Abstract
Objective
With emergence and geographically expanding of antimalarial resistance worldwide, molecular markers are essential tool for surveillance of resistant Plasmodium parasites. Recently, single-nucleotide polymorphisms (SNPs) in the PF3D7_1343700 kelch propeller (K13-propeller) domain are shown to be associated with artemisinin (ART) resistance in vivo and in vitro. This study aims to investigate the ART resistance-associated polymorphisms of K13-propeller and PfATPase6 genes in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea (EG).
Methods
A total of 172 samples were collected from falciparum malaria patients on Bioko Island between 2013 and 2014. The polymorphisms of K13-propeller and PfATPase6 genes were analyzed by Nest-PCR and sequencing.
Results
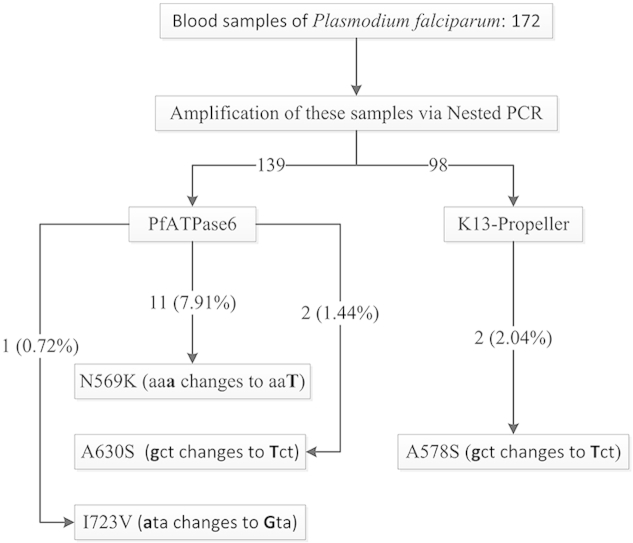
Sequences of K13-propeller and PfATPase6 were obtained from 90.74% (98/108) and 91.45% (139/152) samples, respectively. The 2.04% (2/98) cases had non-synonymous K13-propeller A578S mutation but no found the mutations associated with ART resistance in Southeast Asia. For PfATPase6, the mutations were found at positions N569K and A630S with the mutation prevalence of 7.91% (11/139) and 1.44% (2/139), respectively. In addition, a sample with the mixed type at position I723V was discovered (0.72%, 1/139).
Conclusions
This study initially offers an insight of K13-propeller and PfATPase6 polymorphisms on Bioko Island, EG. It suggests no widespread ART resistance or tolerance in the region, and might be helpful for developing and updating guidance for the use of ART-based combination therapies (ACTs).
Keywords: Plasmodium falciparum, Polymorphism, Artemisinin resistance, K13-propeller
Graphical abstract
Highlights
-
•
The PfATPase6 mutations were found at positions N569K (7.91%), A630S (1.44%) and I723V (0.72%).
-
•
Only one non-synonymous K13-propeller mutation A578S (2.04%) was observed.
-
•
Limited SNPs of PfATPase6 and K13-propeller indicated no widespread ART on Bioko Island, EG.
1. Introduction
The emergence of Plasmodium falciparum resistance to antimalarial drugs has been threatening the world's malaria control and elimination efforts (Young et al., 1963). Currently, P. falciparum have developed resistance/tolerance to antimalarial drugs chloroquine (CQ) and sulfadoxine-pyrimethamine (SP). Although World Health Organization (WHO) has recommended artemisinin (ART)-based combination therapies (ACTs) as the first-line treatment for uncomplicated P. falciparum malaria, P. falciparum are becoming insensitive to ART and its derivatives (Harinasuta et al., 1965, Wongsrichanalai et al., 2002, Amaratunga et al., 2014). At present, virtually all malaria endemic countries in sub-Saharan Africa are adopting either Artemether-Lumefantrine (AL) or Artesunate-Amodiaquine (AS-AQ) as the front-line ACTs. AS-AQ and the ART derivative dihydroartemisinin-piperaquine (DP) are used in Equatorial Guinea (EG) (Barrette and Ringwald, 2010).
Emerging evidence indicate that P. falciparum has been developing the resistance to ART and its derivatives. In Pailin of western Cambodia and other Southeast (SE) Asia area, parasite clearance was delayed following the treatment with ART monotherapy or ACTs (Noedl et al., 2008, Dondorp et al., 2009, Amaratunga et al., 2012, Miotto et al., 2013, Ashley et al., 2014). Our recent study showed the presence of high prevalent mutations in Pfmdr1 (91.39%) and Pfcrt (98.67%) which markers for antimalarial drug resistence in P. falciparum clinical isolates on Bioko Island, EG (Li et al., 2015). It is globally threatening for malaria prevention and treatment (Wootton et al., 2002, Roper et al., 2004). It is imperative to conduct surveillances to identify areas that are potentially developing drug resistance.
