Abstract
We studied the kinetics and structural determinants of closed-state inactivation (CSI) in Kv4.2 channels, considering a multistep process and the possibility that both intra- and intersubunit dynamic binding (i.e., loss and restoration of physical contact) may occur between the S4-S5 linker, including the initial S5 segment (S4S5), and the S6 gate. We expressed Kv4.2 channels in Xenopus oocytes and measured the onset of low-voltage inactivation under two-electrode voltage clamp. Indicative of a transitory state, the onset kinetics were best described by a double-exponential function. To examine the involvement of individual S4S5 and S6 amino acid residues in dynamic binding, we studied S4S5 and S6 single alanine mutants and corresponding double mutants. Both transitory and steady-state inactivation were modified by these mutations, and we quantified the mutational effects based on apparent affinities for the respective inactivated states. Double-mutant cycle analyses revealed strong functional coupling of the S6 residues V404 and I412 to all tested S4S5 residues. To examine whether dynamic S4S5/S6 binding occurs within individual α-subunits or between neighboring α-subunits, we performed a double-mutant cycle analysis with Kv4.2 tandem-dimer constructs. The constructs carried either an S4S5/S6 double mutation in the first α-subunit and no mutation in the second (concatenated) α-subunit or an S4S5 point mutation in the first α-subunit and an S6 point mutation in the second α-subunit. Our results support the notion that CSI in Kv4.2 channels is a multistep process that involves dynamic binding both within individual α-subunits and between neighboring α-subunits.
Introduction
Voltage-gated potassium (Kv) channels are key players in the fine-tuning of nerve and muscle excitation (1). They are activated when the membrane is depolarized, and the resultant potassium efflux counteracts depolarization or supports repolarization. However, sustained membrane depolarization may also drive Kv channels into a nonconducting refractory state called inactivation (2). This inactivation may require channel opening or may occur from preopen closed states, and some Kv channels exhibit both open- and closed-state inactivation (CSI) in parallel, with different weights and kinetics (3).
The initial cloning of potassium channel genes from the fruit fly Drosophila melanogaster identified a group of Kv channels (Shaker, Shab, Shaw, and Shal) with high structural homology and high conservation across species (4). Notably, Shaker, the first potassium channel gene to be cloned (5), mediates rapidly inactivating, so-called A-type currents (6). Therefore, using the newly available cloned potassium channel DNA, researchers subjected not only the mechanism of voltage sensing and voltage-dependent activation but also the mechanism(s) of inactivation to molecular investigations early on. It turned out that Shaker inactivation mainly occurs from the open state and follows two different mechanisms. The channel can be inactivated by an N-terminal inactivation domain, which plugs the open pore from the inside (N-type inactivation) (7, 8), and by a conformational change of the external pore vestibule (P/C-type inactivation) (9, 10). Members of the Kv4 subfamily, which are related to the Shal gene of Drosophila, also mediate prominent A-type currents that are hardly distinguishable from the ones mediated by Shaker. However, the mechanism of Kv4 channel inactivation differs substantially from the classical Shaker N- and P/C-type inactivation. Although these mechanisms are detectable in Kv4 channels in vestigial form (11, 12), Kv4 channels exhibit preferential CSI (13). In fact, Kv4 channels accumulate in closed-inactivated states at all relevant membrane potentials, which is why they have become an attractive model for demonstrating and studying CSI in Kv channels (14).
In contrast to N- and P/C-type inactivation, the mechanism of CSI has remained elusive for a long time. More recently, however, gating-current measurements have shown that conformational changes of the voltage sensor must be tightly associated with conformational changes during CSI in Kv4.2 channels (15). Furthermore, scanning mutagenesis along the S4-S5 linker, including the initial S5 segment (S4S5), and the distal S6 segment (S6) combined with double-mutant cycle analysis has suggested that dynamic binding between the voltage sensor and the activation gate is involved in Kv4.2 channel CSI (16). It is now thought that movement of the voltage sensor is accompanied by its temporary unbinding from the activation gate, leaving Kv4.2 channels in a closed and inactivated state (14). The proposed structural model of dynamic binding implies that CSI and Kv channel activation share molecular determinants (14, 16, 17, 18, 19, 20). Consequently, the main interaction partners that are critically involved in the dynamic binding during Kv4.2 channel CSI are found at sites homologous to the ones identified for Shaker channel activation (16, 21) (see also Fig. 11 A and Discussion). The process of voltage-dependent activation in potassium channels has been kinetically studied in great detail, and early experiments have shown that all four voltage sensors must move before the pore opens in a single cooperative step (24, 25). From a structural point of view, cooperativity implies that neighboring α-subunits must communicate during gating, and, in fact, structural correlates of such intersubunit communication during Kv channel activation have been identified (26) (see Discussion). However, previous studies of dynamic binding in CSI did not consider transitory states or intersubunit communication via the identified functional coupling sites (16). We conducted this study to address these issues. We performed a detailed kinetic and steady-state analysis of low-voltage inactivation (at −65 mV) for wild-type (wt) and mutant Kv4.2 channels. In particular, we used Kv4.2 tandem-dimer constructs with point mutations located in either the same or different α-subunits. Our results support the notion that CSI in Kv4.2 channels is a multistep process that involves dynamic binding both within individual α-subunits and between neighboring α-subunits.
Figure 11.
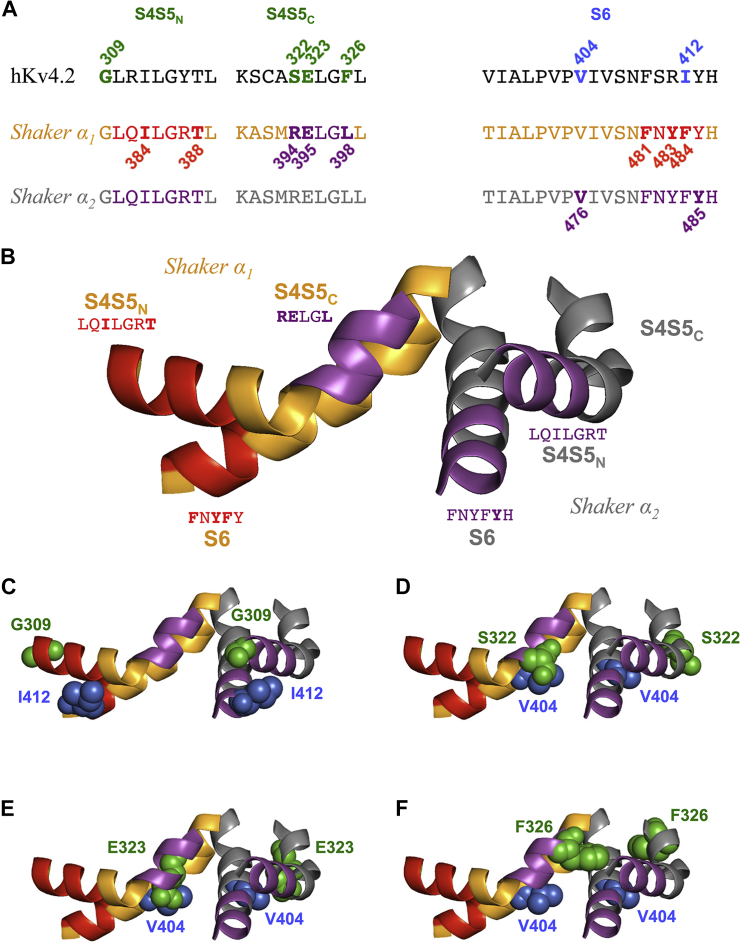
S4S5 and S6 domains and amino acids involved in electromechanical coupling in Kv4.2 and Shaker channels. (A) The previously studied S4S5N, S4S5C, and S6 domains in human Kv4.2 (16) are aligned with the corresponding Shaker domains (Shaker α1 (orange) and Shaker α2 (gray) represent two neighboring α-subunits). The numbered Kv4.2 residues in S4S5 (green) and S6 (blue) were tested for intra- and/or intersubunit interactions in this study. In Shaker, the S4S5N residues I384 and T388 are thought to insert into a hydrophobic pocket formed by the S6 residues F481, Y483, and F484 of the same α-subunit (22). The S4S5C residues R394, E395, and L398 of Shaker α1, and the S6 residue Y485 of Shaker α2 are involved in the RELY intersubunit interaction under the control of V476 (23). (B) S4S5 and S6 of two neighboring α-subunits in the Kv1.2-2.1 chimera crystal structure (19) illustrate putative intra- and intersubunit interactions in the Shaker channel. Domains involved in intrasubunit interactions between S4S5N and S6 in Shaker α1 are shown in red. The S6 domain of Shaker α2 is nestled between S4S5N of Shaker α2 (intrasubunit interaction) and S4S5C of Shaker α1 (intersubunit interaction, magenta). Amino acid sequences and color coding correspond to (A). (C–F) Homology modeling of the Kv4.2 S4S5 (green) and S6 (blue) residues tested for intra- and/or intersubunit interactions in this study. The relevant portions of two neighboring α-subunits are shown, and the ribbon representation is colored according to (B). Kv4.2 coupling pairs: G309/I412 (C), S322/V404 (D), E323/V404 (E), and F326/V404 (F). The Kv4.2 residues are modeled on both α-subunits. They are shown in space-filling representation and the color coding corresponds to (A).
Materials and Methods
Channel constructs
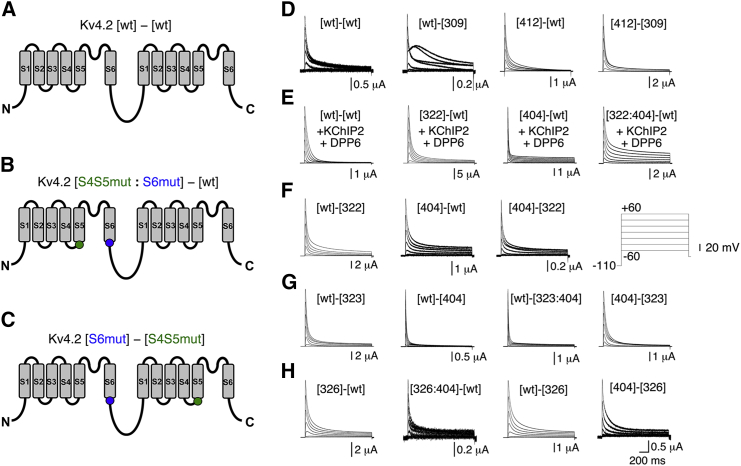
All constructs used in this study are based on the human Kv4.2 clone (27) in a pGEM-HE bacterial expression vector. Mutations were introduced in the S4-S5 linker, including the initial part of S5 (hereafter referred to as S4S5; S4S5N refers to the actual linker helix between the voltage sensor and pore module, and S4S5C refers to the initial S5 segment), in the distal S6 segment (hereafter referred to as S6; see Fig. 1 A), or in both. In particular, we focused on amino acid residues, the alanine mutation of which had resulted in ln (Kci/Kci,wt) magnitudes (see below) of >1 in a previous study (16). Thus, in this study, we reused the following previously characterized Kv4.2 constructs: the S4S5 single mutants G309A, R311A, L313A, S322A, E323A, and F326A; the S6 single mutants L400A, V404A, S407A, N408A, and I412A; and the double mutants S322A:V404A, S322A:S407A, S322A:N408A, S322A:I412A, E323A:V404A, E323A:S407A, E323A:N408A, and E323A:I412A (16). We extended our previous mutational analysis by creating the double mutants G309A:V404A, G309A:S407A, G309A:N408A, G309A:I412A, R311A:L400A, R311A:V404A, R311A:S407A, R311A:N408A, R311A:I412A, L313A:L400A, L313A:V404A, L313A:S407A, L313A:N408A, L313A:I412A, S322A:L400A, E323A:L400A, F326A:L400A, F326A:V404A, F326:S407A, F326:N408A, and F326A:I412A. All point mutations were introduced with overlap PCR using Pfu-Turbo DNA polymerase (Fermentas) and verified by automated DNA sequencing. After site-directed mutagenesis was completed, we generated Kv4.2 tandem dimers for selected pairs of amino acids that carried either a double-point mutation and no mutation in the first and second of two concatenated α-subunits, respectively (see Fig. 7 B), or a single-point mutation in each of two concatenated α-subunits (see Fig. 7 C). Furthermore, we created tandem dimers with a single-point mutation in only one of the two concatenated α-subunits and tandem dimers consisting of two concatenated wt α-subunits (see Fig. 7 A). The cloning strategy for the dimer construction is illustrated in Fig. S1 in the Supporting Material. Two protomer plasmids were designed to have a unique six-base restriction site (for ClaI or EcoRV) in addition to the endogenous SpeI site. In protomers 1 and 2, six bases (including the stop codon for the former and the start codon for the latter) were mutated to a ClaІ restriction site (ATCGAT). Alternatively, in some constructs, unique EcoRV restriction sites (GATATC) were introduced in protomers 1 and 2 at these positions. To obtain concatemeric cDNAs, the 5′ end of the protomer 2 coding sequence had to be linked to the 3′ end of the protomer 1 coding sequence. To achieve this, the protomers were first double-digested with ClaІ and SpeІ (New England BioLabs), followed by purification and extraction on agarose (gel extraction kit; QIAGEN). The ClaІ/SpeІ fragment of protomer 2 (containing the Kv4.2 coding sequence 2 with a stop codon, but no start codon) was ligated into the protomer 1 fragment (containing the Kv4.2 coding sequence 1 with a functional start codon, but no stop codon) using the LigaFast Rapid DNA ligation system (Promega). The ligation reaction product was used to transform JM109-competent cells (Promega), and colonies were selected and screened the next day. QIAGEN prep kits were used to extract DNA. For RNA synthesis, Kv4.2 cDNA was linearized using NotІ (New England Biolabs), and in vitro transcription was performed with the RiboMaxLargeScale RNA production system T7 (Promega). Since individual amino acid exchanges were always made in Kv4.2 and always to alanine, we will use a simplified nomenclature for the constructs used in this study. Mutants (and mutations) will only be named by the respective numbers of the mutated amino acid residue (see Fig. 1 A) in square brackets, with dimers hyphenated (e.g., [323] or [404] for monomers and [wt]-[323] or [404]-[323] for dimers). Two mutations in one α-subunit will be denoted by a colon between the respective numbers (e.g., [323:404] for a monomer and [wt]-[323:404] for a dimer). We failed to clone the constructs [309:400], [309:412]-[wt], and [309]-[wt].
