Abstract
N ′-Nitrosonornicotine (NNN) is carcinogenic in multiple animal models and has been evaluated as a human carcinogen. NNN can be metabolized by cytochrome P450s through two activation pathways: 2′-hydroxylation and 5′-hydroxylation. While most previous studies have focused on 2′-hydroxylation in target tissues of rats, available evidence suggests that 5′-hydroxylation is a major activation pathway in human enzyme systems, in non-human primates, and in target tissues of some other rodent carcinogenicity models. In the study reported here, we investigated DNA damage resulting from NNN 5′-hydroxylation by quantifying the adduct 2-(2-(3-pyridyl)-N-pyrrolidinyl)-2′-deoxyinosine (py-py-dI). In rats treated with NNN in the drinking water (7–500 ppm), py-py-dI was the major DNA adduct resulting from 5′-hydroxylation of NNN in vivo. Levels of py-py-dI in lung and nasal cavity were highest, consistent with the tissue distribution of CYP2A3. In rats treated with (S)-NNN or (R)-NNN, the ratios of formation of (R)-py-py-dI to (S)-py-py-dI were not the expected mirror image, suggesting that there may be a carrier for one of the unstable intermediates formed upon 5′-hydroxylation of NNN. Rat hepatocytes treated with (S)- or (R)-NNN or (2′S)- or (2′R)-5′-acetoxyNNN exhibited a pattern of adduct formation similar to live rats. In vitro studies with human liver S9 fraction or human hepatocytes incubated with NNN (2–500 μM) demonstrated that py-py-dI formation was greater than formation of pyridyloxobutyl-DNA adducts resulting from 2′-hydroxylation of NNN. (S)-NNN formed more total py-py-dI adducts than (R)-NNN in human liver enzyme systems, which is consistent with the critical role of CYP2A6 in the 5′-hydroxylation of NNN in human liver. The results of this study demonstrate that the major DNA adduct resulting from NNN metabolism by human enzymes is py-py-dI and provide potentially important new insights on the metabolic activation of NNN in rodents and humans.
Keywords: NNN, metabolism, 5′-hydroxylation, DNA adduct, py-py-dI, carcinogenesis, humans, rats
Graphical Abstract
Introduction
N′-Nitrosonornicotine (NNN, 1, Scheme 1), a tobacco-specific nitrosamine, is one of the most abundant strong carcinogens in tobacco products.1, 2 NNN is a potent carcinogen in animal models, producing tumors in rats (esophagus, oral cavity, and nasal cavity), mice (lung and forestomach), hamsters (trachea and nasal cavity), and mink (nasal cavity and forebrain).3 NNN and the related carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) have been evaluated by the International Agency for Research on Cancer as carcinogenic to humans (Group 1).2, 4
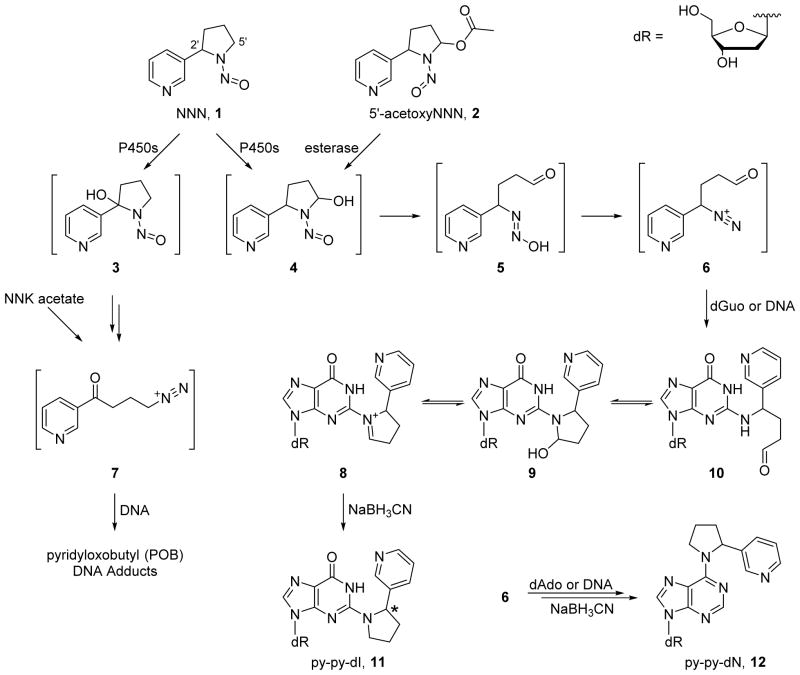
Scheme 1.
Formation of DNA adducts from NNN metabolism. Hydroxylation of the 2′ carbon is known to yield POB-DNA adducts. Hydroxylation of the 5′ carbon of NNN can be modeled in vitro by the hydrolysis of 5′-acetoxyNNN (2). Intermediate 4 spontaneously rearranges to yield the highly reactive diazonium ion 6, which reacts with the N2 position of guanine or the N6 position of adenine. The intermediate adducts are reduced by sodium cyanoborohydride, yielding 11 or 12, which were detected in vivo in this study.
To exert their carcinogenicity, NNN and NNK must be metabolically activated by α-hydroxylation, a process which is catalyzed by cytochrome P450s. NNN activation occurs via one of two pathways: 2′-hydroxylation or 5′-hydroxylation (Scheme 1). The 2′-hydroxylation pathway has been extensively studied in vitro and in target tissues of rats, and it is believed to be the more mutagenic and carcinogenic pathway.5–10 However, data suggest that 5′-hydroxylation of NNN is the more prevalent metabolic pathway in non-human primates.11 Also, in vitro data demonstrate that human liver microsomes and human P450s preferentially 5′-hydroxylate NNN, showing 3-fold to 40-fold selectivity over 2′-hydroxylation.12–14 Studies with ex vivo human esophagus tissue also demonstrate that 5′-hydroxylation is the major metabolic pathway of NNN activation.15, 16 Based on these data, 5′-hydroxylation of NNN is likely to be the major metabolic pathway in humans exposed to tobacco products, and this could be an important source of DNA damage. 5′-Hydroxylation is also the major metabolic pathway in A/J mouse lung and in Syrian golden hamster trachea, which are two important target tissues of NNN tumorigenicity.17–19
The formation of DNA adducts is a key step early in the process of chemical carcinogenesis for many carcinogens.20, 21 In the study reported here, we aimed to characterize and quantitate the DNA damage caused by 5′-hydroxylation of NNN. We have previously identified five DNA adducts (11–15; Scheme 1 and Figure 1) which are formed in vitro from the reaction of 5′-acetoxyNNN (2) and DNA followed by NaBH3CN reduction,22, 23 but the formation of these adducts had not yet been investigated in vivo. One of the major adducts in vitro was 2-(2-(3-pyridyl)-N-pyrrolidinyl)-2′-deoxyinosine (py-py-dI, 11), which forms upon the reaction of the N2 position of guanine with the diazonium ion 6 followed by sodium cyanoborohydride reduction (Scheme 1). Herein we describe studies showing that py-py-dI is by far the major 5′-hydroxylation DNA adduct formed in vivo. We also demonstrate that py-py-dI is the major known DNA adduct formed in vitro by human enzymatic metabolism of NNN. The characterization of DNA adducts resulting from NNN metabolic activation could ultimately lead to a biomarker which could inform cancer risk among tobacco users.
Figure 1.
