Abstract
Background
Circulating microRNA (miRNA) has attractive interests as a non-invasive biomarker of physiological and pathological conditions. Our study aimed to investigate the potential effects of chronic benzene poisoning (CBP) and benzene exposure on miRNA expression, and identify CBP-related miRNAs.
Methods
In the discovery stage, we used a microarray assay to detect the miRNA expression profiles among pooled plasma samples from ten CBP patients, ten healthy benzene-exposed individuals and ten non-benzene exposed individuals. Subsequently, we conducted an expanded validation of six candidate miRNAs in 27 CBP patients- low blood counts, 54 healthy benzene-exposed individuals and 54 non-exposed individuals. Moreover, we predicted the biological functions of putative target genes using a Gene Ontology (GO) function enrichment analysis and KEGG pathway analysis.
Results
In the discovery stage, compared with non-exposures, 36 and 12 miRNAs demonstrated at least a 1.0-fold differential expression in the CBP patients and the benzene exposures, respectively. And compared with benzene exposures, 58 miRNAs demonstrated at least a 1.0-fold differential expression in the CBP patients. In the expanded validation stage, compared with non-exposures as well as exposures, miR-24-3p and miR-221-3p were significantly up-regulated (1.99- and 2.06-fold for miR-24-3p, 2.19- and 3.93-fold for miR-221-3p, P<0.01) while miR-122-5p and miR-638 were significantly down-regulated (−3.45- and −2.60-fold for miR-122-5p, −1.82- and −3.20-fold for miR-638, P<0.001) in the CBP patients; compared with non-exposures, the plasma level of miR-638 was significantly up-regulated (1.38-fold, P<0.01) while the plasma levels miR-122-5p and miR-221-3p were significantly down-regulated (−0.85- and −1.74-fold, P<0.01) in the exposures, which were consistent with the results of microarray analysis.
Conclusions
The four indicated plasma miRNAs may be biomarkers of indicating responses to benzene exposure. Further studies are warranted to verify our findings with a large sample and to confirm the underlying mechanisms.
Keywords: MicroRNA (miRNA), plasma, benzene, chronic benzene poisoning (CBP)
Introduction
Benzene is one of the most common environmental pollutants, and it is still widely used as an industrial solvent in the world, especially in developing countries, representing a significant occupational hazard (1). It was confirmed to be a carcinogen by the International Agency for Research on Cancer. During the benzene related production, benzene was usually inhaled by people as its steamy form. Benzene exposure can lead to numerous non-cancerous health effects associated with functional aberration of vital systems in the body like cardiovascular and respiratory, besides, ambient exposure to benzene was associated with increased risk of lung cancer. Long-term exposure to benzene may result in chronic benzene poisoning (CBP) characterized by hematotoxicity, leading to leucopenia, thrombocytopenia, aplastic anemia, even acute myelogenous leukemia and other forms of leukemia (2-4). According to annual report of occupational diseases in China, CBP is one of the most common chronic occupational poisoning (www.niohp.net.cn/jbjcbg/). The prognosis for restoring CBP remains poor, and increasing numbers of employees facing CBP, especially in developing countries. Therefore, CBP should be highly concerned to explore new screening and therapy strategies.
MicroRNA (miRNA) is an abundant class of small (18–24 nucleotides), endogenous and non-coding RNA, which regulate gene expression at the post-transcriptional level by base pairing to 3’ untranslated region of target messenger RNA (mRNA), and works either by direct cleavage of the target mRNA or inhibition of protein synthesis (5,6). miRNAs play crucial roles in a broad range of physiological and pathological processes, including the processes of normal hematopoiesis and hematopoietic malignancies progression (7-9). Simultaneously, growing evidences have indicated that exposure to hazardous environmental factors may significantly disrupt miRNA expression patterns (10-12). Previous studies also revealed that dysregulated miRNAs are involved in responses to polycyclic aromatic hydrocarbons (PAHs) (13). Further studies suggested that miRNAs and their interactions with environmental factors might affect the effects of PAHs exposure on oxidative DNA damage and lipid peroxidation (14). However, no epidemiological studies have yet demonstrated the potential effects of CBP and benzene exposure on miRNA expression.
Circulating miRNAs in plasma are extremely stable and can be predicted as a noninvasive biomarker of physiological and pathological conditions (15). Thus the aim of current study was to identify differentially expressed CBP-related plasma miRNAs by using microarray analysis and subsequent validation as well as the quantitative reverse-transcription polymerase chain reaction (qRT-PCR). We also performed a predicted target analysis for the indicated miRNAs using bioinformatics, which contributes to gain insights into the potential roles of miRNAs in CBP.
Materials and methods
Ethics statement
This project was approved by the Ethics Committee of Jiangsu Provincial Center for Disease Prevention and Control, each subject gave written informed consent and ethics were respected throughout the whole study period.
Subjects
A total of 27 CBP patients were enrolled in our study. Nineteen of 27 CBP patients came from seven factories in Zhengjiang (Zhengjiang, China), and other 8 CBP patients came from two factories in Yangzhou (Yangzhou, China), where clusters of cases were reported. CBP was diagnosed from 2010 to 2012, And the diagnostic criteria for occupational CBP, according to the Ministry of Health, China, include (I) total white blood cell count <4,000/μL or neutrophil count <2,000/μL or platelet count <80,000/μL, with repeated confirmation of this count in a few months in a peripheral blood examination; (II) the individual with documented benzene exposure has been employed for at least three months in a factory; and (III) exclusion of other causes of abnormal blood counts such as chloromycetin use and ionizing radiation. We chose 54 workers from nine factories, who had been occupationally exposed to benzene, as healthy benzene-exposed controls. Meanwhile, other 54 subjects without occupational benzene exposure were enrolled from rural commercial bank in Lianyungang (Lianyungang, China) as non-exposed controls.