Figure 1.
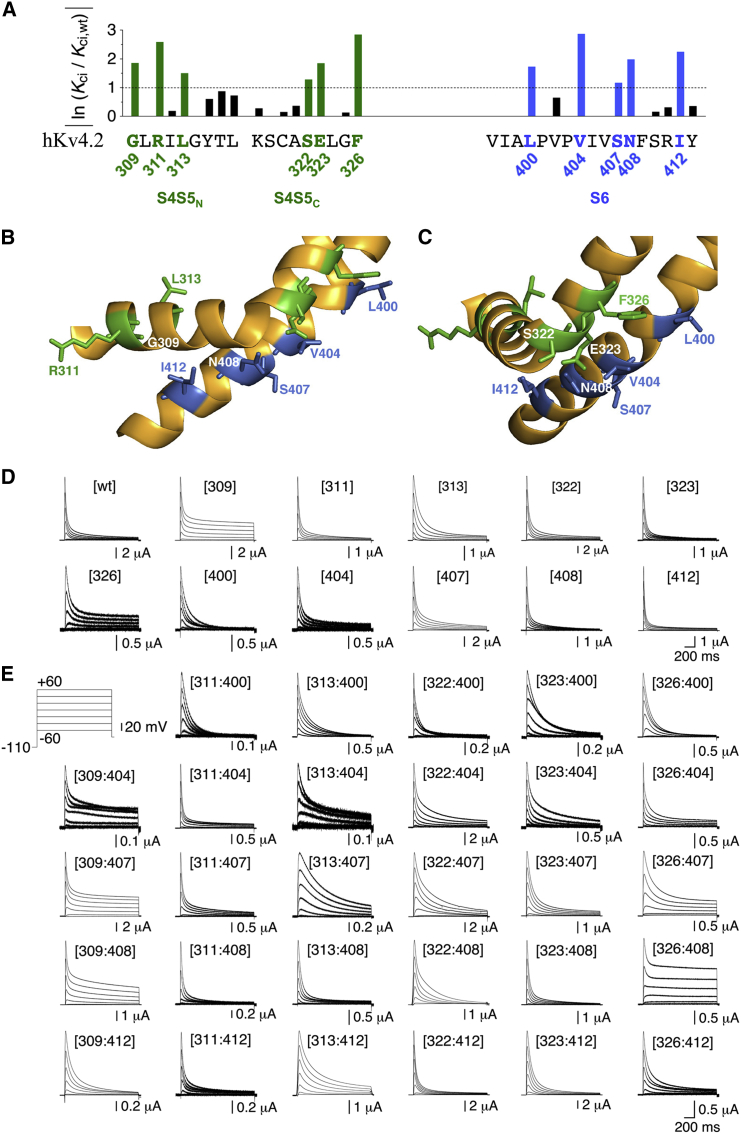
Mutational analysis of S4S5/S6 dynamic binding in Kv4.2 channel CSI with monomeric constructs. (A) Effects of mutating individual S4S5 (left) and S6 (right) residues in Kv4.2 to alanine. The effects refer to low-voltage inactivation as a measure of CSI and are quantified as ln (Kci/Kci,wt) magnitude (Kci refers to a single closed-inactivated equilibrium constant (data based on Barghaan and Bähring (16); see Materials and Methods). The human Kv4.2 S4S5 and S6 native amino acid sequences in single letter code represent the x axis of the bar graph. S4S5 is subdivided in N-terminal (S4S5N, S4-S5 linker helix) and C-terminal (S4S5C, initial S5 segment) portions. An ln (Kci/Kci,wt) magnitude of >1 (dotted line) was arbitrarily chosen as a cutoff value for a significant effect (16). Significant effects on CSI when mutated to alanine were previously obtained for G309, R311, L313, S322, E323, and F326 in S4S5 (green), and for L400, V404, S407, N408, and I412 in S6 (blue) (16). (B and C) Structure of the S4S5/S6 interface in an individual α-subunit. Kv4.2 S4S5 and S6 homology modeling is based on the crystal structure of the Kv1.2-2.1 paddle chimera (19) (green, S4S5 residues; blue, S6 residues). In (B) the S4S5N (harboring G309, R311, and L313) is shown from the side. In (C) the S4S5/S6 interface is rotated by 90°, showing S4S5N from the front and offering a better view of the S4S5C residues S322, E323, and F326. In S6 the residues L400, V404, S407, N408, and I412 are shown. (D) Current families obtained under two-electrode voltage clamp from oocytes expressing monomeric Kv4.2 wt, S4S5 single mutants, and S6 single mutants. (E) Currents mediated by monomeric Kv4.2 S4S5/S6 double mutants (inset: voltage protocol; only test pulses between −60 and +60 mV in 20 mV increments are shown).
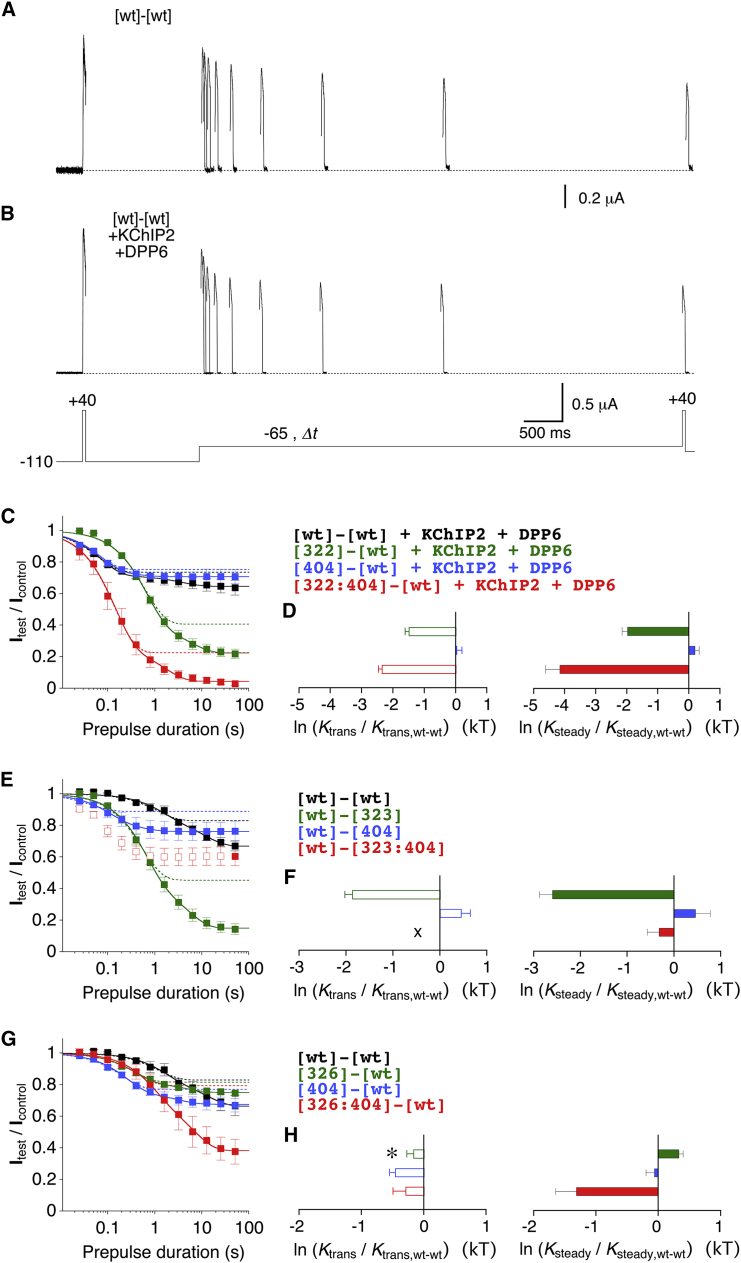
Figure 7.
Kv4.2 tandem-dimer constructs. (A–C) Membrane topology cartoons illustrating different types of the generated tandem dimers. (A) The wt dimer. (B) The construct has a double-point mutation in the first α-subunit and no mutation in the second (concatenated) α-subunit (intrasubunit configuration). (C) The construct has an S6 point mutation in the first α-subunit and an S4S5 point mutation in the second α-subunit (intersubunit configuration). Individual point mutations in S4S5 and S6 are depicted by green and blue dots, respectively. (D–H) A-type currents mediated by the different dimer constructs were generated to test for intra- and intersubunit interactions of the coupling pairs G309/I412, S322/V404, E323/V404, and F326/V404 (inset: voltage protocol; only test pulses between −60 and +60 mV in 20 mV increments are shown). (D) Currents mediated by constructs generated to test for intersubunit interactions of the G309/I412 coupling pair. (E) The constructs generated to test for intrasubunit interactions of the S322/V404 coupling pair were coexpressed with KChIP2 and DPP6 (see Materials and Methods). (F) Currents mediated by constructs generated to test for intersubunit interactions of the S322/V404 coupling pair. (G) Currents mediated by constructs generated to test for intra- and intersubunit interactions of the E323/V404 coupling pair. (H) Currents mediated by constructs generated to test for intra- and intersubunit interactions of the F326/V404 coupling pair.
Heterologous channel expression
Kv4.2 channels were expressed in Xenopus laevis oocytes as previously described (16). Female frogs were anesthetized in ethyl 3-aminobenzoate methanesulfonate (Sigma; 1.2 g/L of tap water), and part of the ovary lobes was surgically removed. The tissue was digested for 3–5 h in a calcium-free solution containing (in mM) 82.5 NaCl, 2 KCl, 1 MgCl2, 5 HEPES, and 1.3 mg/mL collagenase type II (Biochrom), pH 7.5, with NaOH. Defolliculated stage V–VI oocytes were selected the next day, and 50 nL of aqueous cRNA solution was injected per oocyte using a Nanoliter 2000 microinjector (World Precision Instruments). For the purposes of reproducibility and comparability, we expressed most Kv4.2 constructs in the absence of auxiliary subunits, as in a previous study (16), knowing that native Kv4.2 channels usually form ternary complexes with Kv channel interacting proteins (KChIPs) and dipeptidyl aminopeptidase-like proteins (DPPs). Notably, the ternary complexes show modified gating (28), including a KChIP-mediated suppression of vestigial N-type inactivation (11). For expression of Kv4.2 monomers and tandem dimers, 5 ng and 50 ng cRNA, respectively, was injected per oocyte. Some Kv4.2 tandem dimers were expressed together with KChIP2 (human 2b splice variant) (29) and DPP6 (human 6-S splice variant) (30) to enhance the surface expression of channel protein (28). The Kv4.2 tandem dimer, KChIP2, and DPP6 cRNAs were injected at a mass ratio of 1:10:10 (50 ng total cRNA). The dimer construct [323]-[wt] showed no functional expression in the absence and presence of auxiliary subunits. Injected oocytes were incubated at 16°C in a solution containing (in mM) 75 NaCl, 5 Na-pyruvate, 2 KCl, 2 CaCl2, 1 MgCl2, 5 HEPES, and 50 μg/mL gentamicin (Sigma), pH 7.5, with NaOH, and used for recordings after 1–4 days.