Several molecular markers for antimalarial drug resistance has been identified (Wongsrichanalai et al., 2002, Barrette and Ringwald, 2010). Polymorphisms of the sarcoplasmic/endoplasmic reticulum Ca2+-ATPase ortholog in P. falciparum (PfSERCA or PfATPase6) has been associated with ART resistance, although the association of SNPs in PfATPase6 with resistance to ART and the underlying mechanism remains to be confirmed (Jambou et al., 2005, Afonso et al., 2006, Mugittu et al., 2006, Cui et al., 2012).
Single-nucleotide polymorphisms (SNPs) in the PF3D7_1343700 kelch propeller (K13-propeller) have been identified to be a key causal determinant of ART resistance in SE Asia (Mok et al., 2015). From 2009 to 2015, a series of studies associated with K13-propeller has been published particular in Asia (Talundzic et al., 2015, Tun et al., 2015, Wang et al., 2015) and WHO Africa region (Cooper et al., 2015, Ouattara et al., 2015). In Africa, limited mutations of K13-propeller were found in Dakar (Torrentino-Madamet et al., 2014), Uganda (Cooper et al., 2015), Mali (Ouattara et al., 2015), and even 12 countries from sub-Saharan Africa (Kamau et al., 2015). These studies showed the mutational loci in African countries were different from those of SE Asia (Kamau et al., 2015).
Malaria is a serious health problem in EG, especially on Bioko Island. However, whether the ART resistance has been developed on the Bioko Island remains unclear. In this study, we surveyed polymorphisms in K13-propeller and PfATPase6 genes in clinical isolates collected from Bioko Island, EG. Our findings may provide a clue to prevent and treat malaria using ART on Bioko Island, EG.
2. Materials and methods
2.1. Study area
Bioko Island belongs to EG and is located in the Gulf of Guinea, about 100 km off the coast of southern Nigeria and 160 km northwest of continental EG (Fig. 1). The island has a population of 266 000 inhabitants (2001 census) and a humid tropical environment. The launch of the Bioko Island Malaria Control Project (BIMCP) have had a marked impact on malaria transmission, malaria due to P. falciparum is still the major public health problem on the island. The entomological inoculation rates (EIRs) in Bioko Island ranged from 163 to 840, with the outdoor EIRs reaching more than 900 infective mosquito bites yearly and a malaria prevalence of 52% under the age of five years (Overgaard et al., 2012, Rehman et al., 2013).
Fig. 1.
Geographical map of Bioko Island, Equatorial Guinea.
2.2. Samples collection
Blood samples (3 ml) were collected from the confirmed malaria cases between September 2013 and March 2014. Approximately 300 μl of blood was aliquoted on 3 MM Whatman® filter paper (Whatman International Ltd., Maidstone, England), and air dried. These filters were then stored individually in Ziplock bags containing silica desiccant beads and kept at −20 °C. These samples were examined using the ICT malaria P.f. Cassette Test (ICT Diagnostics, South Africa) and Giemsa-stained thick and thin peripheral blood smear examination with microscope. For quality control, archived malaria positive slides were re-examined and parasitaemia was recorded. The Plasmodium spp. was confirmed by Plasmodium malaria real time PCR diagnostic kit (Shanghai Liferiver Bio-Tech Corp, China). This study was approved by the ethics committees of Malabo Regional Hospital. The informed consent was obtained from all participated subjects.
2.3. DNA extraction from blood samples
Genomic DNA (gDNA) was extracted from dried filter blood spots (DBS) by following Chelex-100 extraction procedure described in our previous report (Li et al., 2014). An 18S-rRNA-based RT-PCR was used to evaluate the quality of P. falciparum gDNA.
2.4. Genotyping
Nucleotide and amino-acid sequence of K13-propeller and PfATPase6 used in current study has been reported in PlasmoDB (http://plasmodb.org) under Gene ID: PF3D7_1343700 and PF3D7_0106300. In order to illustrate the mutations of K13-propeller and PfATPase6, one segment of K13-propeller gene and one fragment from PfATPase6 gene were amplified by a nested PCR (Zhang et al., 2008, Li et al., 2014), respectively.
The K13-propeller and PfATPase6 genes were amplified by nested PCR using the primers in Table 1. For first round PCR, 0.5 μl of DNA was amplified with 10 μl 2 × NovoStar Green PCR Mix (1.25 U/μl NovoStar Taq DNA Polymerase, 0.4 mM dNTP Mixture, 2 × PCR Buffer, and 4 mM Mg2+), 0.5 μl forward primer (10 μM), 0.5 μl reverse primer (10 μM), and sterile ultrapure water to a final volume of 20 μl. For the second round PCR, 0.5 μl primary PCR products were amplified with 40 μl reaction system, including 20 μl 2 × NovoStar Green PCR Mix, 1.0 μl forward primer (10 μM), 1.0 μl reverse primer (10 μM), and H2O (up to 40 μl).
Table 1.
