Abstract
Oxidation of guanine by reactive oxygen species and high valent metals produces damaging DNA base lesions like 8-oxo-7,8-dihydroguanine (8-oxoG). 8-oxoG can be further oxidized to form the spiroiminodihydantoin (Sp) lesion, which is even more mutagenic. DNA polymerases preferentially incorporate purines opposite the Sp lesion, and DNA glycosylases excise the Sp lesion from the duplex, although the rate of repair is different for the two Sp diastereomers. To further understand the biological processing of the Sp lesion, differential scanning calorimetry (DSC) studies were performed on a series of 15-mer DNA duplexes. The thermal and thermodynamic stability of each of the Sp diastereomers paired to the four standard DNA bases was investigated. It was found that, regardless of the base-pairing partner, the Sp lesion was always highly destabilizing in terms of DNA melting temperature, enthalpic stability, and overall duplex free energy. We found no significant differences between the two Sp diastereomers, but changing the base pairing partner of the Sp lesion produced slight differences in stability. Specifically, duplexes with Sp:C pairings were always the most destabilized, whereas pairing the Sp lesion with a purine base modestly increased stability. Overall, these results suggest that although the stability of the Sp diastereomers cannot explain the differences in the rates of repair by DNA glycosylases, the most stable base pairing partners do correspond with the nucleotide preference of DNA polymerases.
Graphical Abstract
INTRODUCTION
Reactive oxygen species (ROS) and oxidative stress have been linked to a number of diseases, including cancer and neurological disorders.1,2 ROS can be produced endogenously during cellular respiration1,3 or by neutrophils,4 as well as by exposure to outside agents such as ionizing radiation5,6 and certain transition metals.7,8 These species can oxidize DNA bases, with the reduction potential of guanine making it the most easily oxidized base.9 The oxidation of guanine creates 8-oxo-7,8-dihydroguanine (8-oxoG, Figure 1A), which can be further oxidized to a number of other lesions. These oxidative lesions can create mutations in the DNA, potentially leading to genotoxic effects.10,11 For example, 8-oxoG has been found in elevated concentrations in the brains of Parkinson’s patients12 and the blood of Alzheimer’s patients,13 while a reduced ability to repair 8-oxoG has been associated with head and neck cancer.14 Other oxidative DNA base lesions have been detected in elevated concentrations in mice prior to the onset of colon cancer.15
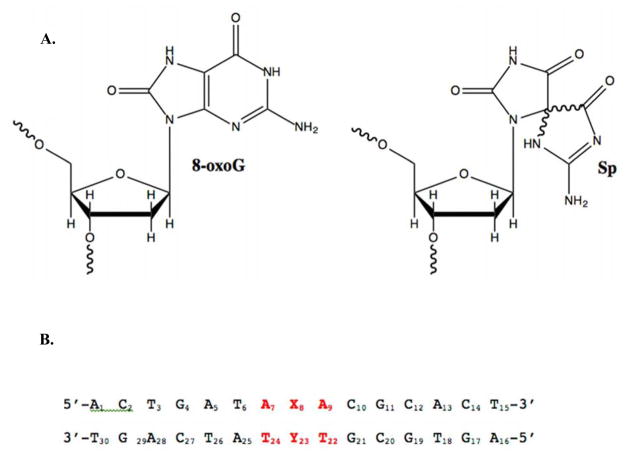
Figure 1.
A. Structures of the oxidized guanine lesions 7,8-dihydro-8-oxo-2′-deoxguanosine (8-oxoG) and spiroiminodihydantoin (Sp). B. DNA sequence used in DSC studies, where X8 is the location of the Sp lesion (or G in the case of the control) and Y23 was varied to be either C, T, G, or A.
One product of the hyperoxidation of oxidation of guanine is the spiroiminodihydantoin (Sp) lesion (Figure 1A). This adduct contains two rigid, mostly flat 5-membered rings that are oriented roughly perpendicular to each other.16–22 The chiral carbon at the spiro center in the nucleobase creates R and S enantiomers that become a pair of diastereomers when bonded to the furanose ring of DNA. The presence of the orthogonal rings as well as the ability to introduce different stereochemistry give this lesion a unique structure that has the potential to greatly impact both the stability and biological processing of duplex DNA.