Recording technique and pulse protocols
Currents were recorded at room temperature under two-electrode voltage clamp using a TurboTec-10CX amplifier (npi Electronic Instruments, Tamm, Germany) controlled by PULSE software (HEKA). Glass microelectrodes, tip-sealed with agar (2% in 3 M KCl) and filled with 3 M KCl, had bath resistances between 500 and 800 kΩ. Oocytes were superfused with a solution containing (in mM) 91 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, and 5 HEPES, pH 7.4, with NaOH. The holding voltage was −80 mV. For all constructs, the voltage dependencies of peak conductance activation and isochronal inactivation were examined. For this purpose, depolarizing voltage jumps of different amplitudes (test potentials between −90 and +70 mV in 10 mV increments) were applied from a prepulse voltage of −110 mV (see Figs. 1 and 7), or different prepulse voltages (between −120 and 0 mV in 5 mV increments) were applied for 30 s before a +40 mV test pulse (not shown), respectively. To assess CSI, the onset of low-voltage inactivation was measured via a protocol in which a prepulse voltage of −65 mV was applied for variable durations (between 25 ms and 51.2 s) before a +40 mV test pulse. The prepulse to −65 mV was preceded by a control pulse from −110 to +40 mV and a 1.5 s recovery period at −110 mV (see Figs. 2 and 8). Capacitive current transients were not compensated, and a P/5 routine was used for leak subtraction.
Figure 2.
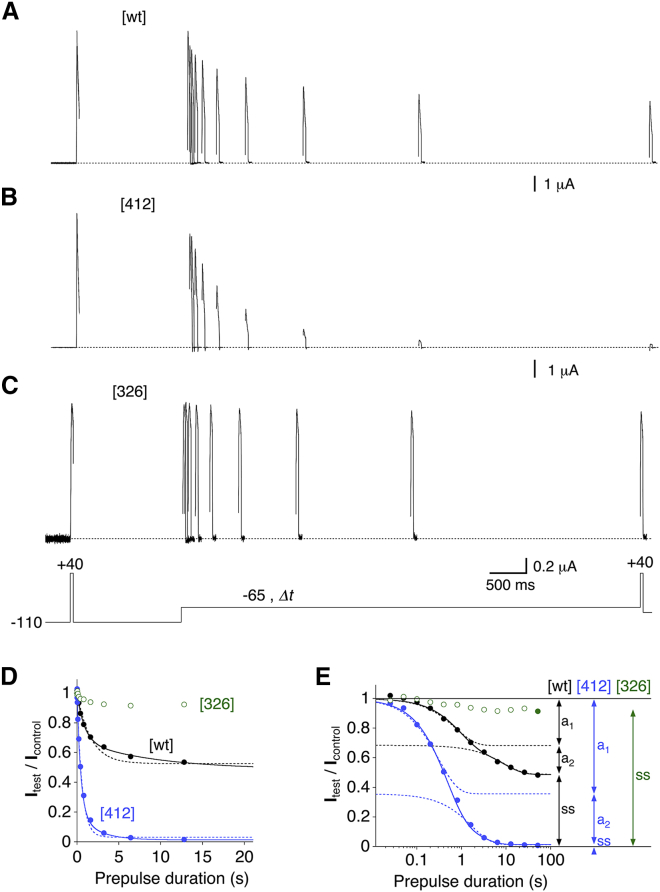
Onset of low-voltage inactivation as a measure of CSI in Kv4.2 channels. Inactivation was induced by a prepulse protocol (control pulse and test pulse to +40 mV before and after, respectively, a −65 mV conditioning prepulse of variable duration, Δt; residual inactivation was removed by voltage steps to −110 mV). (A–C) Currents obtained with the prepulse protocol (only prepulses from 25 ms to 6.4 s are shown) from oocytes expressing Kv4.2 wt ([wt], A), the S6 single mutant [412] (B), and the S4S5 single mutant [326] (C). Dotted lines represent zero current. (D) Relative current amplitudes (Itest/Icontrol) obtained after the conditioning prepulse was plotted against the prepulse duration and the data were subjected to exponential fitting (except for [326]; see Materials and Methods; individual oocyte data from the recordings shown in (A)–(C), including the data points obtained with a 12.8 s prepulse). Dotted lines represent single-exponential fits, and continuous lines represent double-exponential fits to the data points. (E) Individual oocyte data from (A)–(C), including all prepulse durations (25 ms to 51.2 s) were plotted on a semi-log scale and fitted by a double-exponential function for [wt] (black) and [412] (blue, continuous lines). Dotted lines represent the individual components of the double-exponential fits, illustrating the amplitudes of τ1 (a1) and τ2 (a2). The fit yields the steady-state noninactivating current fraction (ss) and a1; all other parameters are calculated (see Materials and Methods). Based on a1, a transitory noninactivating current fraction (1 − a1) was determined. Double-exponential fitting was not possible for the [326] data, and ss was determined from the data point obtained after the longest prepulse (solid green symbol).
Figure 8.
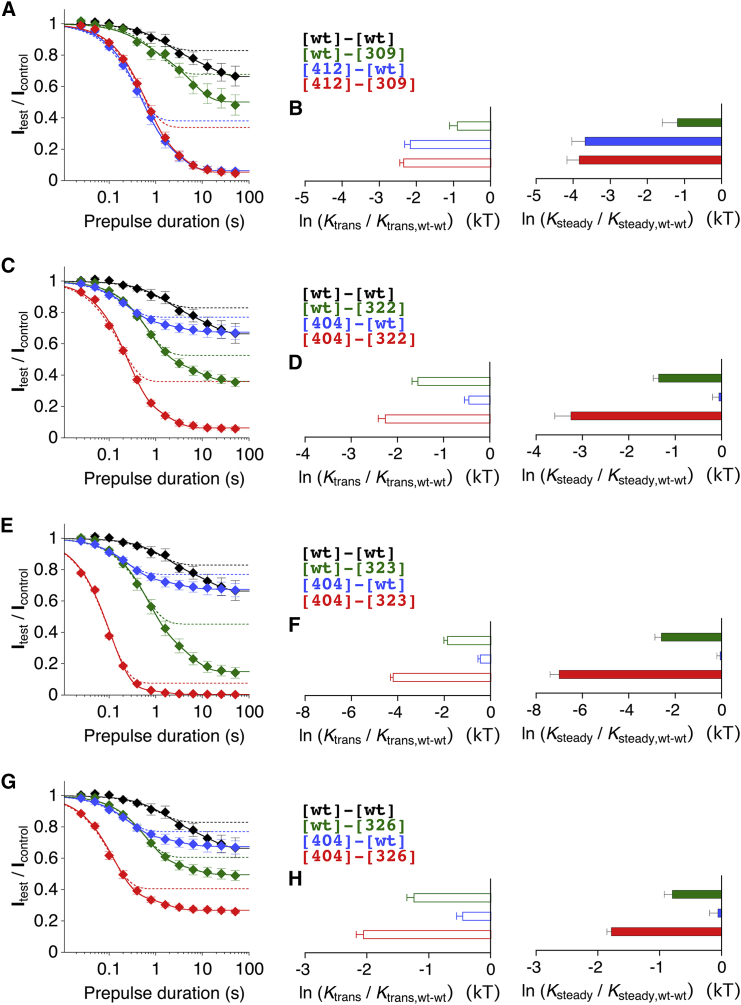
Onset of low-voltage inactivation and analysis of intrasubunit dynamic binding in Kv4.2 tandem-dimer constructs. (A and B) Currents obtained with the prepulse protocol (only prepulses from 25 ms to 6.4 s are shown) from oocytes expressing the Kv4.2 wt tandem dimer ([wt]-[wt]) in the absence (A) and presence (B) of auxiliary subunits (KChIP2 and DPP6). Dotted lines represent zero current. (C, E, and G) Low-voltage inactivation analyses for the tandem-dimer constructs designed to probe intrasubunit S4S5/S6 dynamic binding. Relative current amplitudes (Itest/Icontrol) were plotted against prepulse duration on a semi-log scale and the data were subjected to exponential fitting (continuous lines, double-exponential fits leveling off at ss; dotted lines, initial component indicating the transitory noninactivating current fraction 1 − a1). Black squares, [wt]-[wt]; green squares, single S4S5 mutant dimers; blue squares, single S6 mutant dimers; red squares, intrasubunit double-mutant dimers. (D, F, and H) Mutation-induced changes in apparent affinity for a transitory state (left, open bars) and steady-state inactivation (right, solid bars) expressed as ln (Ktrans/Ktrans,wt-wt) and ln (Ksteady/Ksteady,wt-wt), respectively (green, dimers with a single S4S5 mutation; blue, dimers with a single S6 mutation; red, dimers with a double mutation in one of the two concatenated α-subunits). Separate graphs show data for the coupling pairs S322/V404 (D), E323/V404 (F), and F326/V404 (H), respectively. Note that, similarly to the monomeric constructs, the mutation-induced effects on Ktrans and Ksteady are qualitatively identical (except for [326]-[wt], asterisk). X: double-exponential fitting was not possible, and therefore Ktrans data are not available for [wt]-[323:404] (see Materials and Methods).
Data analysis
Data were analyzed with the use of PULSEFIT (HEKA), Kaleidagraph (Synergy Software), Excel (Microsoft), and Prism (GraphPad Software). The voltage dependencies of activation and isochronal inactivation were analyzed using fourth- and first-order Boltzmann-functions, respectively (see Table S1 for details). The onset kinetics of inactivation at −65 mV were described by a double-exponential function of the form Itest/Icontrol = ss + a1 × exp (− t/τ1) + (1 − ss − a1) × exp (− t/τ2), where Icontrol and Itest are respectively the peak current amplitudes before and after a conditioning pulse to −65 mV of duration t. The equation yielded the low-voltage inactivation time constants τ1 and τ2, the amplitude of τ1 (a1), and the steady-state noninactivating current fraction ss. The amplitude of τ2 (a2) and the corresponding percentage of the total decay (i.e., to reach ss) were calculated from a1 and ss, which are fractions of one (Fig. 2 E and Table S2). Further data analysis was largely adapted from a previous study (16). From the noninactivating current fractions 1 − a1 and ss, the apparent affinities for the transitory state and steady-state inactivation (expressed as equilibrium constants Ktrans and Ksteady, respectively) were calculated. The calculations were based on the following definitions: equilibrium constant K = reverse rate/forward rate; reverse rate = fraction/τ; and forward rate = 1/τ − reverse rate. From this, it follows that Ktrans = (1 − a1)/a1 and Ksteady = ss/(1 − ss) (Fig. 3). For some constructs (usually with very little inactivation at −65 mV), the onset could not be fitted with a double-exponential function. In these cases, 1 − a1 (and Ktrans) could not be obtained, and ss was determined based on the relative current obtained after the longest prepulse (51.2 s; for instance, see [326] in Fig. 2). Note that the equilibrium constants K used here do not reflect the binding affinity of protein domains, but rather the apparent affinity of the channel for closed-inactivated states (see Discussion). Large and small K values denote low and high likelihoods, respectively, for entering the respective inactivated state. In experiments that produced complete inactivation at −65 mV, ss was arbitrarily set to 10−6. Mutation-induced deviations from wt affinity were quantified as ln (Ktrans/Ktrans,wt) and ln (Ksteady/Ksteady,wt) (16). Negative and positive ln values respectively denote an increase and decrease in the affinity for (i.e., the likelihood of entering) the respective inactivated state relative to the wt. For the analysis of the tandem-dimer data, the wt dimer ([wt]-[wt]) was used as a reference. To quantify the nonadditivity of two individual point mutations (indicative of the degree of functional coupling between the native amino acid residues), separate double-mutant cycle analyses (16) were performed based on Ktrans and Ksteady values, respectively, for both monomeric and tandem-dimer constructs. The coupling coefficients for pairs of mutations were calculated as follows: Ωtrans = (Ktrans,wt:wt × Ktrans,S4S5mut:S6mut)/(Ktrans,S4S5mut:wt × Ktrans,wt:S6mut), and Ωsteady = (Ksteady,wt:wt × Ksteady,S4S5mut:S6mut)/(Ksteady,S4S5mut:wt × Ksteady,wt:S6mut), where wt:wt denotes the Kv4.2 wt (or the wt dimer), and S4S5mut:S6mut denotes the double mutant. In a tandem-dimer construct, the two mutations may be located in the same α-subunit (intrasubunit configuration; Fig. 7 B) or in different α-subunits (intersubunit configuration; see Fig. 7 C). S4S5:wt and wt:S6 denote the single mutants with a point mutation in S4S5 or S6, respectively (corresponding tandem dimers carry a mutation in only one of the two concatenated α-subunits). We assumed linear propagation of independent errors through the equations used to calculate Ω. For Ω < 1, the reciprocal was used, and all values are plotted as lnΩ. A Ω value of 1 (lnΩ = 0) means that the two native amino acid residues under study are not functionally coupled (i.e., they are not bound to each other or their binding status does not change) in the respective CSI gating transition. Pooled data are presented as mean ± SEM unless stated otherwise. We examined the interdependence of two measured parameters within a data pool by performing a Pearson correlation analysis (see Figs. S2 and S3). To illustrate the orientation of side chains of the amino acids mutated in this study, we performed structural homology modeling based on the crystal structure coordinates of the Kv1.2-2.1 paddle chimera (PDB accession number 2R9R) (19) using PyMol software (Schrödinger).