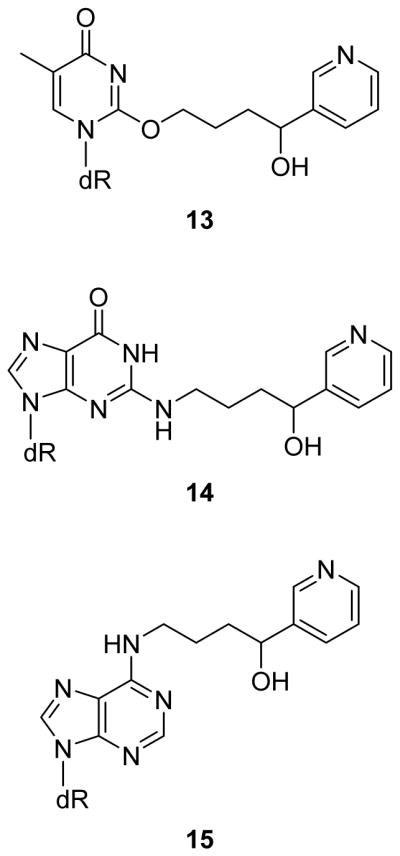
DNA adducts other than 11 and 12 previously identified in vitro as products of the reaction of 5′-acetoxyNNN (2) with DNA followed by treatment with NaBH3CN. The mechanism of formation has been detailed previously.22, 23 These adducts were not detected in vivo in these studies.
Experimental Procedures
CAUTION: NNN is a strong carcinogen and should be handled with extreme care and personal protective equipment. The chemically activated form, 5′-acetoxyNNN, is a potent mutagen and should also be handled with extreme care.
Chemicals and Reagents
Enantiopure (S)- and (R)-NNN,24, 25 analytical standards of (R)- and (S)-py-py-dI,22 and 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK acetate)26 were synthesized as described previously. Hepatocytes, human liver S9 fraction, and corresponding incubation media were obtained from XenoTech LLC (Kansas City, KS). DNA isolation solutions were purchased from Qiagen (Valencia, CA). 2′-Deoxy[15N5]guanosine was procured from Cambridge Isotope Laboratories (Tewksbury, MA). All other chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO).
dGuo was incubated with NNK acetate essentially as described previously.27 Half of the reaction mixture was reduced with NaBH3CN, and both halves were analyzed for the presence of DNA adducts.
Synthesis and Analysis of Enantiopure (2′S)- and (2′R)-5′-AcetoxyNNN
This synthetic procedure was adapted from a previously published synthesis of racemic 5′-acetoxyNNN.28 The previous method employed (±)-tert-butylsulfinamide, but here we utilized enantiopure (S)-(−)- or (R)-(+)-tert-butylsulfinamide. The synthetic procedure is detailed in the Supporting Information (Scheme S1 and Table S1). The final products, (2′S)- and (2′R)-5′-acetoxyNNN, were analyzed by chiral HPLC-UV. The diastereomers were separated on a 250 × 4.6 mm (R,R) Whelk-O1 Pirkle-style HPLC column (Regis Technologies, Morton Grove, IL) under isocratic conditions of 75% isopropanol and 25% hexanes, 1 mL/min flow rate. The two mixtures of diastereomers were dissolved in CH2Cl2 and analyzed by polarimetry on an Autopol V polarimeter (Rudolph Research Analytical, Hackettstown, NJ).
Internal Standards (R)- and (S)-[15N5]Py-py-dI
The internal standards were synthesized by reacting 2′-deoxy[15N5]guanosine (5 mg) with racemic 5′-acetoxyNNN (30 mg) in pH 7.4 phosphate buffer at 37 °C in the presence of porcine liver esterase (5 mg). The reaction was allowed to proceed for 24 h, then the samples were filtered through a 30 kDa molecular weight cutoff filter (Centrifree; Millipore, Jaffrey, NH) and treated with NaBH3CN. The resulting product mixture was purified by solid-phase extraction, then HPLC (linear gradient from 5% to 95% MeOH in H2O over 45 min, 1 mL/min, on a Luna C18(2), 5 μm, 4.6 × 250 mm column), yielding separate stock solutions of (R)- and (S)-[15N5]py-py-dI diastereomers.
Metabolism of NNN by Human Liver S9 Fraction
The human liver S9 fraction (Xenotech) was a pooled mixture isolated from 200 donors (100 male, 100 female, 11–83 years of age, and predominantly Caucasian). The S9 preparation included a recommended protocol for optimal P450 activity, which was adopted in these experiments. Briefly, a buffer was prepared to final concentrations of 50 mM potassium phosphate (pH 7.4), 3 mM MgCl2, 1 mM EDTA, 1 mM NADPH, 5 mM glucose-6-phosphate, 1 mg/mL calf thymus DNA, 1 U/mL glucose-6-phosphate dehydrogenase, 1 mg/mL human liver S9 fraction, and the appropriate concentration of NNN (2–500 μM). The DNA and all salts were added first and vortexed to ensure full dissolution before enzymes or NNN were added. The final solution (5 mL) was incubated for 24 h at 37 °C. Ice cold IPA (7.5 mL) was added to precipitate the DNA. The DNA was then purified by extraction and analyzed as detailed below.
Incubations of Human and Rat Hepatocytes with NNN or 5′-AcetoxyNNN
Hepatocytes (XenoTech) were thawed according to the included protocol. Cells (4–5 million) were suspended in Hepatocyte Incubation Media (4–5 mL, XenoTech), transferred to a 25 cm2 Nunc flask with filter cap, and placed in a humidified incubator at 37 °C with 5% CO2. After 30 min of acclimation, the cells were treated with (S)-, (R)-, or racemic NNN or (2′S)-, (2′R)-, or racemic 5′-acetoxyNNN in the presence of porcine liver esterase (5 mg). After 4 h, the cells were pelleted at 500g for 10 min. The medium was removed and the DNA was isolated and purified as described below.
Treatment of Rats with racemic NNN, (S)-NNN, or (R)-NNN
These studies were approved by the University of Minnesota Institutional Animal Care and Use Committee. Thirty-six male F-344 rats (Charles River Laboratories, Frederick, MD), 6 weeks of age, were housed two per cage (20–24 °C, 12 h light/dark cycle). After a 2-week acclimation period, the rats were treated with either 0, 50, 100, or 500 ppm racemic NNN in their drinking water (9 rats per group). After 3 weeks of treatment, the rats were euthanized by CO2 overdose, and the lung, liver, oral cavity mucosa, esophageal mucosa, nasal respiratory mucosa, and nasal olfactory mucosa were collected and stored at −80 °C until the DNA was isolated. The oral cavity, nasal cavities, and esophageal mucosa yield small amounts of DNA, so tissues from three rats were combined for a single DNA adduct analysis. Liver and lung samples from individual rats were analyzed for DNA adducts. We quantified N = 3 replicate analyses from each tissue except esophageal mucosa, where N = 2.
Under similar conditions, 81 rats were treated with 7, 14, or 28 ppm (S)-NNN or (R)-NNN, or 14, 28, or 56 ppm racemic NNN (9 rats per group) for 5 weeks.29 The same tissues were collected, but only lung, nasal respiratory mucosa, and nasal olfactory mucosa were analyzed for 5′-hydroxylation DNA adduct formation.
DNA Isolation and Purification Procedures
DNA was isolated from tissues and cells following an adapted Gentra Puregene protocol (Qiagen), as described previously.30 The Puregene protocol was modified such that the samples were never heated above 37 °C. The isolated DNA was then dissolved in 10 mM Tris, 1 mM EDTA buffer (pH 7). An equal volume of 24:1 CHCl3:isoamyl alcohol was added, the samples were mixed vigorously and centrifuged at 3,000g for 15 min. The aqueous layer was transferred to a clean tube and the extraction was repeated until there were no visible solids at the solvent interface. The DNA was then precipitated with ice cold EtOH and washed once with 70% (v/v) EtOH and twice with 100% EtOH.
DNA Adduct Analyses by LC-MS/MS
Purified DNA was dissolved in 10 mM sodium succinate buffer containing 5 mM CaCl2 (1 mL, pH 6.5). Four fmol (R)-[15N5]py-py-dI and 5 fmol (S)-[15N5]py-py-dI were added as internal standards. Micrococcal nuclease (30 U/mg DNA) and phosphodiesterase II (250 mU/mg DNA) were added and the samples were incubated at 37 °C for 5 h. Alkaline phosphatase (75 U/mg DNA) was added and the samples were incubated at 37 °C overnight. Samples were filtered through a 30 kDa molecular weight cutoff filter, a 10 μL aliquot was removed for dGuo quantitation, then NaBH3CN was added for 1 h to convert the precursor adduct 8 into py-py-dI, 11. The samples were purified by solid-phase extraction (Strata-X polymeric reversed phase, 30 mg, Phenomenex, Torrance, CA). The cartridges were preconditioned with MeOH and H2O (1 mL each). The samples were loaded and the cartridges were washed with H2O and 25% (v/v) MeOH (1 mL each). The analytes were eluted with 1 mL 50% (v/v) MeOH, the solvent was evaporated in vacuo, and the samples were reconstituted in 20 μL 5% MeOH in H2O for LC-MS/MS analysis.
