Summary
The T-cell receptor (TCR) controls the cellular adaptive immune response to antigens, but our understanding of TCR repertoire diversity and response to challenge is still incomplete. For example, TCR clones shared by different individuals with minimal alteration to germline gene sequences (public clones) are detectable in all vertebrates, but their significance is unknown. Although small in size, the zebrafish TCR repertoire is controlled by processes similar to those operating in mammals. Thus, we studied the zebrafish TCR repertoire and its response to stimulation with self and foreign antigens. We found that cross-reactive public TCRs dominate the T-cell response, endowing a limited TCR repertoire with the ability to cope with diverse antigenic challenges. These features of vertebrate public TCRs might provide a mechanism for the rapid generation of protective T-cell immunity allowing a short temporal window for the development of more specific private T-cell responses.
Graphical abstract

Introduction
The T-cell repertoire, constituted by the pool of T-cell receptor (TCR) specificities, governs the ability of the immune system to respond to both foreign and self-derived immune challenges (Linnemann et al., 2013; Newell and Davis, 2014; Nikolich-Zugich et al., 2004; Turner et al., 2009). Ninety five percent of the TCRs are composed of an α and a β protein chain. The antigen specificity of the TCR is primarily determined by the CDR3 of α and β chains (Rudolph et al., 2006), which interacts with the peptide MHC complex (Davis and Bjorkman, 1988). Indeed, the majority of TCR variation is localized in the third complementarity-determining region (CDR3) as a result of the recombination of variable (V), diversity (D) and joining (J) segments and the incorporation of multiple nucleotide insertions and deletions. Thus, the study of CDR3 sequences provides information about the fraction of the TCR repertoire relevant for antigen recognition. However, the complexity and dynamics of the TCR repertoire remain unknown because of the limited power of the tools used for its investigation.
Previous studies estimated CDR3 diversity based on the analysis of a relatively small number of T cells. These studies are based on a solution for the “unseen species problem” developed to estimate the total number of species in a given population based on random samples of species (Efron and Thisted, 1976; Fisher et al., 1943). This method assumes that the number of TCR clones follows a Poisson distribution, however recent studies found a power law distribution instead (Weinstein et al., 2009). Indeed, studies based on the sequencing of small T-cell samples produced estimates of TCR diversity that were directly proportional to the number of sequences analyzed, suggesting that these methods do not capture the complete TCR repertoire diversity (Freeman et al., 2009). Even when advanced methods are used to study the TCR repertoire, these methods are still limited by their lack of consideration of tissue resident T cells (Burzyn et al., 2013; Park and Kupper, 2015). Because of these limitations, it is still unclear what fraction of the potential T-cell repertoire is expressed, and how similar are the repertories of different individuals in the quiescent state and during the course of an immune response. In addition, TCR sequences shared by different individuals (termed public TCR sequences) are detected in all vertebrates in multiple biological contexts, a surprising finding when the number of potential unique CDR3 sequences generated by VDJ recombination is considered (McBerry et al., 2012; Venturi et al., 2008). However, the significance of public TCRs on the repertoire, as well as their response to stimulation is unknown.
Zebrafish (Danio rerio) is an ideal immunological model system to study the TCR repertoire because its adaptive immune system shares important features with its mammalian counterpart. Examples of these shared elements are the presence of a recombination activating gene (RAG), a combinatorial rearrangement of V, D and J gene segments, junctional diversity during recombination and somatic hypermutation (Lieschke and Trede, 2009; Trede et al., 2004). In addition, the number of T-cells in the zebrafish has been approximated to about 2×10 5 cells, a 106 fold lower number compared to the T-cell numbers found in mice. Therefore, in contrast to TCR sequencing studies performed in mammals using isolated T-cell populations, the zebrafish offers the possibility to perform far more complete immune repertoire studies. In this work we combine the experimental advantages offered by the zebrafish with high-coverage sequencing and computational approaches to investigate the full diversity of the TCR repertoire under homeostatic conditions and its response to challenge with self and non-self antigens.
Results
The TCR β-chain repertoire provides an accurate representation of TCR diversity (Miles et al., 2011). Moreover, although two C-region TCR β-chain genes have been identified in the zebrafish, Cβ1 and Cβ2, transcripts of the Cβ2 segment are very rare (Meeker et al., 2010). Thus, we focused our efforts on the analysis of the zebrafish TCRβ1 repertoire. To analyze the TCR repertoire in zebrafish we developed a method for TCR library generation from whole zebrafish mRNA based on 5′RACE amplification from a single primer annealing to the constant TCR region (Douek et al., 2002) (Fig. 1A). This method uses a single constant region (C-region) and a 5′-anchor primer rather than multiple J or V region primers to avoid differential PCR amplification efficiencies and subsequent library bias (Boyd et al., 2009; Robins et al., 2009; Wang et al., 2010). To confirm the specificity of the method, we cloned and sequenced the amplification products. We found that 100% of the amplification products correspond to TCRβ1 sequences, demonstrating that this amplification method utilizing a single C-region primer is specific (Fig. 1B).
Figure 1. Sequencing of the zebrafish TCR repertoire.
