Abstract
We introduce Photon-HDF5, an open and efficient file format to simplify exchange and long-term accessibility of data from single-molecule fluorescence experiments based on photon-counting detectors such as single-photon avalanche diode, photomultiplier tube, or arrays of such detectors. The format is based on HDF5, a widely used platform- and language-independent hierarchical file format for which user-friendly viewers are available. Photon-HDF5 can store raw photon data (timestamp, channel number, etc.) from any acquisition hardware, but also setup and sample description, information on provenance, authorship and other metadata, and is flexible enough to include any kind of custom data. The format specifications are hosted on a public website, which is open to contributions by the biophysics community. As an initial resource, the website provides code examples to read Photon-HDF5 files in several programming languages and a reference Python library (phconvert), to create new Photon-HDF5 files and convert several existing file formats into Photon-HDF5. To encourage adoption by the academic and commercial communities, all software is released under the MIT open source license.
Introduction
Single-molecule fluorescence techniques have facilitated groundbreaking developments in biophysics, as recently recognized by the Nobel committee (1). Techniques based on acquisition of long series of images obtained with ultrasensitive cameras are the most popular among those, with numerous published software tools (2, 3, 4). A related set of single-molecule fluorescence techniques relying on photon-counting devices is used in studies of freely diffusing single molecules (5, 6, 7), studies of single molecules in microfluidic mixers (8), optical or ABEL traps (9, 10, 11), or on immobilized molecules (12), where high temporal resolution (<1 ms) is required. Among these techniques, single-molecule fluorescence resonant energy transfer (smFRET) (13), microsecond alternating laser excitation (14), nanosecond alternating laser excitation (15), also known as pulse interleaved excitation (16), two-color coincident detection (17, 18), polarization anisotropy (19, 20), fluorescence correlation and cross-correlation spectroscopy (21, 22) or multiparameter fluorescent detection (23) are but a few in a constantly growing list.
Contrary to camera-based techniques, where most manufacturers provide a way to save data in a standardized format such as TIFF images, there is no such standardization for photon-counting hardware, where manufacturers of integrated systems have their own binary formats, and most systems are custom-built, including custom software generating oftentimes poorly documented binary files.
This situation creates a number of issues. First, it is rarely possible for an outsider (for instance a reviewer) to access the raw data of a published study without considerable coding work. Moreover, this assumes that the authors have provided a comprehensive description of their format. Worse, it is often difficult for different generations of students in a single laboratory to decipher data file formats created by their predecessors. In summary, the proliferation of file formats impedes interoperability between different software, creates barriers to data sharing among researchers, and undermines results reproducibility and long-term preservation of data. Moreover, with the growing trend by funding agencies to request data sharing and documentation, having multiple file formats will inevitably result in significant duplication of efforts by each group.
To help address these issues, we developed Photon-HDF5 (24), an open file format for single-molecule fluorescence experiments using photon-counting devices, including time-correlated single-photon counting (TCSPC) data. The file format description comes accompanied with a reference Python library (phconvert) (25) to create Photon-HDF5 files from raw data or existing file formats, as well as examples (26) on how to read Photon-HDF5 files in Python, MATLAB, and LabVIEW. Both format documentation and software are hosted on GitHub, a popular collaborative platform (27), with the aim of allowing the biophysics community to contribute to this project with comments, suggestions, and code (28).
The article is organized as follows. In the Current Situation section, we briefly review the current situation (characterized by a myriad of formats) and showcase a few successful examples of standardization in other scientific domains. In the Proposal of a New File Format section, we examine requirements for an efficient, maintainable file format, and introduce HDF5, a format on which Photon-HDF5 is based. We then describe the overall structure of Photon-HDF5 files and support software. In the Scenarios of Photon-HDF5 Usage section, we illustrate the use of this format with a few scenarios, and in the Conclusions and Perspectives section, we conclude with a summary of Photon-HDF5 key features and with a call for the community to participate in shaping the evolution of the format.
Current Situation
Photon-counting based experiments
Photon-counting based single-molecule fluorescence experiments can rapidly generate large amounts of data: in a typical experiment, count rates from a few kHz to several 10 kHz per detector and measurement durations of hours are not uncommon and, increasingly, a large number of individual detectors are encountered (29, 30, 31). Because each photon’s information (timestamp, channel number, etc.) may occupy 64 or more bits per photon, most experiments store data in binary formats, which minimize disk space usage, at the expense of human readability. Because there is no natural or standardized structure for such a file format, each research group or company designing or putting together photon-counting instrumentation for single-molecule fluorescence experiments usually ends up developing its own custom binary file format.