The subjects were interviewed by trained personnel, and administered a structured questionnaire for information on demographic characteristics, lifetime occupational history, lifestyle habits (smoking and alcohol consumption), disease history, environmental benzene exposures, history of hematotoxicity drug utilization, physical and chemical factor exposure. Venous blood sample of each subject was collected with BD Vacutainer® Venous Blood Collection Tubes (cat. no. 367525) containing ethylene diamine tetraacetic acid (EDTA), and the plasma was isolated within one hour and stored at −80 °C until RNA extraction. Subjects with previous or present treatment on hematotoxicity drugs (e.g., chloromycetin, sulfonamides), habitual exposure to other chemical or physical factors causing hematotoxicity (e.g., ionizing radiation, arsenic) and recent infection were excluded from the study. Exposure estimation was based on benzene concentration monitoring data at the work place provided by Jiangsu Provincial Center for Disease Prevention and Control.
The subjects were classified into three groups: CBP (chronic benzene poisoning patients), exposure (healthy benzene-exposed individuals) and non-exposure (healthy non-benzene exposed individuals). We firstly selected the CBP patients, healthy benzene-exposed subjects and non-benzene exposed subjects who were frequency-matched (1:2:2) to cases by gender, age (±5 years), nationality (Han Chinese), smoking status and drinking status. Then the healthy benzene-exposed subjects were frequency-matched to cases by exposure duration (±3 years) and exposure level. We intentionally frequency-matched the distribution of important general characteristics among these three groups to minimize their confounding effects on miRNA expression profiles. All the subjects were Han Chinese, which indicated that the subjects are homogeneous on nationality. Those who smoked at least one cigarette per day for more than one year were considered as regular smokers. Alcohol consumption was defined as drinking at least seven standard units of alcohol on average per week [1 standard unit =10 g of alcohol equivalent; e.g., a glass/can/bottle (330 mL) of regular beer (5%), a measure (40 mL) of liquor, a glass (120 mL) of wine] for more than 6 months.
Purification of total RNA
In preparation for microarray analysis, three plasma pools were produced by mixing ten samples per group (120 μL per sample) for the LC Sciences microfluidic microarray analysis. The pooled plasma was then subjected to total RNA extraction using Norgen’s Total RNA Purification Kit (Norgen Biotech, Thorold, Canada) following the manufacturer’s instructions provided by Lianchuan Biotechnology Limited (subsidiary of LC Sciences, Hangzhou, China). In the validation stage, RNA was extracted from individual plasma sample (200 μL per sample) using a miRNeasy Serum/Plasma Kit (QIAGEN, cat. no. 217184, Germany). A synthetic caenorhabditis elegans miRNA [cel-miR-238 (25 fmol); Takara, Dalian, China] was added to each plasma sample as an internal control prior to the isolation procedure. The quality and quantity of isolated RNAs were measured by using an ND-1000 NanoDrop spectrophotometer (ThermoScientific, Wilmington, DE, USA).
MicroRNA (miRNA) microarray assay
miRNA expression profiles of three plasma pools were examined using the microarray assay (version 19.0) performed by service provider (subsidiary of LC Sciences, Hangzhou, China) as described previously (16). Briefly, about 0.5 µg total RNA samples from plasma were 3’-extended with a poly(A) tail, to which an oligonucleotide tag was ligated for later fluorescent dye staining. On the microfluidic chip, each detection probe consists of a chemically modified nucleotide coding segment complementary to target miRNA or other RNA (control sequences) and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. After hybridization, tag-conjugating Cy3 dye was circulated through the microfluidic chip for dye staining. Microarray was then scanned with a laser scanner (GenePix 4000B, Molecular Device), and raw data were imput into Array-Pro image analysis software (Media Cybernetics). The data were analyzed by firstly subtracting the background, and then normalizing the signals using a LOWESS filter (Locally-weighted Regression) (17). The signal values, higher than the background value by three folds of standard deviation, were considered valuable. The fold changed values of any two out of three sets of detected signals (log2 transformed and balanced) were calculated.
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) validation
To focus on most likely CBP-related miRNAs in the validation stage, we selected candidate miRNAs from the microarray analysis based on the following criteria: demonstrating at least 1.0 fold higher or −1.0 fold lower expression between any two out of three groups, expressing more than 500 copies in any group, and giving priority to choosing molecules that may be associated with a benzene response based on the literature. The expression levels of candidate miRNAs selected from the array analysis were validated with the use of TaqMan miRNA qRT-PCR primers and regents. As described previously (18), Purified total RNA (1.67 μL) from the individual plasma sample was subjected to reverse transcription in a 5-μL reaction mixture using miRNA-specific stem-loop primers and a TaqMan® miRNA Reverse Transcription Kit (Applied Biosystems, USA) following the manufacturer’s protocol. The real-time PCR assay was performed in a 10-μL reaction mixture that contained 5 μL of 2× TaqMan Universal PCR Master Mix (No AmpErase UNG), 4.5 μL of diluted cDNA (1:15) and 0.5 μL of TaqMan miRNA Assay primers (Applied Biosystems, USA). The thermal cycling procedure was set as follows: an initial denaturation step at 95 °C for 10 min, 40 cycles of PCR amplification at 95 °C for 15 s, and 60 °C for 1 min. Each extracted total RNA sample for each candidate miRNA was run in triplicate. The expression level of each miRNA was individually normalized to cel-miR-238.
