Abstract
Current in vitro optical studies of microtubule dynamics tend to rely on fluorescent labeling of tubulin, with tracking accuracy thereby limited by the quantum yield of fluorophores and by photobleaching. Here, we demonstrate label-free tracking of microtubules with nanometer precision at kilohertz frame rates using interferometric scattering microscopy (iSCAT). With microtubules tethered to a glass substrate using low-density kinesin, we readily detect sequential 8 nm steps in the microtubule center of mass, characteristic of a single kinesin molecule moving a microtubule. iSCAT also permits dynamic changes in filament length to be measured with <5 nm precision. Using the arbitrarily long observation time enabled by label-free iSCAT imaging, we demonstrate continuous monitoring of microtubule disassembly over a 30 min period. The ability of iSCAT to track microtubules with nm precision together with its potential for label-free single protein detection and simultaneous single molecule fluorescence imaging represent a unique platform for novel approaches to studying microtubule dynamics.
Introduction
Microtubules (MTs) are tubular filaments with an outer diameter of 24 nm built via head-to-tail polymerization of tubulin heterodimers. Polymerization is coupled to GTP hydrolysis, such that newly polymerized GTP-tubulin forms a cap at the ends of the MT, whereas the older core of the filament is composed of less stable GDP-tubulin. As a result, MTs built from pure tubulin grow steadily provided the stabilizing GTP-tubulin caps remain intact, whereas breaches in the cap expose the GDP core of the MT and trigger rapid shrinkage. The cycle of growth, catastrophe, shrinkage, and regrowth (rescue) is termed dynamic instability and is critical to the cellular function of MTs (1).
In addition, MTs play numerous important roles by acting as a scaffold for intracellular structure and serving as molecular tracks for a variety of motor proteins (2). Biogenesis and subsequent motility of cilia and flagella requires MT assembly. The tight control of MT dynamics is essential for MT function in cell proliferation and differentiation (3, 4).
Direct imaging of MT assembly and kinesin-driven MT gliding motility is possible using dark-field and a number of interference-based microscopy techniques (5, 6, 7, 8). More recently, it has been shown that MTs can also be visualized using computer-enhanced bright-field microscopy (9). The intrinsic background and noise characteristics of these three traditional contrast modalities, however, limit the localization precision and therefore the ability to visualize nanometer-scale dynamics.
Fluorescent labeling of tubulin enables superresolution imaging of MTs but has several limitations: fluorescent dyes photobleach, which limits the number of photons that can be collected from a single fluorophore and, thereby, the spatiotemporal resolution (10, 11). The localization precision is further affected by residual background originating from free tubulin. For nanometer-precise imaging, more sophisticated labeling schemes have been applied, such as the use of quantum dots (12). Even for quantum dots, however, the temporal resolution in tracking MTs is limited to several tens of milliseconds. Further difficulties include the potential for structural perturbations due to the label, and blinking.
Here, we show that interferometric scattering (iSCAT) microscopy (13, 14) can be used to track the position and length of individual, unlabeled MTs with simultaneous millisecond temporal and subnanometer lateral precision for arbitrarily long observation times.
Materials and Methods
iSCAT microscope
Our experimental setup is similar to that reported recently (15). Briefly, we image a 6 × 6 μm region at 1000 frames/s using a 445 nm diode laser scanned across the sample by two acoustooptic deflectors. The beam deflections generated by the acoustooptic deflectors are imaged into the back focal plane of an oil immersion objective (PLAPON 1.42 NA, 60×; Olympus, Center Valley, PA) after passing through a polarizing beam splitter. A quarter wave plate before the objective causes any light reflected and scattered by the sample to be reflected by the polarizing beam splitter before being imaged onto a complementary metal-oxide semiconductor camera (MV-D1024-160-CL-8, Photonfocus, Lachen, Switzerland) at 333× magnification. The incident power was 17 kW/cm2 at the sample adjusted to achieve near saturation of the camera at an exposure time of 0.56 ms.
Sample preparation
The kinesin used was a truncated dimeric kinesin-1, consisting of the first 430 residues of the rat kinesin heavy chain.
MT preparation
MTs were polymerized from 20 μg porcine tubulin (Cytoskeleton) suspended in 20 μL GBRB80 buffer (80 mM PIPES, pH 6.9, 1 mM EGTA, and 5% glycerol) with 1 mM NaGTP and 2.5 nM MgCl2 and incubated for 30 min at 37°C before adding taxol to 20 μM. Fresh aliquots were used every day.