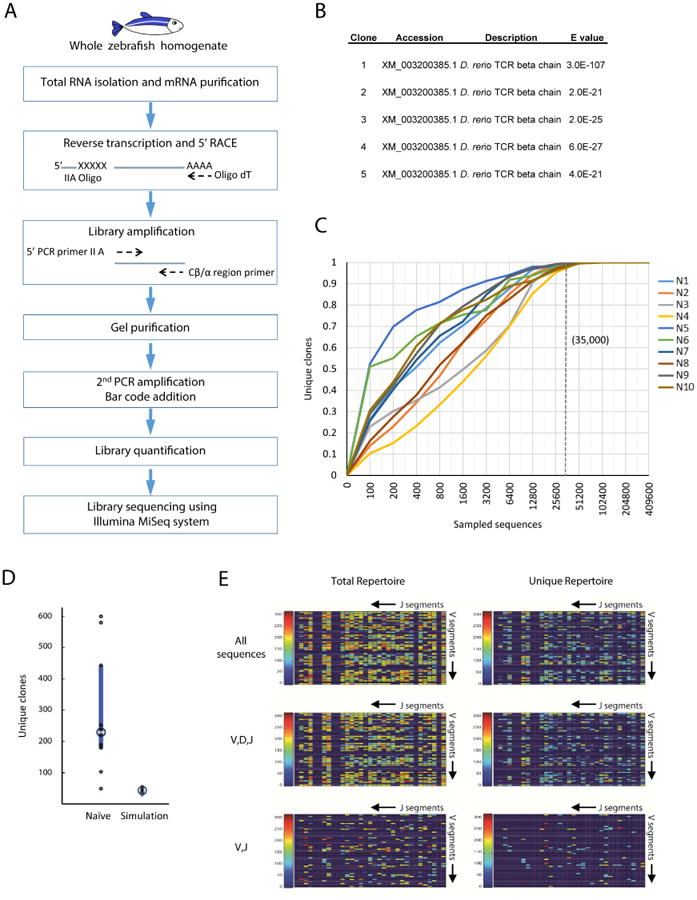
(A) Flow diagram of the experimental protocols used to sequence the zebrafish TCRβ1 (and TCRα) repertoire. mRNA was purified from whole zebrafish, each fish separately. Reverse transcription was performed using an oligo dT primer and 5′RACE was obtained using oligonucleotide IIA (IIA Oligo). Library amplification was done using a primer specific for the Cβ1 or Cα region (Cβ1/α region primer) and a primer complementary to IIA Oligo (5′ PCR primer IIA). The library was gel purified and bar codes were added. The library was quantified and sequenced. (B) Verification of TCRβ1 sequences obtained after the library amplification step. Five clones were sequenced and the top hits of the blasted sequences are presented. (C) Rarefaction analysis of TCRβ1 diversity. Each curve gives the fraction of the observed repertoire as a function of the number of obtained sequences in each of 10 naïve (non-immunized) fish. The dotted line indicates the point at which all fish reach 99% coverage of the total unique sequences, corresponding to 35,000 sequences. (D) Number of unique clones in 10 naïve fish and 10 simulated repertoires. The circle indicates the median. The small number of sequences in the simulated repertoire results form adherence to the distribution of clones within each fish. The abundance of specific clones (learned from the natural occurring repertoire in the naïve fish) dictates a distribution with a few highly represented clones. (E) Heat maps of V,J combination abundance in the total and the unique repertoire of naïve zebrafish. The data are segregated based on the usage of the D segment.
To analyze the number of sequences required to provide a comprehensive coverage of the TCRβ1 repertoire with our sequencing strategy we performed rarefaction studies using partial samples of the full TCRβ1 sequencing data. We found that the total number of V(D)J combinations detected was asymptotic towards saturation, with all of the expressed V(D)J species predicted to be detected by sampling 35,000 sequences or more (Fig. 1C).
Small TCRβ1 repertoire in adult zebrafish
The number of potential TCR combinatorial possibilities exceeds the total number of peripheral T cells in an individual (Davis and Bjorkman, 1988), suggesting that only a fraction of the potential TCR repertoire is actually expressed. Indeed, the repertoire of unique αβ TCRs has been estimated at ∼107 clones in the human (Arstila et al., 1999) and ∼106 in the mouse (Casrouge et al., 2000), a small fraction of the 1015-1020 unique αβ TCRs repertoire that could be potentially generated by these mammalian immune systems. To determine the fraction of the potential TCRβ1 repertoire expressed by zebrafish, we first constructed a computational model of TCR recombination in the zebrafish based on available sequences and our own data on V(D)J recombination, deletions, insertions and substitutions in TCRβ1 sequences (Fig. S1). This simulation, which considers the biophysical properties of recombination, asymptotically estimates an upper limit of 400,000 unique sequences. Because of the limitations imposed by the biophysical features of the recombination process, this estimate is smaller than the 108-1020 sequences that could result from all the potential V(D)J combinations (Benichou et al., 2012).
We then used our computational model to simulate the TCRβ1 repertoire for 10 individual fish, considering not only the number of unique sequences detected, but also, the frequency of these sequences in the total TCRβ1 repertoire. Surprisingly, the model predicted a TCRβ1 repertoire consisting of only 40 unique TCRβ1 clones per fish. This low predicted number is comparable to the zebrafish TCR repertoire detected in our sequencing efforts, in which we detected 49 - 599 unique TCRβ1 CDR3 sequences per individual fish (Fig. 1D). This small number of unique T-cell clones is in agreement with previous studies of the zebrafish B cell repertoire, which has been estimated to harbor 9-200 unique V(D)J sequences generated by recombination, expanded to 1200-6000 antibody clones by somatic hypermutation (Weinstein et al., 2009). Taken together, these data suggest that only a small fraction of the potential TCRβ1 repertoire is actually expressed in adult zebrafish, a fraction significantly lower than the one estimated for the murine and human immune system.
Biased TCRβ1 repertoire in naïve zebrafish
The V(D)J recombination system generates a diverse TCRβ1 repertoire based on the stochastic use of V, D and J gene segments and the deletion and insertion of nucleotides (Davis and Bjorkman, 1988; Fujimoto and Yamagishi, 1987; Malissen et al., 1986; Okazaki et al., 1987; Tonegawa, 1983). Although each V, D, J segment has a theoretically equal chance of being incorporated into a mature TCR, the murine and human TCR repertoires are not evenly distributed and specific V,D,J genes are used more often (Argaet et al., 1994; Cibotti et al., 1994; Moss et al., 1991). Thus, we studied the TCRβ1 repertoire in 10 naïve zebrafish for V,D,J usage bias. We analyzed between 97,503 and 232,193 sequences per fish, a number significantly higher than the 35,000 sequences required to cover the whole TCRβ1 repertoire. Each sequence was aligned with reference sequences (Howe et al., 2013; Kettleborough et al., 2013) to identify specific V, D and J genes, and then the frequency of each V,J combination, including or not the D segment, was estimated (Turner et al., 2006). We found that almost all possible V, J pairs were used in the zebrafish TCRβ1 repertoire when sequences containing the D segment were analyzed (Fig. 1E). However, only a subset of pairs was utilized when the D segment was not included, suggesting that only a limited set of available VJ combinations overcomes the limitations imposed by the 12/23 rule (Akira et al., 1987; Yancopoulos et al., 1986). These data suggest that biases in gene segment usage characterize the zebrafish TCRβ1 repertoire. In addition, the analysis of the total repertoire, that is the collection of TCRβ1 unique sequences adjusted for their frequency, revealed the over-representation of specific V, J pairs (Fig. 1E) suggesting that their expansion results from antigenic stimulation.
