Summary
Here we provide evidence that RBBP4 modulates temozolomide (TMZ) sensitivity through coordinate regulation of 2 key DNA repair genes critical for recovery from TMZ-induced DNA damage: methylguanine-DNA-methyltransferase (MGMT) and RAD51. Disruption of RBBP4 enhanced TMZ sensitivity, induced synthetic lethality to PARP inhibition and increased DNA damage signaling in response to TMZ. Moreover, RBBP4 silencing enhanced TMZ-induced H2AX phosphorylation and apoptosis in GBM cells. Intriguingly, RBBP4 knockdown suppressed the expression of MGMT, RAD51 and other genes in association with decreased promoter H3K9 acetylation (H3K9Ac) and increased H3K9 tri-methylation (H3K9me3). Consistent with these data, RBBP4 interacts with CBP/p300 to form a chromatin modifying complex that binds within the promoter of MGMT, RAD51 and perhaps other genes. Globally, RBBP4 positively and negatively regulates genes involved in critical cellular functions including tumorigenesis. RBBP4/CBP/p300 complex may provide an interesting target for developing therapy sensitizing strategies for GBM and other tumors.
Keywords: RBBP4, temozolomide, resistance, MGMT, glioblastoma
Introduction
GBM is the most common and aggressive primary brain tumor, which is routinely managed by surgery, radiation (RT) and temozolomide (TMZ) chemotherapy. TMZ cytotoxicity is predominantly mediated by O6-methylguanine (O6-MG) DNA lesions, which are repaired by the DNA repair protein MGMT (Drablos et al., 2004). Consequently, GBM patients whose tumors express low level MGMT, due to promoter hypermethylation, are more responsive to TMZ based therapy (Hegi et al., 2004; Stupp et al., 2009). Unfortunately, even these patients with MGMT promoter hypermethylation develop TMZ resistance, and the majority die within 2 years (Hegi et al., 2005). We and others have shown that TMZ resistance can be induced by MGMT re-expression in a subset of MGMT hypermethylated patients (Cahill et al., 2008; Gaspar et al., 2010; Kitange et al., 2012; Yip et al., 2009) and that MGMT-dependent and - independent resistance mechanisms can emerge in the same patient-derived xenograft model. In this model, co-administration of TMZ and a histone deacetylase dramatically shifts the mechanism of resistance emergence towards MGMT over-expression (Kitange et al., 2012). These latter data suggest that emergence of TMZ resistance can be mediated at an epigenetic level, and to identify underlying epigenetic mechanisms associated with TMZ resistance, we used an shRNA library screen to identify genes regulating TMZ responsiveness. Using this approach we identified retinoblastoma binding protein 4 (RBBP4) as a potential modulator of TMZ response.
The RBBP4 gene encodes a protein that is a component of several chromatin modifying protein complexes with varying effects on gene expression (reviewed in (Wolffe et al., 2000)). RBBP4 contributes to repression of gene transcription as a key member of the nucleosome remodeling and deacetylation (NURD) complex, polycomb repressor complex 2 (PRC2), and SIN3- chromatin modulating complexes (Kuzmichev et al., 2002; Todd and Picketts, 2012; Vermaak et al., 1999). As a member of the chromatin assembly factor 1 (CAF1) complex, RBBP4 regulates chromatin assembly in normal replication and during repair of DNA damage (Furuyama et al., 2006; Zhang et al., 2013). Finally, in a complex with p300/CBP, RBBP4 activates gene transcription through histone acetylation (Zhang et al., 2000). To date, we are unaware of any studies that have directly implicated this protein in the modulation of chemosensitivity. Thus, this paper reports that RBBP4 is a negative modulator of TMZ sensitivity and that disruption of this protein enhances TMZ sensitivity through down-regulation of MGMT and genes involved in HR, including RAD51.
Results
Genome wide shRNA screen identified RBBP4 as modulator of TMZ sensitivity
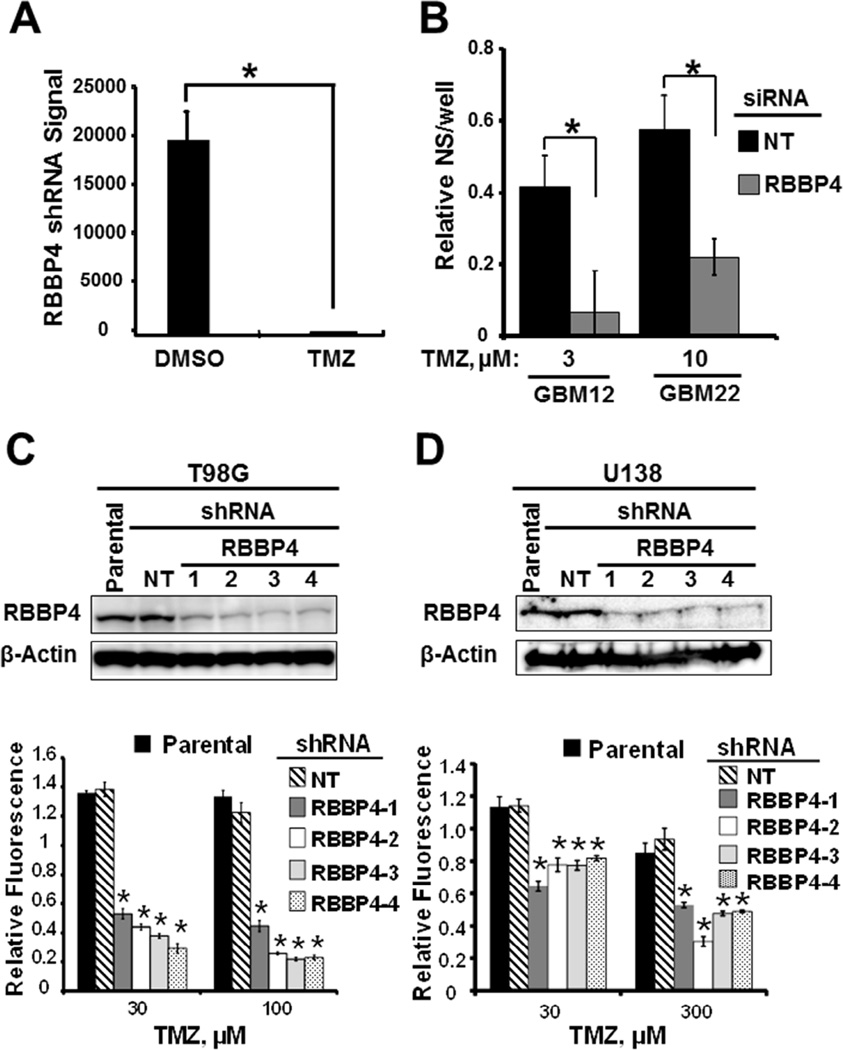
Since resistance is a major hurdle to successful TMZ therapy in GBM, we conducted a high throughput screening to identify potential drivers of TMZ sensitivity in GBM. To that end, we used a pooled shRNA library to identify genome wide modulators of TMZ response in short-term cultured cells from the GBM22 patient-derived xenograft model (Carlson et al., 2011). While this approach identified several candidate genes (data not shown), RBBP4 was particularly interesting because of our ongoing work focused on understanding the epigenetic mechanisms of TMZ resistance (Kitange et al., 2009a; Kitange et al., 2012). The RBBP4 shRNA amplification signal enrichment was significantly higher in the DMSO-treated than in TMZ-treated GBM22 cells (p<0.0001) (Figure 1A), suggesting enhanced TMZ efficacy in cells expressing RBBP4 shRNA. To confirm these results, we evaluated whether siRNA disruption of RBBP4 would sensitize MGMT hypermethylated patient-derived GBM12 and GBM22 cells to TMZ in vitro. Consistently, RBBP4 siRNA, but not the control NT siRNA, induced TMZ sensitization in GBM22 cells following treatment with 10 µM TMZ (p<0.01) and even more profound sensitizing effects were seen in GBM12 (p=0.002) (Figure 1B). Thus, RBBP4 can modulate TMZ response in GBM cells lacking MGMT expression.
Figure 1. RBBP4 modulates TMZ sensitivity in GBM cells.
(A) Absolute levels of RBBP4 shRNA in GBM22 cells following treatment with either DMSO or TMZ. (B) Effect of RBBP4 siRNA, with and without TMZ treatment, on the primary neurosphere (NS) formation in GBM12 and GBM22 cells. (C) RBBP4 expression in T98G cells stably expressing shNT and 4 different RBBP4 shRNA lentiviral constructs (upper panel) and in vitro TMZ cytotoxicity CyQuant assay in the same cell constructs (lower panel). (D) Effect of control shNT and 4 different RBBP4 shRNA on RBBP4 levels in U138 GBM cells (upper panel) and in vitro cytotoxicity. Shown is the data from 3 independent experiments conducted in triplicate (error bar =S.E.M; * = p <0.05).
