Abstract
Background
The key role of egfr in thymoma pathogenesis has been questioned following the failure in identifying recurrent genetic alterations of egfr coding sequences and relevant egfr amplification rate. We investigated the role of the non-coding egfr CA simple sequence repeat 1 (CA-SSR-1) in a thymoma case series.
Methods
We used sequencing and egfr-fluorescence in situ hybridization (FISH) to genotype 43 thymomas; (I) for polymorphisms and somatic loss of heterozygosity of the non-coding egfr CA-SSR-1 microsatellite and (II) for egfr gene copy number changes.
Results
We found two prevalent CA-SSR-1 genotypes: a homozygous 16 CA repeat and a heterozygous genotype, bearing alleles with 16 and 20 CA repeats. The average combined allele length was correlated with tumor subtype: shorter sequences were significantly associated with the more aggressive WHO thymoma subtype group including B2/B3, B3 and B3/C histotypes. Four out of 29 informative cases analysed for somatic CA-SSR-1 loss of heterozygosity showed allelic imbalance (AI), 3/4 with loss of the longer allele. By egfr-FISH analysis, 9 out of 33 cases were FISH positive. Moreover, the two integrated techniques demonstrated that 3 out of 4 CA-SSR-1-AI positive cases with short allele relative prevalence showed significantly low or high chromosome 7 “polysomy”/increased gene copy number by egfr-FISH.
Conclusions
Our molecular and genetic and follow up data indicated that CA-SSR-1-allelic imbalance with short allele relative prevalence significantly correlated with EGFR 3+ immunohistochemical score, increased egfr Gene Copy Number, advanced stage and with relapsing/metastatic behaviour in thymomas.
Keywords: Thymoma, thymic epithelial tumors (TET), egfr microsatellite CA-SSR-1, allelic imbalance (AI), loss of heterozygosity, egfr-fluorescent in situ hybridization (egfr-FISH)
Introduction
Among thymic epithelial tumors (TET), thymomas are heterogeneous in biology and clinical behaviour, whereas carcinomas (C) are clearly aggressive from the beginning (1). EGFR overexpression has been linked with advanced stage (2). However, in a recent report, progression free survival (PFS) and overall survival (OS) appeared to be improved in EGFR positive thymomas (3). It is important to note that egfr mutations are exceedingly rare in thymomas (4) and egfr amplification rate is low (30%) and associated with EGFR protein overexpression by immunohistochemistry (5). Recent data reported increased egfr Gene Copy Number (GCN) as revealed by FISH in rare thymic carcinomas but not in thymomas (6). Moreover, in a bright-field in situ hybridization (BISH) study, egfr GCN was found to be increased in 39/138 TET oriental cases and cases with egfr gains showed better outcome than cases with egfr native status (7). Relevance of egfr gene has also been suggested by the efficacy of EGFR-targeted therapies in sporadic cases of pretreated relapsing thymomas (8,9). However, molecular mechanisms underlying EGFR overexpression have been largely enigmatic and epigenetic mechanisms could play a role. Our aim was to investigate whether a key regulatory mechanism that controls egfr transcription and expression in breast carcinomas (10,11)—namely a polymorphism and loss of heterozygosity (LOH) of the functional CA-SSR-1 microsatellite—may also be involved and characterized in a clinically annotated thymoma cohort. The CA simple sequence repeat 1 (CA-SSR-1) microsatellite lies inside egfr intron 1 and occurrence of shorter variants is associated with higher pre-m-RNA transcription levels in several tumour systems (10,12). Therefore, we performed CA-SSR-1 length and LOH analyses and correlated the results with EGFR-immunohistochemistry (EGFR-IHC), with egfr-fluorescent in situ hybridization (egfr-FISH) and with the clinicopathological and follow up (FU) data.
Materials and methods
Patient series
A total of 43 thymoma cases surgically treated between 1990 and 2009 at the Thoracic Surgeries of Regina Elena National Cancer Institute and of Catholic University of Rome were included in the study. The main demographical and clinicopathological features are shown in Table 1. The availability of formalin-fixed paraffin-embedded (FFPE) tumor tissue as well as of normal peritumoral thymic tissue or other normal matched tissue were major criteria in case selection. Primary tumors were considered; in three cases (no. 26, 41 and 43, Table 2) only tissue from the relapsed neoplasia was available. Diagnoses were reviewed according to the WHO classification (1) by two independent pathologists (MM, LL). Masaoka’s system was applied as staging system (14). This retrospective study was reviewed and approved by the Institutional Ethics Committee of our Institute. The need for individual patient consent was waived because individuals were not identified in the study.
Table 1. Main demographical and clinicopathological features of the cases included in the study.
| Patient characteristics | No. | % |
|---|---|---|
| Total patient No. | 43 | 100 |
| Male | 19 | 44 |
| Female | 24 | 56 |
| Age of diagnosis (yr) | ||
| Median | 50 | |
| Age range | 37-76 | |
| Thymoma histotype (WHO 2004) | ||
| A | 1 | 2.3 |
| AB | 6 | 13.9 |
| B1 | 5 | 11.6 |
| B1/B2 | 1 | 2.3 |
| B2 | 15 | 35 |
| B2/B3 | 7 | 16,3 |
| B3 | 4 | 9.3 |
| B3/C | 4 | 9.3 |
| Masaoka’s staging | ||
| Stage I | 10 | 23.2 |
| Stage II | 20 | 46.4 |
| Stage III | 13 | 30.4 |
Table 2. Comparison of clinicopathological and molecular and genetic data.