The sample preparation for POB-DNA adducts was slightly different, as described previously.10 For samples where py-py-dI and POB adducts were both quantified, the DNA was dissolved in 2 mL buffer and half of the solution was analyzed for py-py-dI as described above, while half was analyzed for POB adducts. Briefly, the POB analysis requires heating to 100 °C for 30 min prior to enzyme hydrolysis, no NaBH3CN was added, and the solid-phase extraction wash step was only 10% (v/v) MeOH, while the elution step was 95% MeOH.
The DNA hydrolysate was analyzed for py-py-dI by liquid chromatography-positive electrospray ionization-tandem mass spectrometry (LC-ESI+-MS/MS) for samples with relatively high adduct formation. LC was carried out on a 0.5 × 150 mm Zorbax SB-C18 5 μm column (Agilent, Santa Clara, CA) with a linear gradient and flow rate of 15 μL/min. After holding initial conditions at 10% B from 0–2 min, the composition was increased to 60% B from 2–17 min, followed by washout and re-equilibration, where solvent A was 10 mM NH4OAc and solvent B was MeOH. MS was performed on a Finnigan TSQ Quantum Discovery MAX triple quadrupole mass analyzer (Thermo Scientific, Waltham, MA). Selected reaction monitoring mass transitions were m/z 399.2 → 283.1 for py-py-dI and m/z 404.2 → 288.1 for [15N5]py-py-dI at 23 eV collision energy, 0.5 amu isolation width. The other four DNA adducts arising from 5′-hydroxylation of NNN (12–15) were analyzed similarly.23
For samples with lower levels of adduct formation, a high-resolution, accurate mass Orbitrap Fusion Tribrid instrument (Thermo Scientific) was employed with LC-positive nanoelectrospray ionization-high-resolution tandem mass spectrometry (LC-NSI+-HRMS/MS). LC was performed on a hand-packed 75 μm × 15 cm, 15 μm orifice, hydro-RP 4 μm, 80 Å HPLC column (Phenomenex). Initial conditions utilized 5% B at 900 nL/min from 0–6 min to load the sample onto the column. Flow was decreased to 300 nL/min and a linear gradient was employed from 7–24 min from 5% to 95% B before re-equilibration, where A was 10 mM NH4OAc and B was MeOH. Precursor ions were isolated using the quadrupole (1.5 amu isolation width) and fragmented by higher-energy collisional dissociation (HCD) at 20%. Fragment ions (100–500 m/z) were analyzed by the Orbitrap detector at 120,000 resolution and 5.05 automatic gain control (AGC) target with maximum injection time of 750 ms. Exact mass chromatograms corresponding to the neutral loss of deoxyribose (the major fragment observed) were extracted at 3 ppm tolerance for py-py-dI (m/z 399.2 → 283.1302) and [15N5]py-py-dI (m/z 404.2 → 288.1154) with minor calibration adjustments if necessary. Developmental work with py-py-dI demonstrated that MS3 fragmentation at 40% HCD yielded a major fragment ion at m/z 132.0808 in both the py-py-dI and [15N5]py-py-dI standards which corresponds to loss of deoxyribose followed by loss of guanine and observation of the adduct fragment (Figure S2). Minor MS3 fragments for py-py-dI were m/z 204.0880 and 130.0651 and for [15N5]py-py-dI were m/z 209.0732 and 130.0651. The MS2 method was used for quantitation. The calibration curve for py-py-dI was linear (R2 > 0.999) between 1.5 amol and 1.5 fmol on column, which covered the range observed in samples. The limit of detection was determined by serial dilution of a standard in H2O and found to be 1 amol on column. We were able to sporadically detect 300 zmol on column, but the response was not linear. Each set of samples included a positive control sample with 2.5 fmol of py-py-dI standard added to dissolved calf thymus DNA. Eleven replicates analyzed over 6 months were used to assess accuracy and interday precision, with an average accuracy of 115% and a coefficient of variation of 11%.
The POB adducts were quantified on the same Orbitrap Fusion Tribrid system, monitoring the mass transition at 20% HCD of m/z 390.2 → 148.0757 for O2-POB-thymidine and m/z 394.2 → 152.1008 for O2-[D4]POB-thymidine. O2-POB-thymidine is generally the major POB adduct in rats treated with NNN under these conditions,10 and it was the only quantifiable adduct in these experiments. The limit of detection was 500 zmol for a dilute standard in H2O.
Adduct formation was normalized to dGuo content of each sample. Quantitation of dGuo was performed on an Agilent 1100 series HPLC with a UV diode array detector set at 254 nm. LC was performed on a 0.5 × 250 mm Luna C18(2) 5 μm, 100 Å column (Phenomenex) with a linear gradient at 10 μL/min from 5% to 20% MeOH in H2O over 22 min with subsequent washout and re-equilibration.
Results
Synthesis of Enantiopure 5′-AcetoxyNNN
The final products of the synthesis were analyzed by chiral stationary phase HPLC (Figure S3). The two diastereomers resulting from the synthesis of (2′S)-5′-acetoxyNNN eluted at 15.5 and 17.9 min, while the two (2′R)-5′-acetoxyNNN diastereomers eluted at 17.6 and 19.6 min. The peaks were equal in area, suggesting that the addition of acetate in the final step of the synthesis is nonselective. Co-injection of the two mixtures of diastereomers yielded three peaks in a 1:2:1 ratio. The two syntheses of 5′-acetoxyNNN yielded inversely optically active products by polarimetry. The (2′S)-5′-acetoxyNNN diastereomers gave a specific rotation of (1.0 g/100 mL, CH2Cl2), and the (2′R)-5′-acetoxyNNN diastereomers gave a specific rotation of (1.0 g/100 mL, CH2Cl2). By NMR, we did not observe distinct E and Z isomers, as are commonly seen in most NMR spectra of N-nitrosamines. The 1H and 13C chemical shifts of the 2′ position are more consistent with an (E) rotamer, with the nitroso oxygen pointing toward the pyridine ring, by comparison with NMR data for NNN.31
Reaction between dGuo or DNA and 5′-acetoxyNNN
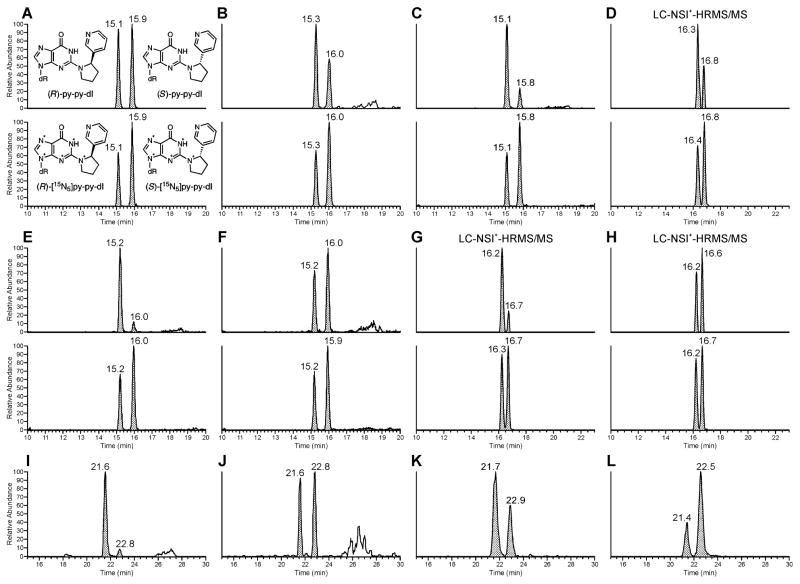
To investigate the formation of (R)- and (S)-py-py-dI in vitro, we exposed either dGuo or calf thymus DNA to (2′S)- or (2′R)-5′-acetoxyNNN dissolved in phosphate buffer under conditions that have been described previously.22 For the reactions with (2′S)-5′-acetoxyNNN, we observed that (R)-py-py-dI was formed to a greater extent than (S)-py-py-dI (Figure 2K), demonstrating that the reaction likely proceeded via an SN2 mechanism, but with some racemization (Scheme 2). This ratio of py-py-dI formation was the same for the reaction with dGuo as for the reaction with DNA. The reactions with (2′R)-5′-acetoxyNNN gave the expected mirror image in vitro, where (S)-py-py-dI was the major peak (Figure 2L), and this held true for the reactions with either dGuo or DNA.