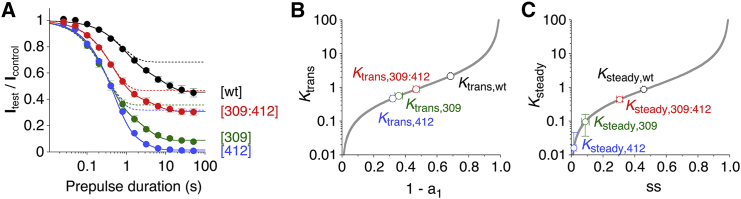
Figure 3.
Onset of low-voltage inactivation and calculation of transitory and steady-state equilibrium constants. (A) Relative current amplitudes (Itest/Icontrol) plotted against prepulse duration on a semi-log scale. Pooled data are shown for Kv4.2 wt ([wt], black), the S4S5 and S6 single mutants [309] (green) and [412] (blue), and the corresponding double mutant [309:412] (red). Continuous lines represent double-exponential fits (leveling off at ss), and dotted lines represent the corresponding fast components in isolation (illustrating the transitory noninactivating current fraction 1 − a1). (B and C) Equilibrium constants Ktrans (B) and Ksteady (C) were calculated from the noninactivating current fractions 1 − a1 and ss, respectively (see Materials and Methods) and plotted logarithmically. The gray lines represent the function f(x) = x/(1 − x). Note that the mutation-induced effects on Ktrans and Ksteady are not additive for the coupling pair G309/I412.
Results
Extended analysis of Kv4.2 channel CSI: kinetics and structural determinants
In a previous study using double-mutant cycle analysis based on low-voltage inactivation parameters, we showed that there is significant functional coupling of E323 and S322 in S4S5 to I412, N408, and V404, but not to an adversely oriented S407, in S6 of Kv4.2 (16). However, although other Kv4.2 residues showed significant effects on CSI when individually mutated to alanine (as deduced from ln (Kci/Kci,wt) magnitudes; Fig. 1 A), no double-mutant cycle analyses had been performed (16). Here, we set out to extend our coupling analyses to those residues (G309, R311, L313, and F326 in S4S5, and L400 in S6; Fig. 1, A–C), and to explore whether dynamic binding within individual α-subunits or between neighboring α-subunits, or both, may be involved in Kv4.2 channel CSI. The S4S5 and S6 amino acid residues of interest were mutated both individually (single mutant) and pairwise (S4S5/S6 double mutant) to alanine, and the constructs were electrophysiologically characterized (see Materials and Methods). All monomeric Kv4.2 constructs produced functional channels that mediated A-type currents in response to depolarizing voltage pulses (Fig. 1, D and E). We analyzed the voltage dependencies of peak conductance activation and isochronal inactivation for the majority of our constructs (see Table S1) and compared the mutation-induced shift in activation with the mutation-induced shift in inactivation. This analysis revealed a significant correlation for monomeric Kv4.2 constructs (p = 0.0020; Fig. S2 A). This finding was not expected for a channel with preferential CSI, and may be due to the vast number of mutations studied, including ones that do not influence dynamic binding (see below and Discussion).
For our functional-coupling analyses, we induced low-voltage inactivation at a fixed prepulse voltage of −65 mV (see Materials and Methods). Representative examples of such experiments are illustrated in Fig. 2 for monomeric Kv4.2 wt ([wt]) and the single mutants [412] and [326]. Obviously, steady-state inactivation is enhanced in [412] and attenuated in [326]. For a more detailed analysis, we plotted the relative current amplitudes that remained after the prepulse against the prepulse duration, and subjected the data to exponential fitting (except for [326]; see Materials and Methods; Fig. 2, D and E). For both the wt and most of the mutants, the onset of inactivation at −65 mV was best described by the sum of two exponential functions (Fig. 2 D). For wt channels, we obtained time constants of 0.86 s (τ1) and 8.12 s (τ2). The fast and slow components of inactivation had amplitudes of 0.315 (a1) and 0.228 (a2), respectively, resulting in a steady-state noninactivating current fraction (ss) of 0.457 (Fig. 2 E). The time constants obtained by double-exponential fitting and the noninactivating current fractions are summarized in Tables S2 and S3, respectively. Notably, with very few exceptions, the mutations caused a decrease of both τ1 and τ2, and the two time constants were equally affected for a given construct (see Table S2). The appearance of two components in the onset of inactivation at −65 mV may be due to a contamination of CSI by a low open probability at this potential that is not detectable macroscopically but causes a delay in the onset of CSI and/or allows open-state inactivation. In this case, one would expect the inactivation time constants and/or their relative contribution to the total decay (i.e., to reach ss) to correlate with the voltage dependence of activation. However, neither the time constants (τ1 and τ2) nor their relative contributions (expressed as % τ2) showed a correlation with V1/2,act (see Fig. S3). Thus, it is reasonable to assume that the observed biphasic onset kinetics of low-voltage inactivation represent two gating transitions related to CSI. Based on this assumption, we used both the transitory (1 − a1) and steady-state (ss) noninactivating current fractions for our functional-coupling analyses.
Analysis of S4S5/S6 coupling pairs in Kv4.2 channels formed by monomeric constructs
Central to the issue of functional coupling between individual amino acid residues in a protein complex is the question of whether the effects of mutating these residues individually are additive or not. Therefore, we performed low-voltage inactivation experiments with pairs of single mutants and the corresponding double mutants, and compared the results with those obtained for the wt. Fig. 3 A shows pooled data for monomeric wt ([wt]), the single mutants [309] and [412], and the double mutant [309:412]. Obviously, the [309:412] double mutant showed smaller degrees of transitory and steady-state inactivation than either of the two single mutants compared with the wt. Thus, the G309A and I412A amino acid exchanges cause nonadditive effects on low-voltage inactivation, and it is likely that the native amino acid residues G309 and I412 are functionally coupled during CSI in Kv4.2 channels. To pursue a more quantitative approach, we calculated affinities for the transitory state and steady-state inactivation based on our double-exponential fitting results (see Materials and Methods). By plotting the equilibrium constants Ktrans (Fig. 3 B) and Ksteady (Fig. 3 C) obtained for [wt], [309], [412], and [309:412] against the respective noninactivating current fraction, we found that for the coupling pair G309/I412, nonadditivity applies to both transitory and steady-state inactivation. The Ktrans and Ksteady values of low-voltage inactivation are summarized in Table S3.
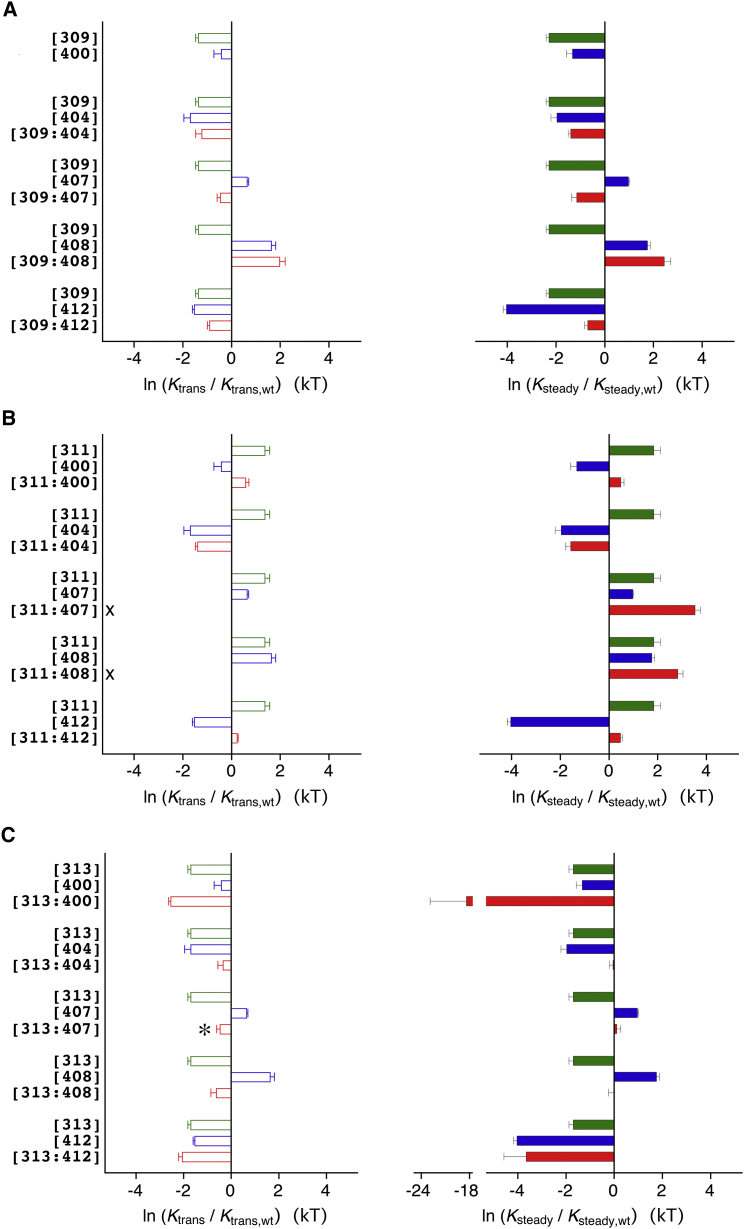
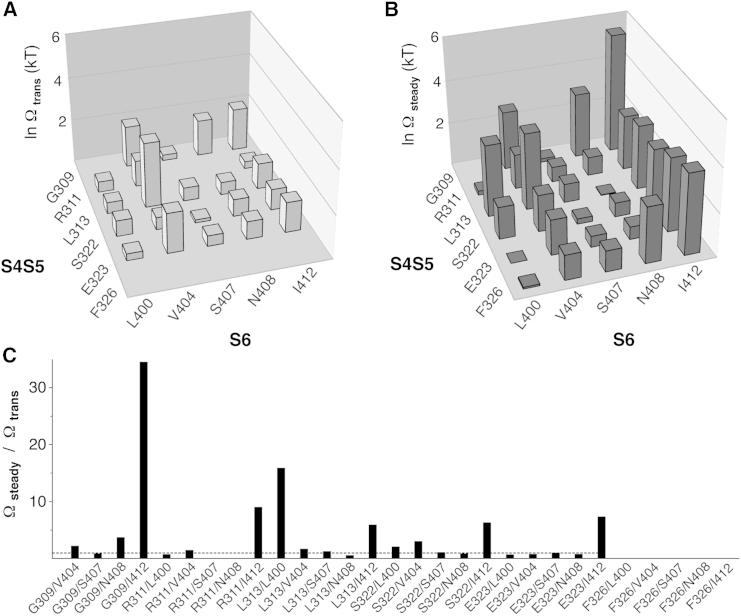
For all mutants, we calculated the ln (Ktrans/Ktrans,wt) and ln (Ksteady/Ksteady,wt) values to quantify their effects (see Materials and Methods). As expected, these values directly and most significantly correlated with the corresponding shifts in the voltage dependence of isochronal inactivation (p < 0.0001; Fig. S2, C and E). Figs. 4 and 5 show the ln (Ktrans/Ktrans,wt) and ln (Ksteady/Ksteady,wt) values for monomeric single mutants and their corresponding double mutants. The mutation-induced effects on Ktrans and Ksteady were qualitatively identical for all mutants (except for [313:407], where the effect was small; Fig. 4 C). The additivity (or nonadditivity) of mutation-induced effects may be assessed by visual inspection of the data. The effects observed for the double mutants [309:407] (Fig. 4 A), [311:400] and [311:407] (Fig. 4 B), [323:407] (Fig. 5 B), and [326:400] and [326:407] (Fig. 5 C) are likely to be caused by additive effects of the corresponding single-point mutations. The results obtained for [311:408] and [311:412] (Fig. 4 B); [313:400], [313:407], and [313:408] (Fig. 4 C); [322:407] and [322:408] (Fig. 5A); and [323:400] and [323:408] (Fig. 5 B) are somewhat ambiguous; however, for the remaining double mutants ([309:404], [309:408], [309:412], [311:404], [313:404], [313:412], [322:400], [322:404], [322:412], [323:404], [323:412], [326:404], [326:408], and [326:412]), the effects are certainly not based on additive effects of the corresponding single-point mutations (Figs. 4 and 5). Of the tested S4S5 mutations, none was exclusively involved in additive or nonadditive effects, but the result depended on the S6 counterpart. Similarly, for the S6 mutations [400], [407], and [408], additivity (or nonadditivity) depended on the S4S5 counterpart. By contrast, the S6 mutations [404] and [412] were exclusively involved in nonadditive effects. Based on these results, one may expect strong functional coupling of the S6 residues V404 and I412 to all tested S4S5 residues. We quantified the magnitude of nonadditivity (i.e., functional coupling) by performing a double-mutant cycle analysis based on the obtained Ktrans and Ksteady values (see Materials and Methods). Fig. 6 shows the results for all 29 S4S5/S6 coupling pairs tested with monomeric constructs (see also Table S3). The coupling coefficients determined based on Ktrans (lnΩtrans in Fig. 6 A) were on average smaller than those determined based on Ksteady (lnΩsteady in Fig. 6 B). The largest Ktrans-based coupling coefficient (lnΩtrans = 2.99) was obtained for the coupling pair L313/V404, and the largest Ksteady-based coupling coefficient (lnΩsteady = 5.52) was obtained for the coupling pair G309/I412. As expected, consistently large lnΩsteady values were obtained for pairings with the S6 residues V404 and I412 (Fig. 6 B). Fig. 6 C illustrates how functional coupling between individual S4S5 and S6 amino acid residues may change when the channel goes from a transitory state to steady-state inactivation. For I412, functional coupling to all S4S5 residues tested was much stronger at steady state. The same applied to the coupling pair L313/L400, whereas S4S5 coupling of the S6 residue V404 remained largely unchanged (except for G309/V404 and S322/V404, which showed a small increase). Other coupling pairs, including L313/N408, showed weaker functional coupling at steady state. Taken together, the results of our extended mutational analysis of Kv4.2 channel CSI reveal a previously unrecognized involvement of the S4-S5 linker helix (S4S5N) in the dynamic binding to distal parts of S6 (Fig. 1 B). Moreover, residue F326 in the initial S5 segment (S4S5C) is identified as a further dynamic binding partner for S6 residues (Fig. 1 C). In principle, dynamic S4S5/S6 binding may occur within individual α-subunits or between neighboring α-subunits. Our analysis results based on double-exponential fitting raise a question as to whether the multistep kinetics correlate with the existence of these two configurations.