RBBP4 disruption sensitizes MGMT-expressing GBM cells to TMZ
Next, we evaluated whether RBBP4 can modulate TMZ response in MGMT unmethylated T98G and U138 GBM cell lines using 4 RBBP4 shRNA constructs (RBBP4-1 to -4) and a single non-specific targeting (NT) shRNA. All 4 shRNA constructs significantly silenced RBBP4 in both T98G and U138 cells (Figure 1C–D; upper panels). Moreover, all 4 different RBBP4 shRNA constructs significantly sensitized T98G and U138 cells to TMZ as compared to NT shRNA (Figure 1C–D; lower panels). The relative fluorescence for T98G expressing NT shRNA (T98GNT) treated with 100 µM TMZ was significantly higher compared with all 4 RBBP4 shRNA clones (Figure 1C, lower panel; p<0.001). Similar significant sensitization was observed in TMZ-treated (300 µM) U138shRBBP4 clones compared with the control U138shNT cells (Figure 1D, lower panel; p<0.001) and GBM6shRBBP4 cells (p<0.05, see Figure S6C). Therefore, RBBP4 is a negative modulator of TMZ response, and silencing of this protein enhances the sensitivity in both MGMT-expressing and non-expressing GBM cells.
RBBP4 regulates TMZ sensitivity by modulating MGMT expression
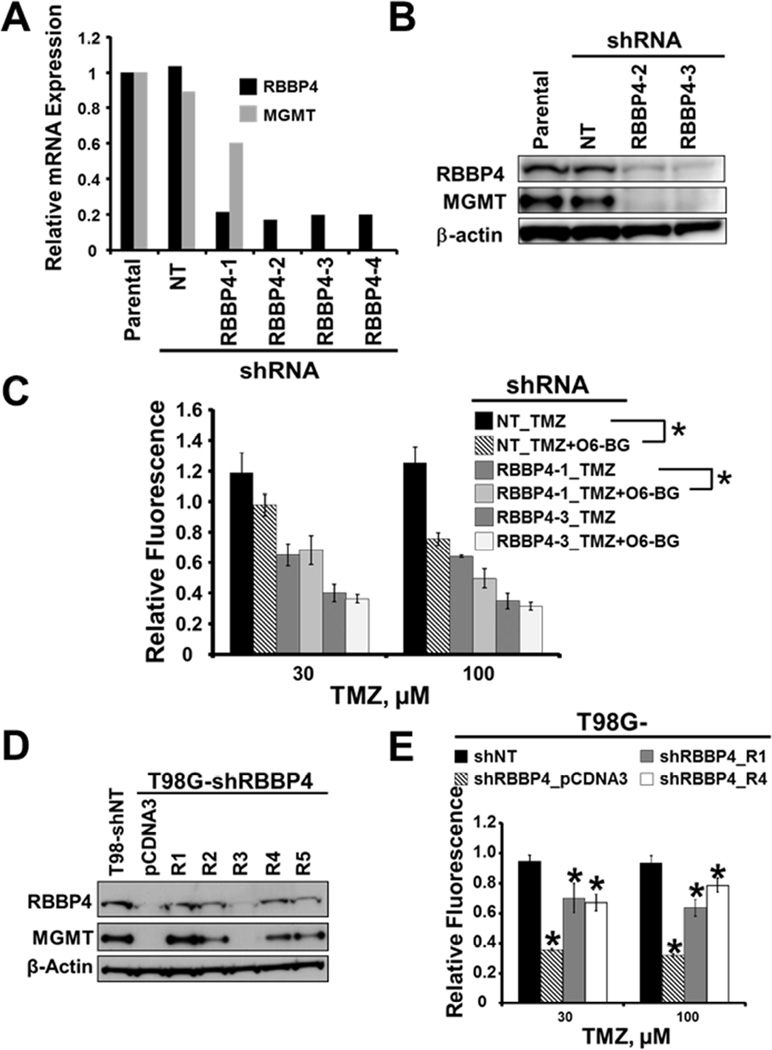
The above finding that RBBP4 is involved in the regulation of TMZ response in MGMT-expressing GBM cells has not been reported previously. Because MGMT is critical for TMZ response (Hegi et al., 2005; Kitange et al., 2009a; Kitange et al., 2009b), we evaluated whether RBBP4 modulates TMZ response through regulation of MGMT expression in unmethylated GBM cells. All 4 RBBP4 shRNAs significantly suppressed MGMT mRNA level in T98G cells compared to control T98GshNT cells (Figure 2A). Consistently, suppression of MGMT mRNA was coupled with decreased MGMT protein levels (Figure 2B), and knockdown of RBBP4 also suppressed MGMT expression in U138 (Figure S1A–B) and GBM6 cells (see Figure S6A). To test whether suppression of MGMT is responsible for the TMZ sensitivity associated with RBBP4 knockdown, we treated T98GshNT and T98G cells with 2 different RBBP4 shRNA constructs (T98GshRBBP4-1 and shRBBP4-3) with TMZ alone or in combination with the MGMT inhibitor O6-benzylguanine (O6-BG). As expected, O6-BG sensitized T98GNT cells to TMZ (p<0.001) but in contrast, addition of O6-BG did not significantly enhance TMZ sensitivity in T98GshRBBP4-3 (p=0.237). Consistent with incomplete suppression of MGMT mRNA expression (see Figure 2A), addition of O6-BG further sensitized T98GshRBBP4-1 cells (p<0.05; Figure 2C). In subsequent sub-cloning, we found heterogeneity in T98GshRBBP4-1 sub-clones with more robust RBBP4 suppression associated with very low MGMT levels (data not shown). Thus, RBBP4 negatively modulates TMZ sensitivity through transcriptional regulation of MGMT in GBM cells. To solidify this view, we first evaluated whether re-expression of an shRNA-resistant RBBP4 construct in T98GshRBBP4-3 clone (from here onward referred to as T98GshRBBP4) would rescue the expression of MGMT and reverse TMZ sensitivity. As expected, re-expression of RBBP4 restored MGMT expression in T98GshRBBP4 cells (Figure 2D). Two of the reconstituted clones (T98GshRBBP4_R1 and R4) were selected for further testing. In comparison to the T98GshNT cells, T98GshRBBP4 cells expressing an empty vector (T98GshRBBP4_pcDNA3) were significantly sensitive to 30 and 100 µM TMZ (p < 0.001; Figure 2E). Conversely, re-expression of RBBP4 significantly restored TMZ resistance in T98GshRBBP4_R1 and R4 clones (p < 0.001 for each clone compared to T98GshRBBP4_pcDNA3; Figure 2E). Therefore, RBBP4 modulates TMZ response through transcriptional regulation of MGMT in unmethylated GBM cells.
Figure 2. RBBP4 regulates TMZ sensitivity through modulation of MGMT expression in GBM cells.
(A) Real time PCR evaluating RBBP4 and MGMT expression using total RNA extracted from T98G cells expressing control shNT and 4 different constructs of RBBP4 shRNA (B) A representative western blot depicting RBBP4 and MGMT protein levels in T98G cells expressing NT-shRNA and two RBBP4 shRNA constructs (C) Effects of MGMT inhibitor O6-BG on TMZ sensitivity in T98G cells expressing control shNT and two RBBP4 shRNA (RBBP4-1 and RBBP4-3) constructs as measured in a CyQuant assay. (D) Western blot for RBBP4 and MGMT in T98G shNT compared with T98G-shRBBP4 cells stably expressing pCDNA3 vector or an shRNA resistant pCDNA3-RBBP4 expression construct (clones R1-R5). (E) TMZ sensitivity in T98G-shNT compared with T98G-shRBBP4 reconstituted with either pCDNA3 or shRNA resistant RBBP4 (clones R1 and R4). Shown in each bar graph is the data from 3 independent experiments conducted in triplicate (error bar = S.E.M; * = p <0.05).