Figure 2.
LC-MS/MS traces obtained upon analysis of py-py-dI in hydrolysates of DNA from experiments carried out in vitro and in vivo. In all sections, the top chromatogram is the mass transition for (R)-py-py-dI (eluting first), and (S)-py-py-dI (eluting second). The bottom chromatogram in each section is the mass transition for the [15N5]-labeled internal standards, which were not available at the time samples (I–L) were analyzed. Asterisks denote 15N isotopes. Chromatograms are from the following sources (treatment): (A) Analytical standards in H2O (5 fmol/uL), (B) Rat liver (100 ppm racemic NNN), (C) Rat olfactory mucosa (50 ppm racemic NNN), (D) Human liver S9 (5 μM racemic NNN), (E) Rat hepatocytes (50 μM (2′S)-5′-acetoxyNNN), (F) Rat hepatocytes (50 μM (2′R)-5′-acetoxyNNN), (G) Human liver S9 (5 μM (S)-NNN), (H) Human liver S9 (5 μM (R)-NNN), (I) Rat respiratory mucosa (28 ppm (S)-NNN), (J) Rat respiratory mucosa (28 ppm (R)-NNN), (K) Calf thymus DNA exposed to (2′S)-5′-acetoxyNNN in buffer. (L) Calf thymus DNA exposed to (2′R)-5′-acetoxyNNN in buffer. All traces except (D), (G), and (H) were from the triple quadrupole LC-ESI+-MS/MS method.
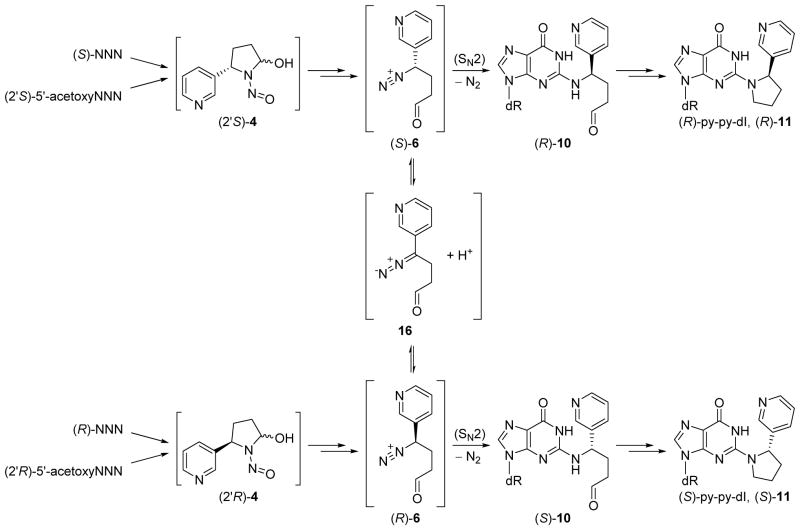
Scheme 2.
Proposed mechanism for the observed stereochemistry of py-py-dI formation. Intermediate 4 can be generated either from hydroxylation of NNN (1) or hydrolysis of 5′-acetoxyNNN (2). Spontaneous rearrangement to the diazonium ion 6, followed by reaction with DNA in an SN2 manner causes inversion of the stereocenter. Interconversion of 6 with the diazo species 16 will partially racemize the stereocenter before reaction with DNA.
Reaction between dGuo and NNK Acetate
To test the specificity of py-py-dI as a marker of 5′-hydroxylation of NNN, we incubated dGuo with NNK acetate. Upon hydrolysis, NNK acetate forms the same reactive species 7 (Scheme 1) as is formed by 2′-hydroxylation of NNN. Intermediate 7 can react with the N2 position of dGuo to form N2-pyridyloxobutyl-2′-deoxyguanosine (N2-POB-dGuo), which has been observed in vitro.27 We hypothesized that if this DNA adduct were to cyclize by reaction of the −N2-H with the carbonyl adjacent to the pyridine ring, then reduction with NaBH3CN could be an alternate route toward formation of py-py-dI. Formation of O6-POB-dGuo in this reaction was clearly detected and confirmed by comparison to a synthetic standard, but there was no detectable formation of py-py-dI in this reaction after reduction with NaBH3CN. This demonstrates that py-py-dI is a specific marker of DNA damage arising exclusively from 5′-hydroxylation of NNN.
Adduct Formation in Rats
Py-py-dI (11) was the major DNA adduct arising from 5′-hydroxylation of NNN in vivo. DNA from rats treated with relatively high doses of racemic NNN (50, 100, or 500 ppm) was analyzed for the presence of five types of 5′-hydroxylation DNA adducts (adducts 11–15), and py-py-dI formation predominated in all tissues analyzed. Representative chromatograms of py-py-dI in hydrolysates of DNA from the liver and olfactory mucosa are shown in Figure 2B and C, respectively. As discussed in our previous publication,22 the samples were treated with NaBH3CN because the equilibrium among 8–10 interferes with chromatography and makes quantitation difficult. There are 8 separable species present among the diastereomers of 8, 9, and 10, whereas py-py-dI elutes as two easily-separable peaks. We have previously established that (R)-py-py-dI elutes first and (S)-py-py-dI elutes second when chromatographed on C18-based stationary phases (Figure 2A).22 A clear dose-response relationship was observed for py-py-dI formation in all tissues (Figure 3). The DNA adduct 6-(2-(3-pyridyl)-N-pyrrolidinyl)-2′-deoxynebularine (py-py-dN, 12) arising from the analogous reaction with adenosine was also detected in rat tissues, but at lower levels. Py-py-dN formation was assessed by LC-MS/MS peak area and was found to be ~10-fold lower than py-py-dI in the lung, ~20-fold lower in the nasal cavity, and py-py-dN was not detected in the liver, esophagus, or oral cavity. The remaining three adducts (13–15, Figure 1) arising from 5′-hydroxyNNN previously characterized in vitro23 were not detected in vivo.
Figure 3.
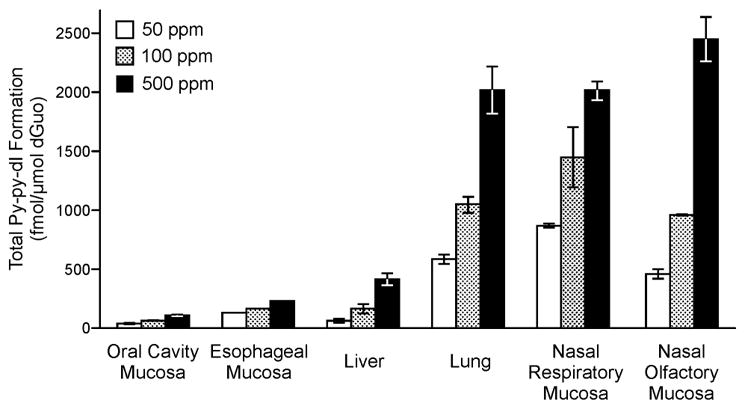
Total py-py-dI adduct formation in rats treated with racemic NNN. Mean and standard deviation data are shown from rats treated with 50, 100, or 500 ppm NNN in drinking water for 3 weeks. Triplicate samples were analyzed except in the esophageal mucosa, where duplicate samples were analyzed. Adducts were not detected in any tissues of the control rats (0 ppm).