Convergent recombination drives the generation of zebrafish public TCR clones
Public T-cell clones express TCR sequence motifs shared by different individuals, and are often expanded by immunization, infection, or autoimmunity (McBerry et al., 2012; Venturi et al., 2008). To study the role of public repertoires in zebrafish, we defined a public sequence as one appearing in at least two different individuals, as previously defined in other studies (Li et al., 2012). We found that public clones represent 36% of the total TCRβ1 CDR3 nucleotide sequences and 40% of the amino acid sequences (Fig. 2A). Conversely, our computational model predicted no sharing of TCR sequences between individual fish (p<10−9). Thus, the zebrafish TCRβ1 repertoire contains a relatively low number of unique sequences, many of which are shared between different individuals.
Figure 2. Convergent recombination characterizes public TCR clones that dominate the zebrafish TCRβ1 repertoire.
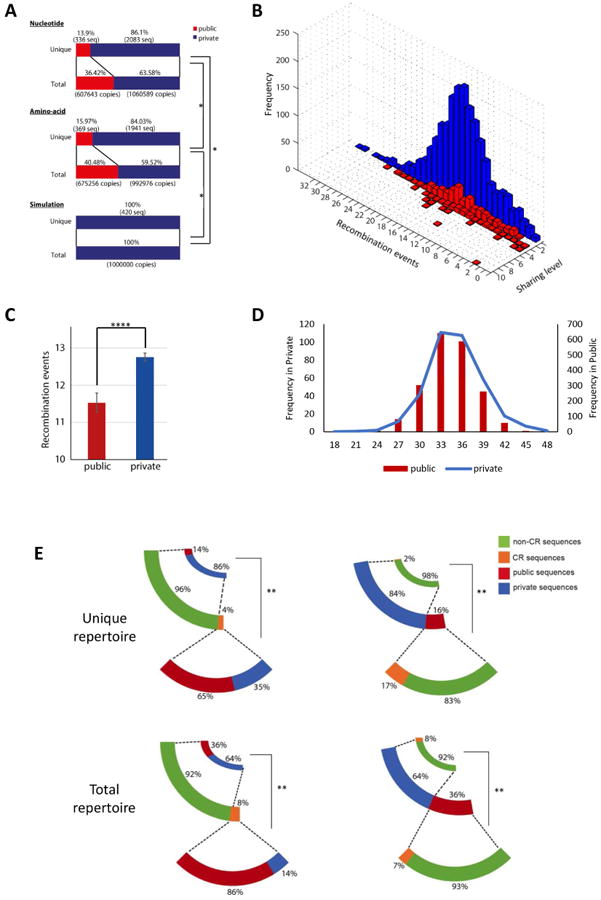
(A) Observed and predicted public and private TCRβ1 sequences in naïve fish. (B) Recombination events in the private and in the public TCRβ1 repertoires, and their association with their sharing. The height of the histogram bars represents the frequency of the clone in the pooled repertoire. Red, public; blue, private. (C) Statistical analysis of recombination events in public and private TCRβ1 clones. **** p<1E-12. (D) Distribution of CDR3 lengths in the private and public TCRβ1 clones. (E) Fraction of public and private TCRβ1 clones within the Unique TCRβ1 repertoire. The fraction of sequences generated by convergent recombination is shown. ** p<0.01.
To study the genetic mechanisms involved in the generation of public T-cell clones we analyzed the frequency of recombination events in public and private TCRβ1 sequences. We found significantly fewer recombination events in public TCRβ1 sequences (Figs. 2B and C), in agreement with previous reports of public TCRs being closer to germline configurations (Ishizuka et al., 2008; Miles et al., 2010; Vermijlen et al., 2010). Of note, public and private clones do not differ in their CDR3 length (Fig. 2D), suggesting that the reduction in recombination events in public clones is not a byproduct of shorter CDR3 sequences.
Convergent recombination, the process by which multiple recombination events produce the same nucleotide sequence and multiple nucleotide sequences encode the same amino-acid sequence, is considered an important driving force in the generation of public T-cell responses (Quigley et al., 2010; Venturi et al., 2006). To study the contribution of convergent recombination in zebrafish public T-cell responses we searched for identical TCRβ1 amino acid sequences originating from different nucleotide sequences in naïve zebrafish. We found a significant contribution of convergent recombination to the public TCR repertoire of naive fish. Four percent of the amino acid sequences in the unique TCRβ1 repertoire are produced by convergent recombination (Fig. 2E, left panel). Strikingly, 65% of TCRβ1 sequences generated by convergent recombination are public (Fig. 2E, left panel), suggesting that convergent recombination plays a significant role during the generation of public TCRβ1 sequences. Indeed, 17% of the public TCRβ1 sequences in the unique repertoire were generated by convergent recombination, as opposed to 2% in private sequences (Fig. 2E right panel). Taken together, these data show that convergent recombination drives the generation of the public repertoire in naïve zebrafish.
Antigenic stimulation expands public TCRβ1 clones
The frequency of a specific TCR in the total repertoire reflects the number of T cells bearing that specific TCR and the amount of mRNA produced by each T cell, both of which are controlled by the stimulation of T cells by their cognate antigen. The size of public clones in the naïve total TCRβ1 repertoire was directly correlated with their usage by different individuals (Fig. 3A), suggesting that the same clones are expanded in different individuals in response to antigenic stimulation. Thus, to study the effect of antigenic stimulation on private and public T-cell responses we analyzed the TCRβ1 repertoire 21 days after immunization of naïve zebrafish with the self-antigen calmodulin (CALM, Fig. S2A), the non-self antigen keyhole limpet hemocyanin (KLH) or administration of the common polyclonal stimulus lectin from Phaseolus vulgaris (PHA). Only 16% of the TCRβ1 clones expanded by PHA administration were expanded by immunization with KLH or CALM, suggesting that PHA activates a larger number of TCRβ1 bearing T cells than protein antigens (not shown).
Figure 3. Public clones dominate the TCRβ1 repertoire.