| Case No. | WHO | Stage | AD | FU | EGFR IHC | CA-SSR-1 genotype | (CA)n sum | CA-SSR-1 LOH | egfr-FISH |
|---|---|---|---|---|---|---|---|---|---|
| 1 | AB | I | MG | N/A | 1+ | 16/17 | 33L | (−) | NA |
| 2 | AB | I | NO | 7 yr, NED | 2+ | 20/20 | 40H | NI | NA |
| 3 | AB | I | NO | 7 yr, NED | 2+ | 20/21 | 41H | (−) | NA |
| 4 | B1 | I | ND | N/A | 2+ | 16/18 | 34L | (−) | ND |
| 5 | B1/B2 | I | NO | 10 yr, relapse | 2+ | 16/16 | 32L | NI | ND |
| 6 | B2 | I | MG | 5 yr, NED | 1+ | 16/19 | 35L | (−) | NA |
| 7 | B2 | I | NO | N/A | 2+ | 15/19 | 34L | (−) | NA |
| 8 | B2 | I | MG | 5 yr, NED | 3+ | 18/20 | 38H | (−) | ND |
| 9 | B2/B3 | I | MG | N/A | 2+ | 16/16 | 32L | NI | NA |
| 10 | A | II | NO | 6 yr, ET metastasis | 2+ | 16/20 | 36L | (−) | Trisomy |
| 11 | AB | II | MG | N/A | 2+ | 16/18 | 34L | (−) | Trisomy |
| 12 | AB | II | NO | N/A | 2+ | 16/16 | 32L | NI | NA |
| 13 | B1 | II | MG | N/A | 2+ | 18/18 | 36L | NI | NA |
| 14 | B1 | II | NO | 3 yr, NED | 2+ | 20/20 | 40H | NI | NA |
| 15 | B1 | II | NO | N/A | 1+ | 16/20 | 36L | (−) | Trisomy |
| 16 | B1 | II | MG | N/A | 1+ | 16/20 | 36L | (−) | NA |
| 17 | B2 | II | Other | 4 yr; death for AD | 1+ | 18/18 | 36L | NI | NA |
| 18 | B2 | II | MG | Postoperative death | 2+ | 16/16 | 32L | NI | ND |
| 19 | B2 | II | MG | 9 yr, ET metastases | 2+ | 16/18 | 34L | (−) | NA |
| 20 | B2 | II | NO | N/A | 2+ | 20/20 | 40H | NI | ND |
| 21 | B2 | II | NO | 8 yr, NED | 3+ | 16/17 | 33L | (−) | ND |
| 22 | B2 | II | MG | 4 yr, NED | 3+ | 16/21 | 37L | (−) | NA |
| 23 | B2 | II | ND | N/A | 3+ | 17/20 | 37L | (−) | NA |
| 24 | B2 | II | MG | N/A | 3+ | 18/20 | 38H | (−) | NA |
| 25 | B2/B3 | II | MG | 3 yr, NED | 2+ | 16/17 | 33L | (−) | NA |
| 26 | B2/B3 | II | MG | 6 yr, relapse | 3+ | 16/17 | 33L | (−) | NA |
| 27 | B3 | II | MG | 11 yr; NED | 2+ | 15/21 | 36L | (−) | Trisomy |
| 28 | B3/C | II | ND | Early death of disease | 3+ | 13/14 | 27L | (−) | ND |
| 29 | B3/C | II | NO | 13 yr; relapses, ET metastasis, death | 3+ | 16/21 | 37L | (−) | ND |
| 30 | B3/C | II | MG | 18 yr, relapses, death for C HT complications | 3+ | 16/16 | 32L | NI | NA |
| 31 | AB | III | NO | 3 yr, NED | 3+ | 16/20 | 36L | (−) < | Low polysomy |
| 32 | B2 | III | MG | 7 yr, NED | 2+ | 14/18 | 32L | (−) | NA |
| 33 | B2 | III | ND | 2 yr, NED | 3+ | 16/19 | 35L | (+) < | Low polysomy |
| 34 | B2 | III | MG | N/A | 2+ | 16/22 | 38H | (−) | NA |
| 35 | B2 | III | MG | 4 yr, NED | 2+ | 20/22 | 42H | (−) | NA |
| 36 | B2/B3 | III | MG | 15 yr; relapses; death of disease | 3+ | 16/- | 16L | NI | Monosomy |
| 37 | B2/B3 | III | NO | 4yr; relapses, death of disease | 3+ | 16/17 | 33L | (−) | NA |
| 38 | B2/B3 | III | NO | N/A | 3+ | 20/20 | 40H | NI | ND |
| 39 | B2/B3 | III | ND | N/A | 2+ | 20/20 | 40H | NI | NA |
| 40 | B3 | III | MG | 13 yr; relapse; alive | 3+ | 16/18 | 34L | (+) > | ND |
| 41 | B3 | III | MG | 20 yr; relapse; death for complications | 3+ | 16/20 | 36L | (+) < | High polysomy |
| 42 | B3 | III | NO | 6 yr; relapses; IT metastasis; lost to FU | 3+ | 16/20 | 36L | (−) | Trisomy |
| 43 | B3/C | III | MG | 20 yr; relapse; death for complications | 3+ | 16/21 | 37L | ND | Low polysomy |
N/A, not available follow up (<2 years); NI, not informative (homozygous); NA, not amplified; ND, not determined; (+) <, LOH of the longer allele; (+) >, LOH of the shorter allele; (CA)n sum, sum of CA of 2 alleles: low ≤37; H=, high ≥38 (13); NED, no evidence of disease; AD, autoimmune disease; FU, follow up; MG, Myasthenia gravis; NO, no autoimmune disease; IT, intrathoracic; ET, extrathoracic; CHT, chemiotherapy.
EGFR immunohistochemistry (EGFR-IHC)
EGFR immunostains were performed on FFPE Tumor slides with the Dako Cytomation PharmDx EGFR kit and evaluated by two independent pathologists (SS, MM) according to recommended scores, as reported in Supplementary material.
Genomic DNA extraction, PCR amplification and mutation analysis of EGFR exon 21 by directed sequencing; EGFR CA-SSR-1 length microsatellite analysis
Manually microdissected epithelial cell-rich areas from representative FFPE tumor tissue slides as well as peritumoral thymic tissue or other normal tissue were selected for genomic DNA extraction and the molecular and genetic investigations. Peritumoral thymic tissues were selected in cases with a well-represented epithelial cell compartment or from involuted thymus with predominance of epithelial cells, as previously reported (15). Other normal FFPE tissue included matched mediastinal lymph nodes or lung parenchyma. After proteinase K digestion, genomic DNA was isolated with the QIAamp DNA FFPE Tissue Kit (Qiagen) and was used for PCR reaction to amplify egfr exon 21 and the egfr CA-SSR-1 microsatellite by specifically designed primers (Table S1). The PCR reactions were performed by using AmpliTaq Gold; sequencing reactions were analysed on a 3130 Genetic Analyzer (Applied Biosystems, Life technologies, Thermo Fisher) for egfr mutation and egfr CA-SSR-1 microsatellite length analyses. Methods were detailed in Supplementary material.