Figure 4.
Quantification of the effect on CSI caused by single- and double-point mutations in S4S5N and S6 of monomeric Kv4.2 constructs. The mutation-induced changes in apparent affinity for a transitory state (left, open bars) and steady-state inactivation (right, solid bars) are expressed as ln (Ktrans/Ktrans,wt) and ln (Ksteady/Ksteady,wt), respectively (green, S4S5N single mutations; blue, S6 single mutations; red, S4S5N/S6 double-point mutations). Positive (i.e., mutation-induced increases in K) and negative (i.e., mutation-induced decreases in K) values indicate a decreased and increased likelihood, respectively, for entering the respective closed-inactivated state. The data for individual S4S5/S6 coupling pairs are grouped in bar triplets to illustrate additivity or nonadditivity. (A–C) Separate graphs show the data for the S4S5/S6 coupling pairs with the S4S5N residues G309 (A), R311 (B), and L313 (C). Inclusion of data with ss = 10−6 (three of four experiments with [313:400]; see Materials and Methods) resulted in a mean ln (Ksteady/Ksteady,wt) value of −18.4 kT. Note that the mutation-induced effects on Ktrans and Ksteady are qualitatively identical (except for [313:407], asterisk). X: double-exponential fitting was not possible, and therefore Ktrans data are not available for [311:407] and [311:408] (see Materials and Methods).
Figure 5.
Quantification of the effect on CSI caused by single- and double-point mutations in S4S5C and S6 of monomeric Kv4.2 constructs. Mutation-induced changes in apparent affinity for a transitory state (left, open bars) and steady-state inactivation (right, solid bars) are expressed as ln (Ktrans/Ktrans,wt) and ln (Ksteady/Ksteady,wt), respectively (green, S4S5C single mutations; blue, S6 single mutations; red, S4S5C/S6 double-point mutations). (A–C) Separate graphs show the data for the coupling pairs with the S4S5C residues S322 (A), E323 (B) and F326 (C). X: Double-exponential fitting was not possible, and therefore Ktrans data are not available for [326], [326:407], [326:408], and [326:412] (see Materials and Methods).
Figure 6.
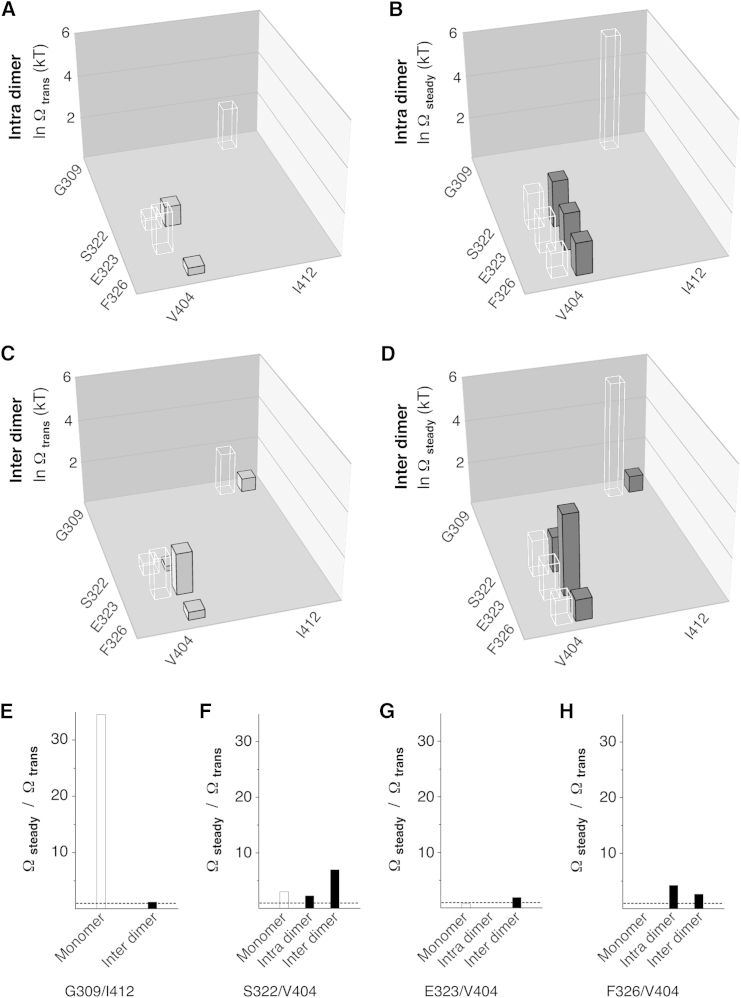
Functional-coupling analysis for S4S5/S6 amino acid pairs based on monomeric Kv4.2 constructs. (A and B) Coupling coefficients for all S4S5/S6 pairings studied were determined by double-mutant cycle analysis based on Ktrans and Ksteady values and plotted as lnΩtrans (A) and lnΩsteady (B), respectively (see Materials and Methods). The three-dimensional plots in (A) and (B) show the native S4S5 amino acid residues on the left x axis and the native S6 amino acid residues on the right x axis. The mean lnΩtrans and lnΩsteady values are shown as vertical cuboids (common y axis, error bars omitted). Note the homogeneous and strong functional coupling during steady-state inactivation for pairings with the S6 residues V404 and I412. (C) The ratio Ωsteady/Ωtrans shows how functional coupling may change when the channel goes from transitory to steady-state inactivation (dotted line: Ωsteady/Ωtrans = 1, no change).
Dynamic binding analysis of CSI in Kv4.2 channels formed by tandem-dimer constructs
We decided to study the S4S5/S6 coupling pairs G309/I412, S322/V404, E323/V404, and F326/V404 (see also Fig. 1, B and C) in more detail. We chose the G309/I412 coupling pair because these two amino acids demarcate the most distal border of the region studied. We chose pairings with V404 because an amino acid exchange at this position appears to play a role in neuropathophysiology (31). In particular, we chose S322/V404 and E323/V404 because homologous pairs appear to be gating relevant in Shaker (21, 23), and F326/V404 because the aromatic phenylalanine demarcates the proximal border of the region studied in S4S5C (see also Fig. 11 and Discussion). To answer the question concerning intra- and intersubunit dynamic binding of these coupling pairs, we made appropriate Kv4.2 tandem-dimer constructs (Fig. 7, B and C). The constructs with a double-point mutation in the first α-subunit and no mutation in the second (concatenated) α-subunit, or vice versa, were expected to specifically perturb (i.e., to probe) putative intrasubunit interactions. The constructs with an S6 point mutation in the first α-subunit and an S4S5 point mutation in the second α-subunit, or vice versa, were expected to probe putative intersubunit interactions. All dimer constructs included in our dynamic-binding analysis produced channels that mediated A-type currents in response to depolarizing voltage pulses (Fig. 7, D–H). The [wt]-[309] current kinetics were unusual and showed a strong voltage dependence (see Discussion). The construct [322]-[wt] alone did not express voltage-dependent currents, which were clearly distinguishable from endogenous oocyte currents. However, functional [322]-[wt] expression was sufficiently increased by coexpression with KChIP2 and DPP6 (see Materials and Methods). Therefore, all constructs involved in the S322/V404 intrasubunit analysis (i.e., [wt]-[wt], [322]-[wt], [404]-[wt], and [322:404]-[wt]) were studied in the presence of these auxiliary subunits. The larger peak current amplitudes and the modulation of macroscopic inactivation kinetics of [wt]-[wt] and [404]-[wt] caused by the coexpression with KChIP2 and DPP6 (not analyzed, but see Fig. 7, D–F), as well as the negative shift of the voltage dependence of activation (see Table S1), indicate that the dimer constructs were capable of interacting with auxiliary subunits. The order of α-subunit concatenation had a big impact on macroscopic inactivation in one case ([404]-[wt] versus [wt]-[404], Fig. 7, F and G), but not so much in the other case ([326]-[wt] versus [wt]-[326]; Fig. 7 H; see Discussion). As opposed to the data set obtained with all channels composed of monomers, the shift in the voltage dependence of inactivation did not significantly correlate with the shift in the voltage dependence of activation for the Kv4.2 tandem-dimer constructs (p = 0.7412; Fig. S2 B).
Fig. 8, A and B, show low-voltage inactivation experiments with oocytes expressing the wt dimer ([wt]-[wt]) in the absence and presence of auxiliary subunits, respectively. The onset kinetics for [wt]-[wt] were also biphasic (Fig. 8 E) but slower (τ1 = 1.41 s, τ2 = 12.1 s), and the transitory and steady-state noninactivating current fractions were larger (1 − a1 = 0.830, ss = 0.664) than the ones obtained for channels composed of monomeric [wt]. Coexpression of [wt]-[wt] with KChIP2 and DPP6 preserved the biphasic nature (Fig. 8 C) but caused an acceleration of onset kinetics (τ1 = 0.06 s, τ2 = 2.71 s), whereas the transitory and steady-state noninactivating current fractions changed only moderately (1 − a1 = 0.735, ss = 0.646; see also Tables S2 and S3). Analogously to the effects on A-type currents, the modulation of low-voltage inactivation kinetics of [wt]-[wt] and [404]-[wt] caused by coexpression with KChIP2 and DPP6 (Fig. 8; Table S2) demonstrates that the dimer constructs were capable of interacting with auxiliary subunits. Moreover, the preservation of double-exponential onset kinetics in the ternary complexes does not support the notion that the biphasic onset kinetics reflects the involvement of N-type inactivation. The order of α-subunit concatenation had an impact on low-voltage inactivation in one case ([326]-[wt] versus [wt]-[326]; Figs. 8 G and 9 G), but to a smaller extent in the other case ([wt]-[404] versus [404]-[wt]; Fig. 8, E and G; see Discussion).
Figure 9.
Probing intersubunit dynamic binding with Kv4.2 tandem-dimer constructs. (A, C, E, and G) Low-voltage inactivation analyses for the tandem-dimer constructs designed to probe intersubunit dynamic binding. Black diamonds, [wt]-[wt]; green diamonds, single S4S5 mutant dimers; blue diamonds, single S6 mutant dimers; red diamonds, intersubunit double-mutant dimers; continuous lines, double-exponential fits; dotted lines, initial component. (B, D, F, and H) Mutation-induced changes in apparent affinity for the transitory (left, open bars) and steady-state inactivation (right, solid bars) expressed as ln (Ktrans/Ktrans,wt-wt) and ln (Ksteady/Ksteady,wt-wt), respectively (green, dimers with a single S4S5 mutation; blue, dimers with a single S6 mutation; red, dimers with one mutation in the first α-subunit and another mutation in the second α-subunit). Separate graphs show the data for the coupling pairs G309/I412 (B), S322/V404 (D), E323/V404 (F), and F326/V404 (H), respectively.