Primers and PCR conditions for genotyping.
| Gene | Primer | Sequence (5′–3′) | Size (bp) | Mutation | PCR condition |
|---|---|---|---|---|---|
| K13-propeller | 1st round PCR | ||||
| K13-1 | CGGAGTGACCAAATCTGGGA | 2097 | 95 °C 3 min; followed by 30 cycles (95 °C 30 s, 55 °C 30 s, 72 °C 30 s); 72 °C 5 min; then store 12 °C. | ||
| K13-4 | GGGAATCTGGTGGTAACAGC | ||||
| 2nd round PCR | |||||
| PfK13_inF2 | TCAACAATGCTGGCGTATGTG | 501 | T474I, M476I, A481V, Y493H, T508N, P527T, G533S, N537I, R539T, I543T, P553L, R561H, V568G, P574L, A578S, and C580Y | 95 °C 3 min; followed by 30 cycles (95 °C 30 s, 55 °C 30 s, 72 °C 30 s); 72 °C 5 min; then store 12 °C. | |
| PfK13_inR2 | TGATTAAG GTAATTAAAAGCTGCTCC | ||||
| PfATPase6 | 1st round PCR | ||||
| PfATPase6-N1F | AATATTGTTATTCAGAATATGATTATAA | 896 | 95 °C 3 min; followed by 30 cycles (95 °C 30 s, 55 °C 30 s, 72 °C 50 s); 72 °C 5 min; then store 12 °C. | ||
| PfATPase6-N1R | TGGATCAATAATACCTAATCCACCTA | ||||
| 2nd round PCR | |||||
| PfATPase6-N2F | AGCAAATATTTTCTGTAACGATAATA | 798 | K561N, N569K, A623E, A630S, G639D, N683K, I723V, and S769N | 95 °C 3 min; followed by 30 cycles (95 °C 30 s, 58 °C 30 s, 72 °C 45 s); 72 °C 5 min; then store 12 °C. | |
| PfATPase6-N2R | TGTTCTAATTTATAATAATCATCTGT | ||||
PCR reaction conditions were listed in Table 1. All PCR products were analyzed using 1.0% agar gel electrophoresis and DNA sequencing using a ABI 3730×L automated sequencer (PE Biosystems, CT, USA). The data was analyzed using the DNAstar (DNASTAR Inc., Madison, WI, USA). The 3D7 K13-propeller and PfATPase6 sequences were used as the references.
2.5. Data analysis
The data was analyzed using SPSS 17.0 (SPSS Inc., Chicago, IL). The mutant and wild-type alleles of the collected clinical samples were used to generate the prevalence of the alleles. A two-tailed P-value is less than 0.05 was considered statistically significant. The 95% confidence intervals (95% CI) was calculated as described previously (Li et al., 2014).
3. Results
3.1. K13-propeller polymorphisms
Sequence of a total of 98 (90.74%, 98/108) K13-propeller nested PCR products was obtained from 108 (62.79%, 108/172) PCR-positive samples out of the 172 isolates. The K13-propeller SNPs were analyzed by comparing with the reference 3D7 strain (PF3D7_1343700) and listed in Table 2. There was a mutation at position 578. The frequency of A578S was 2.04% (2/98) (Table 2). No K13-propeller mutation was detected at positions 474, 476, 493, 508, 527, 537, 539, 543, 553, 561, 568, 574, and 580. Notably, the C580Y, R539T, and Y493H substitutions that were associated with ART resistance in vitro or delayed P. falciparum parasite clearance in vivo in SE Asia were not detected on the samples from Bioko Island, EG.
Table 2.
K13-propeller and PfATPase6 polymorphisms in Plasmodium falciparum isolates on Bioko Island, Equatorial Guinea.
| Gene | Reference |
No. of isolates |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Codon position | AA | Codon | AA | Codon | Base position | PCR positive | Sequencing | Mutation | Prevalence (%, 95% CI) | |
| K13-propeller | 578 | A | gct | S | Tct | 1732 | 108 | 98 | 2 | 2.04, −0.76 to 4.84 |
| PfATPase6 | 569 | N | aaa | K | aaT | 1707 | 152 | 139 | 11 | 7.91, 3.42 to 12.4 |
| 630 | A | gct | S | Tct | 1888 | 152 | 139 | 2 | 1.44, −0.54 to 3.42 | |
| 723 | I | ata | V | Gta | 2167 | 152 | 139 | 1 | 0.72, −0.69 to 2.13 | |
Bold, AA and No. represent single nucleotide polymorphism mutation, amino acid residue and number.
3.2. PfATPase6 polymorphisms
Sequence of a total of 139 (91.45%, 139/152) PfATPase6 nested PCR products was obtained from 152 (88.37%, 152/172) PCR-positive samples out of the 172 isolates. The PfATPase6 SNPs were analyzed by comparing with the reference 3D7 strain (PF3D7_0106300) and listed in Table 2. The PfATPase6 SNPs were found at positions N569K and A630S with the mutation prevalence of 11 (7.91%, 11/139), and 2 (1.44%, 2/139), respectively (Table 2). In addition, a sample with the mixed type at position I723V was also discovered with the mutation prevalence of 1 (0.72%, 1/139) (Table 2). The remaining mutations of K561N, A623E, G639D, N683K, and S769N, all the investigated samples verified the wild-type genotype. Notably, the S769N substitution that was connected with ART resistance was not found from these samples.