Bioinformatic analysis
To understand the potential molecular function of indicated miRNAs, their putative targets were predicted with the use of Targetscan (http://www.targetscan.org), Pictar (http://pictar.mdc-berlin.de) and miRanda (http://www.microrna.org/microrna/home.do) software. Results from the target databases were collected, and only the common targets of three software programs were evaluated. GO term enrichment and KEGG pathway analysis were performed based on David 6.7 (http://david.abcc.ncifcrf.gov/home.jsp) (P<0.01).
Statistic analysis
The ratios among three sets (CBP, exposure and non-exposure) from array analysis were caculated (as log2 value). In the validation stage, the relative expression level of each miRNA in an individual sample was calculated according to the equation 2−△Ct, in which △Ct = Ct miRNA − Ct cel-miR-238. Then the relative quantification value underwent a Log2-transformation to compare the expression levels of candidate miRNAs between groups. Statistical analysis was performed with SPSS software and P value <0.05 was considered statistically significant.
Results
Subject characteristics
A total of 135 participants were enrolled in our study, including 54 non-exposure controls (36 men and 18 women, mean age: 38.0±6.3), 54 healthy exposure subjects (36 man and 18 women, mean age: 38.1±6.5) and 27 cases (18 men and 9 women, mean age: 38.4±5.5), of which 10 participants from each group were selected for the microarray analysis. The detailed information about research subjects was listed in Table 1. In this study, distributions of sex, age, smoking and drinking status did not significantly differ (all P>0.05) among the subjects, benzene-exposed years and benzene-exposed intensity also were not significantly different (all P>0.05) between the exposure and CBP.
Table 1. The basic characteristics of all subjects.
| Sample characteristic | Microarray stage |
Expanded validation stage |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-exposure | Exposure | CBP | P | Non-exposure | Exposure | CBP | P | ||
| Number | 10 | 10 | 10 | − | 54 | 54 | 27 | − | |
| Sex (male/female) | 7/3 | 7/3 | 7/3 | 1.000 | 36/18 | 36/18 | 18/9 | 1.000 | |
| Age (mean ± SD) | 39.0±4.0 | 39.1±4.6 | 39.4±4.5 | 0.978 | 38.0±6.3 | 38.1±6.5 | 38.4±5.5 | 0.955 | |
| Smoking (n, %) | 4 (40.0) | 4 (40.0) | 4 (40.0) | 1.000 | 22 (40.7) | 25 (46.3) | 13 (48.1) | 0.769 | |
| Drinking (n, %) | 2 (20.0) | 2 (20.0) | 2 (20.0) | 1.000 | 20 (37.0) | 22 (40.7) | 12 (44.4) | 0.806 | |
| Exposure years | − | 11.1±4.5 | 11.0±4.5 | 0.961 | − | 11.9±9.9 | 12.3±4.9 | 0.841 | |
| Exposure intensity (mg/m3) | − | 12.1±9.3 | 13.9±6.9 | 0.627 | − | 10.5±9.2 | 14.2±8.1 | 0.076 | |
Global analysis of the plasma microRNA (miRNA) expression profile
The expression levels of 2019 human miRNAs listed in Sanger miRBase Release 19.0 (http://www.sanger.ac.uk/Software/Rfam/mirna/) were tested with a microarray assay. In the discovery stage, 287 and 272 miRNAs were found to be expressed in the plasma pool of the non-exposed subjects and the healthy exposure subjects, respectively, whereas only 236 miRNAs were detected in that of CBP patients. Further analysis revealed that 219 miRNAs were commonly detected in all three groups with the remaining 8, 18 and 30 molecules exclusively expressed in CBP patients, exposures and non-exposures, respectively (Figure 1).
Figure 1.

Global analysis of the plasma miRNA expression profile among the CBP patients, the healthy benzene exposures and the non-exposures. The results indicated that 1,703 of 2,019 miRNAs were not expressed in either research group. 316 miRNAs were detected; 219 miRNAs were commonly detected in three groups, 3, 6 and 32 miRNAs were commonly expressed in only two out of three groups, with the remaining 8, 18 and 30 molecules were exclusively expressed in the CBP patients, the exposures and the non-exposures, respectively. miRNA, microRNA; CBP, chronic benzene poisoning.
Notably, compared with the expression levels in the non-exposure group, 27 miRNAs demonstrated at least a 1-fold higher expression, and 9 miRNAs showed at least a −1-fold lower expression in the CBP group (Table 2), meanwhile four miRNAs were significantly up-regulated, and eight miRNAs were significantly down-regulated in the exposure group (Table 2). In addition, compared with the exposure group, 58 miRNAs (40 up-regulated and 18 down-regulated) were differentially expressed in the CBP patients (Table 2). Based on the miRNA selection criteria (see “Materials and methods”), we selected six significantly differentially expressed miRNAs at the stage of validation. The expression levels and the related functions of six candidate miRNAs are shown in Table 3.
Table 2. Differential expressed miRNAs by microarray (differentially expressed transcripts with fold-change >1 or < −1).