The functional disruption of RBBP4 enhances TMZ-induced DNA damage
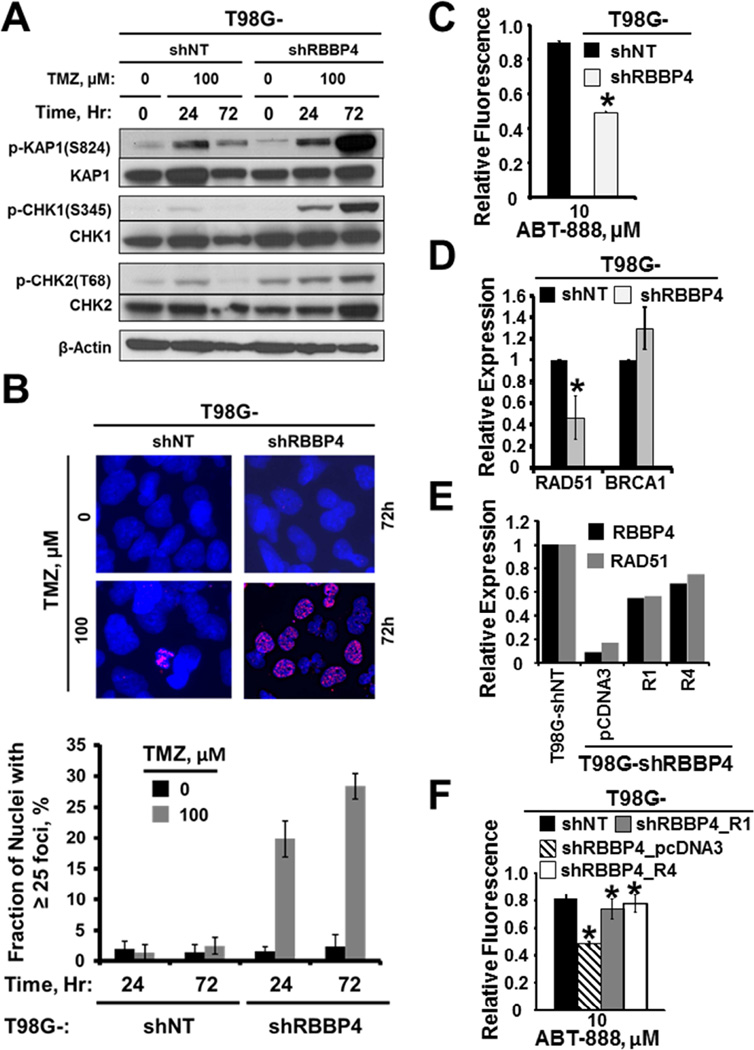
To further evaluate potential mechanisms of enhanced TMZ cytotoxicity in the RBBP4 knock-down cells, we tested the impact on DNA damage signaling and repair. As shown in Figure 3A, TMZ treatment induced a similar level of damage signaling 24 hours later in both T98G-shNT- and -shRBBP4 cells, but by 72 hours, there was a marked increase in damage signaling to KAP1, CHK1, and CHK2 only in the T98G-shRBBP4 cells. In contrast, the DNA damage response signaling was almost back to basal levels by 72 hours in the T98G-shNT cells, suggesting that disruption of RBBP4 suppressed repair of TMZ-induced DSBs. Consistent with a role in the repair of DSBs, TMZ-induced γH2AX foci were significantly higher in T98G-shRBBP4 than in the T98-shNT cells (p<0.001; Figure 3A–B). Since TMZ-induced DSBs are primarily repaired through homologous recombination (HR) (Roos et al., 2009), we next evaluated whether RBBP4 disruption can compromise HR proficiency in T98G cells. For this, we employed a recently established paradigm whereby HR deficiency results in synthetic lethality with PARP inhibition (Clark et al., 2012; Dedes et al., 2011). Consistent with HR deficiency, T98GshRBBP4 cells were significantly sensitized to the PARP inhibitor ABT-888 while sensitization was not observed in T98GshNT cells (p< 0.0001; Figure 3C). Next, we evaluated the possible mechanism by examining the effect of RBBP4 disruption on the expression of RAD51 and BRCA1, 2 key components of the HR pathway (Stark et al., 2002). As shown in Figure 3D, RAD51 expression was significantly down-regulated in T98GshRBBP4 relative to the T98GshNT (p=0.01). In contrast, RBBP4 disruption did not significantly alter the expression of BRCA1 relative to T98GshNT cells (Figure 3D). Similar levels of RAD51 suppression were observed with shRBBP4 in U138, GBM22 (Figure S2A–B) and U251 cells (see Figure 7A). Moreover, re-expression of an shRNA-resistant RBBP4 construct restored RAD51 expression (Figure 3E) and reversed ABT-888 sensitivity in T98GshRBBP4_R1 and R4 clones (p < 0.001; Figure 3F). Collectively, these findings suggest that RBBP4 modulates repair of TMZ-induced DNA damage, possibly through regulation of RAD51. Interestingly, RBBP4 disruption also significantly sensitized T98G cells to lomustine (CCNU) and the radiomimetic bleomycin but did not affect T98G response to hydroxyurea or cisplatin (Figure S1C–E), suggesting that RBBP4 knockdown does not indiscriminately sensitize cells to DNA damaging agents.
Figure 3. RBBP4 modulates repair of TMZ-induced DNA damage and regulates RAD51 expression level in GBM cells.
(A) Western blot depicting the time-course activation of DNA damage signaling following TMZ comparing T98G shNT and shRBBP4-3 clone (B) Nuclear p-H2AX foci in response to 72 hour exposure of T98G cells expressing shNT compared with shRBBP4-3 clone (upper panel), while the lower panel shows the fraction of cells with mean γ-H2AX foci of 3 experiments conducted in triplicate (C) Effect of RBBP4 disruption by shRNA clone shRBBP4 on PARP inhibitor ABT-888 sensitization in T98G GBM cells measured in a CyQuant assay (D) Real time PCR showing RAD51 and BRCA1 expression in T98G-shNT cells compared to T98G-shRBBP4. (E) Real time PCR evaluating RAD51 expression comparing T98G-shRBBP4_pCDNA3 with the RBBP4 reconstituted T98G-shRBBP4_R1 and _R4 cells (F) CyQuant evaluation of ABT-888 sensitivity after RBBP4 reconstitution in T98G-shRBBP4 clone compared with the T98G-shRBBP4_pcDNA3 and T98G-shNT cells Error bar = S.E.M.; * = p<0.05).
RBBP4 mediates epigenetic regulation of MGMT and RAD51
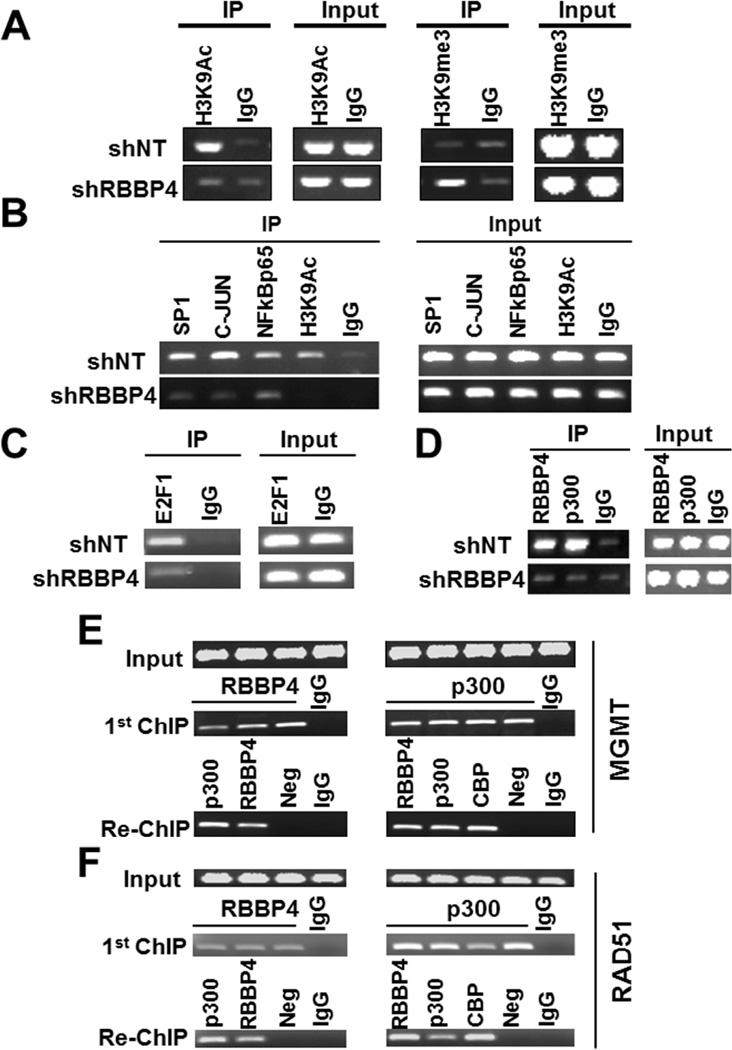
Even though the majority of studies have associated RBBP4 with transcriptional repression (Kuzmichev et al., 2002; Todd and Picketts, 2012; Vermaak et al., 1999), this protein also has been implicated in transcriptional activation in a complex with the histone acetyltransferase p300 (Zhang et al., 2000). Since our findings suggest that RBBP4 may play a role in the transcriptional activation of MGMT and RAD51, we hypothesized that RBBP4 is recruited to MGMT and RAD51 promoters as part of this complex to enhance transcription through histone modification, including acetylation of lysine 9 of histone H3 (H3K9Ac). To test this hypothesis, we evaluated the impact of RBBP4 knockdown on the prevalence of H3K9Ac within the MGMT promoter. As shown by chromatin immunoprecipitation (ChIP), MGMT expression in T98G-shNT was associated with high H3K9Ac levels within the MGMT promoter region (Figure 4A - left upper panel), while suppression of MGMT expression in the T98G-shRBBP4 cells was accompanied by loss of H3K9Ac from the MGMT promoter (left lower panel). Loss of H3K9Ac observed in T98G-shRBBP4 cells was coupled with an increase of H3K9me3 mark within the MGMT promoter region (Figure 4A, right panel). H3K9Ac-based ChIP-sequencing (ChIP-seq) comparing T98G-shNT with T98G-shRBBP4 cells revealed similar changes in H3K9Ac marks within the RAD51 promoter (Figure S2C). Beside H3K9Ac and H3K9me3, RBBP4 disruption also suppressed other histone marks associated with open chromatin within MGMT promoter region (see Figure S5B). Thus, RBBP4 is involved in the epigenetic regulation of both MGMT and RAD51.
Figure 4. RBBP4 regulates H3K9Ac and binds MGMT and RAD51 promoters in complex with p300.