egfr CA-SSR-1 loss of heterozygosity (LOH)
egfr CA-SSR-1 LOH analysis was performed in 29/30 informative cases by comparing tumor and matched peritumoral thymus/other normal tissues. Case no. 43 was not analysed for LOH as no normal tissue was available. Genomic DNA was analysed for CA-SSR-1-LOH by specifically designed primers (Table S1) (16). PCR reactions were done as reported in section 2.3; fragment separation was performed as shown in Supplementary material. The scans for constitutional heterozygous patients were analysed by comparison of the peak area of the two alleles in normal and tumour tissue by the following equation (17): LOH score = T1 X N2/T2 X N1, where T is tumour, N is normal, 1 is the area under the peak corresponding to the shorter allele, and 2 is the area under the peak corresponding to the longer allele. The result was considered statistically significant different from the ratio of the normal allele peak areas when the LOH score was <0.79 (loss of the longer allele) or >1.27 (loss of the shorter allele) (16). The reliability of peak area and lengths for allelic imbalance (AI) score determination was tested by two runs for each independent PCR.
egfr-fluorescent in situ hybridization (egfr-FISH analysis)
egfr-FISH analysis was performed in 33 cases by using the EGFR/CEN-7 Probe Mix (Dako) as reported in Supplementary material. H&E-pre-selected histological areas were identified and evaluated by FISH. In ten cases FISH analysis could not be performed (among them case no. 40, an AI positive case with increase of the long allele) due to unavailability of left over material or for technical reasons. The egfr-FISH evaluation was performed by three independent observers (ADB, EG, MM) in a cell-by-cell approach, by counting at least in 100 non-overlapping interphase nuclei red and green signals respectively for egfr gene and chromosome 7 centromere. Samples were grouped as follows: normal disomy: ≤2 gene copies in >90% of cells; “trisomy”: 3 gene copies in >10% of cells and ratio gene/chromosomes ≤2; “low polysomy”: ≥4 gene copies in >10% but <40% of cells and ratio gene/chromosomes ≤2; “high polysomy”: ≥4 gene copies in >40% cells and ratio gene/chromosomes ≤2; gene amplification: ratio gene/chromosome >2 or 15 gene copies in ≥10% of cells (18,19). Trisomy, low polysomy, high polysomy, and/or gene amplification were considered egfr-FISH positive (19).
Statistical analyses
Associations among genetic variables and clinicopathological characteristics were assessed by Spearman χ2 test or Fisher test, as appropriate. Differences in mean values were evaluated by the Student t-test. The overall threshold of significance was 0.05. The SPSS (version 18.0) statistical program (SPSS Inc., Chicago, IL, USA) was used for analyses.
Results
Clinical data
In our study 43 Caucasian Thymoma patients were included (Table 1); Table 2 shows in detail the clinicopathological features, the FU data and the molecular and genetic variables investigated. In Stage I (9 cases) all thymoma subtypes were represented but the A subtype; in stage II (21 cases) there were: one A, two AB subtype cases and most cases were B subtype. Among these last there were three B3 thymomas with isolated squamous cell carcinoma foci (B3/C). In stage III with one notable exception (an AB case) all cases were B2 and B3 subtypes or “combined” B2/B3; one case was a B3/C case. Among the 38 cases with available informations, 22 patients had myasthenia gravis (MG), one patient had a severe form of Good’s syndrome and 15 patients were free of autoimmunity. FU data, available in 28 patients (65.12%), including two early death, one for surgical complications and one for disease, ranged 2 to 20 years, median 7 years. FU < to 2 years was not considered.
EGFR-immunohistochemistry (EGFR-IHC)
All cases were positive by EGFR-IHC (Table 2). All AB and B1 thymoma cases showed score 1+ or 2+, with a single exception of 3+ (AB case, no. 31, in stage III). All B2 cases except two and all B2/B3 and B3 cases showed a score 2+ or 3+, irrespective of clinical stage (I to III). B3/C cases received score 3+. The single A case (No. 10) had a 2+ score and was in Stage II. A significant statistical correlation by χ2 test was found between EGFR expression score and histological subtype (P=0.009) but only a trend with clinical stage (P=0.07).
EGFR mutation status and EGFR-CA-SSR-1 microsatellite analyses
By tumor genotyping, no exon 21 egfr activating mutations were found. CA-SSR-1 allele length distribution and frequencies were shown in Figure 1. Alleles 13 to 17 dinucleotide long repeat constituted about 50%, the 16 long repeat being the most frequently represented. One case (“monosomic”) had only one CA-SSR-1 allele, 16 CA repeat long. Therefore the distribution of 85 alleles was shown in Figure 1. Thirty out of 43 cases were heterozygous (informative rate 70%) for CA-SSR-1 allele length. The prevalent genotype was heterozygous 16/20 CA (6 cases) followed by a homozygous genotype 16 CA (5 cases) and a heterozygous 16/17 CA repeats (5 cases). The allele length sum for (CA)n ((Ca)n Sum) (Sum of CA repeats of two alleles, or “combined” allele length) was calculated in each single case (Table 2) as well as by histotype (Table 3). Mean length values in the AB, B1, B2 group was 36.1, while in the B2/B3, B3 and B3/C group was 33.5, the differences in these two groups showed statistical significance by the Student t-test (P=0.05). In our series 33 cases, including the “monosomic” case, had an allele length sum ≤37 (Low repeats, “L”), and 10 cases had an allele length ≥38 (High repeats, “H”) (13).
Figure 1.