The Sp lesion has been shown to significantly affect genomic integrity in a number of studies. Kornyushyna et al. showed that the Klenow fragment of E. coli DNA polymerase I incorporates dAMP and dGMP, but not dCMP opposite the Sp lesion.11 The Sp lesion blocked DNA elongation and affected Klenow’s proofreading ability.11,23 They suggested that the proofreading efficiency of the enzyme may depend upon the thermodynamic stability of the lesion containing duplex.23 In another study, when Sp modified bacteriophage DNA was transfected into E. coli cells, the lesion was bypassed efficiently enough during replication to produce progeny phage.10 The mutation frequency for both Sp diastereomers was ~100%, at least an order of magnitude higher than what was observed with 8-oxoG. The stereoisomers are known to display different mutation spectra, with one study showing one Sp isomer generating 72% G to C and 27% G to T transversions, while the other produced 57% G to C and 41% G to T.10 The Sp diastereomers also displayed different mutational signatures and responded differently to knock-outs of polymerase V in studies of the role of SOS inducible E. coli DNA polymerases in the bypass of oxidative DNA lesions.24
Computational studies have suggested that the S- stereoisomer of Sp is less thermodynamically stable than the R- isomer due to steric effects in the larger context of duplexed DNA.25 Both the hydrogen bonding and stacking interactions are optimized in duplexes where Sp is placed opposite a guanine or adenine, rather than the cytidine that is generally opposite the parent base guanine. It was conjectured that Sp acts as a pseudothymine or pseudopyrimidine in these cases, which would contribute to the high rate of G to T and G to C mutations observed with this lesion.
The recognition and repair of Sp lesions by base excision repair (BER) enzymes represents another critical component of Sp lesion mutagenicity. The Sp adduct is repaired by the E. coli glycosylases Fpg (MutM), Nth, and Nei;26,27 yeast enzymes yOGG1 and yOGG2;28 and both murine and human NEIL1.29,30 Some of these enzymes, such as Fpg (MutM), Nth, Nei, and hNEIL1, exhibit preferences for particular base pairing partners, while others like yOGG1 and yOGG2 do not. In addition, studies have shown that the two Sp diastereomers can be distinguished by hNEIL1, which repairs one isomer much more quickly than the other.30
In this work, the stability of both Sp diastereomers paired with the four standard DNA bases was examined in the context of a 15-mer duplex using differential scanning calorimetry (DSC). These results demonstrate how changing the base pairing partner affects the thermal and thermodynamic stability of the Sp diastereomers, providing insight into the processing of this lesion by key biological enzymes.
EXPERIMENTAL PROCEDURES
Reagents
Oligonucleotides containing standard DNA bases were purchased from Integrated DNA Technologies (IDT), and 8-oxoG containing oligonucleotides were obtained from Midland Certified Reagent Company. Acetonitrile (HPLC grade), ammonium acetate, sodium phosphate dibasic heptahydrate (Na2HPO4•7H2O) and sodium chloride were obtained from Fisher Scientific. Sodium hexachloroiridate(IV) hexahydrate (Na2IrCl6•6H2O), sodium phosphate monobasic monohydrate (NaH2PO4•H2O), and EDTA were purchased from Sigma Aldrich.
Synthesis of the Sp Lesion and Annealing of Duplexes
The Sp lesion was synthesized by oxidizing an 8-oxoG containing oligonucleotide with sodium hexachloroiridate(IV) hexahydrate (Na2IrCl6•6H2O) according to literature procedures.11 Briefly, the 8-oxoG containing oligonucleotide (12 μM) was reacted with 100 μM Na2IrCl6•6H2O in 10 mM sodium phosphate buffer (pH 7) and 100 mM sodium chloride. The reaction mixture was heated at 65°C for 30 minutes and quenched with 20 mM EDTA (pH 8.0). After quenching, the reaction solution was desalted and concentrated by using Amicon Ultra Centrifugal Filters (3K MWCO, Millipore).
The Sp oligonucleotide strands were purified on an HP Series 1100 HPLC by using a Dionex DNAPac PA-100 9 × 250 mm anion exchange column with a linear gradient from 60% mobile phase A (10% aqueous acetonitrile) and 40% mobile phase B (90% 1.5 M ammonium acetate, pH 6, 10% acetonitrile), to 100% of mobile phase B over the course of 30 minutes with a flow rate of 2.5 mL/min.31 The two diastereoisomers of the Sp lesion eluted at approximately 20 (peak 1, Sp1) and 21 minutes (peak 2, Sp2), respectively. The two peaks were collected separately and desalted and concentrated by using Amicon Ultra Centrifugal Filters (3K MWCO, Millipore). The samples were characterized by ESI mass spectrometry.
Each of the Sp lesion oligonucleotide strands was annealed to its corresponding complementary strand before performing DSC experiments. The concentrations of single-stranded oligonucleotides were determined spectrophotometrically using molar extinction coefficients calculated with nearest neighbor parameters.32–35 Strands were mixed in a 1:1 ratio in sodium phosphate buffer (10 mM sodium phosphate, pH 7, 100 mM NaCl and 0.1 mM EDTA), heated to 90°C for ~5 min, and slow cooled to room temperature.
