Tripodi D, Friedman TB, Fananapazir L, Morell RJ. Novel association of hypertrophic cardiomyopathy, sensorineural deafness, and a mutation in unconventional myosin VI (MYO6) Journal of medical genetics. 2004;41:309–314. doi: 10.1136/jmg.2003.011973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SM, Arden SD, Roberts RC, Kendrick-Jones J, Cooper JA, Luzio JP, Buss F. Myosin VI binds to and localises with Dab2, potentially linking receptor-mediated endocytosis and the actin cytoskeleton. Traffic. 2002;3:331–341. doi: 10.1034/j.1600-0854.2002.30503.x. [DOI] [PubMed] [Google Scholar]
- Morriswood B, Ryzhakov G, Puri C, Arden SD, Roberts R, Dendrou C, Kendrick-Jones J, Buss F. T6BP and NDP52 are myosin VI binding partners with potential roles in cytokine signalling and cell adhesion. Journal of cell science. 2007;120:2574–2585. doi: 10.1242/jcs.007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- Sahlender DA, Roberts RC, Arden SD, Spudich G, Taylor MJ, Luzio JP, Kendrick-Jones J, Buss F. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. The Journal of cell biology. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. The EMBO journal. 2009;28:3903–3909. doi: 10.1038/emboj.2009.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR. Myosins: a diverse superfamily. Biochimica et biophysica acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- Shen WC, Li HY, Chen GC, Chern Y, Tu PH. Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy. 2015;11:685–700. doi: 10.4161/auto.36098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Molecular cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich G, Chibalina MV, Au JS, Arden SD, Buss F, Kendrick-Jones J. Myosin VI targeting to clathrin-coated structures and dimerization is mediated by binding to Disabled-2 and PtdIns(4,5)P2. Nature cell biology. 2007;9:176–183. doi: 10.1038/ncb1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Sivaramakrishnan S. Myosin VI: an innovative motor that challenged the swinging lever arm hypothesis. Nature reviews Molecular cell biology. 2010;11:128–137. doi: 10.1038/nrm2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagishi Y, Murata Y. Myosin Va mutation in rats is an animal model for the human hereditary neurological disease, Griscelli syndrome type 1. Annals of the New York Academy of Sciences. 2006;1086:66–80. doi: 10.1196/annals.1377.006. [DOI] [PubMed] [Google Scholar]
- Tomatis VM, Papadopulos A, Malintan NT, Martin S, Wallis T, Gormal RS, Kendrick-Jones J, Buss F, Meunier FA. Myosin VI small insert isoform maintains exocytosis by tethering secretory granules to the cortical actin. The Journal of cell biology. 2013;200:301–320. doi: 10.1083/jcb.201204092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello DA, Kendrick-Jones J, Buss F. Myosin VI and its cargo adaptors - linking endocytosis and autophagy. Journal of cell science. 2013;126:2561–2570. doi: 10.1242/jcs.095554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello DA, Manna PT, Allen M, Bycroft M, Arden SD, Kendrick-Jones J, Buss F. The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy. PLoS pathogens. 2015;11:e1005174. doi: 10.1371/journal.ppat.1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello DA, Waxse BJ, Arden SD, Bright NA, Kendrick-Jones J, Buss F. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nature cell biology. 2012;14:1024–1035. doi: 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Molecular cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Vreugde S, Ferrai C, Miluzio A, Hauben E, Marchisio PC, Crippa MP, Bussi M, Biffo S. Nuclear myosin VI enhances RNA polymerase II-dependent transcription. Molecular cell. 2006;23:749–755. doi: 10.1016/j.molcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Walters KJ, Ferentz AE, Hare BJ, Hidalgo P, Jasanoff A, Matsuo H, Wagner G. Characterizing protein-protein complexes and oligomers by nuclear magnetic resonance spectroscopy. Methods in enzymology. 2001;339:238–258. doi: 10.1016/s0076-6879(01)39316-3. [DOI] [PubMed] [Google Scholar]
- Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Cheng W, Hung J, Montell D, Geisbrecht E, Rosen D, Liu J, Naora H. Lessons from border cell migration in the Drosophila ovary: A role for myosin VI in dissemination of human ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8144–8149. doi: 10.1073/pnas.0400400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorca CE, Kim LK, Kim YJ, Krause MR, Zenklusen D, Spilianakis CG, Flavell RA. Myosin VI regulates gene pairing and transcriptional pause release in T cells. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1587–1593. doi: 10.1073/pnas.1502461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.