(A) ChIP evaluating the H3K9Ac and H3K9me3 levels within MGMT promoter region comparing T98G-shNT versus T98G-shRBBP4 cells. (B) ChIP evaluating recruitment of SP1, C-JUN, NF-kB (p65) and H3K9Ac to bind MGMT promoter region comparing T98G-shNT versus T98G-shRBBP4 cells. (C) ChIP assessing the recruitment of E2F1 to the RAD51 promoter in T98G-shNT compared with the T98G-shRBBP4. (D) ChIP evaluating RBBP4 and p300 recruitment to the MGMT promoter in T98G-shNT comparing with T98G-RBBP4 cells. (E) RBBP4 and p300 ChIP re-ChIP depicting co-occupancy of RBBP4 within the MGMT promoter region and (F) the RAD51 promoter.
We hypothesized that chromatin condensation modulated by RBBP4 loss resulted in the exclusion of transcription factors from MGMT and RAD51 promoters. MGMT expression is mainly regulated by SP1, C-JUN and NF-kB transcription factors (Bhakat and Mitra, 2000; Kitange et al., 2012), while RAD51 transcription is modulated by E2F transcription factors (Ogiwara and Kohno, 2012). In comparing T98GshNT with T98GshRBBP4 cells, SP1, C-JUN and NF-kB (p65) transcription factors were all recruited to bind the MGMT promoter in the control T98GshNT cells whereas transcription factor binding was suppressed in T98G-shRBBP4 cells (Figure 4B). Similarly, the binding of E2F1 within the RAD51 promoter was exclusively observed in the T98GshNT cells (Figure 4C). Thus, RBBP4 facilitates acetylation of histones to maintain opened chromatin within promoter regions that allows transcription factors to bind and drive MGMT and RAD51 expression.
RBBP4 and p300 form a complex that binds MGMT and RAD51 promoters
A previous study has shown that RBBP4 interacts with CBP/p300 to form a complex involved in histone acetylation (Zhang et al., 2000), however, whether this specific complex binds the MGMT promoter region is unknown. We hypothesized that if RBBP4 and p300 are recruited to MGMT in a common complex, knockdown of RBBP4 should result in loss of p300 binding to the MGMT promoter region. Consistently, as shown by ChIP assay, both RBBP4 and p300 bind the MGMT promoter in the T98shNT cells (Figure 4D, left upper panel) whereas in T98G-shRBBP4 cells, loss of RBBP4 was accompanied with a loss of p300 enrichment within the MGMT promoter region (Figure 4D, left lower panel). Together, these findings suggest that RBBP4 and p300 are likely recruited to bind the MGMT promoter region in a conjoined complex. To support this view, we performed sequential ChIP (ChIP re-ChIP). As shown in Figure 4E, an initial RBBP4 and p300 ChIP demonstrated recruitment of these protein to the MGMT promoter in the T98G-shNT cells (middle panel). Subsequently, p300-bound chromatin was re-ChIPed from the original RBBP4 ChIP (Figure 4E, left lower panel) and conversely, RBBP4-bound chromatin was re-ChIPed from the original p300 ChIP (Figure 4E, right lower panel). Since CBP is known to interact with p300, an antibody against this protein was included as an additional positive control for the re-ChIP performed on p300 bound chromatin (Figure 4E, left lower panel). Similar results were observed for RAD51 promoter (Figure 4F). Consistent with our hypothesis that RBBP4 functions in a complex with p300, the p300 inhibitor C646 suppressed MGMT expression and sensitized cells to TMZ (see Figure S5C–D). Collectively, these findings strongly support the idea that RBBP4/p300 complex binds the MGMT and RAD51 promoters to drive expression in GBM cells.
Dual effects of RBBP4 on global gene transcription in GBM
Even though many studies have linked RBBP4 protein with transcriptional repression (Kuzmichev et al., 2002; Todd and Picketts, 2012; Vermaak et al., 1999), to the best of our knowledge, we are only aware of a single study that has shown that RBBP4 can interact with CBP/p300 and enhance transcription through histone acetylation (Zhang et al., 2000). Furthermore, understanding of the effect of RBBP4 on global gene expression is lacking. Thus, we used RNAseq to globally identify genes significantly altered by the disruption of RBBP4 in T98G-shNT compared with T98G-shRBBP4. RNAseq revealed that after silencing RBBP4, the expression of 1065 genes was significantly altered with 671 (63%) genes up-regulated and 394 (37%) suppressed (Figure S3A and Tables S1–2). RBBP4-knockdown affected genes involved in variety of cellular functions (Tables S3–4). The top 20 ontologies enriched within the under-expressed genes are shown in Figure S3B and those enriched in the overexpressed genes are shown in Figure S3C. Consistent with the previously established role of RBBP4 in chromatin regulation (reviewed in (Wolffe et al., 2000), 6 (30%) of the top 20 down-regulated ontologies are involved in chromatin and/or nucleosome assembly (Figure S3B, arrows). Importantly, RBBP4 disruption suppressed genes encoding for histones and histone chaperones including NASP, H2AFY2 and H3F3A (Table S1).