Differential Scanning Calorimetry (DSC)
Differential scanning calorimetry experiments were conducted on a Model 6300 NanoDSCIII differential scanning calorimeter (TA Instruments). A sample of each duplex (~50 μM) was prepared in sodium phosphate buffer (10 mM sodium phosphate, pH 7, 100 mM NaCl and 0.1 mM EDTA) and dialyzed (Slide-A-Lyzer MINI dialysis cassettes, 3K MWCO) against excess sodium phosphate buffer to equilibrate the buffer and sample solution. The concentration of each duplex sample was determined spectrophotometrically both before and after DSC analysis. DSC samples were heat denatured by equilibrating them in the sample cell of the spectrometer at 75°C for at least 15 minutes. Absorbance readings at 260 nm. were measured, and the concentration of the duplex was calculated using the sum of the extinction coefficients for the two relevant oligonucleotide strands.
Solutions were degassed for ~10 minutes, and excess heat capacity (Cpex) versus temperature curves were measured from 10 to 90°C with a heating/cooling rate of 1°C/min. For each sample, buffer versus buffer baseline scans were also determined under the same conditions. The program CpCalc was used to calculate the thermodynamic parameters, ΔHcal and ΔScal. For each sample, the buffer scan was subtracted and the oligonucleotide scan was normalized for concentration by CpCalc in order to calculate the molar heat capacity. A linear baseline, drawn from the pre- to the post-transition region, was used to determine the values for ΔHcal and ΔScal in CpCalc. The standard free energy change for duplex formation at 25°C (ΔG25) was calculated by using the equation: ΔG = ΔHcal − TΔScal.
RESULTS AND DISCUSSION
The sequence of the 15-mer oligonucleotide duplex used in this study is shown in Figure 1B. The control sequence contains a central G:C base pair (G8:C23). DSC experiments were performed with the control and eight different Sp lesion duplexes, where X8 was either Sp1 or Sp2 and Y23 was C, T, G, or A. The data obtained from the DSC experiments are presented in Table 1.
Table 1.
Thermodynamic parameters for the formation of the Sp 15-mer duplexes obtained from DSC experiments.
| Sequence (X8:Y23) | Tm (°C) | ΔHcal (kcal/mol) | ΔScal (cal/mol•K) | ΔG25 (kcal/mol) |
|---|---|---|---|---|
| G:C Control | 63.6 ± 0.1 | −101 ± 7 | −301 ± 20 | −11.6 ± 0.8 |
|
| ||||
| Sp1:C | 44.8 ± 0.1 | −80 ± 4 | −253 ± 12 | −5.0 ± 0.2 |
| Sp1:T | 48.0 ± 0.2 | −76 ± 9 | −236 ± 30 | −5.5 ± 0.6 |
| Sp1:G | 48.5 ± 0.1 | −83 ± 2 | −257 ± 7 | −6.1 ± 0.2 |
| Sp1:A | 47.5 ± 0.1 | −86 ± 5 | −269 ± 15 | −6.1 ± 0.3 |
|
| ||||
| Sp2:C | 44.8 ± 0.6 | −77 ± 4 | −241 ± 13 | −4.8 ± 0.4 |
| Sp2:T | 47.8 ± 0.1 | −77 ± 4 | −240 ± 13 | −5.5 ± 0.4 |
| Sp2:G | 47.8 ± 0.1 | −80 ± 4 | −250 ± 13 | −5.7 ± 0.3 |
| Sp2:A | 47.2 ± 0.1 | −86 ± 5 | −267 ± 15 | −6.0 ± 0.3 |
There has been a considerable amount of debate in the literature concerning the assignment of the absolute stereochemistry for the Sp diastereomers. Broyde and Geacintov used experimental and computational optical rotatory dispersion (ORD) and electronic circular dichroism (ECD) to assign absolute configurations to Sp nucleobases.36,37 Their findings were supported by Sugden et al.38 However, Cadet’s research group, using NMR methods, reached the opposite conclusion.39 The Burrows lab has demonstrated that these results can be reconciled by including water molecules in the calculations supporting the ORD and ECD assignments,40 and their results were consistent with the recent crystal structure of DNA polymerase β complexed with a DNA duplex containing an Sp lesion.41
One thing that has become clear as various groups have studied this issue is that that the order of elution for the Sp diastereomers changes based on the type of HPLC column used. With an ion exchange column or amino-silica column the S-Sp isomer elutes first, but the order switches when a Hypercarb HPLC column is used.40 Following this most recent assignment, Sp1 would be the S isomer and Sp2 would be the R isomer. However, since we did not do a full digestion analysis to confirm the absolute stereochemistry of the Sp diastereomers in our oligonucleotides, the duplexes studied here are simply named Sp1 and Sp2 based on their elution order from the Dionex anion exchange column.