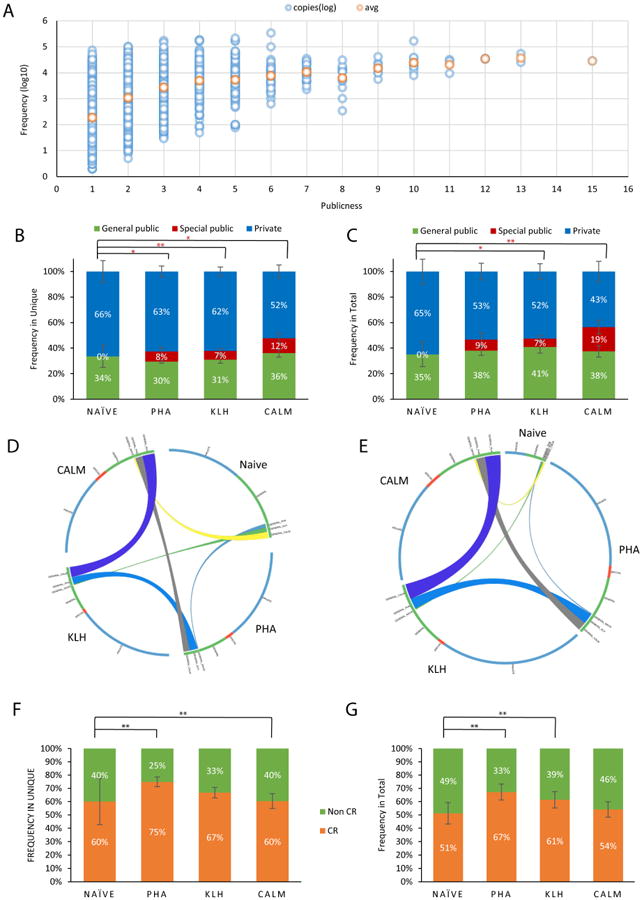
(A) Relationship between sequence sharing between different individuals and the number of copies of each TCRβ1 clone. (B, C) Contribution of private, general public and special public sequences to the unique (B) and total (C) TCRβ1 repertoire following immunization with KLH, CALM or polyclonal stimulation with PHA. (D, E) Sharing of TCRβ1 sequences between the different groups. In panel (D) each group occupies the same fraction of the circle, regardless of repertoire size, while in panel (E) each sequence occupies the same portion of the circle. The circle is colored based on whether the TCRβ1 clone is private (blue), special public (red) or general public (green). Edges represent sequences shared between each 2 groups. (F, G) Fraction of TCRβ1 clones generated by convergent recombination in the unique repertoire (F) and the total (G) TCRβ1 public repertoire.
We then investigated the effect of immunization on the public TCRβ1 repertoire. Immunization with KLH or CALM, or polyclonal activation with PHA expanded public clones in the unique and the total TCRβ1 repertoire (Figs. 3B and C). Indeed, our analyses identified two groups within the public clones: general public clones, consisting of TCRβ1 sequences shared by all immunization groups, and special public clones, consisting of TCRβ1 sequences shared only by fish that received the same antigenic stimulation. Special public clones were detected in the unique and total TCRβ1 repertoire following KLH and CALM immunization and also PHA treatment (Figs. 3B and C). However, immunization with the self-antigen CALM resulted in higher special public responses, suggesting that public TCRβ1 clones are enriched for self reactive elements. Immunization did not affect TCR diversity as indicated by the analysis of the Gini coefficient (Fig. S3A). Thus, immunization with self and non-self antigens stimulates public T-cell responses, which are partially cross-reactive because 41% of the public TCRβ1 expanded by CALM were also expanded by KLH immunization.
The expansion of the public repertoire in response to immunization resulted in part from the expansion of TCRβ1 T-cell clones identified as public in naïve zebrafish, and also from the sharing of TCRβ1 sequences previously identified as private in naïve zebrafish (Figs. 3D and E). Interestingly, most of the public T-cell clones in the unique and the total TCRβ1 repertoire, both general and specific, were generated by convergent recombination (Figs. 3F, G and S3B). Taken together these data identify convergent recombination as an important mechanism for the generation of public clones responsive to self and foreign antigens.
We then investigated the origin of public clones. We found that public clones in KLHimmunized fish or those treated with PHA originated mostly from low frequency clones in naïve zebrafish (Figs. 4A and B). However, public clones in CALM-immunized fish originated from both high and low frequency clones in naïve zebrafish, suggesting that self-reactive public T clones are major components of the adult T-cell repertoire in naïve fish.
Figure 4. Origin of general public TCRβ1 clones.
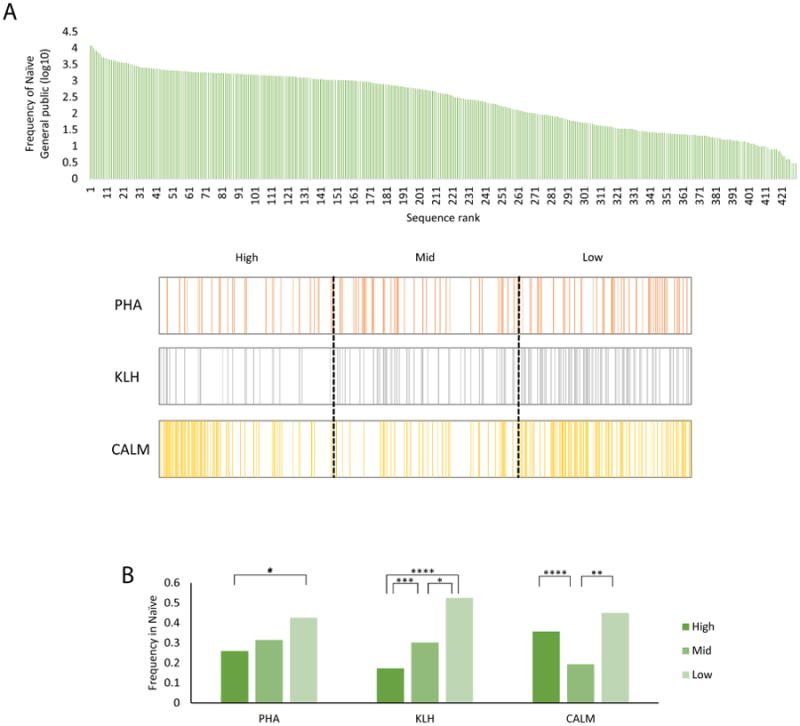
(A) TCRβ1 general public sequences in the naïve repertoire, ranked according to their frequency. The lower panels show general public sequences in the TCRβ1 repertoire of fish in the PHA, KLH and CALM groups. (B) To analyze the origin of general public sequences detected in the immunized repertoire, sequences were classified in the naïve TCRβ1 repertoire into low-, mid- and high-abundant clones and then quantified for the fraction of these groups in the immunized repertoire.