Py-py-dI was readily detected in the lung and nasal cavity of rats treated with lower doses of (S)-, (R)-, or racemic NNN (7, 14, 28, or 56 ppm) by the triple quadrupole LC-ESI+-MS/MS method. Interestingly, rats treated with (S)-NNN showed a different ratio of formation of (R)- to (S)-py-py-dI than rats treated with (R)-NNN. (S)-NNN-treated rats formed primarily (R)-py-py-dI (Figure 2I), while (R)-NNN-treated rats showed nearly equal formation of (R)- and (S)-py-py-dI (Figure 2J). This divergent pattern of adduct formation from (S)-NNN treatment versus (R)-NNN treatment was observed in all tissues analyzed from these rats.
The ratio of (R)- to (S)-py-py-dI formation was variable among tissues of rats treated with racemic NNN (Table S2). The ratios in oral cavity, esophagus, liver, and lung were 1.5:1 to 2.2:1 (Figure 2B). The difference was greater in the nasal respiratory and nasal olfactory mucosa, where the ratio ranged from 3.5:1 to 4.7:1 (R):(S)-py-py-dI (Figure 2C). As indicated above, the 5′-hydroxylation of (S)-NNN yields primarily (R)-py-py-dI in vivo, while 5′-hydroxylation of (R)-NNN yields both (R)- and (S)-py-py-dI. If this holds true for the metabolism of each enantiomer in racemic NNN-treated rats, then we can conclude that (S)-NNN is more extensively 5′-hydroxylated than (R)-NNN in the nasal cavity, while (R)-NNN is more extensively 5′-hydroxylated in other tissues.
Adduct Formation by Human Liver S9 Fraction with Added Calf Thymus DNA
We based our NNN concentration range on previous studies of NNN metabolism with human liver microsomes and expressed human P450 enzymes.12, 14 At these low concentrations, analysis of the DNA required the more sensitive LC-NSI+-HRMS/MS method which allows quantitation of low levels of py-py-dI (6–52 amol on column, Figure 2D, G, and H). Quantifiable and reproducible levels of py-py-dI were observed in incubations with concentrations of (S)-NNN as low as 2 μM (the reported Km value for (S)-NNN 5′-hydroxylation by CYP2A6), and concentrations of (R)-NNN or racemic NNN as low as 5 μM (Figure 4 and Table S3). Human liver enzymes were more efficient at 5′-hydroxylation of (S)-NNN than of (R)-NNN, demonstrating 3- to 6-fold higher adduct formation at every concentration tested here (5–250 μM, Table S3). Levels of POB-DNA adducts were only detected at relatively high concentrations of NNN. Triplicate samples of 20 μM racemic NNN yielded POB-DNA adducts near the LOD (~4:1 signal-to-noise ratio), but we could not reliably quantify adduct formation at this level. At 50 μM NNN or higher, we were able to reliably detect formation of POB-DNA adducts by human liver enzymes. The limit of detection for py-py-dI was 1 amol on column for a dilute standard in H2O, and for O2-POB-thymidine the LOD was 0.5 amol on column. For DNA samples, additional information must be considered, including matrix suppression effects, analyte recovery, and total amount of DNA in each sample. For the human liver S9 incubations, which provided a large amount of DNA, the limit of detection for py-py-dI was 0.8 adducts per 1010 nucleotides, and for O2-POB-thymidine it was 1 adduct per 1010 nucleotides. The intraday reproducibility of the S9 incubations was highly consistent, as shown in Figure 4. The interday reproducibility was more variable, showing variation up to two-fold for a given concentration of NNN (data not shown). The S9 incubations were performed for 24 h to maximize adduct formation, but we have observed that the majority of the adducts are formed within the first 4–8 hours. After this time, the NADPH regenerating system loses activity, which could explain the variability in adduct formation between different preparations of these enzymes. All triplicate incubations at all concentrations shown here were performed on the same day with the same stock of enzymes.
Figure 4.
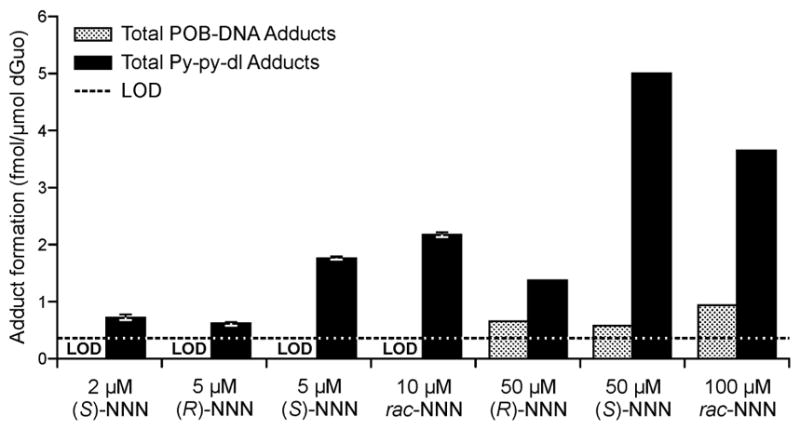
Total py-py-dI and POB adduct formation in vitro from incubation of NNN with human liver S9 enzymes and added calf thymus DNA. Black bars represent py-py-dI formation and gray textured bars represent POB-DNA adduct formation, by quantifying O2-POB-thymidine. Mean and standard deviation data are shown for triplicate incubations at ≤10 μM NNN. Single analyses were performed at ≥50 μM. The black dotted line is the limit of detection for this analysis. LOD denotes the incubations where POB adduct formation was below the limit of detection.
Adduct Formation in Rat and Human Hepatocytes
The hepatocytes were more difficult to analyze since the yield of DNA was low (5–27 μg per incubation) because cell viability decreased rapidly after 3 h. Shorter incubation times yielded less metabolism of NNN and thus adduct formation was near or below the LOD. We were only able to quantify py-py-dI in hepatocytes at higher concentrations of NNN (≥50 μM). At these concentrations, human hepatocytes formed 2-fold to 6-fold more py-py-dI than rat hepatocytes (Table S4).
Rat hepatocytes yielded the same type of stereoselective formation of py-py-dI as we observed in rats treated with NNN, where the (S) intermediates 4–6 generated from (2′S)-5′-acetoxyNNN almost exclusively form (R)-py-py-dI while the (R) intermediates 4–6 generated from (2′R)-5′-acetoxyNNN produce a ratio closer to 1:1 (R):(S)-py-py-dI (Figure 2E, F and Table S4). We observed similar ratios of (R):(S)-py-py-dI formation in rat and human hepatocytes treated with (S)- or (R)-NNN (Table S4). This suggests that the chirality of intermediates 4, 5, and/or 6 is determining the reactivity toward DNA in cells and the effect is similar across species. The observed difference in adduct formation is not due to stereospecific activation by cytochrome P450s or hydrolysis of 5′-acetoxyNNN by porcine liver esterase.
We also used rat hepatocytes to investigate DNA repair processes. After 1 h of exposure to 5′-acetoxyNNN, the media was removed and fresh media was supplied to the cells. DNA was isolated and analyzed at 0, 1, 2, and 3 h after initial exposure, and both (R)- and (S)-py-py-dI adduct levels remained constant over this time period. This confirms that the difference in observed (R)- and (S)-py-py-dI is not due to stereoselective repair processes, and it also suggests that these adducts are not rapidly repaired within live cells.
Discussion
The research reported here demonstrates for the first time that py-py-dI is the major 5′-hydroxylation DNA adduct formed from NNN in vivo in rats. In previous studies, DNA adduct formation resulting from metabolic activation of NNN has primarily been investigated in tissues of rats in which 2′-hydroxylation is often the predominant pathway.8 Our results are consistent with these previous studies; py-py-dI arising from 5′-hydroxylation is a minor constituent of the total known DNA adducts in all rat tissues tested here (Table 1). The 5′-hydroxylation of NNN in rat tissues is primarily catalyzed by CYP2A3,14 which is highly expressed in the lung and nasal cavity, but is minimally expressed in the esophagus and not detectable in the liver.32 The formation of py-py-dI reflects the tissue distribution of CYP2A3.