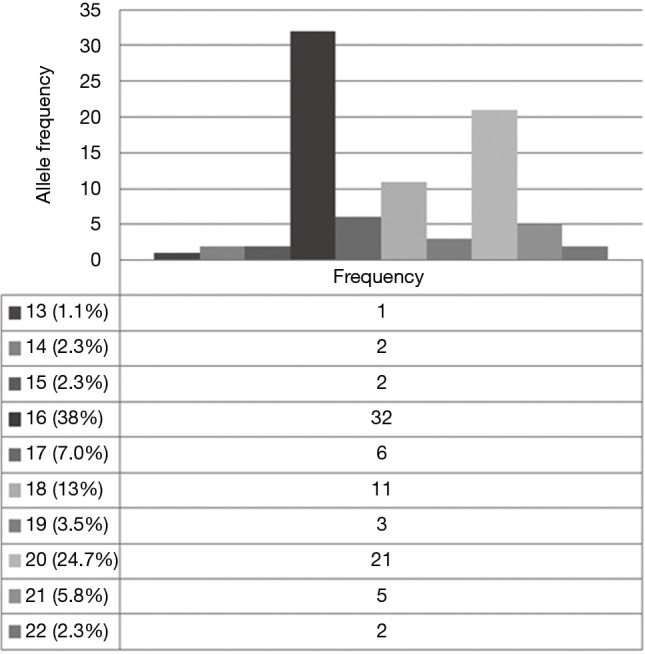
Distribution of CA-SSR-1 allele length among Thymoma patients. The most frequent allele in our caucasian patient group was (CA)n =16 long (38%), followed by (CA)n =20 (24.7%), (CA)n =17 (7%), (CA)n =18 (13%), (CA)n =21 (5.8%), (CA)n =19 (3.5%), (CA)n =14 (2.3%), (CA)n =15 (2.3%), (CA)n =22 (2.3%), (CA)n =13 (1.1%). A total of 85 alleles were reported, as in case no. 36 one allele was absent. This finding was confirmed by egfr-FISH analysis, as the tumor was chromosome 7” monosomic” (case no. 36, Figure S1).
Table 3. Comparison between thymoma histotypes and their frequency with the average “combined” allele length sum for (CA)n for each subtype.
| Thymoma histotype | No. of cases | (%) | (CA)n allelic sum average |
|---|---|---|---|
| A | 1 | 2.3 | 36 |
| AB | 6 | 13.9 | 36 |
| B1 | 5 | 11.6 | 36.4 |
| B1/B2 | 1 | 2.3 | 32 |
| B2 | 15 | 35 | 36.2 |
| B2/B3 | 7 | 16.3 | 32.4 |
| B3 | 4 | 9.3 | 35.5 |
| B3/C | 4 | 9.3 | 33.2 |
egfr CA-SSR-1-LOH analysis
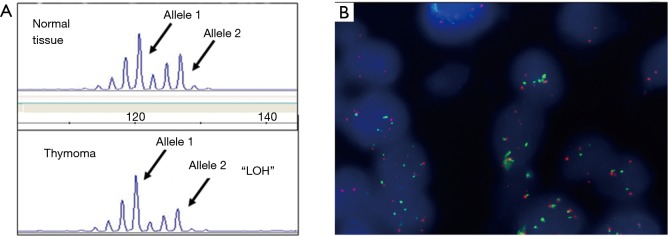
By LOH analysis of 29/30 informative cases, 4/29 (13.8%), all in Stage III, showed AI at the CA-SSR-1 locus, 3/4 cases with loss of the long allele compared to the short allele (Table 2 and Figure 2A). Three out of 4 cases CA-SSR-1-AI positive were B (one B2 and two B3) and one case was an AB subtype. LOH data were comparable when thymus or a different tissue (a lymph node or lung parenchyma) was used as “normal” tissue source.
Figure 2.
Relationship between CA-SSR-1-allelic imbalance (AI) and egfr-Gene Copy Number (GCN) increase/chromosome 7 “polysomy”. (A) Fragment analysis of the CA-SSR-1 locus of normal thymus (top) in comparison with thymoma (bottom). In the tumor tissue it is remarkable the loss of the longer with respect to shorter allele (allelic imbalance, AI); (B) the panel shows the egfr-FISH positive status (high polysomy; ≥4 gene copies in >40% cells) in the same case (case No. 41) (18,19).
egfr-fluorescent in situ hybridization (egfr-FISH) analysis
Twenty two out of 33 cases were disomic; nine cases were egfr-FISH positive showing increased GCN and case no. 36 was chromosome 7 “monosomic” (reported as having only one CA-SSR-1 microsatellite in the previous section on egfr mutation status and egfr-CA-SSR-1 microsatellite analyses; Table 2; Figure S1). Among FISH positive cases, 5 cases were “trisomic”, 3 cases showed “low-polysomy” and one case showed “high polysomy” according to reported criteria (18,19) (Table 2 and Figure 2B). A disomic status was found in 100% of cases tested in stage I, in 75% of cases tested in stage II and in 45% of cases tested in stage III. “Trisomy” was observed in 25% of cases tested in stage II (4/16). In stage III, FISH positivity was found in 5/11 cases (45%) tested, trisomy being found in 9% (1/11 cases tested); both low (27%, 3/11) and high (9%, 1/11) polysomy were observed only in stage III. The single monosomic case was also in stage III.
Integrated correlation of molecular genetic/immunohistochemical data and clinicopathological features
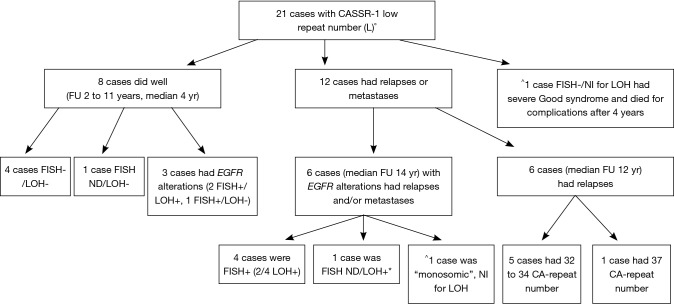
The egfr-FISH analysis showed that three AI positive cases with relative increase of the shorter CA-SSR-1 allele were FISH positive with low or high “chromosome 7 polysomy”/increased GCN, thus suggesting that the increased gene copies carried the CA-SSR-1 shorter allele (cases no. 31, 33 and 41; Table 2, Figure 2A and B). Other five FISH positive cases (“trisomic” for chromosome 7) showed no detectable AI. Statistical analysis revealed a nearly significant correlation between sum length and FISH positivity, short allele sum (“L” status) being associated to positivity (9/9) (P=0.06). Moreover, among the 23 cases analyzed both by LOH and FISH, a statistically significant correlation among associated FISH and AI positivity (observed in three cases) was found with respect to AI negative cases (P=0.01). Further findings derived by correlating the egfr status with FU data, available in 28 patients: no neoplastic adverse events were recorded among the five cases with CA-SSR-1 “H” status, with FU 3 to 7 years. Among the CA-SSR-1 cases with “L” status (21 cases) (Figure 3), eight, including three FISH positive cases, did well (median FU 4 ys). Adverse events (relapses or metastases) were recorded in 12/21 CA-SSR-1 “L” cases (57.14%). Among these “L” cases, six had a range of egfr alterations (FISH positivity, AI, or chromosome 7 “monosomy”). Among FISH positive cases, it is worthy to note that two patients, one with “low” and the other with “high” egfr polysomy, had multiple relapses, were long survivors and died after 20 years because of complications. Our “monosomic” case experienced long survival (15 years) with multiple relapses and ultimately death because of complications. Among the other six relapsing or metastatizing patients with “L” CA-SSR-1, 5/6 cases had very short combined CA-SSR-1 allele length (32 to 34). Moreover, the patient with the shortest combined allele length (case No. 28, 27 Ca repeat sum) died shortly after surgery due to the disease. Case no. 26, included in the study as an example of early relapse, had also a very short [33] Ca repeat sum.