In contrast with a relatively well self-documenting text-based format, binary formats require detailed documentation of the file structure to be properly read (or written). Furthermore, extending a binary format to handle a new piece of information (such as modifications to the setup hardware) usually results in a new, incompatible format requiring specific handling and new, time-consuming code development. Because most researchers are, understandably, not inclined to invest significant resources in developing and documenting file formats, it is common to end up with a collection of poorly documented ad hoc binary formats within each individual research group. The obvious risk is that, over the years, information about those file formats gets lost, effectively leading to data loss.
Although vendors of photon-counting hardware and software have usually done a better job at designing and documenting file formats, their formats are nonetheless tightly linked to specific hardware and therefore, generally not suitable to store data from a competitor’s systems or from custom hardware used in individual laboratories. Additionally, although these formats can store some predefined metadata (such as for instance author, date of creation, and comment), they are not generally extensible to accept additional metadata, necessary to describe a measurement in details.
The result of this situation is that it is difficult, if not impractical, for research groups not using identical hardware and software to exchange data to compare methods and results, and within each group, migration to new hardware or software is discouraged or results in incompatible data formats.
Examples of other formats
The need for standard file formats for domain-specific data sets is a common occurrence in many scientific fields. Most in the biophysics community will be familiar with the PDB file format for biomolecular structures maintained by the Protein Data Bank (32), or with the already mentioned TIFF format for images. The latter format, copyrighted by Adobe Systems (San Jose, CA), is only marginally adapted to the complexity of scientific data, but is used as a least common denominator export format by camera vendors in the absence of a better solution. The FITS format developed by the astronomy community (33) (and supported by some camera manufacturers) is a good example of a powerful alternative, although its design principles are dated and it has not been adopted by the biophysics or microscopy communities.
In the single-molecule community, a text-based file format (single-molecule dataset (SMD)) was recently proposed to store data from a variety of surface-immobilized, camera-based experiments (34). These measurements consist of a series of images, from which time traces (time binned data) are extracted for individual emitters identified in the series. These time traces have relatively short duration (limited by fluorophore bleaching) and therefore have minimal space requirements. The proposed SMD format based on JavaScript Object Notation (JSON) (34) is therefore an appropriate choice for these structured data sets of relatively small size. On the other end, using SMD for timestamp-based experiments, would be inefficient (due to its text-based nature) and result in irreversible information loss (due to the time binning).
Proposal of a New File Format
Design principles
The design of a standard file format for long-term data preservation and to facilitate interoperability and data sharing, presents several (often contradicting) requirements.
Flexibility
The format must be capable of efficiently storing a variety of numeric data types and strings to represent both raw data and all metadata associated with an experiment. Moreover, the structure must be general enough to accommodate different types of timestamp-based single-molecule experiments. For instance, it should support experiments using continuous wave or pulsed excitation, one or many detection channels, one or more excitation wavelengths, etc. Because it would be impossible to predict all possible future technical developments, it should be easily extensible and customizable to accommodate them.
Ease of use
To encourage adoption and facilitate long-term maintainability, the file structure and complexity should be kept as low as possible. It should be possible for a user to quickly write a reader for the file format in any programming language and operating system of choice. Furthermore, there should be tools to facilitate creation of data files that complies with the specifications.
Small size
Although disk storage is increasingly inexpensive, the advantages of a compact file format should not be overlooked. Smaller files are easier to backup, archive online, and share as well as generally faster to read. The format must therefore support smart compression allowing fast read and write, and partial loading of large data sets.
The parent HDF5 format
HDF5 is an existing open hierarchical file format satisfying all requirements listed previously. In the following, we provide a brief overview of the HDF5 format. Detailed information can be found on the HDF Group website (35).
HDF5 is a general-purpose binary format designed to store numerical data sets, widely used in industry and in several scientific communities. HDF5 development is led by the nonprofit HDF Group, which also maintains open-source platform-independent libraries to read and write HDF5 files in multiple languages.