4. Discussion
For EG, both the AS-AQ and DP are considered as the front-line treatment for uncomplicated falciparum malaria. It is essential to understand molecular mutation profiles of P. falciparum parasite for ART resistance and use this initial information for molecular assessment under antimalarial drug pressure. In current study, we only find limited mutation of K13-propeller and low frequency of PfATPase6 mutation of the clinical isolates from Bioko Island, EG. It is the initial report focusing on the molecular markers of K13-propeller and PfATPase6 for ART resistance on this Island.
The ART resistance considers as a major risk to public health, with the most rigorous potential effect in Sub-Saharan Africa, where the burden is seriously. Furthermore, the international investments and domestic investments, the prevention and elimination system for malaria are also insufficient in the region. The hazard of ART-resistant parasites scattering from western Cambodia to the Greater Mekong Subregion and then to Africa particularly Sub-Saharan Africa, as happened previously with CQ and SP-resistant parasites (Wootton et al., 2002, Roper et al., 2004), is worrying (Ariey et al., 2014). A significant action of the WHO Global Plan for Artemisinin Resistance Containment is to increase monitoring and molecular surveillance (Talisuna et al., 2012). However, it is difficult to evaluate how long or when ART resistance mutations will appear in Africa including EG, and molecular detection can offer a profile to speedily discover for the appearance or importation of resistance alleles (Taylor et al., 2014). In SE Asia, the mutations of K13-propeller are found in both western Cambodia (Ariey et al., 2014, Straimer et al., 2015) and Bangladesh (Mohon et al., 2014). In Africa, only limited polymorphisms of K13-propeller are detected (Conrad et al., 2014, Taylor et al., 2014, Torrentino-Madamet et al., 2014). However, the K13-propeller polymorphism in returned migrant workers from Ghana is found in Shanglin of China (Feng et al., 2015). Thus, it is very necessary to strengthen Plasmodium parasites genotypic resistance surveillance with K13-propeller polymorphism. If there is no sufficient attention to long-term survey, it will be a disaster for human health.
The survey of P. falciparum K13-propeller polymorphisms primarily explore a diversity of mutations across on Bioko Island, EG. Limited polymorphisms associated with ART resistance from SE Asia are observed in the clinical isolates. Recent study reports that the SNP mutations at Y493H, I543T, R539T, and C580Y are powerfully connected with prolonging P. falciparum parasite survival time ex vivo; ART resistance in vitro has to be M476I mutation-related, which indicates that mutations of K13-propeller can generate an ART-resistance phenotype in vitro with genetic background from African parasite (Ariey et al., 2014). Although the five mutations play crucial role during ART-resistance to P. falciparum parasite in vitro and in vivo, we observe none of these mutations in the survey parasite samples. Only one K13-propeller A578S mutation (2.04%, 2/98) that previously reported from Cambodia is discovered in the six K13-propeller blades. This mutation is previously found in 0.75% (1/133) of the isolates tested in Uganda and also presents in parasites from DRC, Gabon, Ghana, Kenya, and Mali (Ariey et al., 2014, Conrad et al., 2014, Kamau et al., 2015). Although the prevalence of A578S mutant allele from Bioko Island is lower than Kenya at 2.7%, it is still higher in parasites compared to 1% in the other four countries of Sub-Saharan Africa. This unusual polymorphism also merits further characterization (Taylor et al., 2014). Mutations of A481V, G533C and A578S are confirmed and adjacent to the Y493H, R539T, C580Y mutation, and propose the mutations may have a significant effect on three-dimensional structure of the K13-propeller. Furthermore, the mutations of S522S, Y558H report in Ugandan children and A557S in Congo have not detected on Bioko Island (Conrad et al., 2014, Taylor et al., 2014). The V520A mutation identified from West, Central and East Africa is also not found on the Island, Dakar and Uganda (Sylla et al., 2013, Taylor et al., 2014). These results encourage and suggest ART resistance is not yet established in Africa particular on Bioko Island, EG. Although none of the mutations associated with ART resistance in SE Asia are detected in Africa (Conrad et al., 2014, Taylor et al., 2014), numerous novel K13-propeller coding substitutions bothers in the whole continent of Africa. The phenotypes of these coding polymorphisms remain unclear and will require further characterization to better characterize the clinical impact on ART resistance in Africa. Further analysis of phylogenetic tree or haplotype network is needed to trace the origin of the K13-propeller mutations and to determine whether the ART resistance is widely spreaded from SE Asia or emerged independently in Bioko Island, EG (Nyunt et al., 2014).