Figure 3.
Integrated correlation of molecular and genetic data and clinicopathological features in 21 cases with low-number (“L”) CA-SSR-1 CA repeats (FU ≥2 yr). All WHO histotypes were represented with B2 to B3 cases representing the majority. Cases “L” which did well: no. 6, 21, 22, 25, 32; cases which did well but had egfr gene alterations: no. 27, 31, 33. Cases “L” which had relapse or metastases and had egfr alterations: no. 10, 40, 41, 42, 43; case no. 36 was “monosomic”. Cases “L” which had relapse or metastases, no detectable egfr gene alterations but had short CA-SSR-1 microsatellite: no. 5, 19, 26, 30, 37. Case “L” which had relapse or metastases, with no detectable egfr alterations and CA-SSR-1 microsatellite 37 repeat long: no. 29. The case “L”, FISH-/Not Informative for LOH with Good’s syndrome which had combined CA-SSR-1 microsatellite length of 36 was no. 17. *In this LOH+ case “(+) >”, with LOH of the shorter allele, the FISH status could not be determined (case No. 40, Table 2); ^NI for LOH = not Informative for LOH study (cases No. 17 and 36); Cases 18 and 28 (early deaths) were not reported here. LOH, loss of heterozygosity; FISH, fluorescent in situ hybridization.
Discussion
Thymomas are slow-growing tumors, requiring long term FU; the average time to relapse was reported to be 8.5 years in a recent case series (20). We investigated the egfr polymorphic dinucleotide repeat microsatellite locus CA-SSR-1 length and AI and the egfr-FISH status in a clinically annotated Caucasian Thymoma series with on average long FU. By tumor genotyping, our thymoma series was found to bear similar CA dinucleotide repeat length (13 to 22 CA) frequency than reported in Caucasian populations (21). By considering the combined medium allele length, we showed that the shorter sequences were significantly mostly associated with the B2/B3, B3 and B3/C histotypes. The shorter egfr CA-SSR-1 alleles and combined shorter CA-SSR-1 sequences have been related to increased egfr gene expression/mRNA synthesis (22). In other tumor systems, Caucasian patients with Head & Neck Squamous Cell Carcinomas (HNSCC) bearing short (<17) repeat number of egfr CA-SSR-1 were found to have poorer prognosis in comparison to patients with CA-SSR-1 repeat number ≥17 in both alleles (23). Increased risk was found in esophageal cancer patients with CA-SSR-1 short alleles (cut off ≤18) (24). No mutations were found in our series in exon 21, as already reported (4); mutations were neither found in CA-SSR-1 AI positive cases, at variance with lung cancer (22). The finding of egfr-CA-SSR-1 microsatellite AI in 4/29 cases represents an aberrant genomic somatic signature previously unreported in thymoma. The occurrence of AI with respect to peritumoral thymic tissue suggested that there was no “zone” phenomenon as reported in breast cancer (25). Among our FISH positive cases only those cases with “polysomy”, both low or high, were associated to CA-SSR-1-AI, probably because the slight increase in GCN occurring in “trisomic” cases was not sufficient to be detected as LOH. Moreover, we cannot exclude that the long history and previous treatments had induced secondary egfr genetic changes in one (case no. 41) of the four cases bearing AI. Recently, increased FISH-determined egfr GCN gain was found to have prognostic significance in Head and Neck Squamous Cell Carcinomas (HNSCC) (26). In thymomas a predictive value of egfr GCN gain, could also be argued, as the case AI positive no. 41 in our series was treated in second line with Cetuximab with partial response (8). In other tumor systems, the integrated assessment of egfr polymorphisms, histopathological features and staging factors were shown to contribute to prognostic estimates in breast cancer (27). Our pilot study data indicated that egfr short CA-SSR-1 sequences were significantly associated with the more aggressive B2/B3, B3 and B3/C WHO histotype groups (28) Furthermore, egfr-FISH positivity was associated with malignant behaviour in thymomas in 5 out of the 7 cases with available FU. Moreover, the occurrence of adverse events in patients with the previous mentioned egfr “alterations” appears to be of note both in limited stages (I-II) and in advanced stage (III). Finally, by integrating LOH and egfr-FISH data we showed here that the increased egfr GCN carried on the shorter microsatellite allele in 3 out of 4 cases, a finding previously not described in thymoma.
Conclusions
Our pilot study showed that an egfr-related “signature”, mostly but not exclusively B-histotype related, with short CA-SSR-1, AI for the intron 1 microsatellite CA-SSR-1 and increased egfr GCN could be relevant both for progression and therapeutical purposes in thymoma. The role of egfr-CA-SSR-1 microsatellite and of similar (TG/CA)n repeats occurring in intron 1 of other cancer-related genes has been recently questioned as a class of universal gene expression regulators (29). Therefore a larger prospectively collected thymoma series should be investigated to acquire further insight in thymoma-specific egfr transcription regulatory mechanisms.
Acknowledgements
This work was partially supported by the Italian Ministry of Health, Italy-US program Grants for Rare Diseases to M Marino. Moreover, S Conti received a Research fellowship of the Ministry of Health (Current Research Grants) for this study. The authors thank Prof. Alexander Marx and Dr. Marcella Mottolese for suggestions and critical reading and Dr. Tania Merlino for English language editing. The authors thank the International Thymic Malignancy Interest Group (ITMIG) for providing fruitful discussion.