| miRNA | Fold-changea |
|---|---|
| CBP/non-exposure | |
| miR-4707-5p | 1.76 |
| miR-663a | 1.76 |
| miR-6724-5p | 1.62 |
| miR-1237-5p | 1.51 |
| miR-24-3p | 1.50 |
| miR-5095 | 1.40 |
| miR-4467 | 1.31 |
| miR-5001-5p | 1.28 |
| miR-2861 | 1.24 |
| miR-6722-3p | 1.24 |
| miR-144-3p | 1.22 |
| miR-652-3p | 1.17 |
| miR-1587 | 1.17 |
| miR-6125 | 1.15 |
| miR-221-3p | 1.15 |
| miR-4800-3p | 1.11 |
| miR-125b-5p | 1.10 |
| miR-4734 | 1.10 |
| miR-1227-5p | 1.04 |
| miR-4466 | 1.03 |
| miR-1915-3p | 1.03 |
| hsa-miR-5787 | 1.03 |
| miR-4459 | 1.02 |
| miR-192-5p | 1.02 |
| miR-3196 | 1.01 |
| miR-371b-5p | 1.01 |
| miR-6090 | 1.01 |
| miR-379-5p | −1.06 |
| miR-638 | −1.07 |
| miR-199a-3p | −1.11 |
| let-7d-5p | −1.21 |
| let-7e-5p | −1.32 |
| miR-98-5p | −1.34 |
| miR-625-5p | −1.49 |
| miR-122-5p | −2.05 |
| miR-6073 | −3.16 |
| Exposure/non-exposure | |
| hsa-miR-652-3p | 2.30 |
| hsa-miR-30c-1-3p | 1.16 |
| hsa-miR-4301 | 1.13 |
| hsa-miR-638 | 1.01 |
| hsa-miR-221-3p | −1.01 |
| hsa-miR-122-5p | −1.02 |
| hsa-miR-6088 | −1.03 |
| hsa-miR-4632-5p | −1.03 |
| hsa-miR-3162-5p | −1.26 |
| hsa-miR-4281 | −1.31 |
| hsa-miR-4433-3p | −1.33 |
| hsa-miR-6073 | −1.42 |
| CBP/exposure | |
| miR-221-3p | 2.16 |
| miR-6724-5p | 2.08 |
| miR-2861 | 2.05 |
| miR-5095 | 2.04 |
| miR-663a | 2.01 |
| miR-24-3p | 1.87 |
| miR-4632-5p | 1.83 |
| miR-4707-5p | 1.77 |
| miR-6088 | 1.74 |
| miR-4459 | 1.70 |
| miR-6090 | 1.65 |
| miR-5787 | 1.55 |
| miR-4466 | 1.54 |
| miR-3185 | 1.49 |
| miR-5001-5p | 1.48 |
| miR-6125 | 1.45 |
| miR-5096 | 1.43 |
| miR-6089 | 1.42 |
| miR-6722-3p | 1.41 |
| miR-4800-3p | 1.38 |
| miR-1234-5p | 1.34 |
| miR-371b-5p | 1.32 |
| miR-3665 | 1.31 |
| miR-149-3p | 1.30 |
| miR-1227-5p | 1.29 |
| miR-4516 | 1.28 |
| miR-3162-5p | 1.26 |
| miR-4467 | 1.25 |
| miR-194-5p | 1.24 |
| miR-192-5p | 1.22 |
| miR-4486 | 1.20 |
| miR-3656 | 1.20 |
| miR-4433-3p | 1.18 |
| miR-3196 | 1.18 |
| miR-1915-3p | 1.16 |
| miR-4488 | 1.15 |
| miR-502-3p | 1.15 |
| miR-4687-3p | 1.10 |
| miR-376a-5p | 1.08 |
| miR-3940-5p | 1.07 |
| miR-130b-3p | −1.01 |
| miR-744-5p | −1.03 |
| miR-122-5p | −1.03 |
| miR-139-5p | −1.04 |
| miR-199a-3p | −1.04 |
| miR-30e-3p | −1.05 |
| miR-199a-5p | −1.08 |
| miR-361-5p | −1.10 |
| miR-4267 | −1.11 |
| miR-652-3p | −1.13 |
| miR-223-3p | −1.17 |
| miR-4454 | −1.29 |
| miR-625-5p | −1.39 |
| miR-425-3p | −1.41 |
| miR-4301 | −1.47 |
| miR-4286 | −1.57 |
| miR-6073 | −1.74 |
| miR-638 | −2.08 |
a, the fold change value underwent a Log2-transformation. miRNA, microRNA; CBP, chronic benzene poisoning.
Table 3. Expression levels and related functions of the six selected human miRNAsa.
| miRNA | Discovery stage: FCb |
Validation stagec |
Relevant functions | Sources | |||||
|---|---|---|---|---|---|---|---|---|---|
| CBP/Non | CBP/Exp | Exp/Non | CBP/Non | CBP/Exp | Exp/Non | ||||
| let-7d-5p | −1.21 | −0.71 | −0.50 | 0.25 (0.12, 0.55) | 0.30 (0.15, 0.67) | 0.49 (0.15,0.97) | RAS regulation; regulation of iron metabolism in erythroid cells | (19,20) | |
| miR-24-3p | 1.50 | 1.87 | −0.37 | 3.52 (1.85, 5.94) | 3.31 (1.74, 5.59) | 0.69 (0.35, 1.22) | PAHs response; DNA damage response; ARNT regulation | (13) | |
| miR-122-5p | −1.99 | −1.22 | −0.76 | 0.07 (0.03, 0.09) | 0.12 (0.06, 0.16) | 0.32 (0.18, 0.51) | Heme oxygenase-1 regulation | (21) | |
| miR-221-3p | 1.15 | 2.16 | −1.01 | 3.96 (1.77, 8.05) | 11.92 (5.33, 24.23) | 0.23 (0.14, 0.45) | low-level hydroquinone response | (22) | |
| miR-223-3p | −0.83 | −1.17 | 0.35 | 0.52 (0.18, 1.37) | 0.34 (0.11, 0.88) | 0.59 (0.36, 1.77) | Environmental cigarette smoke (ECS) response | (21) | |
| miR-638 | −1.07 | −2.08 | 1.01 | 0.20 (0.13, 0.42) | 0.06 (0.04, 0.12) | 2.20 (0.89, 4.98) | Benzo(a)pyrene response; regulation of differentiation and proliferation in leukemic cells | (23,24) | |
a, Non = non-exposure and Exp = Exposure; b, the fold change value underwent a Log2-transformation; c, values are median (25th percentile, 75th percentile) qRT-PCR expression relative to cel-miR-238. qRT-PCR, quantitative reverse-transcription polymerase chain reaction.
