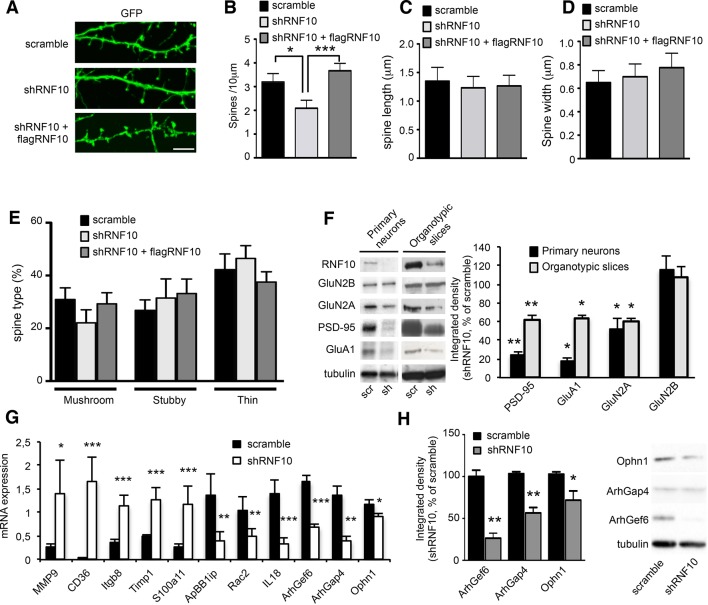
Figure 4. RNF10 silencing induces molecular and morphological modifications of the glutamatergic synapse.
(A) Confocal images of primary hippocampal neurons (DIV14) transfected at DIV7 with pGIPZ-scramble, shRNF10 and shRNF10 plus flagRNF10 and immunolabeled for GFP (green); scale bar: 5 μm. (B-E) Histograms showing the quantification of dendritic spine density (B) (n=6–10; *p<0.05, scramble vs shRNF10; ***p<0.001, shRNF10 vs shRNF10 + flagRNF10; one-way ANOVA, followed by Tukey post-hoc test), dendritic spine length (C), dendritic spine head width (D) and dendritic spine type (E). (F) WB analysis from homogenates of primary hippocampal neurons (DIV14) and organotypic hippocampal slices (DIV14) lentivirally infected with pGIPZ-scramble sequence (scramble) as control or with pLKO-shRNF10 (shRNF10). The histogram shows the quantification of the expression levels of GluN2A, GluA1, PSD-95 and GluN2B in shRNF10-infected neurons and slices, normalized on tubulin and expressed as % of scramble (n=6; *p<0.05; **p<0.01; unpaired Student’s t-test). (G) mRNA expression levels of genes associated with synaptic transmission or dendritic spine morphology by real-time PCR from DIV14 organotypic hippocampal slices lentivirally infected (DIV4) with pGIPZ-scramble sequence (scramble) as control or with pLKO-shRNF10 (shRNF10) (n=4, ***p<0.001; **p<0.01; *p<0.05; unpaired Student’s t-test). (H) WB for ArhGef6, ArhGap4, Ophn1 and tubulin from cell lysates of organotypic hippocampal slices infected with pGIPZ-scramble or with pLKO-shRNF10. The histogram shows the quantification of protein levels from shRNF10 samples with respect to pGIPZ-scramble, following normalization on tubulin (n=3, *p<0.05; **p<0.01; unpaired Student’s t-test).