Time course analysis of the TCRβ1 repertoire in response to immunization
To further elucidate the effect of antigenic stimulation on the T-cell response, we analyzed the TCRβ1 repertoire 14, 21 and 28 days after immunization of naïve zebrafish with PHA, KLH and CALM. In these experiments the zebrafish were boosted by immunization at day 14. Special public TCRβ1 clones were identified at all time points after immunization or PHA administration (Fig. 5). In agreement with our previous findings, the T-cell response to PHA stimulation or immunization with KLH or CALM was dominated by public TCRβ1 clones generated by convergent recombination (Figs. S3C-F). However, the clonal responses induced by the different stimuli showed differences in their kinetics. In KLH-immunized and PHA-treated fish, the frequency of special public TCRβ1 clones peaked 14 days after treatment. Immunization with CALM resulted in higher frequencies of both general and special public clones (Fig. 5). However, the peak in the number of special public clones expanded by CALM immunization was delayed and was only observed 1 week after boosting probably reflecting the need for additional antigenic stimulation needed to break self-tolerance (Fig. 5). Of note, immunizations over time had no effect on TCRβ1 diversity, as measured by the Gini coefficient (Fig. S3G).
Figure 5. Time course analysis of the TCRβ1 repertoire.

Changes in the private, general public and special public fractions of the TCRβ1 repertoire following treatment with PHA or immunization with KLH or CALM.
Antigen stimulation expands public clones in the TCRα repertoire
A diverse repertoire has also been described for the zebrafish TCRα (Haire et al., 2000). Thus, we analyzed the TCRα repertoire using a primer specific for the C-region of the TCRα-chain as described in Fig. 1A. This method was specific because 100% of the amplification products corresponded to TCRα sequences (Fig. 6A). Similarly to our observations on the TCRβ1 repertoire, we found that the size of public clones in the naïve total TCRα repertoire was directly correlated with their usage by different individuals (Fig. 6B).
Figure 6. The zebrafish TCRα repertoire.
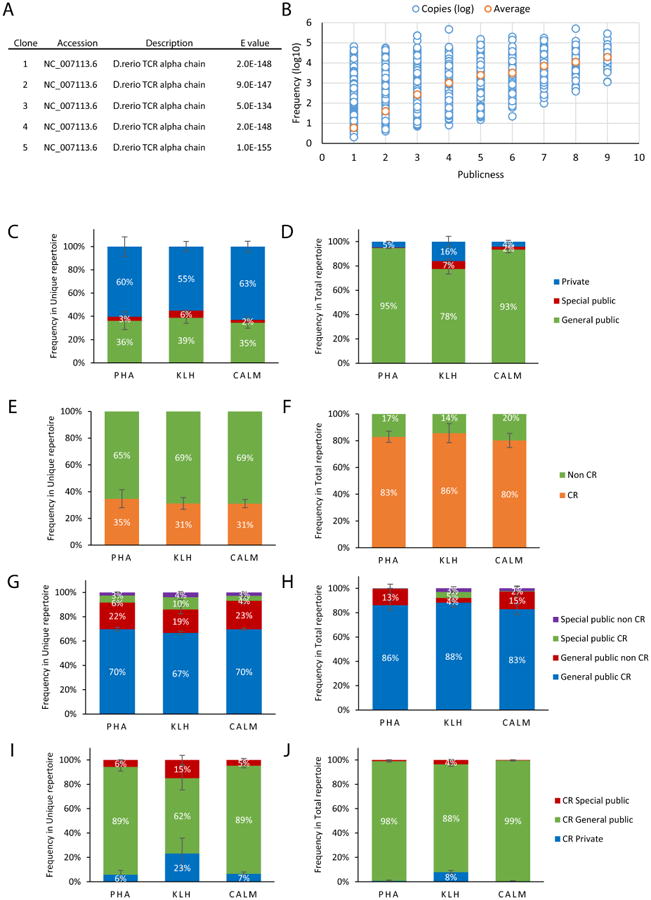
(A) Verificaton of TCRα sequences obtained after library amplification, five clones were sequenced and the top hits of the blasted sequences are presented in the table. (B) Relationship between sequence sharing and the number of copies of each TCRα clone. The orange dot for each sharing level gives the average frequency of clones at that specific sharing level. (C, D) Effect of PHA administration or immunization with KLH or CALM in the private, general public and special public fractions of the unique (C) and total (D) TCRα repertoire. (E, F) Frequency of clones generated by CR in the unique (E) and total (F) TCRα repertoire following PHA administration or immunization with KLH or CALM. (G, H) Fraction of CR and non-CR clones in the general public and special public unique(G) and total TCRα repertoire (H). (I, J) Fractions of private, general public and special public clones within CR and non-CR clones in the unique (I) and total (J) TCRα repertoire.
We then investigated the response of the TCRα repertoire to immunization with KLH or CALM, or to PHA administration. Immunization with CALM or KLH or PHA administration did not affect the number of TCRα unique sequences (Fig. S4). Sequence sharing in the TCRα repertoire, however, is at least as strong as that detected in TCRβ1 repertoire, with most TCRα clones being general public sequences (Figs. 6C and D). We also identified special public TCRα clones following immunization, which were more prevalent in the KLH-immunized fish (Fig. 6D). Taken together, these observations suggest that public TCRα sequences in the naive repertoire are expanded in response to antigenic stimulation.
We detected a large percentage of clones generated by convergent recombination in the unique TCRα repertoire across all the immunization and treatment groups (Fig. 6E), while in the total repertoire the percentages were even higher, suggesting that T-cell clones bearing TCRα public sequences are larger (Fig. 6F). Similar to our previous observations with TCRβ public clones, TCRα public clones were mostly generated by CR (Figs. 6G-J). Collectively, these observations suggest that the TCRα repertoire follows the same rules that we described for the TCRβ, being dominated by public clones some of which are expanded by antigenic stimulation.
Finally, based on the frequency of each TCRα and TCRβ zebrafish clone detected in our sequencing efforts we constructed a probabilistic model of all potential TCRαβ combinations (Fig. 7); a list of the most frequent TCRα and TCRβ sequences used in the construction of the model is provided in Table S2. Based on this model, we estimate that the 2×10 5 T cells present in an adult zebrafish contain at least unique 1.5×10 4 TCRαβ pairs, present in low frequency in the zebrafish TCRαβ repertoire. It should be noted that this is a lower estimate and TCRαβ diversity may be higher, for example as a result of the expression of more than one a chain by T cells described in humans and other vertebrates (Padovan et al., 1993).
Figure 7. A computational model of the TCRαβ repertoire.