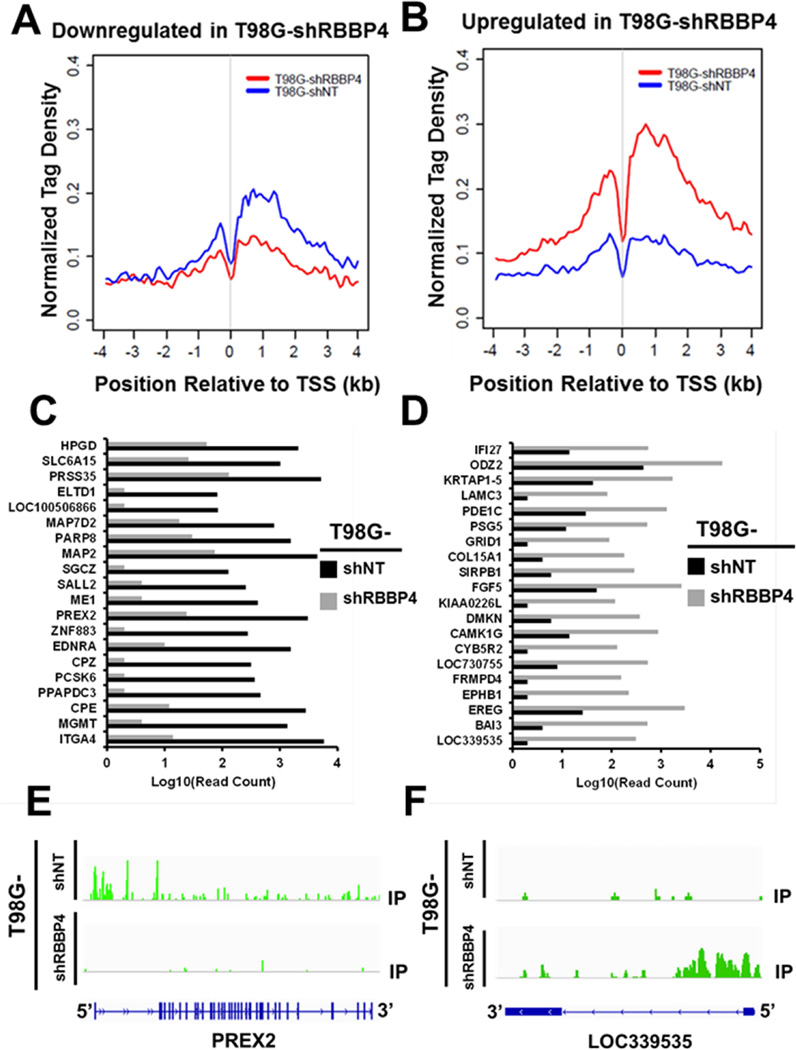
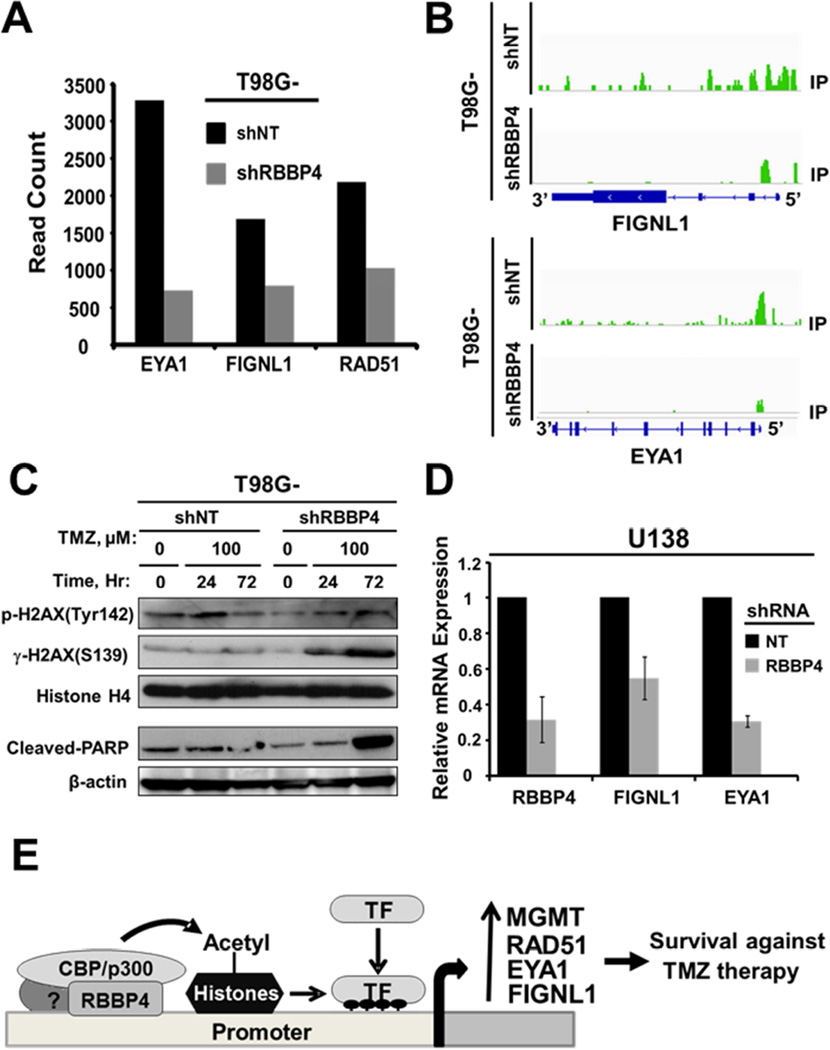
Next, we performed ChIPseq to gain an insight on how RBBP4 regulates genes identified through RNAseq. The H3K9Ac ChIPseq analysis demonstrated that of the 394 downregulated genes, 162 (41%) genes had reduced H3K9Ac with an average H3K9Ac normalized tag density (NTD) within 8-kb windows of 0.069 ± 0.084 (red line Figure 5A), whereas 301 of 671(45%) upregulated genes had an increased H3K9Ac mark with the average H3K9Ac NTD of 0.168 ± 0.066 (red line Figure 5B). The average binding density around TSS (TSS +/− 4kb) is significantly different between T98G-shRBBP4 and T98G-shNT cells for genes that were up- or down-regulated (p-value < 2.2e-16). Therefore, RBBP4 regulation of gene expression is mediated through histone modifications favoring chromatin decondensation (for upregulated genes) or condensation (for suppressed genes). The top 20 suppressed or upregulated genes by RNAseq are shown in Figures 5C–D, respectively. A representative example of H3K9Ac mark alteration by ChIPseq within the 5-prime region of a downregulated gene (PREX2) and an upregulated gene (LOC339535) is shown in Figure 5E–F, respectively. Interestingly, Fidgetin like 1 (FIGNL1) and Eye absence homolog 1 (EYA1), which are genes involved in DNA repair (Cook et al., 2009; Stucki, 2009; Yuan and Chen, 2013), were among the genes suppressed by RBBP4 disruption in association with loss of H3K9Ac marks within their promoters (Figure 6A–B). In contrast, the expression of other key DNA repair genes was not affected by the RBBP4 status (Figure S2D). Since EYA1 plays a key role in DNA repair through de-phosphorylation of tyrosine 142 of histone H2AX (H2AX-Tyr142) (Cook et al., 2009), we next examined whether TMZ can induce de-phosphorylation of H2AX-Tyr142 and whether this effect was abrogated by the RBBP4 disruption. As shown in the upper panel of Figure 6C, TMZ treatment reduced the phosphorylation of H2AX Tyr142 in T98G-shNT cells while the opposite effect was observed in T98G-shRBBP4. Increased phosphorylation of H2AX-Tyr142 and H2AX-S139 paralleled an increase in apoptosis as indicated by elevated cleaved PARP (Figure 6C, lower panel). Similarly, suppression of EYA1 and FIGNL1 was observed in U138 (Figure 6D) and U251 (see Figure 7A) GBM cells with silenced RBBP4 expression. Thus, RBBP4 can epigenetically regulate transcription of genes involved in multiple cellular functions, including EYA1 and FIGNL1 that may be additional DNA damage repair target genes. Both RNA- and ChIP-seq data are available for public access through GSE72477.
Figure 5. RBBP4 upregulates and downregulates gene transcription in GBM.
Representative line graphs displaying the H3K9Ac binding tags within 8-kb region surrounding the transcriptional start site (TSS) of (A) under-expressed and (B) over-expressed genes associated with shRBBP4 (C) Top 20 genes suppressed by RBBP4 shRNA (positively regulated by nativeRBBP4) (D) Top 20 genes elevated by RBBP4 shRNA (negatively regulated by native RBBP4). Representative H3K9Ac ChIP-seq display of a gene that was (E) suppressed and (F) elevated by RBBP4 shRNA.
Figure 6. RBBP4 regulates the expression of FIGNL1 and EYA1.
(A) RNAseq was performed using RNA extracted from T98G-shNT and T98G-shRBBP4 constructs and shown are the gene counts for FIGNL1 and EYA1. (B) ChIPseq display of H3K9Ac marks within FIGNL1 (upper panel) and EYA1 (lower panel) promoter regions. (C) Upper panel shows the effect of shRBBP4 on TMZ induced p-H2AX (Y142) and p-H2AX (S139), whereas the lower panel depicts the effect on cleaved-PARP. (D) RBBP4, FIGNL1 and EYA1 expression in U138 cells expressing shNT compared with shRBBP4 (E) A proposed model of RBBP4 associated with CBP/p300, which leads to histone acetylation and an open chromatin structure that facilitates transcription factor binding and expression of target genes.
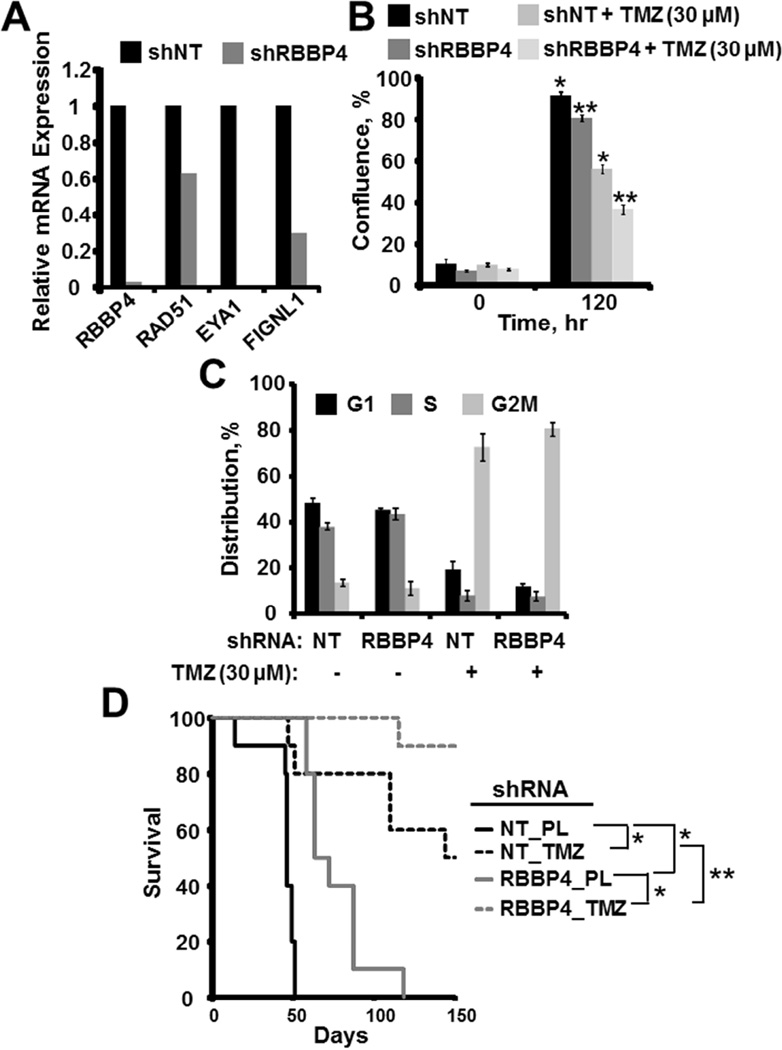
Figure 7. RBBP4 disruption in U251suppresses growth and enhances TMZ sensitivity in vivo.
(A). qRT-PCR showing expression of RAD51, EYA1 and FIGNL1 in U251 cells expressing RBBP4shRNA. (B). U251shRBBP4 and control U251shNT cells were treated with 0 or 30 µM TMZ and confluence was monitored using Incucyte live cell imaging. The bar graph displays the mean of 3 experiments conducted independently in triplicate (Error bar=S.E.M., Asterisks=p<0.05). (C). Cell cycle evaluation was performed using U251shRBBP4 and control U251shNT following 72 hours exposure to 0 or 30 µM TMZ. (Error Bar= S.E.M). (D) Kaplan Meier survival plots depicting the in vivo growth and TMZ sensitivity of control U251shNT compared with the U251RBBP4 cells (PL=placebo, *= p < 0.001, ** =p < 0.05).