In Fig. 8, C–H, results from the analysis of low-voltage inactivation and the quantification of mutation-induced effects as ln (Ktrans/Ktrans,wt-wt) and ln (Ksteady/Ksteady,wt-wt) are shown for the dimer constructs that were generated to probe putative intrasubunit interactions. Fig. 9 shows analogous analysis results for the dimer constructs that were generated to probe putative intersubunit interactions. As expected, and similar to the data obtained with the monomeric constructs, ln (Ktrans/Ktrans,wt-wt) and ln (Ksteady/Ksteady,wt-wt) values for the dimer constructs directly and significantly correlated with the corresponding shifts in the voltage dependence of inactivation (p ≤ 0.0003; Fig. S2, D and F). As all coupling pairs chosen for the tandem-dimer experiments (G309/I412, S322/V404, E323/V404, and F326/V404) showed strong functional coupling in monomeric form (see Figs. 4, 5, and 6), we now looked for additivity of mutation-induced effects in their intra- (Fig. 8) or intersubunit (Fig. 9) configuration to exclude a preferential intra- or intersubunit interaction, respectively. Analysis of low-voltage inactivation (Fig. 8 C) and effect quantification (Fig. 8 D) suggested no additivity of the S322A and V404A mutations in the intrasubunit configuration for both transitory and steady-state inactivation. Also, analysis of the E323/V404 coupling pair did not support additivity in the intrasubunit configuration, although there was an additive trend for steady-state inactivation (Fig. 8, E and F). The discrepancy between single- and double-mutant effects for the coupling pair F326/V404 (Fig. 8 G) is also illustrated by the effect quantification, which shows that F326A and V404A are not additive in the intrasubunit configuration for transitory and steady-state inactivation (Fig. 8 H). Thus, none of the tested coupling pairs showed clearly additive effects in the intrasubunit configuration for either the transitory state or steady-state inactivation. For the coupling pair G309/I412, analysis of low-voltage inactivation (Fig. 9 A) and effect quantification suggested that there was also no additivity in the intersubunit configuration for either transitory or steady-state inactivation (Fig. 9 B). Also for the coupling pairs S322/V404 (Fig. 9, C and D), E323/V404 (Fig. 9, E and F), and F326/V404 (Fig. 9, G and H), effect quantification did not support additivity in the intersubunit configuration concerning steady-state inactivation. However, in the effect quantification for the transitory state, the coupling pairs S322/V404 (Fig. 9 D) and F326/V404 (Fig. 9 H) show clear additivity in the intersubunit configuration, and a similar trend is seen for the coupling pair E323/V404 (Fig. 9 F).
To quantify the magnitude of nonadditivity of mutations in the intra- and intersubunit configurations, we applied double-mutant cycle analysis to the Ktrans and Ksteady values obtained with the dimer constructs. Fig. 10 shows the calculated lnΩtrans and lnΩsteady values for the intradimer (Fig. 10, A and B, except for G309/I412) and interdimer (Fig. 10, C and D) constructs together with the lnΩtrans and lnΩsteady values obtained for the corresponding monomeric constructs (data from Fig. 6) for comparison (see also Table S3).
Figure 10.
Functional-coupling analysis for S4S5/S6 amino acid pairs in Kv4.2 tandem-dimer constructs. Coupling coefficients for the S4S5/S6 pairings G309/I412, S322/V404, E323/V404, and F326/V404 were determined by double-mutant cycle analysis based on Ktrans and Ksteady values and plotted as lnΩtrans (left) and lnΩsteady (right), respectively (see Materials and Methods). (A and B) Coupling coefficients obtained with the tandem-dimer constructs designed to probe intrasubunit dynamic binding (intradimer). (C and D) Coupling coefficients obtained with the tandem-dimer constructs designed to probe intersubunit dynamic binding (interdimer). The solid vertical cuboids representing the tandem-dimer results (error bars omitted) are accompanied by the corresponding monomer results for comparison (transparent cuboids; data from Fig. 6, A and B). (E–H) The ratio Ωsteady/Ωtrans shows how functional coupling may change for the coupling pairs G309/I412 (E), S322/V404 (F), E323/V404 (G), and F326/V404 (H) when the channel transitions from transitory to steady-state inactivation (open bars, monomer data from Fig. 6C; black bars, intra- and interdimers; dotted line, Ωsteady/Ωtrans = 1, no change). Note that for all coupling pairs (except for G309/I412), functional coupling increases in the intersubunit configuration.
For the G309/I412 coupling pair, the lnΩ values obtained in the intersubunit configuration (Fig. 10, C and D) were smaller than the corresponding values obtained for the monomer (lnΩtrans ∼32%, lnΩsteady ∼14%). According to the data obtained for the monomer, functional coupling increases for G309/I412 when the channel enters steady-state inactivation, which is not the case in the intersubunit configuration (Fig. 10 E; see also Fig. 6 C). For S322/V404, the lnΩ values were smaller in the intersubunit (Fig. 10, C and D) than in the intrasubunit configuration (Fig. 10, A and B). LnΩtrans was larger than the monomer value in the intrasubunit (Fig. 10 A) and smaller than the monomer value in the intersubunit configuration (Fig. 10 C) for S322/V404, whereas lnΩsteady was comparable to the corresponding monomer values in both configurations (Fig. 10, B and D). Notably, functional coupling for S322/V404 increased during CSI, especially for the intersubunit configuration (Fig. 10 F). For E323/V404, lnΩsteady was larger in the intersubunit configuration than in the intrasubunit configuration (Fig. 10, B and D) and larger than the corresponding monomer value (Fig. 10 D), whereas the other lnΩ values were equal to the corresponding monomer values (Fig. 10, B and C). The higher interdimer lnΩsteady value compared with that in the monomer (Fig. 10 D) and the increase in functional coupling for E323/V404 in the intersubunit configuration, which was not seen in the monomer (Fig. 10 G), are intriguing observations (see Discussion). For the coupling pair F326/V404, the lnΩ values were similar in the intra- (Fig. 10, A and B) and intersubunit (Fig. 10, C and D) configurations, and the lnΩsteady values were similar to the corresponding monomer values (Fig. 10, B and D). Functional coupling for F326/V404 increased during CSI in both the intra- and intersubunit configurations (Fig. 10 H). Taken together, our results suggest that intra- and intersubunit interactions are equally plausible for the coupling pairs S322/V404, E323/V404, and F326/V404 in Kv4.2 channel CSI. By contrast, and despite the lack of data for the intradimer configuration, a preferential intersubunit interaction between G309 and I412 is not supported (see Discussion).
Discussion
In this study, we treated Kv4.2 channel CSI as a multistep process in which steady-state inactivation is preceded by one or more transitory states. We defined this multistep process by fitting the onset kinetics of low-voltage inactivation with a double-exponential function. We then used the fitting results as the basis for a double-mutant cycle analysis to examine the degree of functional coupling between individual amino acid residues in S4S5 and S6, respectively, in CSI. Our results show that both the initial S5 segment (S4S5C) and the S4-S5 linker helix (S4S5N) are involved, and that the S6 residues V404 and I412 play a major role in the dynamic binding between S4S5 and S6 in Kv4.2 channel CSI. The results of our experiments with tandem-dimer constructs support the notion that V404 is functionally coupled not only to S4S5 residues within the same α-subunit but also to S4S5 residues of a neighboring α-subunit, whereas I412 is functionally coupled to G309 of the same α-subunit rather than to a neighboring α-subunit.
Studying inactivation coupled to the activation machinery
A conceptual model of Kv4.2 channel CSI is based on dynamic binding between S4S5 and S6, as discussed previously (3, 14, 16). Dynamic binding means that the physical contact between S4S5 and S6 may temporarily be lost during membrane depolarization when the voltage sensor undergoes conformational changes related to activation, and restored when the membrane is repolarized and the voltage sensor returns to its resting conformation. It is obvious that the dynamic-binding model of CSI involves protein domains that are instrumental in voltage-dependent activation (17, 18, 19, 20), which makes it difficult to perform a direct and specific experimental investigation of the proposed CSI mechanism. Voltage-dependent activation consists of two main processes: voltage-sensor movement and gate opening. Previous approaches to identify the structural determinants of Kv4.2 channel CSI were based on a kinetic matching between CSI and voltage sensor movement, and a structural matching between CSI and gate opening, respectively. Dougherty and co-workers (15) were able to show that the kinetics of CSI and recovery from inactivation in Kv4.2 channels match the kinetics of loss and restoration, respectively, of gating charge. The authors concluded that loss of gating charge and CSI are two manifestations of the same process, i.e., the adoption of a relaxed voltage-sensor conformation during membrane depolarization is mechanistically coupled to the process that renders the gate reluctant to open. To provide support for the hypothesis that a loss of contact between S4S5 and S6 is responsible for the reluctance of the gate to open, we examined structural matching and were able to show that pairwise mutations in Kv4.2 protein domains, homologous to those known to be critical for gate control in Shaker (S4S5 and S6) (20), have short-range effects on CSI. Double-mutant cycle analysis combined with structural homology modeling provided support for the notion that identical structures are utilized for voltage-dependent activation and CSI in Kv4.2 channels (16). In this study, we further exploited our strategy of introducing mild mutations (to alanine) that leave activation largely intact, and using low-voltage inactivation parameters as a readout. But can we exclude the possibility that the effects we are seeing are only secondary to a perturbation of activation? To address this problem, we determined the voltage dependence of peak conductance activation (V1/2,act) as a crude measure of opening probability, for all constructs, in addition to the kinetics and voltage dependence of inactivation. As a strong argument against the notion that our mutational effects are secondary to a perturbation of gate opening, we found no correlation when we compared the voltage dependence of activation with the kinetics (time constants and their relative amplitudes) of low-voltage inactivation (Fig. S3). Apparently, mutations to alanine affect gate opening and CSI differently. Alternatively, the interaction points within S4S5 and S6 for gate opening and CSI may overlap to a large degree, but may not be identical. In support of the latter possibility, we found a significant correlation between the mutation-induced shifts in V1/2,act and V1/2,inact for the analysis including all monomeric constructs (Fig. S2 A), but not for the analysis of tandem dimers, in which only selected coupling pairs with a critical involvement in CSI were examined further (Fig. S2 B). Also, in our previous study with a smaller number of monomeric constructs, we found no such correlation (16).
Double-mutant cycle analysis applied to the multistep process of Kv4.2 channel CSI
To study the dynamic S4S5/S6 binding in Kv4.2 channels, we chose an approach based on equilibrium constants and second-order coupling (32), which directly relates to the idea that functionally coupled amino acid residues in the gating-relevant protein domains may dissociate (onset of inactivation) and reassociate (recovery from inactivation) (3, 14, 16). A similar approach (also based on equilibrium constants describing the reversible interaction between protein domains) was previously applied to potassium channel N-type inactivation, where association and dissociation of the inactivation domain represent the onset of inactivation and recovery from inactivation, respectively (11, 33). It should be noted that, in contrast to the binding of the N-type inactivation peptide to the open pore, the dynamic binding between S4S5 and S6 postulated for Kv4.2 channel CSI has not yet been directly shown. Therefore, the equilibrium constants we used for the double-mutant cycle analysis of CSI in this study (Ktrans and Ksteady) do not refer to binding affinities but to apparent affinities for the respective closed-inactivated states (although, for the purpose of double-mutant cycle analysis, calculated equilibrium constants could have been used as their reciprocals with no influence on the resultant Ω values; see Materials and Methods). It should be further noted that the binding of the inactivation particle to the pore (33) can be viewed as a single-step process, whereas more complex gating rearrangements, such as the dynamic S4S5/S6 binding in Kv4.2 channel CSI, may involve multiple steps, with all four subunits participating. In this case, thermodynamic calculations may be influenced by (and therefore reflect) intermediates. Despite this caveat, thermodynamic mutant cycles based on V1/2,act measurements have been widely used to study the structural determinants of gating transitions for different types of Kv channels, including Shaker (21, 34, 35), Kv2.1 (36), Kv7.1 (37), and Kv11.1 (38, 39, 40). Chowdhury and Chanda (41) used a generalized interaction-energy analysis based on the median voltage of charge transfer to obtain more reliable data on pairwise interactions and coupling free energies (see below).