The horizontal axis represents TCRβ1 chains while the vertical axis represents TCRα chains. Each blue square in the figure represents a potential TCRαβ pair.
Discussion
In this work we analyzed the zebrafish TCRβ1 and TCRα repertoire and its response to immunization with self and non-self antigens. We found that the zebrafish TCR repertoire is small and biased towards the use of certain V-J combinations, in a similar manner to what is known from partial repertoire analyses in other vertebrates (Miles et al., 2011). Moreover, the analysis of the TCR repertoire revealed the over-representation of specific V-J pairs, suggestive of clonal expansion in response to antigenic stimulation. These observations suggest that the zebrafish TCR repertoire is shaped by the balance between T-cell expansion in response to self and non-self antigens and T-cell competition for limited growth and survival factors. In addition, since decreased repertoire diversity is linked to impaired T-cell immunity (Yager et al., 2008), these data suggest that compensatory mechanisms operate in zebrafish to provide protective immunity against pathogens.
Public T-cell clones encoded with minimal alteration to germline gene sequences characterize the TCR repertoire of vertebrates (McBerry et al., 2012; Venturi et al., 2008). We detected high frequencies of public TCRαβ sequences in the zebrafish. High frequency sharing of antibody sequences has also been reported in the zebrafish antibody repertoire, which is also characterized by its small size in agreement with our observations on the TCRαβ repertoire (Jiang et al., 2011; Weinstein et al., 2009). Public T-cell clones have been shown to contribute to anti-viral immune responses (Miles et al., 2011). We identified public T-cell clones as major components of the zebrafish response to immunization. Moreover, we detected a significant overlap in the public TCRαβ sequences expanded in response to self and foreign antigens that share no sequence homology, suggesting that public T-cell clones are highly cross-reactive. Cross-reactivity with self antigens plays an important role in the development of the TCR repertoire and T-cell responses to foreign antigens (Birnbaum et al., 2014; Fulton et al., 2015; Krogsgaard et al., 2005; Mandl et al., 2013; Stefanova et al., 2002). Collectively, these observations suggest that public T-cell responses allow a relatively small TCR repertoire to cope with the diverse range of antigens presented by pathogens.
The dominant role of public T-cell responses in zebrafish might represent an early step during the evolution of adaptive immunity (Boehm et al., 2012; Cooper and Herrin, 2010; Flajnik and Kasahara, 2010; Guo et al., 2009). However, while the cross-reactivity of public T-cell clones could potentially compensate for the small size of the TCR repertoire, it might also increase the risk for the development of pathogenic autoimmunity. Interestingly, a FoxP3 homologue is detectable in zebrafish (Quintana et al., 2010), suggesting that the potential for the development of autoimmunity was co-selected with mechanisms of immune regulation.
Public T-cell clones do not constitute a dominant fraction of the TCR repertoire of mice, humans and other mammals (Miles et al., 2011). However, polyfunctional and cross-reactive public T-cell clones are detected in HIV-1 controllers (Chen et al., 2012; Kosmrlj et al., 2010), and similar observations have been made in the context of infection with herpes virus (Zhu et al., 2013), as well as shared self-peptide MHC-specific clones in healthy individuals (Yu et al., 2015). Although it is still unknown whether public TCRs directly control the polyfunctionality and polyreactivity of public T cells (Tubo et al., 2013), these observations suggest that cross-reactive public T-cell clones contribute to pathogen control in organisms with larger TCR repertoires. Their restricted diversity and sharing by different individuals, together with their ability to respond to diverse self and non-self molecules, are features of vertebrate public TCRs that might provide a mechanism for the rapid generation of protective T-cell immunity allowing a short temporal window for the development of more specific private T-cell responses.
Experimental Procedures
Fish maintenance
1 year old male zebrafish (AB strain) were maintained in a 28-30°C system with a 14/10 hrs light/dark cycle in accordance with guidelines by the Institutional Animal Care and Use Committee of Harvard Medical School.
Immunization
Fish were anaesthetized using 0.02% Tricaine methanesulfonate (Sigma-Aldrich) and immunized intra-peritoneally (i.p.) with a 10μl emulsion containing 1:1 Incomplete Freund's Adjuvant (IFA, Difco Laboratories) and 90% PBS (Invitrogen), 0.25μg lipopolysaccharide (ultrapure LPS, Invivogen), 0.7μg CpG Oligonucleotide ODN 1826 (Invivogen) and 2 μg of either PHA (Sigma-Aldrich), KLH (Sigma-Aldrich) or CALM (Creative BioMart, NY, USA). Two weeks later the fish were boosted with PHA, KLH or CALM in 1:1 IFA: 90% PBS.
TCRαβ sequencing and annotation
Total RNA was extracted from whole fish homogenate and cDNA was generated. cDNA from each of fish was used for TCRβ/α chain library amplification using the 5′PCR primer IIA from the SMARTer™ Pico PCR cDNA Synthesis kit (Clontech) and the constant region primer (Table S1). The library was gel-purified and barcodes were added using the same reaction as for the library amplification and the primers listed in Table S1.
TCRβ and TCRα annotation was performed by using NCBI BLAST+ to identify the V and J germline genes of a TCR read, and then the CDR3 was determined by finding the conserved cysteine at the 5′ end of the CDR3 and the conserved Phenylalanine at the 3′ end of the CDR3.
Supplementary Material
Highlights.
We studied the response of the zebrafish TCRαβ repertoire to antigenic stimulation.
The zebrafish TCRαβ repertoire is dominated by cross-reactive public clones.
Public T-cells facilitate the rapid generation of protective T-cell immunity.
The zebrafish provides a model to study the T cell response at a systems levels.
Acknowledgments
This work was supported by grants AI075285, and AI093903 from the National Institutes of Health and a Harry Weaver Scholar Award and grant RG4111A1 from the National Multiple Sclerosis Society to FJQ, grant 2011154 from the BSF to FJQ and SE. FJQ thanks Maria Ethel del Aguila for useful discussions and support. RC is supported by a postdoctoral fellowship from the Swedish Research Council. MJ is supported by Sigrid Juselius fellowship, The Paulo Foundation, The Finnish Multiple Sclerosis Foundation, Orion-Farmos Research Foundation and Saastamoinen Foundation. This work was funded in part through the intramural program of the National Institute of Allergy and Infectious Diseases, NIH.