Identification of candidate microRNAs (miRNAs)
For further exploring the possible influence of CBP or benzene exposure on miRNA expression, six miRNAs (let-7d-5p, miR-24-3p, miR-122-5p, miR-221-3p, miR-223-3p and miR-638) were selected for the expanded validation by qRT-PCR in 27 CBP patients, 54 healthy exposure subjects and 54 non-exposure subjects. Among those, ten CBP patients, ten healthy exposure subjects and ten non-exposed subjects were repeated in microarray analysis stage.
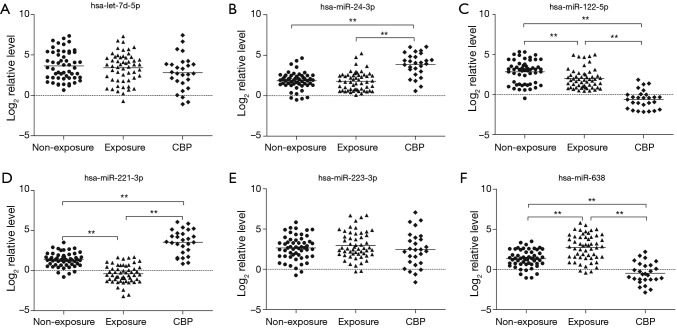
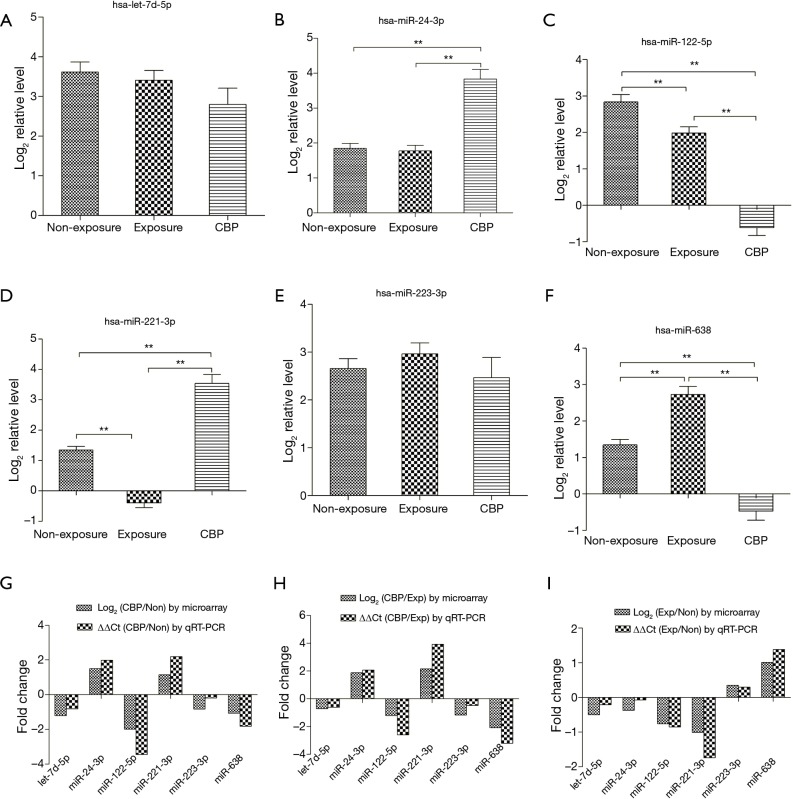
The plasma levels of the six miRNAs are presented in Figures 2 and 3. Compared with the non-exposures and the exposures, miR-24-3p and miR-221-3p were significantly up-regulated (1.99- and 2.06-fold for miR-24-3p, 2.19- and 3.93-fold for miR-221-3p, P<0.01) while miR-122-5p and miR-638 were significantly down-regulated (−3.45- and −2.60-fold for miR-122-5p, −1.82- and −3.20-fold for miR-638, P<0.001) in the CBP patients. And compared with the non-exposures, the plasma level of miR-638 was significantly up-regulated (1.38-fold, P<0.01) while the plasma levels of miR-122-5p and miR-221-3p were significantly down-regulated (−0.85- and −1.74-fold, P<0.01) in the exposures. The validation results of the four miRNAs were in concordance with the microarray analysis (Figure 3).
Figure 2.
The plasma levels of let-7d-5p, miR-24-3p, miR-122-5p, miR-221-3p, miR-223-3p and miR-638 were determined by qRT-PCR individually in the CBP patients (n=27), the healthy exposures (n=54) and the non-exposures (n=54). The expression levels of the six miRNAs were normalized to cel-miR-238 (Log2 relative level). **P<0.01. qRT-PCR, quantitative reverse-transcription polymerase chain reaction; CBP, chronic benzene poisoning.
Figure 3.
Expression of miRNA by qRT-PCR and microarray. (A-F) Relative expression level of differentially expressed miRNAs by qRT-PCR (mean ± SEM); (G-I) comparison of qRT-PCR findings to microarray results by fold-change of eleven-selected miRNAs. Non = non-exposure and exp= exposure. **P<0.01. miRNA, microRNA; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; CBP, chronic benzene poisoning.