Effect of the Sp Lesion on the Thermal Stability of the Duplex
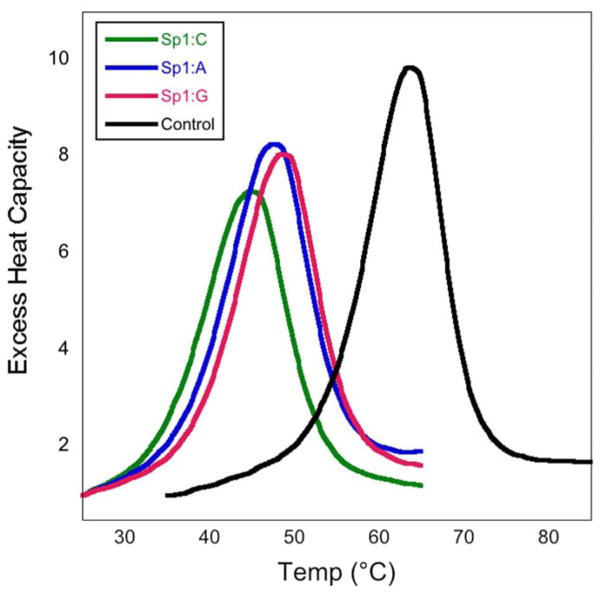
As expected based on our previous studies, the G:C control duplex had a melting temperature that was ~20°C higher than either of the Sp diastereomers paired with a C base.42 In fact, all of the Sp containing duplexes showed a significant thermal destabilization, with melting temperatures at least 15°C lower than the control duplex. By comparison, the presence of an 8-oxoG lesion lowers the melting temperature by only ~1°C, and mispairing either the parent G base or an 8-oxoG lesion with T, G, or A lowers the melting temperature by only ~6–9°C. The base pairing partner modulated the effect of the Sp lesion, with the melting temperatures of the Sp:C duplexes being ~3°C lower than the other base pairing partners examined. However, no notable differences were observed between the two Sp diastereomers in any of the duplexes studied.
Many studies have demonstrated that the presence of an Sp lesion significantly decreases the melting temperature of DNA, although the extent of this effect appears to depend on the sequence, length, and position of the Sp lesion in the duplex. Recent work with an 11-mer duplex containing a central Sp:C base pair showed that the melting temperature was similar for the two Sp diastereomers and decreased by ~30°C relative to the undamaged parent duplex, indicating that this effect may be more severe in shorter duplexes.43 In contrast, studies with 18-mer template/16-mer primer duplexes with the Sp:C lesion located 3 base pairs from the end of the duplex found the melting temperature to decrease by only ~8–13°C depending on the DNA sequence context.11
Changing Sp’s base pairing partner from C to a purine base modestly increased the melting temperature of the duplex in our study. We have obtained similar results with other 15-mer duplexes whose DNA sequence is the same as that used here with the exception of the two bases directly flanking the lesion site (Table 2). A duplex containing a central TSpT sequence (instead of ASpA) yielded almost identical differences in melting temperatures to those reported here, indicating that flipping the A:T base pairs around the lesion site has little to no effect on the melting temperature as might be expected. However, when the central ASpA sequence was changed to CSpC, we found that the values of the melting temperatures for both the control and Sp lesions duplexes increased by a further ~5–10°C, suggesting that having C:G base pairs flanking the lesion site, with their additional hydrogen bond, may help to increase the thermal stability of Sp containing duplexes. With both of these different flanking sequences, we found that pairing the Sp lesion to a purine base increased the melting temperature by a few degrees. Similar results have been observed with the 18-mer template/16-mer primer duplexes mentioned above in a G-C rich sequence, although in an A-T rich sequence only the melting temperature of the Sp:A duplex was increased, with the Sp:G sample having a melting temperature similar to the Sp:C duplex.11
Table 2.
Melting Temperature Data for 15-mer duplexes with central TSpT and CSpC sequences.