HDF5 has a hierarchical (tree-like) data structure similar to files and folders in a file system: groups are the equivalent of folders, whereas data fields (e.g., arrays) are the equivalent of files. The main group of a HDF5 file is called “root” (indicated by the slash character “/”) and can contain data fields or other groups. The HDF5 format is self-describing, meaning that a user does not need to know the hierarchical structure or the data types before reading the file. For example, to access a numerical array in a HDF5 file, the user only needs to specify its path: the array will be automatically loaded into memory using the appropriate data type. In other words, all the byte-level details (such as byte order and data type width) are transparently handled, greatly simplifying data reading and writing. Moreover, HDF5 transparently supports compression, allowing reducing file size while maintaining high reading speed without any added complexity for the user. Finally, each node in a HDF5 file (groups or data fields) can have any number of attributes, such as descriptions, which can be used to embed metadata to a data set.
Libraries to operate on HDF5 files currently exist for the major programming languages used in scientific computing, such as C, Fortran, C++, Java, Python, R, MATLAB, and LabVIEW. Furthermore, a free multiplatform graphical viewer and editor (HDFView (36)) allows users to quickly explore, modify, or export content of HDF5 files. The availability of well-maintained open source libraries allows users to reliably manage HDF5 files without the need to deal with the underlying byte-level structure, effectively overcoming the drawbacks of binary formats highlighted in the Current Situation section.
HDF5 has been widely adopted in industry and in several scientific communities as the basis for specialized data formats. Because of its openness, popularity, and long-term support, it represents a safe choice for long-term data preservation.
Photon-HDF5 file format
HDF5 is the foundation for the proposed format for photon-counting single-molecule fluorescence data, therefore called Photon-HDF5 file format.
Photon-HDF5 is, essentially, a conventional structure to store timestamp-based single-molecule fluorescence data in HDF5 files (Fig. 1). Because Photon-HDF5 files are HDF5 files, the requirements of an open, efficient, well-documented, and platform- and language-independent are automatically fulfilled.
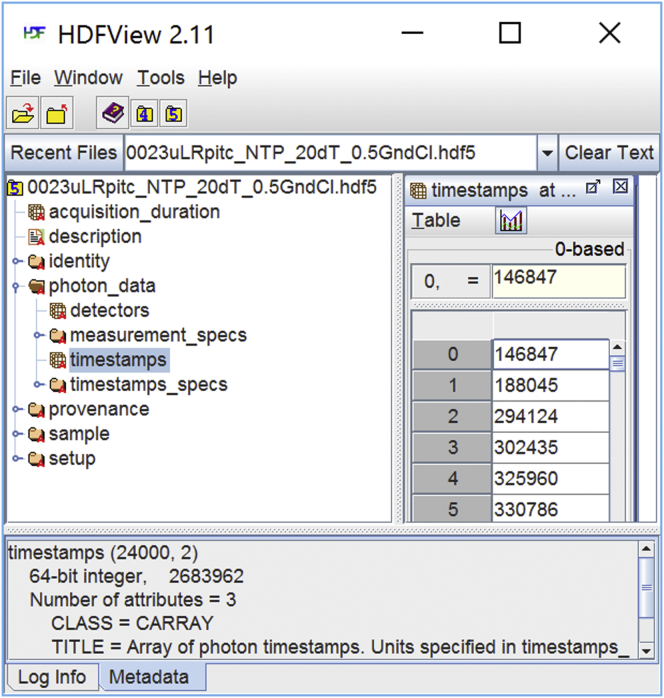
Figure 1.
A Photon-HDF5 file as seen with the HDFView visualizer. In contrast with other binary formats, Photon-HDF5 files (or any other HDF5 file) can be examined with an open-source Java visualizer (HDFView). Note that the file structure (left pane) is immediately intelligible to the user, who can navigate it and select items for display or export. The timestamps array is selected in the left pane and its content displayed in the right pane. In the bottom pane, the TITLE attribute, among other attributes saved in the file, shows a description for the selected item (timestamps). Photon-HDF5 files include descriptions for every field, facilitating interactive file exploration. To see this figure in color, go online.
We now briefly describe the main components of a Photon-HDF5 file. The interested reader is invited to visit the photon-hdf5.org website for further information and a complete description of the format and associated tools.
Photon-HDF5 structure
The Photon-HDF5 file structure contains five main groups (photon_data, setup, sample, identity, and provenance) and two root fields: a general description string (description) and the acquisition duration (acquisition_duration). The general structure is illustrated in Fig. 2 (see also Figs. S1 and S2 in the Supporting Material), whereas Fig. 1 shows a Photon-HDF5 file as it appears when opened with the HDFView visualizer.