Bioinformatic analysis
To further explore the molecular functions of the four miRNAs indicated in CBP, we performed a predicted target analysis. There were 1,140 putative targets for miR-24-3p, 468 for miR-122-5p, 206 for miR-221-3p and 230 for miR-638. GO function enrichment and KEGG pathway analysis indicated that these target genes were involved in 297 enriched biological functions (P<0.01, including regulation of transcription, intracellular signaling cascade, protein amino acid phosphorylation and enzyme linked receptor protein signaling pathway) and 39 signaling transduction pathways (P<0.01, including endocytosis, axon guidance, pathways in cancer and cell adhesion molecules), which were attached as supplementary data.
Discussion
In this study, we identified four indicated miRNAs in plasma that may be potential biomarkers to indicate responses to benzene exposure, which may also provide a new perspective on the pathogenesis of CBP. To date, the mechanism behind benzene poisoning still remains unclear. Benzene has to be metabolized to elicit toxicity to the blood and bone marrow (25), and the benzene poisoning is probably mediated through its metabolites via multiple pathways (26). The proposed hypotheses includs the induction of oxidative stress by reactive oxygen species (ROS) (27), the disruption of cell cycle (28), the DNA damage (29), and the interference in normal cytokine signaling in growth and differentiation of marrow hematopoietic stem cell (30), in which miRNAs were reported to be likely to play crucial roles (5,31). In present study, we first explored the potential effects on occupational benzene exposure and CBP on plasma miRNA expression in Chinese occupational population.
Previous studies have demonstrated that the alteration of miRNAs expression played important roles in cell transformation, particularly in the process of chemical carcinogenesis (32-34). Our microarray analysis and subsequent validation identified four indicated miRNAs in plasma that may affect the responses to benzene exposure, which may also provide a new perspective on the pathogenesis of CBP. And the four miRNAs exhibited distinct patterns in normal subjects and patients. The expression level of miR-24-3p was exclusively elevated in the CBP patients, miR-122-5p progressively decreased from the exposures to the CBP patients, whereas miR-221-3p was first down-regulated in the exposures and then up-regulated in the CBP patients, conversely, the expression of miR-638 was firstly up-regulated in the exposures and then down-regulated in the CBP patients. Actually, previous studies demonstrated that miRNAs may exhibit different functions and expression changes to adapt environmental stress or maintain the state of malignancy (23,35). These results again indicated that miRNA profile implies the distinct mechanisms of chemical which induced biological or pathological consequences (11,36).
The four miRNAs have been reported to regulate genes that could influence the adverse effects of benzene exposures or the progression of CBP. The miR-24-3p can regulate several important target genes which might be mainly involved in oxidative damage regulation. The miR-24-3p can negatively regulate ARNT (aryl hydrocarbon receptor nuclear translocator), and its downstream gene CYP1A1 (cytochrome P450 1A1), which encodes for the main metabolizing enzyme of benzene (37), and thus may reduce the levels of benzene metabolism and oxidative damage to DNA and lipids. In addition, miR-24-3p has been reported to negatively regulate H2AX, which is crucial in double-stranded break repair; thus, reduced expression of this gene might increase cellular sensitivity to DNA-damaging agents and genomic instability (31,38). Furthermore, functional experiments showed that both miR-122 overexpression or siRNA-mediated silencing of its target cyclin G1 determined an increase of p53 transcriptional activity and stability, demonstrating that miR-122 could modulate p53 through the inhibition of cyclin G1 (39), which indicates a cell cycle arrest role for miR-122. Thus, miR-122-5p might play a cell cycle arrest role in DNA-damaging repair to affect the adverse effects of benzene exposures or the progression of CBP. Moreover, it was also noteworthy that miR-122 downregulation was known to induce the expression of heme oxygenase-1 (40), a potent antioxidant activity, that is dramatically up-regulated in the liver of mice transplacentally exposed to environmental cigarette smoke (41).
It has been suggested that miR-221 may be considered as a generalized inhibitor of myelopoiesis by inhibiting molecules involved in myeloid development (42). miR-221 has been reported to be highly expressed in a variety of cancers including leukemia (43), and it has been shown to block the expression of a cell cycle inhibitor and tumor suppressor gene, P27 (44), these observations suggested its oncogenic role. Interestingly, low- level hydroquinone, an important benzene metabolite, could inhibit miR-221 expression and cell apoptosis, while promote cell growth in TK 6 lymphoblastoid cell model (22). Further functional experiments have indicated that miR-221 could regulate PARP-1 inactivation induced by hydroquinone by targeting the 3’ untranslated region (UTR) of MBD2 mRNA (45). miR-638 was down-regulated in the CBP subjects, however, it was up-regulated in the plasma of the benzene exposed subjects. The results were in line with previous findings conducted in BaP-induced human cell transformation experiments (23). Binding to its CDS region, miR-638 could repress the BRCA1 protein level (46), and the functions of BRCA1 were mainly used to repair DNA double-strand breaks and remove bulky DNA adducts through homologous recombination repair and nonhomologous end joining (47,48). The upregulation of miR-638 in PAHs-exposed workers with leading to an inhibition of the cellular repair system, by contrast, the downregulation of miR-638 in BaP-transformed cells implies a chemoresistance role (23). These findings demonstrated that miR-638 might exhibit a dual function in response to environmental stimuli depending upon the state of pathological process. The identification of the miRNA targets that mediate the actions of miRNAs in the CBP pathways can reveal previously unrecognized components of cellular stress responses, which may provide improving diagnosis and treatment options for the clinical conditions (49).
In this study, we used a microarray assay to detect the miRNA expression profiles among pooled plasma samples. Subsequently, we conducted an expanded validation by qRT-PCR. When searching for candidate miRNAs, it would be desirable to select miRNAs whose expression is altered similarly in all individual samples. Theoretically, one should come up with the same candidates when pooling RNA samples from different individuals and hybridizing it on one microarray. Previous studies also revealed that usage of pooled RNA for screening candidates suitable for clinical diagnostic purposes met quality criteria as cited by the MicroArray Quality Control (MAQC) consortium. Moreover, pursuing this RNA pooling approach required unique material, and costs and efforts are reduced about 10-fold (50).