| DNA Sequence | Tm (°C) | |||
|---|---|---|---|---|
| Central TSpT Sequence | ||||
|
| ||||
| G:C Control | 64±1 | |||
| Sp2:C | 43±2 | |||
| Sp2:G | 46±1 | |||
|
| ||||
| Central CSpC Sequence | ||||
|
| ||||
| G:C Control | 73±1 | |||
| Sp2:C | 50±1 | |||
| Sp2:T | 51±1 | |||
| Sp2:G | 55±1 | |||
| Sp2:A | 52±1 | |||
Effect of the Sp Lesion on the Thermodynamic Stability of the Duplex
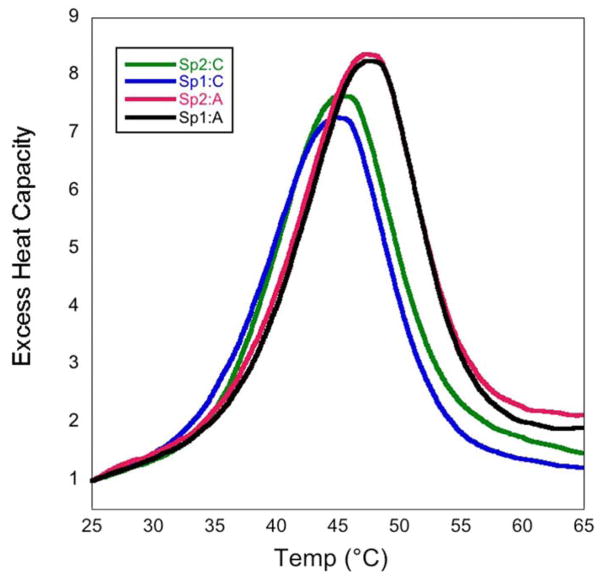
In all of the DNA duplexes studied, the Sp lesion significantly reduced the thermodynamic stability of the duplex, consistent with previous studies.42,43 Overall we saw a increase in the free energy of duplex formation of ~5.5–7 kcal/mol for the Sp lesion containing duplexes. For duplexes with different base pairing partners, changes in the thermodynamic parameters were observed when the Sp lesion was paired to G, A, and T compared to C. In general, the Sp:C duplexes were the least thermodynamically stable, having the most positive ΔG values for helix formation. Pairing the Sp lesion with a purine base produced the most thermodynamically stable lesion containing duplexes, with the Sp:T duplexes falling in between Sp:purines and Sp:C. The DSC thermograms presented in Figure 2 illustrate these differences for the control, Sp:purine, and Sp:C duplexes. Figure 3 shows the DSC thermograms for the Sp1 and Sp2 diastereomers paired with C and A. Just as with the melting temperatures, there were no significant differences observed in the thermodynamic parameters of the two Sp diastereomers in any of the duplexes, in agreement with previous DSC studies.43
Figure 2.
DSC thermograms comparing the control (black), Sp1:C (green), and Sp1:purine (blue and red) 15-mer duplexes.
Figure 3.
DSC thermograms comparing the two Sp diastereomers paired with cytosine (blue and green) and adenine (black and red).
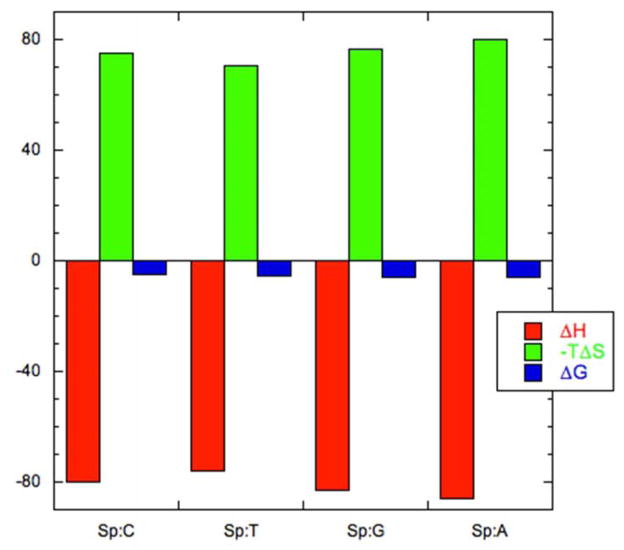
Examination of the thermodynamic data in Table 1 reveals that the destabilization of the Sp lesion duplexes is enthalpic in nature. This destabilization is offset by an entropic stabilization, resulting in an “enthalpy-entropy compensation” that is seen in many biological systems featuring multiple, weak interactions44–46 and is depicted graphically in Figure 4. Our earlier work with the Sp2:C duplex also showed a significant thermodynamic destabilization of the duplex that was enthalpic in nature; however, the absolute magnitudes of the enthalpy and entropy values obtained for the Sp2:C duplex in that study are somewhat different from what was obtained here.42 The reason for these differences is not entirely clear. One possible explanation is that the concentration of the Sp2:C duplex in the earlier study may have been less than initially determined; some sample may have been lost during the dialysis step before DSC. In this work, the concentration of the duplexes was checked both before and after DSC analysis. Reanalyzing that earlier data using a duplex concentration of ~30–35 μM instead of 50 μM yields values that are similar to the ones reported here.
Figure 4.
Enthalpy-entropy compensation for the Sp1 15-mer duplexes. The thermodynamic parameters for the formation of the helices have units of kcal/mol. For each sample, ΔH° values are plotted in red, −TΔS° values are in green, and ΔG° values are in blue.