RBBP4 disruption suppresses growth and TMZ sensitivity in vivo
Finally, we disrupted RBBP4 expression in MGMT-methylated U251 GBM cells to evaluate whether RBBP4 can impact growth and TMZ sensitivity in vivo. Consistent with the above findings, RBBP4 disruption suppressed RAD51, EYA1 and FIGNL1 in U251 cells (Figure 7A). In addition, in vitro evaluation revealed that U251shRBBP4 cells were significantly more sensitive to TMZ (30 µM) compared with the control U251shNT (p < 0.01; Figure 7B). As shown in Figure 7C, flow cytometry revealed an insignificant difference in cell cycle distribution between U251shNT and U251shRBBP4 cells with and without TMZ exposure, suggesting that TMZ sensitization is independent from cell-cycle progression. The U251 shRBBP4 and shNT cells then were used to establish orthotopic tumors in mice. As shown in Figure 7D, placebo-treated U251shRBBP4-bearing mice survived significantly longer compared with the placebo-treated U251shNT (median survival= 47 days vs. 67.5 days; p < 0.001). There also was a corresponding significant survival benefit in TMZ-treated mice implanted with U251shRBBP4 tumors compared with U251shNT (p-value <0.001 with censoring of mice still alive at Day 150; Figure 7D). Intracranial tumors from moribund mice demonstrated that RBBP4 knockdown was maintained for the duration of the in vivo experiment (Figure S7). Similar anti-tumor effects were observed in the patient-derived GBM6 xenograft model in which RBBP4 knockdown suppressed growth of untreated orthotopic tumors (Figure S6D). These data demonstrate that RBBP4 suppression is associated with slower tumor growth and greater TMZ sensitivity in GBM orthotopic xenografts.
Discussion
Epigenetic regulation of chromatin structure is an important modulator of gene expression and can critically influence tumor biology and response to therapies. In many tumor types, mutations or altered expression of the epigenetic regulators have been directly implicated in tumorigenesis through their effects on gene expression (Turcan et al., 2012; Venneti et al., 2013) and reviewed in:(Wilting and Dannenberg, 2012), and this has stimulated significant interest in developing pharmacologic inhibitors of these regulators. The two best examples of this paradigm include BRD4 and the HDAC family of proteins for which there are now several inhibitors that are either clinically approved or in clinical testing (Fiskus et al., 2014b). More recent data also suggest that HDACs or BRD4 are important regulators of therapeutic resistance emergence (Fiskus et al., 2014a; Kitange et al., 2012; Knoechel et al., 2014; Tang et al., 2014). Similar to these two targets, this paper reports the importance of the epigenetic regulator, RBBP4, in the regulation of gene expression patterns in human GBM tumor cells. Knockdown of RBBP4 resulted in marked sensitization of tumor cells to TMZ in conjunction with suppression of several DNA repair genes known to be critical for response to TMZ chemotherapy.
MGMT is a critical mediator of cytotoxicity for DNA alkylating agents such as TMZ (Hegi et al., 2005; Kitange et al., 2009b; Kitange et al., 2012), and either direct MGMT knockdown or treatment with an MGMT inhibitor can markedly sensitize tumors to TMZ therapy (Hirose et al., 2003; Vlachostergios et al., 2013). Thus, the suppression of MGMT associated with RBBP4 shRNA expression can be mechanistically linked to the enhanced TMZ sensitivity seen in T98G and U138 cells. Unrepaired TMZ induced O6-MG lesions are mispaired with thymidine and result in MMR-mediated stalled replication forks that ultimately degenerate into DSBs (Hirose et al., 2001; Sarkaria et al., 2008). The replication-associated DSBs are preferably repaired by the HR system (Chai et al., 2014; Tentori et al., 2014; Yoshimoto et al., 2012), and cells lacking proficient HR are highly sensitive to TMZ-induced damage (Liu et al., 2009; Short et al., 2011). RAD51 is a key component of the HR pathway, and consistent with a significant defect in HR integrity, suppression of RBBP4 resulted both in a significant reduction in RAD51 expression and increased sensitivity to PARP inhibition (Figure 3C). While suppression of additional HR components may contribute to the repair defects, these data demonstrate a similar effect as previous studies, where RAD51 knockdown was associated with an enhanced response to TMZ chemotherapy (Short et al., 2011; Zhang et al., 2012). This effect may be especially important in tumors that lack significant MGMT expression, such as GBM12, GBM22 (Figure 1B) and U251 (Figure 7), where unrepaired O6-MG lesions will result in markedly higher levels of replication stress and efficient processing of replication-induced lesions is critical for cell survival. High levels of TMZ induced replication stress and DSBs are more likely to be observed in highly proliferating cells, while RBBP4 disruption suppressed proliferation potential in GBM cells (Figure 7C, and Figure S6). This effect on reduced proliferation might be mediated through induction of cell differentiation, as suggested by suppressed neurosphere formation in GBM6 shRBBP4 expressing cells (Figure S6B). Delineation of RBBP4 as an important mediator of two major known mechanisms of repair of TMZ-induced DNA damage suggest that the relevant RBBP4 complex mediating these effects may be an attractive therapeutic target.
The influence of RBBP4 on HR may be an important regulatory mechanism that is relevant across multiple tumor types. As described previously, synthetic lethality to PARP inhibition is a hallmark of HR deficiency (Clark et al., 2012; Dedes et al., 2011), and the significant sensitivity of T98G cells to either ABT-888 or BMN673 (Figure 3C and data not shown) are consistent with physiologically meaningful defects in HR. Beyond suppression of RAD51 expression, RBBP4 knockdown also reduced gene expression of EYA1 and FIGNL1, which were recently demonstrated to play a key role in the repair of DSBs and apoptosis (Cook et al., 2009; Yuan and Chen, 2013). Moreover, beyond regulating gene expression, RBBP4 may regulate HR more directly, since this protein can co-associate with a BRCT domain within BRCA1 (Yarden and Brody, 1999). RBBP4 also is an integral component of the NURD complex, and another member of this complex, chromodomain helicase DNA binding protein 4 (CHD4) indirectly regulates BRCA1 recruitment to DNA DSBs and lack of CHD4 promotes sensitivity to PARP inhibitors (Pan et al., 2012). Shown as supplemental data (Figure S1), RBBP4 knockdown was associated with increased sensitivity to the radio-mimetic bleomycin, which is in contrast to a previous study that demonstrated an increased sensitivity to ionizing radiation with RBBP4 overexpression (Torres-Roca et al., 2005). These apparently contradictory results suggest that either suppression or over-expression of RBBP4 can deregulate critical DNA repair capacities and lead to increased cytotoxicity. Future studies will be directed at dissecting the relative contributions to DNA repair deficits conferred by effects of RBBP4 on gene expression versus direct interactions with DNA repair complexes.
The effects of RBBP4 on global gene expression patterns have not been elucidated. Comparison of gene expression in T98G cells expressing RBBP4 shRNA versus non-targeted shRNA revealed that RBBP4 exerts a suppressive effect on almost two-thirds of the genes with altered expression, while expression of the remaining genes are upregulated. Consistent with disruption of a RBBP4/p300/CBP complex by shRBBP4 expression, 41% of the 394 down-regulated genes had specific suppression of H3K9 acetylation within their promoters. This change in the chromatin landscape was most pronounced within an 8-kb region encompassing the TSS. A similar study using direct ChIPseq analysis of CBP/p300 binding in T98G cells demonstrated preferential binding within the TSS and transcript ends (Ramos et al., 2010). Moreover, in their study, a total of 170 genes (including MGMT) with the TSS bound by the p300/CBP were also altered in shRBBP4-expressing T98G cells in our current data set. In conjunction with the ChIP-re-ChIP data demonstrating co-association of RBBP4 and p300/CBP within the same promoter regions of MGMT and RAD51 and the effects of RBBP4 shRNA on p300 within these regions, these data strongly support the concept that RBBP4 and CBP/p300 exist in a complex that can effectively modulate gene expression (Figure 6E). CBP/p300 has important HAT activity and can acetylate several sites within histone tails including H3K9. Overall, histone acetylation mediated by CBP/p300 promotes an open chromatin structure conducive to gene expression. Thus, the association of RBBP4 shRNA with both reduced H3K9Ac and reduced transcription factor binding within the MGMT and RAD51 promoters are all consistent with a model where RBBP4/CBP/p300 promotes an open chromatin structure that allows transcription factor binding and effective gene expression of multiple gene targets. Since the current ChIPseq data are based on a single histone mark, there is a possibility that the identified genes represent only a fraction of genes regulated by RBBP4/CBP/p300 complex. This is particularly notable since MGMT was significantly silenced after RBBP4 disruption despite low baseline levels of H3K9Ac tags within the promoter region (Figure S5A), which suggests other histone marks modulated by RBBP4 also may be important in mediating MGMT regulation. Consistent with this view, RBBP4 disruption elevated multiple histone marks favoring closed chromatin within the MGMT promoter (Figure S5B). Thus, fine mapping of the chromatin landscape regulated by RBBP4/CBP/p300 complex using multiple histone marks is a subject for further investigation.