In this study, we used a double-mutant cycle analysis based on equilibrium constants to quantify the nonadditivity of mutational effects related to Kv4.2 channel CSI, with no reference to thermodynamic free-energy changes. To avoid misinterpretation of our data, we took into consideration the previously neglected double-exponential kinetics of low-voltage inactivation in Kv4.2 channels. Intriguingly, our previous disulfide bonding experiments with a Kv4.2 E323C:V404C double mutant brought these biphasic kinetics to light (16). We treated the initial component of the biphasic decay as the adoption of a transitory state and the second component as the transition into steady-state inactivation, and applied double-mutant cycle analysis to both processes. Validating our coupling analysis, the mutation-induced effects on the two components were qualitatively identical for almost all constructs, and coupling coefficients were on average smaller for the transitory state than for steady-state inactivation. With this analysis method, we confirmed the strong functional coupling of the S4S5C residues S322 and E323 to the S6 residues V404 and I412, and the weaker functional coupling of these residues to the S6 residue S407 in Kv4.2 channel CSI (16). Our data extend the known functional coupling profile to a number of residues, including G309, R311, and L313, located in the S4-S5 linker helix (S4S5N; Fig. 1 B); F326, located in the initial part of S5 (S4S5C) one helical turn above E323; and L400, located in S6 one helical turn above V404 (Fig. 1 C). Our three-dimensional functional coupling profiles for transitory and steady-state inactivation (Fig. 6) suggest that the S6 residues V404 and I412 play a major role in dynamic S4S5/S6 binding during Kv4.2 channel CSI. The steady-state coupling coefficients were on average larger for I412 than for V404, which suggests stronger functional coupling in more distal S6 regions than in regions closer to the PVP motif of the S6 gate (Fig. 1 A; see below). By contrast, L400, which is located beyond the PVP motif in the more proximal part of S6, on average exhibited only weak functional coupling to S4S5. These results are in accordance with the notion that the PVP motif serves as a hinge point for S6 movement (42), and that dynamic binding, as proposed for Kv4.2 channel CSI, may become more manifest in S6 regions with stronger movement. Notably, the largest coupling coefficient was obtained for the coupling pair G309/I412, which suggests that dynamic S4S5/S6 binding also extends well into the linker helix S4S5N, where contact with more distal S6 regions is expected to occur (see below). For the newly identified (to our knowledge) structural determinants of CSI, there is no thermodynamic precedence. The phenylalanine residue in the Shaker S6, homologous to I412 in Kv4.2, has not been studied previously, and S4S5N was not included in a previous Shaker mutagenesis scan combined with double-mutant cycle analysis (Fig. 11 A (21); but see below).
Dynamic S4S5/S6 binding in the Kv4.2 dimer-of-dimers
Double-mutant cycle analysis based on the expression of monomeric constructs is blind regarding the question as to whether functional coupling arises from intra- or intersubunit interactions. Therefore, we applied our double-mutant cycle analysis also to tandem-dimer constructs, which were designed to preferentially perturb either interactions within individual α-subunits (intradimer, i.e., the double mutation is in the intrasubunit configuration) or interactions between neighboring α-subunits (interdimer, i.e., the double mutation is in the intersubunit configuration). The dimer approach relies on proper protein folding and channel formation, which was confirmed electrophysiologically in our study, since the expression of the tandem-dimer constructs produced typical A-type currents. For [wt]-[309], we obtained currents with bimodal macroscopic inactivation. Moderate depolarization activated currents with slow kinetics reminiscent of endogenous oocyte conductances; however, stronger depolarization activated typical A-type currents (see Fig. 7 D), as if two kinetically distinct Kv4.2 channel populations or one Kv4.2 channel population with extremely voltage-dependent inactivation kinetics underlay the macroscopic currents. Bimodal macroscopic inactivation kinetics were, to some extent, also seen with the monomeric construct [309:404] (see Fig. 1 E), but with none of the other constructs. Therefore, we think that mutating G309 in Kv4.2 in an appropriate protein background (e.g., dimer or double mutant) may lead to the expression of channels with bimodal gating. Notably, G309 lies at the N-terminal end of the S4-S5 linker helix, directly adjacent to the S4 segment, and shows strong functional coupling to S6.
We assumed that expressing Kv4.2 dimer constructs led to dimer-of-dimers formation, but can we exclude the possibility that only one of the two concatenated α-subunits contributed to pore formation, leaving the other α-subunit tethered on the outside of the channel complex (43, 44)? Our data show that channels produced by the dimer constructs are able to interact with auxiliary subunits, as deduced from the effects of KChIP2 and DPP6 coexpression on macroscopic current amplitudes and kinetics, as well as on the voltage dependence of activation (28). These results are in accordance with those obtained in a previous study in which KChIP4 coexpression modulated the channels formed by KChIP4-Kv4.2-Kv4.2 concatemers, presumably by occupying two remaining KChIP association sites (45). Although we are confident that dimer-of-dimers formation occurred in our experiments, there is still the question of how the two dimers are oriented with respect to each other (Fig. S4). With random association of the two dimers, the ordering within the dimer construct (AB or BA) should not matter. In this context, it is interesting that [404]-[wt] and [wt]-[404] show similar low-voltage inactivation properties but differ with respect to macroscopic inactivation, whereas [326]-[wt] and [wt]-[326] show similar macroscopic inactivation but differ with respect to low-voltage inactivation (see Figs. 7, 8, and 9). A similar dependence of gating on the ordering within dimers was previously observed for Shaker (44). These findings argue against random association of the dimers. Thus, the constructs must have preferentially adopted either an AABB (the N-termini of the two dimers point in the same direction) or an ABAB (the N-termini of the two dimers point in opposite directions; Fig. S4) conformation. Originally, it was assumed that potassium channel dimers assemble in an ABAB fashion (43), according to the way rotational symmetry is achieved in voltage-gated Na+ and Ca2+ channels. However, a more recent study on Kv1.1/Kv1.2 heteromerization suggested that dimer assembly in an adjacent conformation (AABB) may also be possible (46). The conformation of dimer assembly may have influenced the results obtained in this study. Provided that two residues are able to undergo intersubunit interactions, an isolated investigation of putative intrasubunit interactions (e.g., [wt]-[323:404]) is hampered in the AABB conformation. Putative intersubunit interactions (e.g., [404]-[323]), on the other hand, are perturbed at four sites in the ABAB configuration but at only two sites in the AABB configuration (Fig. S4). Despite the shortcomings of the tandem-dimer approach, the results of our experiments support the notion that dynamic S4S5/S6 binding may occur both within individual α-subunits and between neighboring α-subunits in the multistep process of Kv4.2 channel CSI.
Using our tandem-dimer approach, we searched for additivity of mutational effects for the intra- and interdimer configurations to exclude a significant role of dynamic S4S5/S6 binding in each configuration. However, the results were not mutually exclusive, since for steady-state inactivation, nonadditivity was evident in either configuration (see Figs. 8 and 9). However, concerning the transitory state, the coupling pairs S322/V404 and F326/V404 showed additivity in the intersubunit configuration (Fig. 9, D and H), and there was a similar trend for the coupling pair E323/V404 (Fig. 9 F). These results are reflected by our double-mutant cycle analysis, which leads to the conclusion that dynamic binding of the S4S5 residues S322, E323, and F326 to the S6 residue V404 within the same α-subunit plays a role in both transitory and steady-state inactivation, whereas dynamic binding of these S4S5 residues to V404 in a neighboring α-subunit only plays a role in steady-state inactivation. For the G309/I412 coupling pair, the data are incomplete, and therefore no firm statement can be made concerning an intra- or intersubunit preference. However, considering the small coupling coefficients in the intersubunit configuration compared with those obtained with the monomer, in combination with the absence of an increase in functional coupling from transitory to steady-state inactivation, preferential intersubunit dynamic binding between G309 and I412 appears unlikely. The absence of additivity for G309/V412 in the intersubunit configuration, which one may expect, could be due to effect saturation at ∼−4 kT with almost complete steady-state inactivation (Fig. 9, A and B). For the coupling pair E323/V404, an increase in functional coupling from transitory to steady-state inactivation was seen in the intersubunit configuration, so the coupling coefficient obtained for steady-state inactivation was larger than the corresponding monomer value (Fig. 10 D). These results may be explained by a novel (to our knowledge) intersubunit functional coupling between E323 and V404, which does not exist in tetrameric channels but becomes possible in channels formed by the corresponding interdimer construct. Also, the possibility that E323 and V404 may be involved in an indirect coupling (23) complicates the analysis for this pair of amino acids (see below).
Structural determinants of electromechanical coupling in Kv4.2 and Shaker
Movement of the voltage sensor is thought to be translated into opening and closing of the pore via physical binding between S4S5 and S6, which may be accomplished in two possible ways: S4S5 may keep the S6 gate closed (acting like a bolt) unless the voltage sensor moves, or S4S5 may tear the S6 gate open (acting like a spring) when the voltage sensor moves (26). According to the dynamic-binding model of Kv4.2 channel CSI (3, 14, 16), the S6 gate must adopt a closed conformation if it is disconnected from the voltage sensor (i.e., its default state is closed). This is a reasonable assumption, since a set of point mutations in the S4 segment of Shaker (47) was shown to shift the voltage dependence of the gating charge movement to more negative potentials while shifting that of the open probability to more positive potentials. Such asymmetric effects on the voltage dependence of the gating charge movement and open probability show that the mutations have partly uncoupled the voltage sensor from the gate, such that the former is easier to move and the latter is harder to open. The effects of partial uncoupling support the notion that the default state of the S6 gate is closed and that S4S5 acts like a spring (26, 48). The mechanical properties of a spring also reconcile with the elasticity of the system needed for energy storage, such that the gate will only open if all four voltage sensors have moved (24, 25). We speculate that in Kv channels with prominent CSI, such as Kv4.2, this elasticity is low, and therefore complete uncoupling may occur during preopen-activation steps. Our results lead to the intriguing hypothesis that in Kv4.2 channels, different sites may undergo dynamic binding at different time points. For all coupling pairs involved in CSI, the dynamic binding within individual subunits may occur during the initial phase. During the second phase, intersubunit dynamic binding may occur for some coupling pairs to establish steady-state inactivation (see below).
Alanine mutation of I412 in Kv4.2 led to a tremendous enhancement of CSI (see Fig. 2), and the coupling pairs that included I412, especially G309/I412, yielded the largest coupling coefficients found in this study (see Fig. 6). Both our double-mutant cycle analysis results and structural homology modeling (Fig. 11 C) support the notion that G309 and I412 are functionally coupled within individual Kv4.2 α-subunits. I412 is homologous to F484 in Shaker. The Shaker S4S5N residues I384 and T388 insert into a hydrophobic pocket formed by F481, Y483, and F484 in S6 of the same α-subunit (22) (Fig. 11, A and B), and the same applies to homologous residues in Kv1.5 (49). Notably, great sequence variations exist between Shaker and Kv4.2 in the hydrophobic pocket, with a phenylalanine replaced by isoleucine (I412) and a tyrosine replaced even by a positively charged arginine (R411) in Kv4.2 (Fig. 11 A). Thus, it is quite likely that the strength of binding at these sites in Kv4.2 differs substantially from that observed in Shaker. We did not test the conserved S4S5 residues in Kv4.2 homologous to I384 and T388 in Shaker or the conserved S6 residue in Kv4.2 homologous to Y485 in Shaker (Fig. 11 A), because their individual mutation to alanine does not yield large effects on CSI (see Fig. 1 A) (16).
In this study, we tested the nonconserved S4S5C residues S322 and F326, and the conserved S4S5C residue E323, and focused on V404 (Fig. 11 A) as a coupling partner in S6. Our results show that V404 is involved in strong functional coupling to a number of S4S5 residues (see Fig. 6), and the large coupling coefficients for the respective interdimer (see Fig. 10, C and D) support an intersubunit interaction between E323 and V404 in Kv4.2 channel CSI. E323 and V404 are homologous to E395 and V476 in Shaker. The Shaker S4S5C residues R394, E395, and L398 seem to interact with Y485 in S6 of a neighboring α-subunit (RELY intersubunit interaction) with a special involvement of V476 (Fig. 11, A and B). V476 is thought to normally hold E395 in position in the context of the RELY interaction, and mutation of V476 to alanine leads to a reorientation of the native E395 that allows strong binding between Y485 and R394 (23). Data obtained from a generalized interaction-energy analysis suggest that R394, E395, and Y485 constitute a gating triad that ensures the structural integrity of the intersubunit interface (50). The described intersubunit interactions are thought to play a key role in the cooperativity required for the opening step (23, 26, 50). Notably, drastic sequence variance also exists between Shaker and Kv4.2 in the S4S5C domain: R394 is replaced by serine (S322) and L398 is replaced by phenylalanine (F326; Fig. 11 A).
The apparent inconsistency of a smaller coupling coefficient for the monomer than for the intersubunit configuration reflects the possibility that, similarly to Shaker (23), the E323A and V404A mutations in Kv4.2 add another interaction that occurs elsewhere in the S4S5/S6 interface. Alternatively, E323 and V404 may be involved in indirect interactions via the gating triad (50). In Kv4.2, both E323A and V404A strongly favor CSI (16) (Fig. 5 B), in contrast to the increased S4S5/S6 binding and stabilization of the open state reported for homologous mutations in Shaker (23). The aforementioned sequence variations between Shaker and Kv4.2 may underlie the differences in S4S5/S6 binding. Thus, unlike the positively charged R394 in Shaker, the homologous S322 in Kv4.2 (Fig. 11 A) seems to maintain both intra- and intersubunit interactions, at least with V404, as suggested by our dimer approach (see Fig. 10). Despite the differences in sequence and function, homology modeling based on Kv1.2-2.1 crystal structure data (19) locates all S4S5C residues tested for intra- and/or intersubunit interactions and V404 in Kv4.2 to the RELY interface defined for the Shaker channel (Fig. 11, D–F). It should be noted that the structural data used for homology modeling represent the Kv1.2-2.1 channel in its open state. One may hypothesize that the spatial relationships between the residues of identified coupling pairs represent a situation that exists just before unbinding occurs.