Polymorphisms evaluation of PfATPase6 in Africa have occurred rarely (Mugittu et al., 2006, Legrand et al., 2008, Happi et al., 2009, Menegon et al., 2010, Kamugisha et al., 2011, Zatra et al., 2012). The current study initial describes for the PfATPase6 polymorphism on Bioko Island, EG. Among the three different observed mutations, only one, the N569K mutation, is relatively frequent 7.91% of isolates. The previous studies have also found a high prevalence of this mutation in Zanzibar (36%) and Tanzania (29%) (Dahlstrom et al., 2008), Niger (17.2%) (Ibrahim et al., 2009). The PfATPase6 S769N mutation is absent in all Bioko samples and consistent with previous results in Africa countries (Mugittu et al., 2006, Legrand et al., 2008, Happi et al., 2009, Menegon et al., 2010, Kamugisha et al., 2011, Zatra et al., 2012) and South America (Adhin et al., 2012). It demonstrates that the Africa and South America share the similar molecular pattern of PfATPase6 for ART resistance.
5. Conclusions
The present study shows that the low prevalence polymorphism mutations of PfATPase6 and limited mutations of K13-propeller, potentially associated with ART resistance, are obviously observed on Bioko Island, EG. Continuous molecular surveillance with K13-propeller gene as ART resistance marker is exceedingly recommended on Bioko Island, EG. Furthermore, it might be helpful for developing and updating guidance for the use of ACTs.
Conflicts of interest
We declare that we have no conflict of interest.
Financial support
This study was supported by the China Postdoctoral Science Foundation (M.L. Grant Number 2013M542195); Medical Science Fund of Guangdong Province (M.L. Grant Number A2013780); Scientific Research Foundation for the Returned Overseas Chinese Scholars (J.L. Grant Number JYB201448HBMU01), State Education Ministry; the Natural Science Foundation of Hubei Province of China (J.L. Grant Number 2014CFB648), and the Foundation for Innovative Research Team of Hubei University of Medicine (J.L. Grant Number 2014 CXZ02).
Acknowledgments
The authors thank the Department of Health of Guangdong Province and Department of Aid to Foreign Countries of Ministry of Commerce of People's Republic of China for their help. The authors also thank Dr. Xinsheng Gu for revising the manuscript.
References
- Adhin M.R., Labadie-Bracho M., Vreden S.G. Status of potential PfATP6 molecular markers for artemisinin resistance in Suriname. Malar. J. 2012;11:322. doi: 10.1186/1475-2875-11-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso A., Hunt P., Cheesman S., Alves A.C., Cunha C.V., do Rosario V., Cravo P. Malaria parasites can develop stable resistance to artemisinin but lack mutations in candidate genes atp6 (encoding the sarcoplasmic and endoplasmic reticulum Ca2+ ATPase), tctp, mdr1, and cg10. Antimicrob. Agents Chemother. 2006;50:480–489. doi: 10.1128/AAC.50.2.480-489.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaratunga C., Sreng S., Suon S., Phelps E.S., Stepniewska K., Lim P., Zhou C., Mao S., Anderson J.M., Lindegardh N., Jiang H., Song J., Su X.Z., White N.J., Dondorp A.M., Anderson T.J., Fay M.P., Mu J., Duong S., Fairhurst R.M. Artemisinin-resistant Plasmodium falciparum in Pursat province, western Cambodia: a parasite clearance rate study. Lancet. Infect. Dis. 2012;12:851–858. doi: 10.1016/S1473-3099(12)70181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaratunga C., Witkowski B., Khim N., Menard D., Fairhurst R.M. Artemisinin resistance in Plasmodium falciparum. Lancet. Infect. Dis. 2014;14:449–450. doi: 10.1016/S1473-3099(14)70777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.C., Khim N., Kim S., Duru V., Bouchier C., Ma L., Lim P., Leang R., Duong S., Sreng S., Suon S., Chuor C.M., Bout D.M., Menard S., Rogers W.O., Genton B., Fandeur T., Miotto O., Ringwald P., Le Bras J., Berry A., Barale J.C., Fairhurst R.M., Benoit-Vical F., Mercereau-Puijalon O., Menard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley E.A., Dhorda M., Fairhurst R.M., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J.M., Mao S., Sam B., Sopha C., Chuor C.M., Nguon C., Sovannaroth S., Pukrittayakamee S., Jittamala P., Chotivanich K., Chutasmit K., Suchatsoonthorn C., Runcharoen R., Hien T.T., Thuy-Nhien N.T., Thanh N.V., Phu N.H., Htut Y., Han K.T., Aye K.H., Mokuolu O.A., Olaosebikan R.R., Folaranmi O.O., Mayxay M., Khanthavong M., Hongvanthong B., Newton P.N., Onyamboko M.A., Fanello C.I., Tshefu A.K., Mishra