There also existed some limitations in this study. First, our study is a cross-sectional study, and the discovery could not demonstrate a causal association between the indicated miRNAs and benzene exposure. Moreover, we validated only a subset of the miRNAs found in discovery stage, other miRNAs should be further validated.
Conclusions
The microarray analysis and the expanded validation identified the four indicated plasma miRNAs, which may be associated with the responses to benzene exposure or CBP. Further studies are warranted to verify our findings and determine the underlying mechanisms.
Acknowledgements
Funding: This study was funded by Outstanding Medical Academic Leaders program of Jiangsu Province (LJ201130), National Natural Science Foundation of China (81302466).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Khalade A, Jaakkola MS, Pukkala E, et al. Exposure to benzene at work and the risk of leukemia: a systematic review and meta-analysis. Environ Health 2010;9:31. 10.1186/1476-069X-9-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aksoy M. Hematotoxicity and carcinogenicity of benzene. Environ Health Perspect 1989;82:193-7. 10.1289/ehp.8982193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson TJ. A review of the role of benzene metabolites and mechanisms in malignant transformation: summative evidence for a lack of research in nonmyelogenous cancer types. Int J Hyg Environ Health 2009;212:1-10. 10.1016/j.ijheh.2007.09.013 [DOI] [PubMed] [Google Scholar]
- 4.Zolghadr F, Sadeghizadeh M, Amirizadeh N, et al. How benzene and its metabolites affect human marrow derived mesenchymal stem cells. Toxicol Lett 2012;214:145-53. 10.1016/j.toxlet.2012.08.015 [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 6.Caldas C, Brenton JD. Sizing up miRNAs as cancer genes. Nat Med 2005;11:712-4. 10.1038/nm0705-712 [DOI] [PubMed] [Google Scholar]
- 7.Havelange V, Garzon R. MicroRNAs: emerging key regulators of hematopoiesis. Am J Hematol 2010;85:935-42. 10.1002/ajh.21863 [DOI] [PubMed] [Google Scholar]
- 8.Lv M, Zhang X, Jia H, et al. An oncogenic role of miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by targeting glucocorticoid receptor-α and cAMP/PKA pathways. Leukemia 2012;26:769-77. 10.1038/leu.2011.273 [DOI] [PubMed] [Google Scholar]
- 9.Rommer A, Steinleitner K, Hackl H, et al. Overexpression of primary microRNA 221/222 in acute myeloid leukemia. BMC Cancer 2013;13:364. 10.1186/1471-2407-13-364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jardim MJ, Fry RC, Jaspers I, et al. Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ Health Perspect 2009;117:1745-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bollati V, Marinelli B, Apostoli P, et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect 2010;118:763-8. 10.1289/ehp.0901300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Flora S, Balansky R, D'Agostini F, et al. Smoke-induced microRNA and related proteome alterations. Modulation by chemopreventive agents. Int J Cancer 2012;131:2763-73. 10.1002/ijc.27814 [DOI] [PubMed] [Google Scholar]
- 13.Deng Q, Huang S, Zhang X, et al. Plasma microRNA expression and micronuclei frequency in workers exposed to polycyclic aromatic hydrocarbons. Environ Health Perspect 2014;122:719-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng Q, Dai X, Guo H, et al. Polycyclic aromatic hydrocarbons-associated microRNAs and their interactions with the environment: influences on oxidative DNA damage and lipid peroxidation in coke oven workers. Environ Sci Technol 2014;48:4120-8. 10.1021/es4055516 [DOI] [PubMed] [Google Scholar]
- 15.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Zheng Y, Wang G, et al. Identification of microRNA and bioinformatics target gene analysis in beef cattle intramuscular fat and subcutaneous fat. Mol Biosyst 2013;9:2154-62. 10.1039/c3mb70084d [DOI] [PubMed] [Google Scholar]
- 17.Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185-93. 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- 18.Zhu Z, Qi Y, Ge A, et al. Comprehensive characterization of serum microRNA profile in response to the emerging avian influenza A (H7N9) virus infection in humans. Viruses 2014;6:1525-39. 10.3390/v6041525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635-47. 10.1016/j.cell.2005.01.014 [DOI] [PubMed] [Google Scholar]
- 20.Andolfo I, De Falco L, Asci R, et al. Regulation of divalent metal transporter 1 (DMT1) non-IRE isoform by the microRNA Let-7d in erythroid cells. Haematologica 2010;95:1244-52. 10.3324/haematol.2009.020685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izzotti A, Calin GA, Arrigo P, et al. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J 2009;23:806-12. 10.1096/fj.08-121384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu LH, Liang XH, Ling XX, et al. Influences of biological characteristics and the expression of miR-221 induce by low-level hydroquinone exposure in TK6 lymphoblastoid cells. China Occupational Medicine 2011;38:102-5. [Google Scholar]
- 23.Li D, Wang Q, Liu C, et al. Aberrant expression of miR-638 contributes to benzo(a)pyrene-induced human cell transformation. Toxicol Sci 2012;125:382-91. 10.1093/toxsci/kfr299 [DOI] [PubMed] [Google Scholar]