Significance for Biological Processing
The goal of this study was to perform DSC studies to investigate whether or not thermodynamic differences between the Sp diastereomers paired to different DNA bases could provide insight into how the Sp lesion is processed by biologically important enzymes like DNA polymerases and glycosylases. We did not observe any notable differences in terms of the melting temperatures or free energies of duplex formation between the two Sp diastereomers for any of the duplexes studied. Earlier computational work had predicted a better hydrogen bonding network with R-Sp than with the S stereoisomer, suggesting that R-Sp might display a greater stability in our DSC studies.25 We did not find this to be the case, and in fact, Khutsishvili et al. recently found a similar result with 11-mer duplexes containing the Sp lesion.43 Thus, it appears that even though the two Sp isomers are repaired at different rates by DNA glycosylases,30 this disparity does not stem from differences in either the thermal or thermodynamic stability of the lesions in DNA.
Overall, our results demonstrate that the Sp lesion is more stable when paired with a purine than with a pyrimidine. These results are consistent with findings from computational studies that suggested that the Sp lesion may act as a “pseudothymine” stabilizing pairing with G or A over C.25 Experiments with the Klenow fragment of DNA polymerase I that found that the Sp lesion was paired primarily with G or A.11,23 It is possible that the additional thermal or thermodynamic stability obtained by pairing Sp with a purine is a factor in this enzymatic preference that leads to the types of mutations observed with this lesion. Interestingly, the rate of Sp’s repair by the DNA glycosylase hNEIL1 is influenced by its base pairing partner, with more efficient repair occurring when Sp is opposite T, C, or G than when paired with A.30 In that work, the authors suggest that “less stable base pairing of the hydantoin lesions with T relative to G and A may translate into … more facile recognition and excision by hNEIL1.” Our results provide direct evidence that Sp:purine base pairs are indeed more thermodynamically stable than Sp:T pairs, which may influence the ability of DNA glycosylases to identify and repair these harmful oxidative base lesions.
Finally, while having Sp paired to a purine proved to be the most stable arrangement, our results showed that Sp:T pairs were consistently more stable both thermally and thermodynamically than Sp:C pairs. This result is consistent with previous thermodynamic studies where TT mismatches in DNA duplexes were found to be more stable than CC mismatches.44,47 The increased stability of the TT mismatches has been attributed to their ability to form two hydrogen bonds, whereas CC mismatches are only able to form one.44 If Sp is able to act as a “pseudothymine” then it may be able to form two hydrogen bonds when paired with a thymine base, thus creating more stability than when it is paired to a cytosine, as we observed.
Conclusion
In summary, the results of these studies and others reveal that the Sp lesion is always highly destabilizing to the DNA duplex. There do not appear to be any significant differences between the thermal and thermodynamic stabilities of the two Sp diastereomers, but the base pairing partner of the Sp lesion can impact the amount of destabilization in the duplex. In general, paring the Sp lesion with a purine serves to increase the thermal and thermodynamic stability of the duplex, perhaps influencing to how these lesions are processed during biologically important events like DNA replication and repair.
Acknowledgments
Funding Sources
Funding for this work was provided by a National Institutes of Health AREA grant (National Cancer Institute, 1R15 CA149958-01A1) to ERJ and Smith College.
Smith undergraduate students Melinda Ng ‘09 and Maud Martei ‘10 collected melting temperature data for control DNA sequences. Mass spectral data for these oligonucleotides were obtained by Dr. Kalina Dimova at the Smith College Center for Proteomics. The authors thank Dr. Charles Amass, Dr. Kalina Dimova, Prof. David Gorin, and Prof. Stylianos Scordilis for access to and assistance with instrumentation, Prof. Megan Núñez (Wellesley College) for helpful discussions, and Prof. Sheila R. Smith (U. Michigan, Dearborn) for assistance with the TOC figure.