N., Valecha N., Phyo A.P., Nosten F., Yi P., Tripura R., Borrmann S., Bashraheil M., Peshu J., Faiz M.A., Ghose A., Hossain M.A., Samad R., Rahman M.R., Hasan M.M., Islam A., Miotto O., Amato R., MacInnis B., Stalker J., Kwiatkowski D.P., Bozdech Z., Jeeyapant A., Cheah P.Y., Sakulthaew T., Chalk J., Intharabut B., Silamut K., Lee S.J., Vihokhern B., Kunasol C., Imwong M., Tarning J., Taylor W.J., Yeung S., Woodrow C.J., Flegg J.A., Das D., Smith J., Venkatesan M., Plowe C.V., Stepniewska K., Guerin P.J., Dondorp A.M., Day N.P., White N.J., Tracking Resistance to Artemisinin, C Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrette A., Ringwald P. WHO Press; Switzerland: 2010. Global Report on Antimalarial Drug Efficacy and Drug Resistance 2000-2010. [Google Scholar]
- Conrad M.D., Bigira V., Kapisi J., Muhindo M., Kamya M.R., Havlir D.V., Dorsey G., Rosenthal P.J. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS One. 2014;9:e105690. doi: 10.1371/journal.pone.0105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R.A., Conrad M.D., Watson Q.D., Huezo S.J., Ninsiima H., Tumwebaze P., Nsobya S.L., Rosenthal P.J. Lack of artemisinin resistance in plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob. Agents Chemother. 2015;59:5061–5064. doi: 10.1128/AAC.00921-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Wang Z., Jiang H., Parker D., Wang H., Su X.Z., Cui L. Lack of association of the S769N mutation in Plasmodium falciparum SERCA (PfATP6) with resistance to artemisinins. Antimicrob. agents Chemother. 2012;56:2546–2552. doi: 10.1128/AAC.05943-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrom S., Veiga M.I., Ferreira P., Martensson A., Kaneko A., Andersson B., Bjorkman A., Gil J.P. Diversity of the sarco/endoplasmic reticulum Ca(2+)-ATPase orthologue of Plasmodium falciparum (PfATP6) Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2008;8:340–345. doi: 10.1016/j.meegid.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J., Lwin K.M., Ariey F., Hanpithakpong W., Lee S.J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S.S., Yeung S., Singhasivanon P., Day N.P., Lindegardh N., Socheat D., White N.J. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Li J., Yan H., Feng X., Xia Z. Evaluation of antimalarial resistance marker polymorphism in returned migrant workers in china. Antimicrob. Agents Chemother. 2015;59:326–330. doi: 10.1128/AAC.04144-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happi C.T., Gbotosho G.O., Folarin O.A., Sowunmi A., Hudson T., O'Neil M., Milhous W., Wirth D.F., Oduola A.M. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob. agents Chemother. 2009;53:888–895. doi: 10.1128/AAC.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harinasuta T., Suntharasamai P., Viravan C. Chloroquine-resistant falciparum malaria in Thailand. Lancet. 1965;2:657–660. doi: 10.1016/s0140-6736(65)90395-8. [DOI] [PubMed] [Google Scholar]
- Ibrahim M.L., Khim N., Adam H.H., Ariey F., Duchemin J.B. Polymorphism of PfATPase in Niger: detection of three new point mutations. Malar. J. 2009;8:28. doi: 10.1186/1475-2875-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambou R., Legrand E., Niang M., Khim N., Lim P., Volney B., Ekala M.T., Bouchier C., Esterre P., Fandeur T., Mercereau-Puijalon O. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005;366:1960–1963. doi: 10.1016/S0140-6736(05)67787-2. [DOI] [PubMed] [Google Scholar]
- Kamau E., Campino S., Amenga-Etego L., Drury E., Ishengoma D., Johnson K., Mumba D., Kekre M., Yavo W., Mead D., Bouyou-Akotet M., Apinjoh T., Golassa L., Randrianarivelojosia M., Andagalu B., Maiga-Ascofare O., Amambua-Ngwa A., Tindana P., Ghansah A., MacInnis B., Kwiatkowski D., Djimde A.A. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J. Infect. Dis. 2015;211:1352–1355. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamugisha E., Sendagire H., Kaddumukasa M., Enweji N., Gheysari F., Swedberg G., Kironde F. Detecting adenosine triphosphatase 6 (pfATP6) point mutations that may be associated with Plasmodium falciparum resistance to artemisinin: prevalence at baseline, before policy change in Uganda. Tanzan. J. Health Res. 2011;13:40–47. doi: 10.4314/thrb.v13i1.58580. [DOI] [PubMed] [Google Scholar]