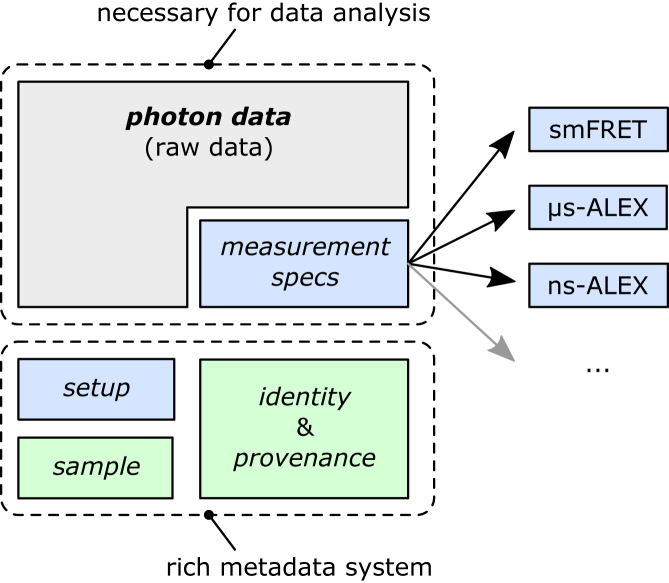
Figure 2.
Photon-HDF5 file structure. The core photon_data group contains the raw data. A file containing only raw data is still a valid Photon-HDF5 file but it cannot be analyzed without additional information on what the data represents. The optional (but recommended) measurement_specs group identifies the type of measurement and specifies the associated metadata (i.e., role of each detector, measurement parameters, etc.). When measurement_spec is present, the user has all the information needed to analyze the data. The other groups enhance the data file legibility, providing important measurement metadata. setup contains information on the system used for acquisition (e.g., excitation wavelengths, laser power(s), CW or pulsed excitation, etc.). sample contains sample information (e.g., dyes, buffer, etc.). The last two groups, identity and provenance, provide author and file information. To see this figure in color, go online.
The photon_data group. As visible in Fig. 1 (and detailed in Fig. S1), the photon_data group contains the raw data (timestamps, detectors, and nanotimes arrays), as well as information needed to analyze it (in the measurement_specs subgroup). All arrays in photon_data have the same length, equal to the number of recorded photons. Information corresponding to the ith photon (its macrotime stamp, detector identity, and when applicable, the microtime measured by TCSPC hardware, or nanotime) is stored in the ith value of each of these arrays.
Except for timestamps, the other arrays are present in a file only when needed, i.e., the detectors array is present only if there is more than one detector and the nanotimes array is present only if TCSPC timing data is recorded.
Inside photon_data, the measurement_specs group contains the measurement type and a type-dependent set of metadata fields. The currently supported measurement types are listed in Table S1. Each measurement type (specified in measurement_type) requires a particular set of fields, which completely describe the experiment. For example, a single-laser single-molecule FRET data file will have a smFRET measurement type, and will include two numeric fields called spectral_ch1 and spectral_ch2 containing the donor and acceptor detector ID.
Defining a new measurement type involves choosing a name and a set of required fields in the measurements_specs group. Defining new measurement types allows gradually extending the file format to support new measurement types, without compromising the legibility of already existing files.
Data contained in the measurement_specs group provides all the necessary information needed to interpret raw experimental data. Files not including a measurement_specs group are still valid Photon-HDF5 files, but a user cannot analyze their content without knowing additional measurement specifications. Such an option is provided only to store incomplete or debugging data and is not recommended for data that need to be shared. For new measurement types, users should submit new measurement_specs definitions to the community for integration in new releases of the specifications (37).
The setup group. The setup group stores setup properties such as laser excitation wavelengths and powers, the number of detected spectral bands (e.g., two-colors or three-colors), the number of detected polarization states (e.g., none = 1 and 2 polarizations = 2) and the number of split channels (i.e., identical spectral or polarization channels split by nonpolarizing beam splitters).
Metadata groups. In addition to the previous groups, a Photon-HDF5 file may contain three other groups (sample, identity, and provenance), which provide additional optional metadata (see Figs. 2 and S2).
The sample group contains a description of the sample (sample_name), the buffer (buffer_name), and the fluorophore names (dye_names). Storing this basic sample information with the data simplifies preservation of experimental details.