Conclusion
Our results do not correspond in all aspects to those obtained with uncoupling mutants in Shaker. This may be due to great sequence variations at critical positions between the two related but differently inactivating channel types. In particular, the nature of the gating triad (50), if present in Kv4.2 channels, may not have evolved to ensure the structural integrity of the intersubunit interface, but rather to allow dynamic binding. It is not clear whether CSI in Kv4.2 channels relies exclusively on intra- and intersubunit dynamic binding between S4S5/S6 interfaces (Fig. 11 B) or alternative mechanism(s) are also involved. The results of our functional-coupling analyses suggest that the double-exponential onset kinetics of low-voltage inactivation represents dynamic binding events between S4S5 and S6 rather than profoundly different mechanisms. Whether the CSI mechanism and its structural determinants that have been postulated for Kv4.2 channels are also applicable to other Kv channels with prominent CSI remains to be shown.
Author Contributions
J.W. made the Kv4.2 constructs, performed the experiments, and analyzed the data. R.B. designed the research, analyzed the data, and wrote the manuscript.
Acknowledgments
We thank Telse Kock for excellent technical assistance and Christiane K. Bauer for discussion.
This work was supported by grant BA 2055/1-3 from the Deutsche Forschungsgemeinschaft to R.B.
Editor: William Kobertz.
Footnotes
Four figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)01124-8.
Supporting Material
References
- 1.Hille B. Sinauer Associates, Inc.; Sunderland, MA: 2001. Ion Channels of Excitable Membranes. [Google Scholar]
- 2.Kurata H.T., Fedida D. A structural interpretation of voltage-gated potassium channel inactivation. Prog. Biophys. Mol. Biol. 2006;92:185–208. doi: 10.1016/j.pbiomolbio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Bähring R., Covarrubias M. Mechanisms of closed-state inactivation in voltage-gated ion channels. J. Physiol. 2011;589:461–479. doi: 10.1113/jphysiol.2010.191965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salkoff L., Baker K., Wei A. An essential ‘set’ of K+ channels conserved in flies, mice and humans. Trends Neurosci. 1992;15:161–166. doi: 10.1016/0166-2236(92)90165-5. [DOI] [PubMed] [Google Scholar]
- 5.Timpe L.C., Schwarz T.L., Jan L.Y. Expression of functional potassium channels from Shaker cDNA in Xenopus oocytes. Nature. 1988;331:143–145. doi: 10.1038/331143a0. [DOI] [PubMed] [Google Scholar]
- 6.Timpe L.C., Jan Y.N., Jan L.Y. Four cDNA clones from the Shaker locus of Drosophila induce kinetically distinct A-type potassium currents in Xenopus oocytes. Neuron. 1988;1:659–667. doi: 10.1016/0896-6273(88)90165-1. [DOI] [PubMed] [Google Scholar]
- 7.Hoshi T., Zagotta W.N., Aldrich R.W. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 8.Zagotta W.N., Hoshi T., Aldrich R.W. Restoration of inactivation in mutants of Shaker potassium channels by a peptide derived from ShB. Science. 1990;250:568–571. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- 9.Choi K.L., Aldrich R.W., Yellen G. Tetraethylammonium blockade distinguishes two inactivation mechanisms in voltage-activated K+ channels. Proc. Natl. Acad. Sci. USA. 1991;88:5092–5095. doi: 10.1073/pnas.88.12.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.López-Barneo J., Hoshi T., Aldrich R.W. Effects of external cations and mutations in the pore region on C-type inactivation of Shaker potassium channels. Receptors Channels. 1993;1:61–71. [PubMed] [Google Scholar]
- 11.Gebauer M., Isbrandt D., Bähring R. N-type inactivation features of Kv4.2 channel gating. Biophys. J. 2004;86:210–223. doi: 10.1016/S0006-3495(04)74097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaulin Y.A., De Santiago-Castillo J.A., Covarrubias M. Mechanism of the modulation of Kv4:KChIP-1 channels by external K+ Biophys. J. 2008;94:1241–1251. doi: 10.1529/biophysj.107.117796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fineberg J.D., Ritter D.M., Covarrubias M. Modeling-independent elucidation of inactivation pathways in recombinant and native A-type Kv channels. J. Gen. Physiol. 2012;140:513–527. doi: 10.1085/jgp.201210869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bähring R., Barghaan J., Wollberg J. Voltage sensor inactivation in potassium channels. Front. Pharmacol. 2012;3:100. doi: 10.3389/fphar.2012.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dougherty K., De Santiago-Castillo J.A., Covarrubias M. Gating charge immobilization in Kv4.2 channels: the basis of closed-state inactivation. J. Gen. Physiol. 2008;131:257–273. doi: 10.1085/jgp.200709938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barghaan J., Bähring R. Dynamic coupling of voltage sensor and gate involved in closed-state inactivation of Kv4.2 channels. J. Gen. Physiol. 2009;133:205–224. doi: 10.1085/jgp.200810073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long S.B., Campbell E.B., MacKinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 18.Long S.B., Campbell E.B., MacKinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 19.Long S.B., Tao X., MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 20.Lu Z., Klem A.M., Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J. Gen. Physiol. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yifrach O., MacKinnon R. Energetics of pore opening in a voltage-gated K+ channel. Cell. 2002;111:231–239. doi: 10.1016/s0092-8674(02)01013-9. [DOI] [PubMed] [Google Scholar]
- 22.Haddad G.A., Blunck R. Mode shift of the voltage sensors in Shaker K+ channels is caused by energetic coupling to the pore domain. J. Gen. Physiol. 2011;137:455–472. doi: 10.1085/jgp.201010573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batulan Z., Haddad G.A., Blunck R. An intersubunit interaction between S4-S5 linker and S6 is responsible for the slow off-gating component in Shaker K+ channels. J. Biol. Chem. 2010;285:14005–14019. doi: 10.1074/jbc.M109.097717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zagotta W.N., Hoshi T., Aldrich R.W. Shaker potassium channel gating. III: Evaluation of kinetic models for activation. J. Gen. Physiol. 1994;103:321–362. doi: 10.1085/jgp.103.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zagotta W.N., Hoshi T., Aldrich R.W. Shaker potassium channel gating. II: Transitions in the activation pathway. J. Gen. Physiol. 1994;103:279–319. doi: 10.1085/jgp.103.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blunck R., Batulan Z. Mechanism of electromechanical coupling in voltage-gated potassium channels. Front. Pharmacol. 2012;3:166. doi: 10.3389/fphar.2012.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu X.-R., Wulf A., Pongs O. Characterization of human Kv4.2 mediating a rapidly-inactivating transient voltage-sensitive K+ current. Receptors Channels. 1999;6:387–400. [PubMed] [Google Scholar]
- 28.Jerng H.H., Pfaffinger P.J. Modulatory mechanisms and multiple functions of somatodendritic A-type K+ channel auxiliary subunits. Front. Cell. Neurosci. 2014;8:82. doi: 10.3389/fncel.2014.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bähring R., Dannenberg J., Isbrandt D. Conserved Kv4 N-terminal domain critical for effects of Kv channel-interacting protein 2.2 on channel expression and gating. J. Biol. Chem. 2001;276:23888–23894. doi: 10.1074/jbc.M101320200. [DOI] [PubMed] [Google Scholar]
- 30.Wada K., Yokotani N., Shimasaki S. Differential expression of two distinct forms of mRNA encoding members of a dipeptidyl aminopeptidase family. Proc. Natl. Acad. Sci. USA. 1992;89:197–201. doi: 10.1073/pnas.89.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H., Lin M.C., Nelson S.F. Exome sequencing identifies de novo gain of function missense mutation in KCND2 in identical twins with autism and seizures that slows potassium channel inactivation. Hum. Mol. Genet. 2014;23:3481–3489. doi: 10.1093/hmg/ddu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yifrach O., Zandany N., Shem-Ad T. Examining cooperative gating phenomena in voltage-dependent potassium channels: taking the energetic approach. Methods Enzymol. 2009;466:179–209. doi: 10.1016/S0076-6879(09)66008-0. [DOI] [PubMed] [Google Scholar]
- 33.Zhou M., Morais-Cabral J.H., MacKinnon R. Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature. 2001;411:657–661. doi: 10.1038/35079500. [DOI] [PubMed] [Google Scholar]
- 34.Shem-Ad T., Irit O., Yifrach O. Inter-subunit interactions across the upper voltage sensing-pore domain interface contribute to the concerted pore opening transition of Kv channels. PLoS One. 2013;8:e82253. doi: 10.1371/journal.pone.0082253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zandany N., Ovadia M., Yifrach O. Direct analysis of cooperativity in multisubunit allosteric proteins. Proc. Natl. Acad. Sci. USA. 2008;105:11697–11702. doi: 10.1073/pnas.0804104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bocksteins E., Ottschytsch N., Snyders D.J. Functional interactions between residues in the S1, S4, and S5 domains of Kv2.1. Eur. Biophys. J. 2011;40:783–793. doi: 10.1007/s00249-011-0694-3. [DOI] [PubMed] [Google Scholar]
- 37.Labro A.J., Boulet I.R., Snyders D.J. The S4-S5 linker of KCNQ1 channels forms a structural scaffold with the S6 segment controlling gate closure. J. Biol. Chem. 2011;286:717–725. doi: 10.1074/jbc.M110.146977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Y.M., Hull C.M., Claydon T.W. Functional interactions of voltage sensor charges with an S2 hydrophobic plug in hERG channels. J. Gen. Physiol. 2013;142:289–303. doi: 10.1085/jgp.201310992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng C.A., Phan K., Perry M.D. Multiple interactions between cytoplasmic domains regulate slow deactivation of Kv11.1 channels. J. Biol. Chem. 2014;289:25822–25832. doi: 10.1074/jbc.M114.558379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perry M.D., Wong S., Vandenberg J.I. Hydrophobic interactions between the voltage sensor and pore mediate inactivation in Kv11.1 channels. J. Gen. Physiol. 2013;142:275–288. doi: 10.1085/jgp.201310975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chowdhury S., Chanda B. Estimating the voltage-dependent free energy change of ion channels using the median voltage for activation. J. Gen. Physiol. 2012;139:3–17. doi: 10.1085/jgp.201110722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster S.M., Del Camino D., Yellen G. Intracellular gate opening in Shaker K+ channels defined by high-affinity metal bridges. Nature. 2004;428:864–868. doi: 10.1038/nature02468. [DOI] [PubMed] [Google Scholar]
- 43.Heginbotham L., MacKinnon R. The aromatic binding site for tetraethylammonium ion on potassium channels. Neuron. 1992;8:483–491. doi: 10.1016/0896-6273(92)90276-j. [DOI] [PubMed] [Google Scholar]
- 44.McCormack K., Lin L., Sigworth F.J. Tandem linkage of Shaker K+ channel subunits does not ensure the stoichiometry of expressed channels. Biophys. J. 1992;63:1406–1411. doi: 10.1016/S0006-3495(92)81703-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kitazawa M., Kubo Y., Nakajo K. The stoichiometry and biophysical properties of the Kv4 potassium channel complex with K+ channel-interacting protein (KChIP) subunits are variable, depending on the relative expression level. J. Biol. Chem. 2014;289:17597–17609. doi: 10.1074/jbc.M114.563452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Sabi A., Shamotienko O., Dolly J.O. Arrangement of Kv1 α subunits dictates sensitivity to tetraethylammonium. J. Gen. Physiol. 2010;136:273–282. doi: 10.1085/jgp.200910398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ledwell J.L., Aldrich R.W. Mutations in the S4 region isolate the final voltage-dependent cooperative step in potassium channel activation. J. Gen. Physiol. 1999;113:389–414. doi: 10.1085/jgp.113.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vardanyan V., Pongs O. Coupling of voltage-sensors to the channel pore: a comparative view. Front. Pharmacol. 2012;3:145. doi: 10.3389/fphar.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Labro A.J., Raes A.L., Snyders D.J. Kv channel gating requires a compatible S4-S5 linker and bottom part of S6, constrained by non-interacting residues. J. Gen. Physiol. 2008;132:667–680. doi: 10.1085/jgp.200810048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chowdhury S., Haehnel B.M., Chanda B. Interfacial gating triad is crucial for electromechanical transduction in voltage-activated potassium channels. J. Gen. Physiol. 2014;144:457–467. doi: 10.1085/jgp.201411185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.