- Legrand E., Volney B., Meynard J.B., Mercereau-Puijalon O., Esterre P. In vitro monitoring of Plasmodium falciparum drug resistance in French Guiana: a synopsis of continuous assessment from 1994 to 2005. Antimicrob. Agents Chemother. 2008;52:288–298. doi: 10.1128/AAC.00263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen J., Xie D., Eyi U.M., Matesa R.A., Obono M.M., Ehapo C.S., Yang L., Yang H., Lin M., Wu W., Wu K., Li S., Chen Z. Molecular mutation profile of Pfcrt and Pfmdr1 in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015 doi: 10.1016/j.meegid.2015.08.039. [DOI] [PubMed] [Google Scholar]
- Li J., Chen J., Xie D., Monte-Nguba S.M., Eyi J.U., Matesa R.A., Obono M.M., Ehapo C.S., Yang L., Lu D., Yang H., Yang H.T., Lin M. High prevalence of pfmdr1 N86Y and Y184F mutations in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Pathogens Glob. Health. 2014;108:339–343. doi: 10.1179/2047773214Y.0000000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegon M., Talha A.A., Severini C., Elbushra S.M., Mohamedani A.A., Malik E.M., Mohamed T.A., Wernsdorfer W.H., Majori G., Nour B.Y. Frequency distribution of antimalarial drug resistance alleles among Plasmodium falciparum isolates from Gezira State, central Sudan, and Gedarif State, eastern Sudan. Am. J. Trop. Med. Hyg. 2010;83:250–257. doi: 10.4269/ajtmh.2010.09-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Almagro-Garcia J., Manske M., Macinnis B., Campino S., Rockett K.A., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J.M., Duong S., Nguon C., Chuor C.M., Saunders D., Se Y., Lon C., Fukuda M.M., Amenga-Etego L., Hodgson A.V., Asoala V., Imwong M., Takala-Harrison S., Nosten F., Su X.Z., Ringwald P., Ariey F., Dolecek C., Hien T.T., Boni M.F., Thai C.Q., Amambua-Ngwa A., Conway D.J., Djimde A.A., Doumbo O.K., Zongo I., Ouedraogo J.B., Alcock D., Drury E., Auburn S., Koch O., Sanders M., Hubbart C., Maslen G., Ruano-Rubio V., Jyothi D., Miles A., O'Brien J., Gamble C., Oyola S.O., Rayner J.C., Newbold C.I., Berriman M., Spencer C.C., McVean G., Day N.P., White N.J., Bethell D., Dondorp A.M., Plowe C.V., Fairhurst R.M., Kwiatkowski D.P. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat. Genet. 2013;45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohon A.N., Alam M.S., Bayih A.G., Folefoc A., Shahinas D., Haque R., Pillai D.R. Mutations in Plasmodium falciparum K13 propeller gene from Bangladesh (2009–2013) Malar. J. 2014;13:431. doi: 10.1186/1475-2875-13-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S., Ashley E.A., Ferreira P.E., Zhu L., Lin Z., Yeo T., Chotivanich K., Imwong M., Pukrittayakamee S., Dhorda M., Nguon C., Lim P., Amaratunga C., Suon S., Hien T.T., Htut Y., Faiz M.A., Onyamboko M.A., Mayxay M., Newton P.N., Tripura R., Woodrow C.J., Miotto O., Kwiatkowski D.P., Nosten F., Day N.P., Preiser P.R., White N.J., Dondorp A.M., Fairhurst R.M., Bozdech Z. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347:431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugittu K., Genton B., Mshinda H., Beck H.P. Molecular monitoring of Plasmodium falciparum resistance to artemisinin in Tanzania. Malar. J. 2006;5:126. doi: 10.1186/1475-2875-5-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noedl H., Se Y., Schaecher K., Smith B.L., Socheat D., Fukuda M.M., Artemisinin Resistance in Cambodia 1 Study, C Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- Nyunt M.H., Hlaing T., Oo H.W., Tin-Oo L.L., Phway H.P., Wang B., Zaw N.N., Han S.S., Tun T., San K.K., Kyaw M.P., Han E.T. Molecular assessment of artemisinin resistance markers, polymorphisms in the K13 propeller, and a multidrug-resistance gene in the eastern and western border areas of Myanmar. Clin. Infect. Dis. off. Publ. Infect. Dis. Soc. Am. 2014 doi: 10.1093/cid/ciu1160. [DOI] [PubMed] [Google Scholar]
- Ouattara A., Kone A., Adams M., Fofana B., Maiga A.W., Hampton S., Coulibaly D., Thera M.A., Diallo N., Dara A., Sagara I., Gil J.P., Bjorkman A., Takala-Harrison S., Doumbo O.K., Plowe C.V., Djimde A.A. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. Am. J. Trop. Med. Hyg. 2015;92:1202–1206. doi: 10.4269/ajtmh.14-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard H.J., Reddy V.P., Abaga S., Matias A., Reddy M.R., Kulkarni V., Schwabe C., Segura L., Kleinschmidt I., Slotman M.A. Malaria transmission after five years of vector control on Bioko Island, Equatorial Guinea. Parasites Vectors. 