ABBREVIATIONS
- ROS
reactive oxygen species
- Sp
spiroiminodihydantoin
- 8-oxoG
7,8-dihydro-8-oxo-2′-deoxyguanosine
- DSC
differential scanning calorimetry
- BER
base excision repair
- ORD
optical rotatory dispersion
- ECD
electronic circular dichroism
References
- 1.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants and the degenerative disease of aging. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckman KB, Ames BN. Oxidative decay of DNA. J Biol Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 3.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 4.Yu H, Venkatarangan L, Wishnok JS, Tannenbaum SR. Quantitation of four guanine oxidation products from reaction of DNA with varying doses of peroxynitrite. Chem Res Toxicol. 2005;18:1849–1857. doi: 10.1021/tx050146h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadet J, Berger M, Douki T, Ravanat JL. Oxidative damage to DNA: Formation, measurement, and biological significance. Rev Physiol, Biochem Pharmacol. 1997;131:1–87. doi: 10.1007/3-540-61992-5_5. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 7.Burrows CJ, Muller JG, Kornyushyna O, Luo W, Duarte V, Leipold MD, David SS. Structure and potential mutagenicity of new hydantoin products from guanosine and 8-oxo-7,8-dihydroguanine oxidation by transition metals. Environ Health Perspect. 2002;110:713–717. doi: 10.1289/ehp.02110s5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 9.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solutions. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 10.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 11.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow Fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Perry G, Smith MA, Robertson D, Olson SJ, Graham DG, Montine TJ. Parkinson’s Disease Is Associated with Oxidative Damage to Cytoplasmic DNA and RNA in Substantia Nigra Neurons. Am J Pathol. 1999;154:1423–1429. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrag M, Mueller C, Zabel M, Crofton A, Kirsch WM, Ghribi O, Squitti R, Perry G. Oxidative stress in blood in Alzheimer’s disease and mild cognitive impairment: A meta-analysis. Neurobiol Dis. 2013;59:100–110. doi: 10.1016/j.nbd.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Paz-Elizur T, Ben-Yosef R, Elinger D, Vexler A, Krupsky M, Berrebi A, Shani A, Schechtman E, Freedman L, Livneh Z. Reduced repair of the oxidative 8-oxoguanine DNA damage and risk of head and neck cancer. Cancer Res. 2006;66:11683–11689. doi: 10.1158/0008-5472.CAN-06-2294. [DOI] [PubMed] [Google Scholar]
- 15.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA. 2012:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 17.Buchko GW, Cadet J, Berger M, Ravanat JL. Photooxidation of d(TpG) by phthalocyanines and riboflavin. Isolation and characterization of dinucleoside monophosphates containing the 4R* and 4S* diastereomers of 4,8-dihydro-4-hydroxy-8-oxo-2′deoxyguanosine. Nucleic Acids Res. 1992;20:4847–4851. doi: 10.1093/nar/20.18.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadet J, Berger M, Decarroz C, Wagner JR, van Lier JE, Ginot YM, Vigny P. Photosensitized reactions of nucleic acids. Biochimie. 1986;68:813–834. doi: 10.1016/s0300-9084(86)80097-9. [DOI] [PubMed] [Google Scholar]
- 19.Cadet J, Decarroz C, Wang SY, Midden WR. Mechanisms and products of photosensitized degradation of nucleic acids and model compounds. Isr J Chem. 1983;23:420–429. [Google Scholar]
- 20.Ravanat JL, Berger M, Benard F, Langlois R, Ouellet R, van Lier JE, Cadet J. Phthalocyanine and naphthalocyanine photosensitized oxidation of 2′-deoxyguanosine: Distinct type I and type II products. Photochem Photobiol. 1992;55:809–814. [Google Scholar]
- 21.Ravanat JL, Cadet J. Reaction of singlet oxygen with 2′-deoxyguanosine and DNA. Isolation and characterization of the main oxidation products. Chem Res Toxicol. 1995;8:379–388. doi: 10.1021/tx00045a009. [DOI] [PubMed] [Google Scholar]
- 22.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Spiroiminodihydantoin lesions derived from guanine oxidation: Structures, energetics, and functional implications. Biochemistry. 2005;44:6043–6051. doi: 10.1021/bi0473657. [DOI] [PubMed] [Google Scholar]
- 23.Kornyushyna O, Burrows CJ. Effect of the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin on proofreading by Escherichia coli DNA polymerase I (Klenow fragment) in different sequence contexts. Biochemistry. 2003;42:13008–13018. doi: 10.1021/bi0350755. [DOI] [PubMed] [Google Scholar]
- 24.Neeley WL, Delaney S, Alekseyev YO, Jarosz DF, Delaney JC, Walker GC, Essigmann JM. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J Biol Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 25.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Structural and thermodynamic features of spiroiminodihydantoin damaged DNA duplexes. Biochemistry. 2005;44:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 26.Hazra TK, Muller JG, Manuel RC, Burrows CJ, Lloyd RS, Mitra S. Repair of hydantoins, one electron oxidation products of 8-oxoguanine, by DNA glycosylases of Escherichia coli. Nucleic Acids Res. 2001;29:1967–1974. doi: 10.1093/nar/29.9.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leipold MD, Muller JG, Burrows CJ, David SS. Removal of hydantoin products of 8-oxoguanine oxidation by the Escherichia coli DNA repair enzyme Fpg. Biochemistry. 2000;39:14984–14992. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- 28.Leipold MD, Workman H, Muller JG, Burrows CJ, David SS. Recognition and removal of oxidized guanines in duplex DNA by the base excision repair enzymes hOGG1, yOGG1, and yOGG2. Biochemistry. 2003;42:11373–11381. doi: 10.1021/bi034951b. [DOI] [PubMed] [Google Scholar]