The identity group contains identification metadata such as data set author (and its affiliation), Photon-HDF5 format version, software used to save the file (and its version), and optional data set identifiers such as DOI or URL. This information is important when sharing data sets online, to keep track of authorship and provide proper credit.
As discussed in the Scenarios of Photon-HDF5 Usage section, a Photon-HDF5 file might be created by converting an original data file saved by an acquisition (or simulation) software outputting some custom file format. The provenance group is the place to store this information, such as the name and creation date of the original file, and the software used to save it.
Additional file format features
Field data types. Leveraging the self-describing nature of HDF5, instead of prescribing specific data types, Photon-HDF5 specifications only indicate whether a field is a scalar, an array, or a string and whether the data type is integer or floating point. For examples, the detectors array (array of detector IDs) typically uses an unsigned 8-bit integer type, which is sufficient for the majority of setups. However, when using single-photon avalanche diode arrays with >256 pixels, a 16- or 32-bit integer data type can be used without any modification to the file format.
Custom user groups. For maximum flexibility, users can store arbitrary amounts of additional (custom) data in any position within the Photon-HDF5 hierarchy. To avoid incompatibilities with future versions of the format, it is required that custom data be stored inside a group (or subgroup) named user.
Future Photon-HDF5 versions. To guarantee long-term accessibility to Photon-HDF5 files, future versions of the file format software will be backward-compatible. Specifically, previously defined fields will never be renamed or have their meaning modified. This ensures that software written to read an old version will be still capable to correctly read a newer version, although it will not be able to interpret newer fields or measurement types.
Reading and writing Photon-HDF5
Reading Photon-HDF5 files does not differ from reading any other HDF5 file and involves using the HDF5 library for the language of choice. A specific advantage is that field names (and their meaning) are defined in the specifications (SM.1 in the Supporting Material). Examples of how to read Photon-HDF5 files in several languages are provided in SM.2 and reference (26) (currently for Python, MATLAB, and LabVIEW). Additional discussion on reading Photon-HDF5 files can be found in the Reading Photon-HDF5 files section of the reference documentation (38).
To create Photon-HDF5 files, users can convert existing files or save them directly from suitably modified acquisition software. Conversion is generally the simplest approach and, when using acquisition software whose source code is unavailable, the only possible one. To simplify saving and converting Photon-HDF5 files, we maintain phconvert, an open-source Python library serving as reference implementation for the Photon-HDF5 format (25). phconvert includes a browser-based interface using Jupyter Notebooks (39, 40) to convert supported file formats into Photon-HDF5. The formats currently supported are HT3 (from PicoQuant TCSPC hardware, Berlin, Germany), SPC/SET (from Becker & Hickl TCSPC hardware, Berlin, Germany) as well as SM (a legacy file format used in S.W’s laboratory ). We provide an online demonstration service to run these notebooks and convert these formats to Photon-HDF5 without software installation on the user’s computer (41).
Additionally, the phconvert library can be used in any Python program to easily create Photon-HDF5 files from scratch (see SM.3). Although Photon-HDF5 files can be created using only a HDF5 library (e.g., pytables or h5py), one advantage of using phconvert is that files are validated and guaranteed to conform to the specifications. Phconvert, in fact, checks that all fields have correct names and types, and adds a description to each field (see Fig. 1 for an example). Furthermore, phconvert automatically computes and fills several fields (e.g., file creation and modification dates, format and software names and versions, acquisition duration, etc.) reducing the amount of metadata the user needs to provide. Programs written in languages different from Python, can benefit from the same advantages of using phconvert for creating Photon-HDF5 files by calling a simple script called phforge (42) whose use is described in SM.3.2.
Other currently unsupported formats can be converted to Photon-HDF5 simply writing a Python function to load the data in memory and using phconvert to save the data to Photon-HDF5. Taking the existing loader functions as examples, this task is relatively easy, even for inexperienced Python programmers. We encourage interested users to contribute to phconvert so that out-of-the-box support for conversion of the largest number of formats can be provided. For a discussion on saving Photon-HDF5 from Python or other languages (see SM.3 and the reference documentation (43).
Scenarios of Photon-HDF5 Usage
In this section, we illustrate a few use cases where employing the Photon-HDF5 format may lead to significant advantages.
Researchers performing (or interested in) photon-counting based single-molecule experiments can be classified in the following three categories. Users using:
-
a.