2012;5:253. doi: 10.1186/1756-3305-5-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman A.M., Mann A.G., Schwabe C., Reddy M.R., Roncon Gomes I., Slotman M.A., Yellott L., Matias A., Caccone A., Nseng Nchama G., Kleinschmidt I. Five years of malaria control in the continental region, Equatorial Guinea. Malar. J. 2013;12:154. doi: 10.1186/1475-2875-12-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper C., Pearce R., Nair S., Sharp B., Nosten F., Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- Straimer J., Gnadig N.F., Witkowski B., Amaratunga C., Duru V., Ramadani A.P., Dacheux M., Khim N., Zhang L., Lam S., Gregory P.D., Urnov F.D., Mercereau-Puijalon O., Benoit-Vical F., Fairhurst R.M., Menard D., Fidock D.A. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylla K., Abiola A., Tine R.C., Faye B., Sow D., Ndiaye J.L., Ndiaye M., Lo A.C., Folly K., Ndiaye L.A., Gaye O. Monitoring the efficacy and safety of three artemisinin based-combinations therapies in Senegal: results from two years surveillance. BMC Infect. Dis. 2013;13:598. doi: 10.1186/1471-2334-13-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talisuna A.O., Karema C., Ogutu B., Juma E., Logedi J., Nyandigisi A., Mulenga M., Mbacham W.F., Roper C., Guerin P.J., D'Alessandro U., Snow R.W. Mitigating the threat of artemisinin resistance in Africa: improvement of drug-resistance surveillance and response systems. Lancet. Infect. Dis. 2012;12:888–896. doi: 10.1016/S1473-3099(12)70241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talundzic E., Okoth S.A., Congpuong K., Plucinski M.M., Morton L., Goldman I.F., Kachur P.S., Wongsrichanalai C., Satimai W., Barnwell J.W., Udhayakumar V. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog. 2015;11:e1004789. doi: 10.1371/journal.ppat.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.M., Parobek C.M., DeConti D.K., Kayentao K., Coulibaly S.O., Greenwood B.M., Tagbor H., Williams J., Bojang K., Njie F., Desai M., Kariuki S., Gutman J., Mathanga D.P., Martensson A., Ngasala B., Conrad M.D., Rosenthal P.J., Tshefu A.K., Moormann A.M., Vulule J.M., Doumbo O.K., Ter Kuile F.O., Meshnick S.R., Bailey J.A., Juliano J.J. Absence of Putative artemisinin resistance mutations among plasmodium falciparum in sub-Saharan africa: a molecular epidemiologic study. J. Infect. Dis. 2014 doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrentino-Madamet M., Fall B., Benoit N., Camara C., Amalvict R., Fall M., Dionne P., Ba Fall K., Nakoulima A., Diatta B., Dieme Y., Menard D., Wade B., Pradines B. Limited polymorphisms in k13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012-2013. Malar. J. 2014;13:472. doi: 10.1186/1475-2875-13-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun K.M., Imwong M., Lwin K.M., Win A.A., Hlaing T.M., Hlaing T., Lin K., Kyaw M.P., Plewes K., Faiz M.A., Dhorda M., Cheah P.Y., Pukrittayakamee S., Ashley E.A., Anderson T.J., Nair S., McDew-White M., Flegg J.A., Grist E.P., Guerin P., Maude R.J., Smithuis F., Dondorp A.M., Day N.P., Nosten F., White N.J., Woodrow C.J. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet. Infect. Dis. 2015;15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang Y., Cabrera M., Zhang Y., Gupta B., Wu Y., Kemirembe K., Hu Y., Liang X., Brashear A., Shrestha S., Li X., Miao J., Sun X., Yang Z., Cui L. Artemisinin resistance at the China-Myanmar border and association with mutations in the K13 propeller gene. Antimicrob. Agents Chemother. 2015;59:6952–6959. doi: 10.1128/AAC.01255-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsrichanalai C., Pickard A.L., Wernsdorfer W.H., Meshnick S.R. Epidemiology of drug-resistant malaria. Lancet. Infect. Dis. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- Wootton J.C., Feng X., Ferdig M.T., Cooper R.A., Mu J., Baruch D.I., Magill A.J., Su X.Z. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- Young M.D., Contacos P.G., Stitcher J.E., Millar J.W. Drug resistance in plasmodium falciparum from Thailand. Am. J. Trop. Med. Hyg. 1963;12:305–314. doi: 10.4269/ajtmh.1963.12.305. [DOI] [PubMed] [Google Scholar]
- Zatra R., Lekana-douki J.B., Lekoulou F., Bisvigou U., Ngoungou E.B., Ndouo F.S. In vitro antimalarial susceptibility and molecular markers of drug resistance in Franceville, Gabon. BMC Infect. Dis. 2012;12:307. doi: 10.1186/1471-2334-12-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.Q., Guan Y.Y., Sheng H.H., Zheng B., Wu S., Xiao H.S., Tang L.H. Multiplex PCR and oligonucleotide microarray for detection of single-nucleotide polymorphisms associated with Plasmodium falciparum drug resistance. J. Clin. Microbiol. 2008;46:2167–2174. doi: 10.1128/JCM.00081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]