EGFR immunohistochemistry (EGFR-IHC)
EGFR immunohistochemistry was performed with the Dako Cytomation PharmDx EGFR kit. Briefly, only tumor cell membrane EGFR expression was defined positive and was graded 0 to 3. An intensity score was applied as follows: negative = no reaction; 1+ if the neoplastic cells displayed an incomplete, weak plasmamembrane/cytoplasmic immunostaining; 2+ if neoplastic cells displayed a complete plasmamembrane immunostaining with moderate intensity; and 3+ if neoplastic cells displayed a complete plasmamembrane strong immunostaining. Score 2+ and 3+ were considered overexpression.
egfr exon 21 PCR Amplification and mutation analysis by direct sequencing
PCR reactions were in a volume of 50 µL which contained; 10 pmol of each primer, 0.25 mM each dNTP, 2.5 mM MgCl2, 3 units of AmpliTaq Gold (Applied Biosystems (AB), Life Technologies, Thermofisher), 1X Taq Gold buffer (AB, Life Technologies, Thermofisher), and the appropriate volume of H2O. A thermocycle touchdown PCR protocol was as follow: 10 minutes at 95 °C, 8 cycles at 95 °C for 30 seconds, 59 °C for 30 seconds, and 72 °C for 30 seconds, followed by 30 cycles at 95 °C for 30 seconds, 57 °C for 30 seconds, and at 72 °C for 30 seconds followed by 72 °C for 10 minutes. PCR products were sequenced directly by using the Big Dye V3.1 Cycle-Sequencing kit (AB, Life Technologies, Thermofisher) with proper reverse primers (Table S1). After sequencing reaction, 1 µL of every mixture was purified by BigDye XTerminator Purification Kit (AB, Life Technologies, Thermofisher) and analyzed on 3130 Genetic Analyzer (Applied Biosystems, Life Technologies, Thermofisher).
egfr CASSRI Loss of heterozygosity (LOH) analysis
About 50–100 ng of genomic DNA was amplified in a volume of 25 µL, 10 pmol of each primer, 0.25 mM each dNTP, 2.5 mM MgCl2, 3 units of AmpliTaq Gold, 1X Taq Gold buffer (AB, Life Technologies, Thermofisher), and the appropriate volume of H2O. The thermocycle PCR protocol as follows: 95 °C for 10 min, 30 cycles of 95 °C for 30 sec, 58 °C for 30 sec, 72 °C for 30 sec, and a final extension at 60°C for 30 min to obtain greater number of fragments with A-tailed ends. From 1 to 2 µL of PCR reaction products were added to a mix containing 0.5 µL GENESCAN 500 LIZ fluorescent size standard and Hi-Di formamide (AB, Life Technologies, Thermofisher), denatured for 5 min at 94 °C and loaded onto the Genetic Analyzer. Products were capillary electrophoresed and the data processed by GeneMapper 4.0 Analysis Software (AB, Life Technologies, Thermofisher). The analyses were performed at least in duplicates of independent PCRs.
EGFR-FISH analysis
The EGFR/CEN-7 Probe Mix (Dako) detects the copy number of the egfr genes located on chromosome 7p11.2 using the chromosome 7 centromere region as a reference (chromosome 7 copy number detection). The Texas Red-labeled DNA probe (EGFR) binds to a 196 kb segment containing the egfr gene on chromosome 7p11.2. The fluoroscein-labelled PNA probe (CEN-7) binds to the centromeric region of chromosome 7. In brief, 2 µm thick paraffin sections were deparaffinized, dehydrated in ethanol, and air-dried. Sections were digested with Pepsin at 37 °C for 25 minutes. Slides were placed on a pre-programmed, humidified slide warmer (Hybridize; DAKO) with the following settings: denaturation at 82 °C for 5 minutes and hybridization at 45 °C for 18 hours. After hybridization, the slides were placed in a staining jar containing diluted Stringent Wash Buffer at room temperature after removing the Coverslips. As soon as coverslips had been removed, slides were transferred from the room temperature, jar was pre-washed to the 65 °C jar in the water bath. Stringent wash was performed for 10 minutes. Subsequently, slides were placed in a wash buffer and dehydrated through a graded series of ethanol: 2 minutes in 70% ethanol, 2 minutes in 85% ethanol, 2 minutes in 96% ethanol. After drying, each slide was covered with 10µL of Fluorescence Mounting Medium containing DAPI and applied a glass coverslip. The FISH results were assessed with an epi-fluorescence microscope (Zeiss, Axioscope 40) equipped with Image Processing analysis software (Media Cybernetics) able to DAPI/specific Texas Red and FITC single filters. Normal thymic tissue sections were included in each FISH assay as negative control.
Table S1. Nucleotide sequence of primers utilized to amplify two DNA regions (egfr exon 21 and egfr CA-SSR-1 locus). Fw (Forward), Rw (Reverse). For CA-SSR-1 locus amplification reverse primer was 5’ end fluorescence-labeled with 6-carboxy-fluorescein (6-FAM).
| egfr exon 21 | Fw | 5’-TTCATGCGCCTTTCCATTCTTT-3’ |
| Rv | 5’-CTGGTCCCTGGTGTCAGGAAA-3’ | |
| CA-SSR-1 locus | Fw | 5’- GTTTGAAGAATTTGAGCCAACC- 3’ |
| Rv | 6-FAM-5’-TTCTGTCTGCACACTTGGCAC-3’ |
Figure S1.
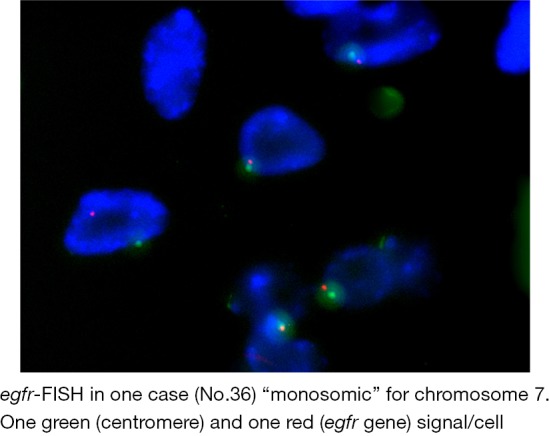
egfr-FISH in one case (No. 36) “monosomic” for chromosome 7. One green (centromere) and one red (egfr gene) signal cell.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare. While performing the study, S Conti was an Italian Ministry of Health Research fellow.