Commercial acquisition hardware and software (Alice)
-
b.
Custom acquisition hardware and software (Bob)
-
c.
Mixture of a and b (Charlie)
In each case, Alice, Bob, and Charlie use one or more custom or proprietary binary formats to store data from their experiments, and will use a custom or commercial software to analyze their data. We now examine three typical situations encountered by researchers.
Example 1 (publication)
Alice wants to publish an article including her data so that as many researchers as possible can look at it (including the reviewers, who may not, however, have time to process it and might just be curious to take a peek at it).
The Photon-HDF5 format and HDFViewer will do the job for the reviewer and researchers using a different system, whereas the original format will allow only readers having the same commercial software to have a look at the data. If she uses one of the two most common commercial formats, she can use phconvert to create the Photon-HDF5 version. If not, writing code to create a Photon-HDF5 version should be simple thanks to the examples provided. Moreover, her code could be added to phconvert for the benefit of the entire community.
Example 2 (collaboration)
Alice and Bob want to collaborate on a project, Alice doing the experiments and Bob performing some custom analysis with his software.
Because Bob does not use Alice’s system, he would have to decipher the proprietary file format description of the vendor and extract the relevant data for his analysis. By asking Alice to first convert her file to the Photon-HDF5 format with phconvert, he will benefit from file size reduction, simple access to the data, and by incorporating the necessary code in his software, he will gain access to a variety of other files generated by other users of Photon-HDF5.
Example 3 (data management)
Charlie works with a student running some experiments on a commercial system, which he wants to compare to similar experiments he is carrying out on a new setup he has built with custom hardware and software.
Because Charlie needs to save new data, he needs a new format. But he also needs to read data generated by the student. The least effort consists in using a format that is efficient, well-documented and supported, and in which his student’s files can be easily converted. Photon-HDF5 naturally fits the bill, and additionally provides him with the same benefits enjoyed by Alice and Bob for publication and sharing.
Summary
Although these scenarios do not directly involve manufacturers of single-photon counting hardware and their associated software, it is clear that manufacturers should take some interest in an effort such as Photon-HDF5. Although it is not expected that they would abandon their proprietary format, their contribution to the definition of future versions of the format or to conversion tools is welcome and encouraged, as is that of anyone interested in the research community.
Conclusions and Perspectives
We have developed the Photon-HDF5 file format as a hardware-agnostic format for single-molecule fluorescence data based on photon-counting detectors. Photon-HDF5 is easy to use, customize, and extend, in addition to inheriting all the advantages of the HDF5 format (open standard, self-described, and efficient).
The format has been designed both for interoperability between different analysis software and long-term data archival. For this purpose, it includes a rich set of metadata to make data files self-contained. Photon-HDF5 facilitates data sharing between research groups by reducing the efforts needed to decode the wide variety of binary formats used across laboratories.
Although we have only discussed standard single-spot confocal experiments in the text, the format supports and is currently used for multispot experiments using multipixel single-photon avalanche diode arrays (24). Further extensions to accommodate future developments or new measurement types can be naturally incorporated.
Currently, the open source FRETBursts software package for smFRET burst analysis supports reading Photon-HDF5 files (44). Therefore, converting smFRET data to Photon-HDF5 already enables researchers to analyze a variety of data with this software. The PyBroMo simulator for smFRET experiments generates simulated smFRET data in Photon-HDF5 format (45), which can be analyzed by FRETBursts or other software supporting Photon-HDF5. Additional software packages supporting Photon-HDF5 files are expected to be released in the near future and links to their websites will be added to the photon-hdf5.org site.
The impact of Photon-HDF5 format will be determined by the extent of its adoption in the single-molecule community and its support by scientific publishers. To maximize the involvement of third parties, we embraced an open development model for both the format specification and the support software. Users are encouraged to send feedback (e.g., on perceived limitations or to add support for new measurement types) or other contributions. To facilitate the process, we include guidelines for contributors in the Photon-HDF5 reference documentation (24). Finally, by adopting an open consensus-based governance, we aim to empower the single-molecule community to determine the future directions of Photon-HDF5 format in order to become a standard widely-useful tool for data sharing and preservation.
Author Contributions
A.I. designed the file format, implemented the supporting software, wrote the reference documentation, websites, and article; T.L. designed the file format, implemented LabVIEW reading examples, and wrote the article; R.B. implemented the MATLAB code for reading and writing Photon-HDF5 files; S.W. wrote the article; X.M. designed the file format, implemented LabVIEW reading and writing examples, and wrote the article.