- 24.Lin Y, Li D, Liang Q, et al. miR-638 regulates differentiation and proliferation in leukemic cells by targeting cyclin-dependent kinase 2. J Biol Chem 2015;290:1818-28. 10.1074/jbc.M114.599191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon BI, Hirabayashi Y, Kawasaki Y, et al. Mechanism of action of benzene toxicity: cell cycle suppression in hemopoietic progenitor cells (CFU-GM). Exp Hematol 2001;29:278-85. 10.1016/S0301-472X(00)00671-8 [DOI] [PubMed] [Google Scholar]
- 26.Sarma SN, Kim YJ, Ryu JC. Differential gene expression profiles of human leukemia cell lines exposed to benzene and its metabolites. Environ Toxicol Pharmacol 2011;32:285-95. 10.1016/j.etap.2011.06.001 [DOI] [PubMed] [Google Scholar]
- 27.Snyder R, Hedli CC. An overview of benzene metabolism. Environ Health Perspect 1996;104 Suppl 6:1165-71. 10.1289/ehp.961041165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irons RD, Neptun DA. Effects of the principal hydroxy-metabolites of benzene on microtubule polymerization. Arch Toxicol 1980;45:297-305. 10.1007/BF00293810 [DOI] [PubMed] [Google Scholar]
- 29.Lévay G, Ross D, Bodell WJ. Peroxidase activation of hydroquinone results in the formation of DNA adducts in HL-60 cells, mouse bone marrow macrophages and human bone marrow. Carcinogenesis 1993;14:2329-34. 10.1093/carcin/14.11.2329 [DOI] [PubMed] [Google Scholar]
- 30.Renz JF, Kalf GF. Role for interleukin-1 (IL-1) in benzene-induced hematotoxicity: inhibition of conversion of pre-IL-1 alpha to mature cytokine in murine macrophages by hydroquinone and prevention of benzene-induced hematotoxicity in mice by IL-1 alpha. Blood 1991;78:938-44. [PubMed] [Google Scholar]
- 31.Wang Y, Taniguchi T. MicroRNAs and DNA damage response: implications for cancer therapy. Cell Cycle 2013;12:32-42. 10.4161/cc.23051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen T. The role of MicroRNA in chemical carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 2010;28:89-124. 10.1080/10590501.2010.481477 [DOI] [PubMed] [Google Scholar]
- 33.Gao SM, Chen C, Wu J, et al. Synergistic apoptosis induction in leukemic cells by miR-15a/16-1 and arsenic trioxide. Biochem Biophys Res Commun 2010;403:203-8. 10.1016/j.bbrc.2010.10.137 [DOI] [PubMed] [Google Scholar]
- 34.Liu L, Jiang Y, Zhang H, et al. Overexpressed miR-494 down-regulates PTEN gene expression in cells transformed by anti-benzo(a)pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Life Sci 2010;86:192-8. 10.1016/j.lfs.2009.12.002 [DOI] [PubMed] [Google Scholar]
- 35.Mascaux C, Laes JF, Anthoine G, et al. Evolution of microRNA expression during human bronchial squamous carcinogenesis. Eur Respir J 2009;33:352-9. 10.1183/09031936.00084108 [DOI] [PubMed] [Google Scholar]
- 36.Rager JE, Smeester L, Jaspers I, et al. Epigenetic changes induced by air toxics: formaldehyde exposure alters miRNA expression profiles in human lung cells. Environ Health Perspect 2011;119:494-500. 10.1289/ehp.1002614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oda Y, Nakajima M, Mohri T, et al. Aryl hydrocarbon receptor nuclear translocator in human liver is regulated by miR-24. Toxicol Appl Pharmacol 2012;260:222-31. 10.1016/j.taap.2012.02.012 [DOI] [PubMed] [Google Scholar]
- 38.Lal A, Pan Y, Navarro F, et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol 2009;16:492-8. 10.1038/nsmb.1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fornari F, Gramantieri L, Giovannini C, et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res 2009;69:5761-7. 10.1158/0008-5472.CAN-08-4797 [DOI] [PubMed] [Google Scholar]
- 40.Shan Y, Zheng J, Lambrecht RW, et al. Reciprocal effects of micro-RNA-122 on expression of heme oxygenase-1 and hepatitis C virus genes in human hepatocytes. Gastroenterology 2007;133:1166-74. 10.1053/j.gastro.2007.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izzotti A, Balansky RM, Cartiglia C, et al. Genomic and transcriptional alterations in mouse fetus liver after transplacental exposure to cigarette smoke. FASEB J 2003;17:1127-9. [DOI] [PubMed] [Google Scholar]
- 42.Georgantas RW, 3rd, Hildreth R, Morisot S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A 2007;104:2750-5. 10.1073/pnas.0610983104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cammarata G, Augugliaro L, Salemi D, et al. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am J Hematol 2010;85:331-9. [DOI] [PubMed] [Google Scholar]
- 44.le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J 2007;26:3699-708. 10.1038/sj.emboj.7601790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu LH, Liang XX, Liang HR, et al. Hydroquinone Induced Down-regulated Expression of PARP-1 and miR-221 in TK6 cells. Journal of Environment and Health 2011;28:485-7. [Google Scholar]
- 46.Nicoloso MS, Sun H, Spizzo R, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res 2010;70:2789-98. 10.1158/0008-5472.CAN-09-3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Page F, Randrianarison V, Marot D, et al. BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesion in human cells. Cancer Res 2000;60:5548-52. [PubMed] [Google Scholar]
- 48.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res 2008;18:134-47. 10.1038/cr.2007.111 [DOI] [PubMed] [Google Scholar]
- 49.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012;148:1172-87. 10.1016/j.cell.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Port M, Seidl C, Ruf CG, et al. Reliable and sample saving gene expression analysis approach for diagnostic tool development. Health Phys 2012;103:159-68. [DOI] [PubMed] [Google Scholar]