Actual address: Salvatore Conti, MS. Sr Field Application Scientist, Life Technologies Brand Thermofisher Scientific, Via Tiepolo 18, 20900 Monza, Italy. Email: salvatore.conti@termofisher.com.
Actual address: Stefano Sioletic, MD, PhD. Department of Pathology, Ospedale Santa Maria Della Misericordia, 33100 Udine, Italy. Email: sioletic.stefano@aoud.sanita.fvg.it.
References
- 1.Travis WD, Brambilla E, Müller-Hermelink HK, et al. editors. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Third Edition ed. IARC 2004. [Google Scholar]
- 2.Suzuki E, Sasaki H, Kawano O, et al. Expression and mutation statuses of epidermal growth factor receptor in thymic epithelial tumors. Jpn J Clin Oncol 2006;36:351-6. 10.1093/jjco/hyl028 [DOI] [PubMed] [Google Scholar]
- 3.Aisner SC, Dahlberg S, Hameed MR, et al. Epidermal growth factor receptor, C-kit, and Her2/neu immunostaining in advanced or recurrent thymic epithelial neoplasms staged according to the 2004 World Health Organization in patients treated with octreotide and prednisone: an Eastern Cooperative Oncology Group study. J Thorac Oncol 2010;5:885-92. 10.1097/JTO.0b013e3181d86a30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoh K, Nishiwaki Y, Ishii G, et al. Mutational status of EGFR and KIT in thymoma and thymic carcinoma. Lung Cancer 2008;62:316-20. 10.1016/j.lungcan.2008.03.013 [DOI] [PubMed] [Google Scholar]
- 5.Ionescu DN, Sasatomi E, Cieply K, et al. Protein expression and gene amplification of epidermal growth factor receptor in thymomas. Cancer 2005;103:630-6. 10.1002/cncr.20811 [DOI] [PubMed] [Google Scholar]
- 6.Weissferdt A, Lin H, Woods D, et al. HER family receptor and ligand status in thymic carcinoma. Lung Cancer 2012;77:515-21. 10.1016/j.lungcan.2012.05.108 [DOI] [PubMed] [Google Scholar]
- 7.Mimae T, Tsuta K, Kondo T, et al. Protein expression and gene copy number changes of receptor tyrosine kinase in thymomas and thymic carcinomas. Ann Oncol 2012;23:3129-37. 10.1093/annonc/mds147 [DOI] [PubMed] [Google Scholar]
- 8.Palmieri G, Marino M, Salvatore M, et al. Cetuximab is an active treatment of metastatic and chemorefractory thymoma. Front Biosci 2007;12:757-61. 10.2741/2098 [DOI] [PubMed] [Google Scholar]
- 9.Farina G, Garassino MC, Gambacorta M, et al. Response of thymoma to cetuximab. Lancet Oncol 2007;8:449-50. 10.1016/S1470-2045(07)70141-9 [DOI] [PubMed] [Google Scholar]
- 10.Gebhardt F, Zänker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem 1999;274:13176-80. 10.1074/jbc.274.19.13176 [DOI] [PubMed] [Google Scholar]
- 11.Buerger H, Gebhardt F, Schmidt H, et al. Length and loss of heterozygosity of an intron 1 polymorphic sequence of egfr is related to cytogenetic alterations and epithelial growth factor receptor expression. Cancer Res 2000;60:854-7. [PubMed] [Google Scholar]
- 12.Gebhardt F, Bürger H, Brandt B. Modulation of EGFR gene transcription by secondary structures, a polymorphic repetitive sequence and mutations--a link between genetics and epigenetics. Histol Histopathol 2000;15:929-36. [DOI] [PubMed] [Google Scholar]
- 13.Han SW, Oh DY, Im SA, et al. Epidermal growth factor receptor intron 1 CA dinucleotide repeat polymorphism and survival of advanced gastric cancer patients treated with cetuximab plus modified FOLFOX6. Cancer Sci 2010;101:793-9. 10.1111/j.1349-7006.2009.01447.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masaoka A, Monden Y, Nakahara K, et al. Follow-up study of thymomas with special reference to their clinical stages. Cancer 1981;48:2485-92. [DOI] [PubMed] [Google Scholar]
- 15.Ganci F, Vico C, Korita E, et al. MicroRNA expression profiling of thymic epithelial tumors. Lung Cancer 2014;85:197-204. 10.1016/j.lungcan.2014.04.008 [DOI] [PubMed] [Google Scholar]
- 16.Tidow N, Boecker A, Schmidt H, et al. Distinct amplification of an untranslated regulatory sequence in the egfr gene contributes to early steps in breast cancer development. Cancer Res 2003;63:1172-8. [PubMed] [Google Scholar]
- 17.Canzian F, Salovaara R, Hemminki A, et al. Semiautomated assessment of loss of heterozygosity and replication error in tumors. Cancer Res 1996;56:3331-7. [PubMed] [Google Scholar]
- 18.Gonçalves A, Esteyries S, Taylor-Smedra B, et al. A polymorphism of EGFR extracellular domain is associated with progression free-survival in metastatic colorectal cancer patients receiving cetuximab-based treatment. BMC Cancer 2008;8:169. 10.1186/1471-2407-8-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li YH, Wang F, Shen L, et al. EGFR fluorescence in situ hybridization pattern of chromosome 7 disomy predicts resistance to cetuximab in KRAS wild-type metastatic colorectal cancer patients. Clin Cancer Res 2011;17:382-90. 10.1158/1078-0432.CCR-10-0208 [DOI] [PubMed] [Google Scholar]
- 20.Harnath T, Marx A, Ströbel P, et al. Thymoma-a clinico-pathological long-term study with emphasis on histology and adjuvant radiotherapy dose. J Thorac Oncol 2012;7:1867-71. 10.1097/JTO.0b013e3182745f73 [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Innocenti F, Chen P, et al. Interethnic difference in the allelic distribution of human epidermal growth factor receptor intron 1 polymorphism. Clin Cancer Res 2003;9:1009-12. [PubMed] [Google Scholar]
- 22.Nomura M, Shigematsu H, Li L, et al. Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers. PLoS Med 2007;4:e125. 10.1371/journal.pmed.0040125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bandrés E, Barricarte R, Cantero C, et al. Epidermal growth factor receptor (EGFR) polymorphisms and survival in head and neck cancer patients. Oral Oncol 2007;43:713-9. 10.1016/j.oraloncology.2006.09.002 [DOI] [PubMed] [Google Scholar]