Analysis of the data for enriched gene ontologies regulated by RBBP4 indicates that several of these ontologies are related to nucleosome assembly, cell cycle progression, angiogenesis and cell survival. Consistently, nucleosome assembly and cell cycle progression also were ontologies shown to be highly enriched upon suppression of CBP/p300 activity in melanoma cells (Yan et al., 2013). In this latter study, the p300 HAT inhibitor C646 also suppressed pathways associated with DNA damage checkpoints and repair. Consistently, p300 inhibitor C646 suppressed MGMT expression and sensitized TMZ in T98G cells in the present study (Figure S5C–D). Since RBBP4 regulates multiple gene pathways involved in gliomagenesis, we queried the Oncomine database (https://www.oncomine.org/resource/) to evaluate whether this gene is overexpressed in gliomas. Intriguingly, RBBP4 is overexpressed in glial tumors with the highest expression in GBM (Figure S4A–B). In a follow up analysis of TCGA mRNA expression data, RBBP4 expression directly correlated with the expression of RAD51 and FIGNL1 in all patients while a significant correlation with EYA1 was specifically observed in unmethylated GBM (Figure S4C). Conversely, RBBP4 expression negatively correlated with MGMT levels in methylated whereas no significant correlation was noted in unmethylated tumors. These findings are consistent with a view that RBBP4 may be a part of multiple epigenetic mechanisms regulating MGMT in GBM. Overall, RBBP4 expression did not correlate with survival of GBM patients within TCGA database (Figure S4D). In an analysis segregated by GBM sub-type, low RBBP4 expression marginally associated with a better survival outcome for mesenchymal and neural GBM subclasses (p=0.05; data not shown). These findings are in part consistent with the current results showing that RBBP4 knockdown in U251 can significantly prolong survival of both placebo and TMZ-treated orthotopic tumors (Figure 7C) and prolongs survival of untreated GBM6 orthotopic tumors (Figure S6D). Thus, besides modulating therapy response, RBBP4 may also control tumor growth, perhaps through cell proliferation, angiogenesis or other unknown mechanisms. Future studies are focused on extending these observations to our panel of patient-derived xenograft models to define the influence of RBBP4 on gene expression and therapy response across a spectrum of GBMs. These studies will provide critical insight into the role of RBBP4 both in tumorigenesis and therapeutic resistance.
In summary, our study provides description of a role for RBBP4 in the regulation of TMZ sensitivity. Of particular translational interest is the finding that RBBP4 interacts with CBP/p300 to form a complex that drives the expression of MGMT, RAD51 and other selected DNA repair genes through histone acetylation. Particularly, since MGMT is overexpressed in approximately 70% of GBM and given the ongoing interest in developing therapies targeting HATs, the RBBP4/CBP/p300 complex poses an interesting target for future therapies in GBM.
Experimental Procedures
Cell culture and drug cytotoxicity assay
Short term primary serum-free cultures were derived from the flank GBM xenografts as described previously by our group (Carlson et al., 2011) and neuro-sphere formation assay were performed as previously reported (Kitange et al., 2012). Additional culture procedures are provided as supplementary information.
Gene knockdown and re-expression
pGIPZ-lentiviral vectors targeting human RBBP4 gene (see supplementary information) and non-specific targeting (NT) pGIPZ control vector were purchased from Mayo Clinic RNA interference shared resource. Lentiviral shRNA pseudo-particles were produced by co-transfection of the Trans-Lentiviral packaging mix (GE Dharmacon, Lafayette, CO) with shRNA transfer vector into HEK 293T packaging cells. Cells were transduced with the lentiviral particles followed by puromycin selection (2–5 µg/mL) for 10 days. The shRNA-resistant Myc3-tagged RBBP4 pCDNA3 construct (Plasmid #20715) was purchased from Addgene (Cambridge, MA). The transfections were conducted as previously reported (Kitange et al., 2010) and selection was performed using 1 mg/ml G418 (Life Technologies, Grand Island, NY).
Western Blotting
Total proteins were isolated by lysing cells in a detergent-containing RIPA buffer (Sigma) supplemented with a cocktail of protease inhibitors (Roche, Indianapolis, ID) while total histone extraction was performed using an Epigenetek kit (Farmingdale, NY). The subsequent steps were performed as previously reported (Kitange et al., 2012).
Real time RT-PCR
Real time PCR was performed as previously reported (Kitange et al., 2012) and additional procedures are included in supplemental information.
Chromatin Immunoprecipitation (ChIP) and Re-ChIP
ChIP was performed using the EZ-ChIP™ kit (Millipore, Billerica, MA). Crosslinking was performed with 1% formaldehyde at room-temperature for 10 minutes and was quenched with 0.1 M glycine for 5 minutes. Subsequent steps were performed as previously reported (Kitange et al., 2012). The ChIP-re-ChIP experiments were performed according to manufacturer’s instructions (Active Motif, Carlsbad, CA).
RNA- and ChIP-sequencing
To identify RBBP4 target genes, RNA- and ChIP-sequencing (RNA- and ChIP-seq) were conducted comparing T98GshNT with T98GshRBBP4 cells. The subsequent steps for RNA-seq and ChIP-seq procedures are described under supplemental data information.
Statistical Analysis
The differences in TMZ sensitivity and mRNA expression in relation to RBBP4 expression status were analyzed using a two-sample t-test and p-value < 0.05 was considered statistically significant. The in vivo survival was evaluated using Kaplan Meir survival plots and log-rank test was used for statistical significance. Additional statistical analyses are described under supplemental experimental procedures.
Supplementary Material
Highlights.
RBBP4 disruption sensitizes GBM cells to temozolomide.
RBBP4 controls temozolomide sensitivity by regulating multiple DNA repair proteins
RBBP4 regulates the expression of MGMT, RAD51, FIGNL1 and EYA1.
RBBP4 interacts with CBP/p300 to enhance chromatin mediated gene expression.
Acknowledgments
We thank Dr. Yuichi Machida, Department of Oncology Research, Mayo Clinic, Rochester, for supplying the shRNA library and Dr. Shiv Gupta, Department of Radiation Oncology Research, Mayo Clinic, Rochester, for reading and critical comments on the manuscript. The project was supported by funding from American Cancer Society (G.J.K.) and National Institute of Health RO1CA176830 (J.N.S.) and the Mayo Brain Tumor SPORE (P50 CA108961).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors Contributions
G.J. K.: Performed experiments, analyzed data and wrote the manuscript.
A.C. M.: Evaluated TMZ sensitivity in RBBP4 reconstituted clones.
M. A. S.: Harvested cells for primary cultures used in this study.
J .L. P.: Performed p-H2AX foci experiments.
B .L. C.: Harvested cells for primary cultures used in this study.
Y. Z.: Analyzed shRNA screening data.
A. A. N.: Analyzed RNA-seq data.
J-H. L.: Performed ChIP-seq
H. Y.: Analyzed ChIP-seq data
P. A. D: Performed TCGA analysis in relation to RBBP4
Z. Z.: Supervised ChIP-seq and edited the manuscript
J. N. S.: Supervised the study and edited the manuscript
Accession numbers
Both RNA- and ChIP-seq data are available for public access through GSE72477.
The authors declare no potential conflicts of interest.