Footnotes
Authors' contribution: RC, MJ, JK and CC performed in vitro and in vivo experiments in zebrafish; JA sequenced the zebrafish TCR libraries; HP, SD, GY, YL, LC and SE performed bioinformatics analysis; RC, MJ, DCD and FJQ conceived the experimental design; RC, MJ, DCD, SE and FJQ wrote the manuscript; RC, MJ, DCD, SE and FJQ conceived the study; DCD and FJQ supervised the TCR sequencing; SE and FJQ supervised the bioinformatic analysis; FJQ supervised the overall project.
Accession Numbers: Sequences have been uploaded to Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) and can be found under the following accession numbers: SAMN04440425, SAMN04440426, SAMN04440427, SAMN04440428, SAMN04440429, SAMN04440430, SAMN04440431, SAMN04440432, SAMN04440433, SAMN04440434, SAMN04440435, SAMN04440436, SAMN04440438, SAMN04440442, SAMN04440443, SAMN04440444, SAMN04440445, SAMN04440446, SAMN04440447, SAMN04440448, SAMN04440449, SAMN04440450, SAMN04440454, SAMN04440455, SAMN04440456, SAMN04440457, SAMN04440458, SAMN04440459, SAMN04440460, SAMN04440461, SAMN04440462, SAMN04440463, SAMN04440464, SAMN04440465, SAMN04440468, SAMN04440469, SAMN04440470, SAMN04440471, SAMN04440472, SAMN04440592, SAMN04440593, SAMN04440594, SAMN04440595, SAMN04440596, SAMN04440597, SAMN04440598, SAMN04440599.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Akira S, Okazaki K, Sakano H. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining. Science. 1987;238:1134–1138. doi: 10.1126/science.3120312. [DOI] [PubMed] [Google Scholar]
- Argaet VP, Schmidt CW, Burrows SR, Silins SL, Kurilla MG, Doolan DL, Suhrbier A, Moss DJ, Kieff E, Sculley TB, Misko IS. Dominant selection of an invariant T cell antigen receptor in response to persistent infection by Epstein-Barr virus. J Exp Med. 1994;180:2335–2340. doi: 10.1084/jem.180.6.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- Benichou J, Ben-Hamo R, Louzoun Y, Efroni S. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology. 2012;135:183–191. doi: 10.1111/j.1365-2567.2011.03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, Ozkan E, Davis MM, Wucherpfennig KW, Garcia KC. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T, Iwanami N, Hess I. Evolution of the immune system in the lower vertebrates. Annual review of genomics and human genetics. 2012;13:127–149. doi: 10.1146/annurev-genom-090711-163747. [DOI] [PubMed] [Google Scholar]
- Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nature immunology. 2013;14:1007–1013. doi: 10.1038/ni.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casrouge A, Beaudoing E, Dalle S, Pannetier C, Kanellopoulos J, Kourilsky P. Size estimate of the alpha beta TCR repertoire of naive mouse splenocytes. Journal of immunology (Baltimore, Md: 1950) 2000;164:5782–5787. doi: 10.4049/jimmunol.164.11.5782. [DOI] [PubMed] [Google Scholar]
- Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nature immunology. 2012;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibotti R, Cabaniols JP, Pannetier C, Delarbre C, Vergnon I, Kanellopoulos JM, Kourilsky P. Public and private V beta T cell receptor repertoires against hen egg white lysozyme (HEL) in nontransgenic versus HEL transgenic mice. J Exp Med. 1994;180:861–872. doi: 10.1084/jem.180.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MD, Herrin BR. How did our complex immune system evolve? Nature reviews Immunology. 2010;10:2–3. doi: 10.1038/nri2686. [DOI] [PubMed] [Google Scholar]
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai KL, Karandikar NJ, Casazza JP, Koup RA. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. Journal of immunology (Baltimore, Md: 1950) 2002;168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- Efron B, Thisted R. Estimating the number of unseen species: How many words did Shakespeare know? Biometrika. 1976;63:435–447. [Google Scholar]
- Fisher RA, Corbet AS, Williams CB. The relation between the number of species and the number of individuals in a random sample of an animal population. The Journal of Animal Ecology. 1943:42–58. [Google Scholar]
- Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nature reviews Genetics. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JD, Warren RL, Webb JR, Nelson BH, Holt RA. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome research. 2009;19:1817–1824. doi: 10.1101/gr.092924.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S, Yamagishi H. Isolation of an excision product of T-cell receptor alpha-chain gene rearrangements. Nature. 1987;327:242–243. doi: 10.1038/327242a0. [DOI] [PubMed] [Google Scholar]
- Fulton RB, Hamilton SE, Xing Y, Best JA, Goldrath AW, Hogquist KA, Jameson SC. The TCR's sensitivity to self peptide-MHC dictates the ability of naive CD8(+) T cells to respond to foreign antigens. Nature immunology. 2015;16:107–117. doi: 10.1038/ni.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Hirano M, Herrin BR, Li J, Yu C, Sadlonova A, Cooper MD. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haire RN, Rast JP, Litman RT, Litman GW. Characterization of three isotypes of immunoglobulin light chains and T-cell antigen receptor alpha in zebrafish. Immunogenetics. 2000;51:915–923. doi: 10.1007/s002510000229. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka J, Stewart-Jones GB, van der Merwe A, Bell JI, McMichael AJ, Jones EY. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its Vbeta domain. Immunity. 2008;28:171–182. doi: 10.1016/j.immuni.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Jiang N, Weinstein JA, Penland L, White RA, 3rd, Fisher DS, Quake SR. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc Natl Acad Sci U S A. 2011;108:5348–5353. doi: 10.1073/pnas.1014277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettleborough RN, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, Sealy I, White RJ, Herd C, Nijman IJ, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, Pereyra F, Carrington M, Walker BD, Chakraborty AK. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogsgaard M, Li QJ, Sumen C, Huppa JB, Huse M, Davis MM. Agonist/endogenous peptide-MHC heterodimers drive T cell activation and sensitivity. Nature. 2005;434:238–243. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- Li H, Ye C, Ji G, Wu X, Xiang Z, Li Y, Cao Y, Liu X, Douek DC, Price DA, Han J. Recombinatorial biases and convergent recombination determine interindividual TCRbeta sharing in murine thymocytes. Journal of immunology (Baltimore, Md: 1950) 2012;189:2404–2413. doi: 10.4049/jimmunol.1102087. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Trede NS. Fish immunology. Curr Biol. 2009;19:R678–682. doi: 10.1016/j.cub.2009.06.068. [DOI] [PubMed] [Google Scholar]