- 24.Vashist YK, Trump F, Gebauer F, et al. EGFR intron-1 CA repeat polymorphism is a predictor of relapse and survival in complete resected only surgically treated esophageal cancer. Target Oncol 2014;9:43-52. 10.1007/s11523-013-0260-2 [DOI] [PubMed] [Google Scholar]
- 25.Kersting C, Tidow N, Schmidt H, et al. Gene dosage PCR and fluorescence in situ hybridization reveal low frequency of egfr amplifications despite protein overexpression in invasive breast carcinoma. Lab Invest 2004;84:582-7. 10.1038/labinvest.3700077 [DOI] [PubMed] [Google Scholar]
- 26.Szabó B, Nelhubel GA, Kárpáti A, et al. Clinical significance of genetic alterations and expression of epidermal growth factor receptor (EGFR) in head and neck squamous cell carcinomas. Oral Oncol 2011;47:487-96. 10.1016/j.oraloncology.2011.03.020 [DOI] [PubMed] [Google Scholar]
- 27.Leite MS, Giacomin LC, Piranda DN, et al. Epidermal growth factor receptor gene polymorphisms are associated with prognostic features of breast cancer. BMC Cancer 2014;14:190. 10.1186/1471-2407-14-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ströbel P, Bauer A, Puppe B, et al. Tumor recurrence and survival in patients treated for thymomas and thymic squamous cell carcinomas: a retrospective analysis. J Clin Oncol 2004;22:1501-9. 10.1200/JCO.2004.10.113 [DOI] [PubMed] [Google Scholar]
- 29.Zhang W, He L, Liu W, et al. Exploring the relationship between polymorphic (TG/CA)n repeats in intron 1 regions and gene expression. Hum Genomics 2009;3:236-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EGFR immunohistochemistry was performed with the Dako Cytomation PharmDx EGFR kit. Briefly, only tumor cell membrane EGFR expression was defined positive and was graded 0 to 3. An intensity score was applied as follows: negative = no reaction; 1+ if the neoplastic cells displayed an incomplete, weak plasmamembrane/cytoplasmic immunostaining; 2+ if neoplastic cells displayed a complete plasmamembrane immunostaining with moderate intensity; and 3+ if neoplastic cells displayed a complete plasmamembrane strong immunostaining. Score 2+ and 3+ were considered overexpression.
PCR reactions were in a volume of 50 µL which contained; 10 pmol of each primer, 0.25 mM each dNTP, 2.5 mM MgCl2, 3 units of AmpliTaq Gold (Applied Biosystems (AB), Life Technologies, Thermofisher), 1X Taq Gold buffer (AB, Life Technologies, Thermofisher), and the appropriate volume of H2O. A thermocycle touchdown PCR protocol was as follow: 10 minutes at 95 °C, 8 cycles at 95 °C for 30 seconds, 59 °C for 30 seconds, and 72 °C for 30 seconds, followed by 30 cycles at 95 °C for 30 seconds, 57 °C for 30 seconds, and at 72 °C for 30 seconds followed by 72 °C for 10 minutes. PCR products were sequenced directly by using the Big Dye V3.1 Cycle-Sequencing kit (AB, Life Technologies, Thermofisher) with proper reverse primers (Table S1). After sequencing reaction, 1 µL of every mixture was purified by BigDye XTerminator Purification Kit (AB, Life Technologies, Thermofisher) and analyzed on 3130 Genetic Analyzer (Applied Biosystems, Life Technologies, Thermofisher).
About 50–100 ng of genomic DNA was amplified in a volume of 25 µL, 10 pmol of each primer, 0.25 mM each dNTP, 2.5 mM MgCl2, 3 units of AmpliTaq Gold, 1X Taq Gold buffer (AB, Life Technologies, Thermofisher), and the appropriate volume of H2O. The thermocycle PCR protocol as follows: 95 °C for 10 min, 30 cycles of 95 °C for 30 sec, 58 °C for 30 sec, 72 °C for 30 sec, and a final extension at 60°C for 30 min to obtain greater number of fragments with A-tailed ends. From 1 to 2 µL of PCR reaction products were added to a mix containing 0.5 µL GENESCAN 500 LIZ fluorescent size standard and Hi-Di formamide (AB, Life Technologies, Thermofisher), denatured for 5 min at 94 °C and loaded onto the Genetic Analyzer. Products were capillary electrophoresed and the data processed by GeneMapper 4.0 Analysis Software (AB, Life Technologies, Thermofisher). The analyses were performed at least in duplicates of independent PCRs.
The EGFR/CEN-7 Probe Mix (Dako) detects the copy number of the egfr genes located on chromosome 7p11.2 using the chromosome 7 centromere region as a reference (chromosome 7 copy number detection). The Texas Red-labeled DNA probe (EGFR) binds to a 196 kb segment containing the egfr gene on chromosome 7p11.2. The fluoroscein-labelled PNA probe (CEN-7) binds to the centromeric region of chromosome 7. In brief, 2 µm thick paraffin sections were deparaffinized, dehydrated in ethanol, and air-dried. Sections were digested with Pepsin at 37 °C for 25 minutes. Slides were placed on a pre-programmed, humidified slide warmer (Hybridize; DAKO) with the following settings: denaturation at 82 °C for 5 minutes and hybridization at 45 °C for 18 hours. After hybridization, the slides were placed in a staining jar containing diluted Stringent Wash Buffer at room temperature after removing the Coverslips. As soon as coverslips had been removed, slides were transferred from the room temperature, jar was pre-washed to the 65 °C jar in the water bath. Stringent wash was performed for 10 minutes. Subsequently, slides were placed in a wash buffer and dehydrated through a graded series of ethanol: 2 minutes in 70% ethanol, 2 minutes in 85% ethanol, 2 minutes in 96% ethanol. After drying, each slide was covered with 10µL of Fluorescence Mounting Medium containing DAPI and applied a glass coverslip. The FISH results were assessed with an epi-fluorescence microscope (Zeiss, Axioscope 40) equipped with Image Processing analysis software (Media Cybernetics) able to DAPI/specific Texas Red and FITC single filters. Normal thymic tissue sections were included in each FISH assay as negative control.