Table 1.
Comparison of DNA adduct formation from 2′-hydroxylation vs 5′-hydroxylation of NNN in rats. Values for py-py-dI have been converted to fmol/mg DNA to match the reported POB-DNA adduct levels. Dosage and treatment length are detailed in the first column. Total POB-DNA adducts are the sum of O2-POB-thymidine, N7-POB-guanine, and O2-POB-cytosine. Adducts arising from 2′-hydroxylation predominate in all rat tissues, after accounting for dosage.
| Oral Cavity Mucosa | Esophageal Mucosa | Liver | Lung | Nasal Respiratory Mucosa | Nasal Olfactory Mucosa | |
|---|---|---|---|---|---|---|
|
|
||||||
| Total py-py-dI Formation, (fmol/mg DNA) 50 ppm Racemic NNN, 3 weeks | 21 | 85 | 37 | 383 | 574 | 302 |
| Total POB-DNA Adduct Formation, (fmol/mg DNA) 10 ppm (R)-NNN, 2 weeks | 205a | 360b | 70b | 310b | 2940a | 851a |
| Total POB-DNA Adduct Formation, (fmol/mg DNA) 10 ppm (S)-NNN, 2 weeks | 644a | 1090b | 340b | 120b | 1580a | 201a |
| Total POB-DNA Adduct Formation, (fmol/mg DNA) 14 ppm (R)-NNN, 10 weeks | 184c | 736c | 121c | 704c | 3250c | 1700c |
| Total POB-DNA Adduct Formation, (fmol/mg DNA) 14 ppm (S)-NNN, 10 weeks | 592c | 2030c | 728c | 327c | 6720c | 431c |
We observed that (R)- and (S)-py-py-dI are formed in different ratios when rats are treated with (S)- or (R)-NNN. (S)-NNN yielded almost exclusive formation of (R)-py-py-dI, while (R)-NNN yielded a nearly 1:1 ratio of (R)- and (S)-py-py-dI. When the diazonium ion 6 reacts with nucleophiles, the chirality is expected to invert via an SN2 mechanism,33 thus (S)-NNN should yield (R)-py-py-dI (Scheme 2). However, previous studies of alkyldiazonium ions at neutral pH suggest that diazonium ions can interconvert with a diazo species.6, 34 If this process occurs to form intermediate 16, we would expect to see some degree of racemization at the 2′ carbon before 6 reacts with DNA. We observed this racemization process in vitro. When (2′S)-5′-acetoxyNNN reacts with calf thymus DNA in pH 7.4 phosphate buffer, we measure a ~2:1 ratio of (R):(S)-py-py-dI formation, with the major peak arising from inversion of the stereocenter (Figure 2K). Similarly, when (2′R)-5′-acetoxyNNN reacts with DNA in buffer, we see a ~1:2 ratio of (R):(S)-py-py-dI formation (Figure 2L).
In an attempt to reproduce the chemistry that we observed in vivo, we incubated rat hepatocytes with either (S)- or (R)-NNN or (2′S)- or (2′R)-5′-acetoxyNNN, and we observed similar ratios of py-py-dI formation that we observed in live rats (Figure 2E, F, I, and J). We observed similar patterns of adduct formation in the hepatocyte incubations with 5′-acetoxyNNN to those with NNN. This demonstrates that it is the stereochemistry of the reactive intermediates 4–6, not the stereospecific activation by P450s or esterase, which is responsible for the observed ratio of (R)- to (S)-py-py-dI formation. We simplified this system further, and we were able to observe ratios of py-py-dI formation similar to those in rats when DNA was dissolved in hepatocyte incubation media (XenoTech, supplemented with fetal bovine serum) and incubated with 5′-acetoxyNNN and esterase. But this phenomenon was not observed for the reaction between 5′-acetoxyNNN and dGuo dissolved in hepatocyte incubation media, nor was it observed for the reaction between 5′-acetoxyNNN and DNA dissolved in buffer (Figure 2K and L). Perhaps there is a protein capable of interacting with double-stranded DNA and with at least one of the chiral reactive intermediates 4, 5, and/or 6, that either facilitates the formation of (R)-py-py-dI or hinders the formation of (S)-py-py-dI. To our knowledge, a carrier protein of this type is unprecedented in studies of DNA alkylation by nitroso compounds. Further investigation is needed to determine the cause of the stereospecific reactivity of the (S)- and (R)-diazonium ions toward DNA in the cell.
Literature reports on the stereospecific formation of DNA adducts by enantiomeric electrophiles are sparse. The first report we are aware of was with an activated benzo[a]pyrene (BP) metabolite,35 wherein (+)-anti-BP diol epoxide readily reacted with DNA to form an N2 adduct with guanine residues in double-stranded DNA while (−)-anti-BP diol epoxide was poorly reactive toward DNA. This selectivity effect was not observed with single-stranded DNA, demonstrating the critical importance of the chiral secondary structure of the DNA double-helix.35 Selective reactivity has also been reported with BP diol epoxide binding to albumin side chains,36 with aflatoxin B1 binding to A-form, B-form, or Z-form DNA,37 and with 2,3-epoxy-4-hydroxynonanal.38 In each of these cases, the secondary structure of the macromolecule guided the stereospecific reactivity of the electrophile. In our work with chiral intermediates 4–6, the reactivity was not solely influenced by the secondary structure of DNA. We were able to observe stereospecific adduct formation when double-stranded DNA was dissolved in cell culture medium, but not when the same DNA was dissolved in phosphate buffer. There remains the possibility that a protein is binding to the DNA and altering its structure, which may affect the interaction with the electrophile. Or the electrophile binds the protein first, and is then guided toward the DNA. A similar three-component system has recently been modeled with the (+)-trans-anti-BP-N2-dGuo DNA adduct and the residues of a histone tail.39
In rats treated with racemic NNN, we observed varying ratios of (R):(S)-py-py-dI formation in different tissues (Table S2). The oral cavity, esophagus, liver and lung showed 1.5:1 to 2.2:1 ratios, while the nasal respiratory and nasal olfactory mucosa showed upwards of 4.7:1 ratios of (R):(S)-py-py-dI formation (Figure 2B and C). We can utilize these ratios to estimate the relative metabolism of (S)- and (R)-NNN in rats that were treated with racemic NNN. As discussed above, the in vivo metabolism of (S)-NNN yields primarily (R)-py-py-dI, while (R)-NNN yields approximately equal amounts of both diastereomers of py-py-dI. Then it should follow that for racemic NNN treatment, if (S)-NNN and (R)-NNN were 5′-hydroxylated to the same extent, we would expect a combined ratio of ~3:1 (R):(S)-py-py-dI. We can use this ratio to determine that (S)-NNN is more extensively 5′-hydroxylated than (R)-NNN in the nasal cavity, where the ratio is higher than 3:1. Likewise, we can conclude that (R)-NNN is more extensively 5′-hydroxylated in the oral cavity, esophagus, liver and lung, where the ratios were lower. This is consistent with previous metabolic studies of rats treated with (S)- or (R)-NNN.8
In human model systems, NNN is preferentially 5′-hydroxylated in vitro,12–14 as was determined by monitoring the major metabolites formed upon hydrolysis of the reactive diazonium ions 6 and 7. However, there is the possibility that these diazonium ions are not equally reactive toward DNA, which is an important factor when considering the genotoxic effects of NNN. The work presented here directly measures the DNA damage resulting from 2′-hydroxylation and 5′-hydroxylation of NNN and demonstrates that the major type of DNA damage in human systems is in the form of py-py-dI. The next important step needing to be evaluated is the mutagenic potential of these adducts. In A/J mouse peripheral lung and in Syrian golden hamster trachea, two target tissues of NNN tumorigenicity, the 5′-hydroxylation of NNN is the predominant metabolic pathway.17–19 It is possible that py-py-dI is playing an important role in the carcinogenesis process in these animal models.