References
- Bhakat KK, Mitra S. Regulation of the human O(6)-methylguanine-DNA methyltransferase gene by transcriptional coactivators cAMP response element-binding protein-binding protein and p300. J Biol Chem. 2000;275:34197–34204. doi: 10.1074/jbc.M005447200. [DOI] [PubMed] [Google Scholar]
- Cahill DP, Codd PJ, Batchelor TT, Curry WT, Louis DN. MSH6 inactivation and emergent temozolomide resistance in human glioblastomas. Clin Neurosurg. 2008;55:165–171. [PubMed] [Google Scholar]
- Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, maintenance and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Curr Protoc Pharmacol. 2011 doi: 10.1002/0471141755.ph1416s52. Chapter 14, Unit 14 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai KM, Wang CY, Liaw HJ, Fang KM, Yang CS, Tzeng SF. Downregulation of BRCA1-BRCA2-containing complex subunit 3 sensitizes glioma cells to temozolomide. Oncotarget. 2014 doi: 10.18632/oncotarget.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CC, Weitzel JN, O'Connor TR. Enhancement of Synthetic Lethality via Combinations of ABT-888, a PARP Inhibitor, and Carboplatin In Vitro and In Vivo Using BRCA1 and BRCA2 Isogenic Models. Mol Cancer Ther. 2012;11:1948–1958. doi: 10.1158/1535-7163.MCT-11-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedes KJ, Wilkerson PM, Wetterskog D, Weigelt B, Ashworth A, Reis-Filho JS. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle. 2011;10:1192–1199. doi: 10.4161/cc.10.8.15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE. Alkylation damage in DNA and RNA--repair mechanisms and medical significance. DNA Repair (Amst) 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Fiskus W, Sharma S, Qi J, Shah B, Devaraj SG, Leveque C, Portier BP, Iyer S, Bradner JE, Bhalla KN. BET protein antagonist JQ1 is synergistically lethal with FLT3 tyrosine kinase inhibitor (TKI) and overcomes resistance to FLT3-TKI in AML cells expressing FLT-ITD. Mol Cancer Ther. 2014a;13:2315–2327. doi: 10.1158/1535-7163.MCT-14-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskus W, Sharma S, Qi J, Valenta JA, Schaub LJ, Shah B, Peth K, Portier BP, Rodriguez M, Devaraj SG, et al. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther. 2014b;13:1142–1154. doi: 10.1158/1535-7163.MCT-13-0770. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc Natl Acad Sci U S A. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar N, Marshall L, Perryman L, Bax DA, Little SE, Viana-Pereira M, Sharp SY, Vassal G, Pearson AD, Reis RM, et al. MGMT-independent temozolomide 23 resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res. 2010;70:9243–9252. doi: 10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S, Otten P, Van Melle G, de Tribolet N, Stupp R. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Berger MS, Pieper RO. p53 effects both the duration of G2/M arrest and the fate of temozolomide-treated human glioblastoma cells. Cancer Res. 2001;61:1957–1963. [PubMed] [Google Scholar]
- Hirose Y, Kreklau EL, Erickson LC, Berger MS, Pieper RO. Delayed repletion of O6-methylguanine-DNA methyltransferase resulting in failure to protect the human glioblastoma cell line SF767 from temozolomide-induced cytotoxicity. J Neurosurg. 2003;98:591–598. doi: 10.3171/jns.2003.98.3.0591. [DOI] [PubMed] [Google Scholar]
- Kitange GJ, Carlson BL, Mladek AC, Decker PA, Schroeder MA, Wu W, Grogan PT, Giannini C, Ballman KV, Buckner JC, et al. Evaluation of MGMT promoter methylation status and correlation with temozolomide response in orthotopic glioblastoma xenograft model. J Neurooncol. 2009a;92:23–31. doi: 10.1007/s11060-008-9737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitange GJ, Carlson BL, Schroeder MA, Decker PA, Morlan BW, Wu W, Ballman KV, Giannini C, Sarkaria JN. Expression of CD74 in high grade gliomas: a potential role in temozolomide resistance. J Neurooncol. 2010;100:177–186. doi: 10.1007/s11060-010-0186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitange GJ, Carlson BL, Schroeder MA, Grogan PT, Lamont JD, Decker PA, Wu W, James CD, Sarkaria JN. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro Oncol. 2009b;11:281–291. doi: 10.1215/15228517-2008-090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitange GJ, Mladek AC, Carlson BL, Schroeder MA, Pokorny JL, Cen L, Decker PA, Wu W, Lomberk GA, Gupta SK, et al. Inhibition of histone deacetylation 24 potentiates the evolution of acquired temozolomide resistance linked to MGMT upregulation in glioblastoma xenografts. Clin Cancer Res. 2012;18:4070–4079. doi: 10.1158/1078-0432.CCR-12-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, Gillespie SM, Fernandez D, Ku M, Wang H, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46:364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Han EK, Anderson M, Shi Y, Semizarov D, Wang G, McGonigal T, Roberts L, Lasko L, Palma J, et al. Acquired resistance to combination treatment with temozolomide and ABT-888 is mediated by both base excision repair and homologous recombination DNA repair pathways. Mol Cancer Res. 2009;7:1686–1692. doi: 10.1158/1541-7786.MCR-09-0299. [DOI] [PubMed] [Google Scholar]
- Ogiwara H, Kohno T. CBP and p300 histone acetyltransferases contribute to homologous recombination by transcriptionally activating the BRCA1 and RAD51 genes. PLoS One. 2012;7:e52810. doi: 10.1371/journal.pone.0052810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan MR, Hsieh HJ, Dai H, Hung WC, Li K, Peng G, Lin SY. Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J Biol Chem. 2012;287:6764–6772. doi: 10.1074/jbc.M111.287037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos YF, Hestand MS, Verlaan M, Krabbendam E, Ariyurek Y, van Galen M, van Dam H, van Ommen GJ, den Dunnen JT, Zantema A, t Hoen PA. Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res. 2010;38:5396–5408. doi: 10.1093/nar/gkq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos WP, Nikolova T, Quiros S, Naumann SC, Kiedron O, Zdzienicka MZ, Kaina B. Brca2/Xrcc2 dependent HR, but not NHEJ, is required for protection against 25 O(6)-methylguanine triggered apoptosis, DSBs and chromosomal aberrations by a process leading to SCEs. DNA Repair (Amst) 2009;8:72–86. doi: 10.1016/j.dnarep.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Kitange GJ, James CD, Plummer R, Calvert H, Weller M, Wick W. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14:2900–2908. doi: 10.1158/1078-0432.CCR-07-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SC, Giampieri S, Worku M, Alcaide-German M, Sioftanos G, Bourne S, Lio KI, Shaked-Rabi M, Martindale C. Rad51 inhibition is an effective means of targeting DNA repair in glioma models and CD133+ tumor-derived cells. Neuro Oncol. 2011;13:487–499. doi: 10.1093/neuonc/nor010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Hu P, Pierce AJ, Moynahan ME, Ellis N, Jasin M. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J Biol Chem. 2002;277:20185–20194. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- Stucki M. Histone H2A.X Tyr142 phosphorylation: a novel sWItCH for apoptosis? DNA Repair (Amst) 2009;8:873–876. doi: 10.1016/j.dnarep.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- Tang Y, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P, Bergthold G, Masoud S, Nguyen B, Vue N, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med. 2014;20:732–740. doi: 10.1038/nm.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tentori L, Ricci-Vitiani L, Muzi A, Ciccarone F, Pelacchi F, Calabrese R, Runci D, Pallini R, Caiafa P, Graziani G. Pharmacological inhibition of poly(ADP-ribose) polymerase-1 modulates resistance of human glioblastoma stem cells to temozolomide. BMC Cancer. 2014;14:151. doi: 10.1186/1471-2407-14-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd MA, Picketts DJ. PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. J Proteome Res. 2012;11:4326–4337. doi: 10.1021/pr3004369. [DOI] [PubMed] [Google Scholar]
- Torres-Roca JF, Eschrich S, Zhao H, Bloom G, Sung J, McCarthy S, Cantor AB, Scuto A, Li C, Zhang S, et al. Prediction of radiation sensitivity using a gene expression classifier. Cancer Res. 2005;65:7169–7176. doi: 10.1158/0008-5472.CAN-05-0656. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. 2013;23:558–564. doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Wade PA, Jones PL, Shi YB, Wolffe AP. Functional analysis of the SIN3-histone deacetylase RPD3-RbAp48-histone H4 connection in the Xenopus oocyte. Mol Cell Biol. 1999;19:5847–5860. doi: 10.1128/mcb.19.9.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachostergios PJ, Hatzidaki E, Papandreou CN. MGMT repletion after treatment of glioblastoma cells with temozolomide and O6-benzylguanine implicates NFkappaB and mutant p53. Neurol Res. 2013;35:879–882. doi: 10.1179/1743132813Y.0000000191. [DOI] [PubMed] [Google Scholar]
- Wilting RH, Dannenberg JH. Epigenetic mechanisms in tumorigenesis, tumor cell heterogeneity and drug resistance. Drug Resist Updat. 2012;15:21–38. doi: 10.1016/j.drup.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Urnov FD, Guschin D. Co-repressor complexes and remodelling chromatin for repression. Biochem Soc Trans. 2000;28:379–386. [PubMed] [Google Scholar]
- Yan G, Eller MS, Elm C, Larocca CA, Ryu B, Panova IP, Dancy BM, Bowers EM, Meyers D, Lareau L, et al. Selective inhibition of p300 HAT blocks cell cycle progression, induces cellular senescence, and inhibits the DNA damage response in melanoma cells. J Invest Dermatol. 2013;133:2444–2452. doi: 10.1038/jid.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden RI, Brody LC. BRCA1 interacts with components of the histone deacetylase complex. Proc Natl Acad Sci U S A. 1999;96:4983–4988. doi: 10.1073/pnas.96.9.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip S, Miao J, Cahill DP, Iafrate AJ, Aldape K, Nutt CL, Louis DN. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15:4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Mizoguchi M, Hata N, Murata H, Hatae R, Amano T, Nakamizo A, Sasaki T. Complex DNA repair pathways as possible therapeutic targets to overcome temozolomide resistance in glioblastoma. Front Oncol. 2012;2:186. doi: 10.3389/fonc.2012.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Chen J. FIGNL1-containing protein complex is required for efficient homologous recombination repair. Proc Natl Acad Sci U S A. 2013;110:10640–10645. doi: 10.1073/pnas.1220662110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wu X, Yang L, Xiao F, Zhang H, Zhou A, Huang Z, Huang S. FoxM1 Inhibition Sensitizes Resistant Glioblastoma Cells to Temozolomide by Downregulating the Expression of DNA Repair Gene Rad51. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Vo N, Goodman RH. Histone binding protein RbAp48 interacts with a complex of CREB binding protein and phosphorylated CREB. Mol Cell Biol. 2000;20:4970–4978. doi: 10.1128/mcb.20.14.4970-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Tyl M, Ward R, Sobott F, Maman J, Murthy AS, Watson AA, Fedorov O, Bowman A, Owen-Hughes T, et al. Structural plasticity of histones H3-H4 facilitates their allosteric exchange between RbAp48 and ASF1. Nat Struct Mol Biol. 2013;20:29–35. doi: 10.1038/nsmb.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.