- Linnemann C, Heemskerk B, Kvistborg P, Kluin RJ, Bolotin DA, Chen X, Bresser K, Nieuwland M, Schotte R, Michels S, et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nature medicine. 2013;19:1534–1541. doi: 10.1038/nm.3359. [DOI] [PubMed] [Google Scholar]
- Malissen M, McCoy C, Blanc D, Trucy J, Devaux C, Schmitt-Verhulst AM, Fitch F, Hood L, Malissen B. Direct evidence for chromosomal inversion during T-cell receptor beta-gene rearrangements. Nature. 1986;319:28–33. doi: 10.1038/319028a0. [DOI] [PubMed] [Google Scholar]
- Mandl JN, Monteiro JP, Vrisekoop N, Germain RN. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity. 2013;38:263–274. doi: 10.1016/j.immuni.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBerry C, Gonzalez RM, Shryock N, Dias A, Aliberti J. SOCS2-induced proteasome-dependent TRAF6 degradation: a common anti-inflammatory pathway for control of innate immune responses. PloS one. 2012;7:e38384. doi: 10.1371/journal.pone.0038384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker ND, Smith AC, Frazer JK, Bradley DF, Rudner LA, Love C, Trede NS. Characterization of the zebrafish T cell receptor beta locus. Immunogenetics. 2010;62:23–29. doi: 10.1007/s00251-009-0407-6. [DOI] [PubMed] [Google Scholar]
- Miles JJ, Bulek AM, Cole DK, Gostick E, Schauenburg AJ, Dolton G, Venturi V, Davenport MP, Tan MP, Burrows SR, et al. Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus infection. PLoS pathogens. 2010;6:e1001198. doi: 10.1371/journal.ppat.1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles JJ, Douek DC, Price DA. Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol. 2011;89:375–387. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- Moss PA, Moots RJ, Rosenberg WM, Rowland-Jones SJ, Bodmer HC, McMichael AJ, Bell JI. Extensive conservation of alpha and beta chains of the human T-cell antigen receptor recognizing HLA-A2 and influenza A matrix peptide. Proc Natl Acad Sci U S A. 1991;88:8987–8990. doi: 10.1073/pnas.88.20.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell EW, Davis MM. Beyond model antigens: high-dimensional methods for the analysis of antigen-specific T cells. Nature biotechnology. 2014 doi: 10.1038/nbt.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nature reviews Immunology. 2004;4:123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Davis DD, Sakano H. T cell receptor beta gene sequences in the circular DNA of thymocyte nuclei: direct evidence for intramolecular DNA deletion in V-D-J joining. Cell. 1987;49:477–485. doi: 10.1016/0092-8674(87)90450-8. [DOI] [PubMed] [Google Scholar]
- Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Expression of two T cell receptor alpha chains: dual receptor T cells. Science. 1993;262:422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- Park CO, Kupper TS. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nature medicine. 2015;21:688–697. doi: 10.1038/nm.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley MF, Greenaway HY, Venturi V, Lindsay R, Quinn KM, Seder RA, Douek DC, Davenport MP, Price DA. Convergent recombination shapes the clonotypic landscape of the naive T-cell repertoire. Proc Natl Acad Sci U S A. 2010;107:19414–19419. doi: 10.1073/pnas.1010586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana FJ, Iglesias AH, Farez MF, Caccamo M, Burns EJ, Kassam N, Oukka M, Weiner HL. Adaptive autoimmunity and Foxp3-based immunoregulation in zebrafish. PloS one. 2010;5:e9478. doi: 10.1371/journal.pone.0009478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annual review of immunology. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Stefanova I, Dorfman JR, Germain RN. Self-recognition promotes the foreign antigen sensitivity of naive T lymphocytes. Nature. 2002;420:429–434. doi: 10.1038/nature01146. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand immunity. Immunity. 2004;20:367–379. doi: 10.1016/s1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- Tubo NJ, Pagan AJ, Taylor JJ, Nelson RW, Linehan JL, Ertelt JM, Huseby ES, Way SS, Jenkins MK. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell. 2013;153:785–796. doi: 10.1016/j.cell.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nature reviews Immunology. 2006;6:883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- Turner SJ, La Gruta NL, Kedzierska K, Thomas PG, Doherty PC. Functional implications of T cell receptor diversity. Current opinion in immunology. 2009;21:286–290. doi: 10.1016/j.coi.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V, Kedzierska K, Price DA, Doherty PC, Douek DC, Turner SJ, Davenport MP. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proc Natl Acad Sci U S A. 2006;103:18691–18696. doi: 10.1073/pnas.0608907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nature reviews Immunology. 2008;8:231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- Vermijlen D, Brouwer M, Donner C, Liesnard C, Tackoen M, Van Rysselberge M, Twite N, Goldman M, Marchant A, Willems F. Human cytomegalovirus elicits fetal gammadelta T cell responses in utero. J Exp Med. 2010;207:807–821. doi: 10.1084/jem.20090348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sanders CM, Yang Q, Schroeder HW, Jr, Wang E, Babrzadeh F, Gharizadeh B, Myers RM, Hudson JR, Jr, Davis RW, Han J. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci U S A. 2010;107:1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager EJ, Ahmed M, Lanzer K, Randall TD, Woodland DL, Blackman MA. Age-associated decline in T cell repertoire diversity leads to holes in the repertoire and impaired immunity to influenza virus. J Exp Med. 2008;205:711–723. doi: 10.1084/jem.20071140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos GD, Blackwell TK, Suh H, Hood L, Alt FW. Introduced T cell receptor variable region gene segments recombine in pre-B cells: evidence that B and T cells use a common recombinase. Cell. 1986;44:251–259. doi: 10.1016/0092-8674(86)90759-2. [DOI] [PubMed] [Google Scholar]
- Yu W, Jiang N, Ebert PJ, Kidd BA, Muller S, Lund PJ, Juang J, Adachi K, Tse T, Birnbaum ME, et al. Clonal Deletion Prunes but Does Not Eliminate Self-Specific alphabeta CD8(+) T Lymphocytes. Immunity. 2015;42:929–941. doi: 10.1016/j.immuni.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Peng T, Johnston C, Phasouk K, Kask AS, Klock A, Jin L, Diem K, Koelle DM, Wald A, et al. Immune surveillance by CD8alphaalpha+ skin-resident T cells in human herpes virus infection. Nature. 2013;497:494–497. doi: 10.1038/nature12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