We were able to detect py-py-dI formation when pooled human liver S9 fraction was incubated with DNA and NNN at concentrations as low as 2 μM (S)-NNN, 5 μM (R)-NNN, or 5 μM racemic NNN. The previously reported Km values for NNN 5′-hydroxylation are 2.3 μM (S)-NNN, 2.1 μM racemic NNN, and 22 μM (R)-NNN by recombinant human CYP2A6, and Km values range from 4.9–45 μM racemic NNN in individual human liver microsome samples.12, 14 In the same studies, the reported Km values for racemic NNN 2′-hydroxylation were 304 μM for CYP3A4 and 312 μM for microsomes, while CYP2A6 had no detectable 2′-hydroxylation activity. When human liver S9 was exposed to higher concentrations of NNN (50–500 μM), we observed that py-py-dI formation did not continue to increase, suggesting that CYP2A6 enzymes are saturated at these concentrations (Table S3). We were only able to quantify POB-DNA adduct formation at 50 μM NNN or higher, which is consistent with the reported Km values for CYP3A4 and suggests that 2′-hydroxylation of NNN is a minor source of DNA damage in human liver enzyme systems at relevant concentrations of NNN.
(S)-NNN is more extensively metabolized by human liver enzymes and produces more 5′-hydroxylation DNA adducts than (R)-NNN. Studies in rats have demonstrated that (S)-NNN is the more carcinogenic enantiomer, and it produces more 2′-hydroxylation DNA adducts in the rat oral cavity, esophagus, and liver.10, 40 Thus, in both the human model and the rat model, (S)-NNN causes more total DNA damage than (R)-NNN. The mutagenicity of py-py-dI has not yet been investigated in human systems. This will be important to determine, as tobacco products contain more (S)-NNN than (R)-NNN.31, 41
Conclusion
We have shown for the first time that py-py-dI forms in vivo and is the major DNA adduct arising from NNN 5′-hydroxylation. In human systems, where 5′-hydroxylation predominates, py-py-dI forms in higher amounts than POB adducts resulting from 2′-hydroxylation. Metabolism of (S)-NNN leads to more total adduct formation than does (R)-NNN in human systems. The ratio of (R)- to (S)-py-py-dI formation differs between the NNN enantiomers, which requires further study on the nature of the chiral reactive intermediates. This work establishes a foundation for a better understanding of intermediates involved in nitrosamine bioactivation and for further biomarker development in humans exposed to tobacco products.
Supplementary Material
Acknowledgments
Funding Information
This work was supported by grant R01-CA-081301 from the National Cancer Institute. Mass spectrometry was carried out in the Analytical Biochemistry Shared Resource at the Masonic Cancer Center, partially supported by grant P30-CA-077598 from the National Cancer Institute.
The authors would like to thank Bob Carlson for editorial assistance. We would also like to thank Dr. Peter Villalta and Xun Ming for mass spectrometry assistance in the Analytical Biochemistry Shared Resource at the Masonic Cancer Center. We thank Anna K. Michel for providing NNK acetate.
Abbreviations list
- AGC
automatic gain control
- dR
2′-deoxyribose
- HCD
higher-energy collisional dissociation
- LC-ESI+-MS/MS
liquid chromatography-positive electrospray ionization-tandem mass spectrometry
- LC-NSI+-HRMS/MS
liquid chromatography-positive nanoelectrospray ionization-high-resolution tandem mass spectrometry
- LOD
limit of detection
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNK acetate
4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNN
N′-nitrosonornicotine
- POB
pyridyloxobutyl
- py-py-dI
2-(2-(3-pyridyl)-N-pyrrolidinyl)-2′-deoxyinosine
- py-py-dN
6-(2-(3-pyridyl)-N-pyrrolidinyl)-2′-deoxynebularine
Footnotes
The supporting information document contains the synthetic details and spectral data for enantiopure (2′S)- and (2′R)-5′-acetoxyNNN, MS fragmentation patterns for py-py-dI, chiral HPLC data for 5′-acetoxyNNN, and (R)- and (S)-py-py-dI formation data in rat tissues, human liver S9, and rat and human hepatocytes. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.IARC. Tobacco smoke and involuntary smoking. IARC Monogr Eval Carcinog Risks Hum. 2004;83:1–1438. [PMC free article] [PubMed] [Google Scholar]
- 2.IARC. Smokeless tobacco and some tobacco-specific N-nitrosamines. IARC Monogr Eval Carcinog Risks Hum. 2007;89:1–592. [PMC free article] [PubMed] [Google Scholar]
- 3.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 4.IARC. Personal habits and indoor combustions. Volume 100 E. A review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum. 2012;100:1–538. [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CB, Hecht SS, Hoffmann D. Metabolic alpha-hydroxylation of the tobacco-specific carcinogen, N′-nitrosonornicotine. Cancer Res. 1978;38:3639–3645. [PubMed] [Google Scholar]
- 6.Hecht SS, Lin D. Comparative mutagenicity of 4-(carbethoxynitrosamino)-4-(3-pyridyl)butanal and 4-(carbethoxynitrosamino)-1-(3-pyridyl)-1-butanone, model compounds for alpha-hydroxylation of N′-nitrosonornicotine. Carcinogenesis. 1986;7:611–614. doi: 10.1093/carcin/7.4.611. [DOI] [PubMed] [Google Scholar]
- 7.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Analysis of pyridyloxobutyl DNA adducts in F344 rats chronically treated with (R)- and (S)-N′-nitrosonornicotine. Chem Res Toxicol. 2007;20:246–256. doi: 10.1021/tx060208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McIntee EJ, Hecht SS. Metabolism of N′-nitrosonornicotine enantiomers by cultured rat esophagus and in vivo in rats. Chem Res Toxicol. 2000;13:192–199. doi: 10.1021/tx990171l. [DOI] [PubMed] [Google Scholar]
- 9.Zhang S, Wang M, Villalta PW, Lindgren BR, Lao Y, Hecht SS. Quantitation of pyridyloxobutyl DNA adducts in nasal and oral mucosa of rats treated chronically with enantiomers of N′-nitrosonornicotine. Chem Res Toxicol. 2009;22:949–956. doi: 10.1021/tx900040j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao L, Balbo S, Wang M, Upadhyaya P, Khariwala SS, Villalta PW, Hecht SS. Quantitation of pyridyloxobutyl-DNA adducts in tissues of rats treated chronically with (R)- or (S)-N′-nitrosonornicotine (NNN) in a carcinogenicity study. Chem Res Toxicol. 2013;26:1526–1535. doi: 10.1021/tx400235x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Upadhyaya P, Zimmerman CL, Hecht SS. Metabolism and pharmacokinetics of N′-nitrosonornicotine in the patas monkey. Drug Metab Dispos. 2002;30:1115–1122. doi: 10.1124/dmd.30.10.1115. [DOI] [PubMed] [Google Scholar]
- 12.Patten CJ, Smith TJ, Friesen MJ, Tynes RE, Yang CS, Murphy SE. Evidence for cytochrome P450 2A6 and 3A4 as major catalysts for N′-nitrosonornicotine alpha-hydroxylation by human liver microsomes. Carcinogenesis. 1997;18:1623–1630. doi: 10.1093/carcin/18.8.1623. [DOI] [PubMed] [Google Scholar]
- 13.Staretz ME, Murphy SE, Patten CJ, Nunes MG, Koehl W, Amin S, Koenig LA, Guengerich FP, Hecht SS. Comparative metabolism of the tobacco-related carcinogens benzo[a]pyrene, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol, and N′-nitrosonornicotine in human hepatic microsomes. Drug Metab Dispos. 1997;25:154–162. [PubMed] [Google Scholar]