Acknowledgments
We thank Dr. Sangyoon Chung and Dr. Eitan Lerner for providing experimental data files.
This work was supported in part by National Institutes of Health (NIH) grant R01-GM95904 and by U.S. Department Energy (DOE) grant DE-FC02-02ER63421-00. Dr. Weiss discloses equity in Nesher Technologies and intellectual property used in the research reported here. The work at UCLA was conducted in Dr. Weiss’s Laboratory. Dr. Laurence’s work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under contract DE-AC52-07NA27344.
Editor: Elizabeth Rhoades.
Footnotes
Supporting Materials and Methods, Supporting Results, Supporting Discussion, five figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)01170-4.
Supporting Material
References
- 1.Lippincott-Schwartz J. Profile of Eric Betzig, Stefan Hell, and W. E. Moerner, 2014 Nobel Laureates in Chemistry. Proc. Natl. Acad. Sci. USA. 2015;112:2630–2632. doi: 10.1073/pnas.1500784112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKinney S.A., Joo C., Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van de Meent J.-W., Bronson J.E., Gonzalez R.L., Jr. Empirical Bayes methods enable advanced population-level analyses of single-molecule FRET experiments. Biophys. J. 2014;106:1327–1337. doi: 10.1016/j.bpj.2013.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenfeld M., Pavlichin D.S., Herschlag D. Single molecule analysis research tool (SMART): an integrated approach for analyzing single molecule data. PLoS One. 2012;7:e30024. doi: 10.1371/journal.pone.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fries J.R., Brand L., Seidel C.A.M. Quantitative identification of different single molecules by selective time-resolved confocal fluorescence spectroscopy. J. Phys. Chem. A. 1998;102:6601–6613. [Google Scholar]
- 6.Dahan M., Deniz A.A., Weiss S. Ratiometric measurement and identification of single diffusing molecules. Chem. Phys. 1999;247:85–106. [Google Scholar]
- 7.Boukobza E., Sonnenfeld A., Haran G. Immobilization in surface-tethered lipid vesicles as a new tool for single biomolecule spectroscopy. J. Phys. Chem. B. 2001;105:12165–12170. [Google Scholar]
- 8.Lipman E.A., Schuler B., Eaton W.A. Single-molecule measurement of protein folding kinetics. Science. 2003;301:1233–1235. doi: 10.1126/science.1085399. [DOI] [PubMed] [Google Scholar]
- 9.Fields A.P., Cohen A.E. Electrokinetic trapping at the one nanometer limit. Proc. Natl. Acad. Sci. USA. 2011;108:8937–8942. doi: 10.1073/pnas.1103554108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q., Goldsmith R.H., Moerner W.E. Probing single biomolecules in solution using the anti-Brownian electrokinetic (ABEL) trap. Acc. Chem. Res. 2012;45:1955–1964. doi: 10.1021/ar200304t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McHale K., Berglund A.J., Mabuchi H. Quantum dot photon statistics measured by three-dimensional particle tracking. Nano Lett. 2007;7:3535–3539. doi: 10.1021/nl0723376. [DOI] [PubMed] [Google Scholar]
- 12.Yang H., Luo G., Xie X.S. Protein conformational dynamics probed by single-molecule electron transfer. Science. 2003;302:262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 13.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 14.Kapanidis A.N., Lee N.K., Weiss S. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laurence T.A., Kong X., Weiss S. Probing structural heterogeneities and fluctuations of nucleic acids and denatured proteins. Proc. Natl. Acad. Sci. USA. 2005;102:17348–17353. doi: 10.1073/pnas.0508584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller B.K., Zaychikov E., Lamb D.C. Pulsed interleaved excitation. Biophys. J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orte A., Clarke R., Klenerman D. Determination of the fraction and stoichiometry of femtomolar levels of biomolecular complexes in an excess of monomer using single-molecule, two-color coincidence detection. Anal. Chem. 2006;78:7707–7715. doi: 10.1021/ac061122y. [DOI] [PubMed] [Google Scholar]
- 18.Clarke R.W., Orte A., Klenerman D. Optimized threshold selection for single-molecule two-color fluorescence coincidence spectroscopy. Anal. Chem. 2007;79:2771–2777. doi: 10.1021/ac062188w. [DOI] [PubMed] [Google Scholar]
- 19.Ha T., Enderle T., Weiss S. Single molecule dynamics studied by polarization modulation. Phys. Rev. Lett. 1996;77:3979–3982. doi: 10.1103/PhysRevLett.77.3979. [DOI] [PubMed] [Google Scholar]
- 20.Schaffer J., Volkmer A., Seidel C.A.M. Identification of single molecules in aqueous solution by time-resolved fluorescence anisotropy. J. Phys. Chem. A. 1999;103:331–336. [Google Scholar]
- 21.Rigler R., Mets Ü., Kask P. Fluorescence correlation spectroscopy with high count rate and low background: analysis of translational diffusion. Eur. Biophys. J. 1993;22:169–175. [Google Scholar]
- 22.Schwille P., Meyer-Almes F.J., Rigler R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 1997;72:1878–1886. doi: 10.1016/S0006-3495(97)78833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eggeling C., Berger S., Seidel C.A.M. Data registration and selective single-molecule analysis using multi-parameter fluorescence detection. J. Biotechnol. 2001;86:163–180. doi: 10.1016/s0168-1656(00)00412-0. [DOI] [PubMed] [Google Scholar]
- 24.Photon-HDF5 Home Page. http://photon-hdf5.org.