- 29.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions of spiroiminodihydantoin and guanidinohydantoin in DNA by mammalian base excision repair glycosylases NEIL1 and NEIL1. DNA Repair. 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slade PG, Hailer MK, Martin BD, Sugden KD. Guanine-specific oxidation of double-stranded DNA by Cr(VI) and ascorbic acid forms spiroiminodihydantoin and 8-oxo-2′-deoxyguanosine. Chem Res Toxicol. 2005;18:1140–1149. doi: 10.1021/tx050033y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devor EJ, Behlke MA. Oligonucleotide yield, resuspension, and storage Integrated DNA Technologies 2005 [Google Scholar]
- 33.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. III Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 34.Warshaw MM, Cantor CR. Oligonucleotide interactions. IV Conformational differences between deoxy- and ribodinucleoside phosphates. Biopolymers. 1970;9:1079–1103. doi: 10.1002/bip.1970.360090910. [DOI] [PubMed] [Google Scholar]
- 35.Vallone PM, Benight AS. Thermodynamic, spectroscopic, and equilibrium binding studies of DNA sequence context effects in four 40 base pair deoxyoligonucleotides. Biochemistry. 2000;39:7835–7846. doi: 10.1021/bi000326k. [DOI] [PubMed] [Google Scholar]
- 36.Durandin A, Jia L, Crean C, Kolbanovskiy A, Ding S, Shafirovich V, Broyde S, Geacintov NE. Assignment of absolute configurations of the enantiomeric spiroiminodihydantoin nucleobases by experimental and computational optical rotatory dispersion methods. Chem Res Toxicol. 2006;19:908–913. doi: 10.1021/tx060078e. [DOI] [PubMed] [Google Scholar]
- 37.Ding S, Jia L, Durandin A, Crean C, Kolbanovskiy A, Shafirovich V, Broyde S, Geacintov NE. Absolute configurations of spiroiminodihydantoin and allantoin stereoisomers: Comparison of computed and measured electronic circular dichroism spectra. Chem Res Toxicol. 2009;22:1189–1193. doi: 10.1021/tx900107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gremaud JN, Martin BD, Sugden KD. Influence of substrate complexity on the diastereoselective formation of spiroiminodihydantoin and guanidinohydantoin from chromate oxidation. Chem Res Toxicol. 2010;23:379–385. doi: 10.1021/tx900362r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karwowski B, Dupeyrat F, bardet M, Ravanat JL, Krajewski P, Cadet J. Nuclear magnetic resonance studies of the 4R and 4S diastereomers of spiroiminodihydantoin 2′-deoxyribonucleotides: absolute configuration and conformational features. Chem Res Toxicol. 2006;19:1357–1365. doi: 10.1021/tx060088f. [DOI] [PubMed] [Google Scholar]
- 40.Fleming AM, Orendt AM, He Y, Zhu J, Dukor RK, Burrows CJ. Reconciliation of Chemical, Enzymatic, Spectroscopic and Computational Data To Assign the Absolute Configuration of the DNA Base Lesion Spiroiminodihydantoin. J Am Chem Soc. 2013;135:18191–18204. doi: 10.1021/ja409254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckenroth BE, Fleming AM, Sweasy JB, Burrows CJ, Doublié S. Crystal Structure of DNA Polymerase β with DNA Containing the Base Lesion Spiroiminodihydantoin in a Templating Position. Biochemistry. 2014;53:2075–2077. doi: 10.1021/bi500270e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chinyengetere F, Jamieson ER. Impact of the oxidized guanine lesion spiroimimodihydantoin on the conformation and thermodynamic stability of a 15-mer DNA duplex. Biochemistry. 2008;47:2584–2591. doi: 10.1021/bi701502t. [DOI] [PubMed] [Google Scholar]
- 43.Khutsishvili I, Zhang N, Marky LA, Crean C, Patel DJ, Geacintov NE, Shafirovich V. Thermodynamic profiles and nuclear magnetic resonance studies of oligonucleotide duplexes conatining single diastereomeric spiroiminodihydantoin lesions. Biochemistry. 2013;52:1354–1363. doi: 10.1021/bi301566v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tikhomirova A, Beletskaya IV, Chalikian TV. Stability of DNA duplexes containing GG, CC, AA, and TT mismatches. Biochemistry. 2006;45:10563–10571. doi: 10.1021/bi060304j. [DOI] [PubMed] [Google Scholar]
- 45.Prabhu NV, Sharp K. Heat capacity in proteins. Annu Rev Phys Chem. 2005;56:521–548. doi: 10.1146/annurev.physchem.56.092503.141202. [DOI] [PubMed] [Google Scholar]
- 46.Dunitz JD. Win some, lose some: Enthalpy-entropy compensation in weak intermolecular interactions. Chem Biol. 1995;2:709–712. doi: 10.1016/1074-5521(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 47.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A•A, C•C, G•G, and T•T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]