- 14.Wong HL, Murphy SE, Hecht SS. Cytochrome P450 2A-catalyzed metabolic activation of structurally similar carcinogenic nitrosamines: N′-nitrosonornicotine enantiomers, N-nitrosopiperidine, and N-nitrosopyrrolidine. Chem Res Toxicol. 2005;18:61–69. doi: 10.1021/tx0497696. [DOI] [PubMed] [Google Scholar]
- 15.Castonguay A, Stoner GD, Schut HA, Hecht SS. Metabolism of tobacco-specific N-nitrosamines by cultured human tissues. Proc Natl Acad Sci U S A. 1983;80:6694–6697. doi: 10.1073/pnas.80.21.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakradeo PP, Nair J, Bhide SV. Metabolism of N′-nitrosonornicotine by adult and fetal human oesophagal cultures. Cell Biol Int. 1995;19:53–58. doi: 10.1006/cbir.1995.1007. [DOI] [PubMed] [Google Scholar]
- 17.Castonguay A, Lin D, Stoner GD, Radok P, Furuya K, Hecht SS, Schut HA, Klaunig JE. Comparative carcinogenicity in A/J mice and metabolism by cultured mouse peripheral lung of N′-nitrosonornicotine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, and their analogues. Cancer Res. 1983;43:1223–1229. [PubMed] [Google Scholar]
- 18.Hecht SS, Reiss B, Lin D, Williams GM. Metabolism of N′-nitrosonornicotine by cultured rat esophagus. Carcinogenesis. 1982;3:453–456. doi: 10.1093/carcin/3.4.453. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann D, Castonguay A, Rivenson A, Hecht SS. Comparative carcinogenicity and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and N′-nitrosonornicotine in Syrian golden hamsters. Cancer Res. 1981;41:2386–2393. [PubMed] [Google Scholar]
- 20.Wiencke JK. DNA adduct burden and tobacco carcinogenesis. Oncogene. 2002;21:7376–7391. doi: 10.1038/sj.onc.1205799. [DOI] [PubMed] [Google Scholar]
- 21.Dipple A. DNA adducts of chemical carcinogens. Carcinogenesis. 1995;16:437–441. doi: 10.1093/carcin/16.3.437. [DOI] [PubMed] [Google Scholar]
- 22.Upadhyaya P, McIntee EJ, Villalta PW, Hecht SS. Identification of adducts formed in the reaction of 5′-acetoxy-N′-nitrosonornicotine with deoxyguanosine and DNA. Chem Res Toxicol. 2006;19:426–435. doi: 10.1021/tx050323e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Upadhyaya P, Hecht SS. Identification of adducts formed in the reactions of 5′-acetoxy-N′-nitrosonornicotine with deoxyadenosine, thymidine, and DNA. Chem Res Toxicol. 2008;21:2164–2171. doi: 10.1021/tx8002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu MWBWE, Hoffmann D. Chemical studies on tobacco smoke XXIII: synthesis of carbon-14 labeled myosmine, nornicotine, and N′-nitrosonornicotine. J Labelled Comp. 1974;10:79–88. [Google Scholar]
- 25.Seeman JI, Chavdarian CG, Secor HV. Synthesis of the enantiomers of nornicotine. J Org Chem. 1985;50:5419–5421. [Google Scholar]
- 26.Spratt TE, Peterson LA, Confer WL, Hecht SS. Solvolysis of model compounds for alpha-hydroxylation of N′-nitrosonornicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone: evidence for a cyclic oxonium ion intermediate in the alkylation of nucleophiles. Chem Res Toxicol. 1990;3:350–356. doi: 10.1021/tx00016a013. [DOI] [PubMed] [Google Scholar]
- 27.Wang M, Cheng G, Sturla SJ, Shi Y, McIntee EJ, Villalta PW, Upadhyaya P, Hecht SS. Identification of adducts formed by pyridyloxobutylation of deoxyguanosine and DNA by 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone, a chemically activated form of tobacco specific carcinogens. Chem Res Toxicol. 2003;16:616–626. doi: 10.1021/tx034003b. [DOI] [PubMed] [Google Scholar]
- 28.Marriner GA, Kerwin SM. An improved synthesis of (+/−)-N′-nitrosonornicotine 5′-acetate. J Org Chem. 2009;74:2891–2892. doi: 10.1021/jo9000417. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Villalta PW, Upadhyaya P, Hecht SS. Analysis of O6-[4-(3-Pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine and other DNA adducts in rats treated with enantiomeric or racemic N′-nitrosonornicotine. Chem Res Toxicol. 2016;29:87–95. doi: 10.1021/acs.chemrestox.5b00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zarth AT, Cheng G, Zhang Z, Wang M, Villalta PW, Balbo S, Hecht SS. Analysis of the benzene oxide-DNA adduct 7-phenylguanine by liquid chromatography-nanoelectrospray ionization-high resolution tandem mass spectrometry-parallel reaction monitoring: application to DNA from exposed mice and humans. Chem Biol Interact. 2014;215:40–45. doi: 10.1016/j.cbi.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carmella SG, McIntee EJ, Chen M, Hecht SS. Enantiomeric composition of N′-nitrosonornicotine and N′-nitrosoanatabine in tobacco. Carcinogenesis. 2000;21:839–843. doi: 10.1093/carcin/21.4.839. [DOI] [PubMed] [Google Scholar]
- 32.Gopalakrishnan R, Morse MA, Lu J, Weghorst CM, Sabourin CL, Stoner GD, Murphy SE. Expression of cytochrome P450 2A3 in rat esophagus: relevance to N-nitrosobenzylmethylamine. Carcinogenesis. 1999;20:885–891. doi: 10.1093/carcin/20.5.885. [DOI] [PubMed] [Google Scholar]
- 33.Brosch D, Kirmse W. Stereochemistry of nucleophilic displacement on 1-alkanediazonium ions. J Org Chem. 1991;56:907–908. [Google Scholar]
- 34.Smith RH, Koepke SR, Tondeur Y, Denlinger CL, Michejda CJ. The methyldiazonium ion in water: competition between hydrolysis and proton exchange. J Chem Soc, Chem Commun. 1985:936–937. [Google Scholar]
- 35.Meehan T, Straub K. Double-stranded DNA steroselectively binds benzo(a)pyrene diol epoxides. Nature. 1979;277:410–412. doi: 10.1038/277410a0. [DOI] [PubMed] [Google Scholar]
- 36.Day BW, Skipper PL, Zaia J, Singh K, Tannenbaum SR. Enantiospecificity of covalent adduct formation by benzo[a]pyrene anti-diol epoxide with human serum albumin. Chem Res Toxicol. 1994;7:829–835. doi: 10.1021/tx00042a017. [DOI] [PubMed] [Google Scholar]
- 37.Raney VM, Harris TM, Stone MP. DNA conformation mediates aflatoxin B1-DNA binding and the formation of guanine N7 adducts by aflatoxin B1 8,9-exo-epoxide. Chem Res Toxicol. 1993;6:64–68. doi: 10.1021/tx00031a010. [DOI] [PubMed] [Google Scholar]
- 38.Sodum RS, Chung FL. Stereoselective formation of in vitro nucleic acid adducts by 2,3-epoxy-4-hydroxynonanal. Cancer Res. 1991;51:137–143. [PubMed] [Google Scholar]
- 39.Fu I, Cai Y, Zhang Y, Geacintov NE, Broyde S. Entrapment of a histone tail by a DNA lesion in a nucleosome suggests the lesion impacts epigenetic marking: a molecular dynamics study. Biochemistry. 2016 doi: 10.1021/acs.biochem.5b01166. Articles ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balbo S, James-Yi S, Johnson CS, O’Sullivan MG, Stepanov I, Wang M, Bandyopadhyay D, Kassie F, Carmella S, Upadhyaya P, Hecht SS. (S)-N′-Nitrosonornicotine, a constituent of smokeless tobacco, is a powerful oral cavity carcinogen in rats. Carcinogenesis. 2013;34:2178–2183. doi: 10.1093/carcin/bgt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stepanov I, Yershova K, Carmella S, Upadhyaya P, Hecht SS. Levels of (S)-N′-nitrosonornicotine in U.S. tobacco products. Nicotine Tob Res. 2013;15:1305–1310. doi: 10.1093/ntr/nts249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.