- 25.Ingargiola, A. phconvert - Library and converter for Photon-HDF5 files. http://photon-hdf5.github.io/phconvert.
- 26.Reading Photon-HDF5 in multiple languages. http://photon-hdf5.github.io/photon_hdf5_reading_examples/.
- 27.Photon-HDF5 project page on GitHub. https://github.com/Photon-HDF5.
- 28.Photon-HDF5. Guidelines for contributors. http://photon-hdf5.readthedocs.org/en/latest/contributing.html.
- 29.Rech I., Marangoni S., Cova S. Multipixel single-photon avalanche diode array for parallel photon counting applications. J. Mod. Opt. 2009;56:326–333. [Google Scholar]
- 30.Laurence T.A., Ly S., Coleman M.A. Fluorescence correlation spectroscopy at micromolar concentrations without optical nanoconfinement. J. Phys. Chem. B. 2014;118:9662–9667. doi: 10.1021/jp505881z. [DOI] [PubMed] [Google Scholar]
- 31.Michalet X., Ingargiola A., Ghioni M. Silicon photon-counting avalanche diodes for single-molecule fluorescence spectroscopy. IEEE J. Sel. Top. Quantum Electron. 2014;20:38044201–380442020. doi: 10.1109/JSTQE.2014.2341568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berman H., Henrick K., Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 33.FITS. The Astronomical Image Table Format. http://fits.gsfc.nasa.gov/.
- 34.Greenfeld M., van de Meent J.-W., Herschlag D. Single-molecule dataset (SMD): a generalized storage format for raw and processed single-molecule data. BMC Bioinformatics. 2015;16:3. doi: 10.1186/s12859-014-0429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.HDF5 Homepage. https://www.hdfgroup.org/HDF5/.
- 36.HDFView Homepage. https://www.hdfgroup.org/products/java/release/download.html.
- 37.Photon-HDF5. Defining new measurement types. http://photon-hdf5.readthedocs.org/en/latest/new_measurement_specs.html.
- 38.Photon-HDF5 Reference. Reading Photon-HDF5 files. http://photon-hdf5.readthedocs.org/en/latest/reading.html.
- 39.Shen H. Interactive notebooks: Sharing the code. Nature. 2014;515:151–152. doi: 10.1038/515151a. [DOI] [PubMed] [Google Scholar]
- 40.Jupyter Project Homepage. https://jupyter.org/.
- 41.Photon-HDF5 Online Converter. http://photon-hdf5.github.io/Photon-HDF5-Converter/.
- 42.phforge: a script for creating Photon-HDF5 files. http://photon-hdf5.github.io/phforge/.
- 43.Photon-HDF5 Reference. Writing Photon-HDF5 files. http://photon-hdf5.readthedocs.org/en/latest/writing.html.
- 44.Ingargiola, A. FRETBursts - Burst analysis software for smFRET experiments. http://tritemio.github.io/FRETBursts/.
- 45.Ingargiola, A. PyBroMo, smFRET simulatotor using 3D Brownian motion and fluorescence emission under confocal excitation. http://tritemio.github.io/PyBroMo/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.