Chamber preparation
A 50 × 25 mm cover glass was cleaned by washing with ultrapure water (Milli-Q; Millipore, Billerica, MA), ethanol, and again with Milli-Q water. After drying, it was passed through a blue flame to remove dust and residual moisture. A channel was created between the cleaned coverslip and a 25 × 25 mm coverslip with double-sided tape. The resulting volume of the chamber was between 15 and 20 μL.
Kinesin gliding assay
The experiments were performed in motility buffer (MB: 20 mM PIPES, pH 7.4, 10 mM K-acetate, 4 mM MgCl2, and 5 mM dithiothreitol). Two volumes of kinesin solution were pipetted into the flow chamber and incubated for 2 min each time. Next, two volumes of 10 mg/mL bovine serum albumin (diluted in MB) were pipetted into the chamber, once again with an incubation time of 2 min each. This was required to block the remaining sites where kinesin motors were not bound. One volume of MT solution (diluted 50 times in MB containing taxol) was pipetted into the chamber; unbound MTs were washed away. At this stage, the iSCAT precision could be tested by moving the piezo stage in distinct steps. Finally, one volume of motility buffer supplemented with 10 μM ATP and 20 μM taxol was added and MT gliding was observed.
MT shrinking
Freshly prepared MTs were attached to the glass surface as described above and washed three times with MB that did not contain taxol.
MT tracking and step detection
Individual MTs were tracked by fitting their PSF to a functional form resulting from the convolution of a 2D Gaussian with a unit box given by the equation
where ; and . A is an amplitude; x and y denote MT position; sx/y is the width of the 2D Gaussian in the x and y directions; L is the long axis of the MT and t its short axis (which was fixed to 24 nm, corresponding to the diameter of the MT); θ refers to a rotation angle; and Erf is the error function.
Step detection was performed by a model- and parameter-free change point-detection algorithm based on the minimization of the Schwartz information criteria (16).
Results and Discussion
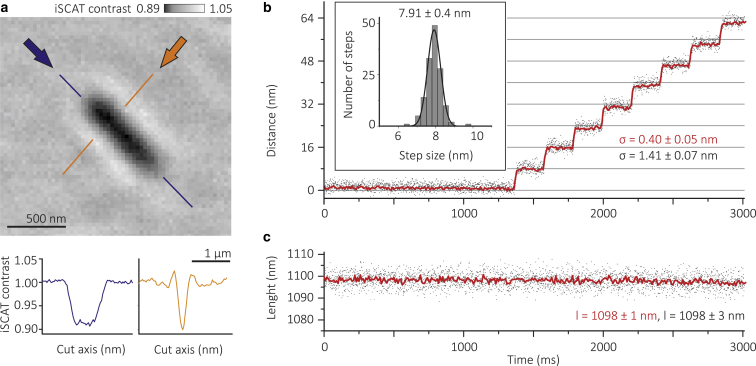
A typical iSCAT image of unlabeled MTs bound to a microscope cover slide is shown in Fig. 1 a. The contrast results from interference between light scattered by the MT and light reflected at the glass-water interface (14). The indicated cross sections along and across the long axis of the MT illustrate the magnitude of the filament signal compared to surface roughness (8% vs. 0.3%). To determine the length and center-of-mass position of such MTs, we fit the image to an elongated 2D Gaussian (see Materials and Methods). The standard deviation of the MT center-of-mass position was on the order of 1.4 nm at 1 kHz frame rate and dropped to 0.4 nm after 10-fold temporal binning (Fig. 1 b). A single kinesin molecule is expected to move an MT in 8 nm steps in a hand-over-hand fashion (17, 18). To test our ability to accurately and precisely measure steps on the order of 8 nm, we translated a surface-bound MT using a nanometric stage. We were able to resolve individual steps with 0.4 nm precision, and MT length could be determined with 1 nm precision at 100 Hz imaging speed (Fig. 1 c).
Figure 1.
Label-free imaging and tracking of a single microtubule (MT) with interferometric scattering microscopy. (a) iSCAT image of a single MT, including cross sections along and across (blue and orange arrows, respectively) its long axis. (b) Artificial steps performed by moving an MT bound to a cover glass with a nanometric stage in 8 nm steps. The tracking precision was determined by calculating the positional fluctuation, σ, which varies with imaging speed, but is independent of MT orientation relative to the motion of the nanometric stage. (c) MT length as determined by fitting the iSCAT images to an elongated Gaussian as described in the text. Raw data were acquired at an imaging speed of 1000 frames/s (gray dots) and averaged to 100 frames/s (red line). To see this figure in color, go online.
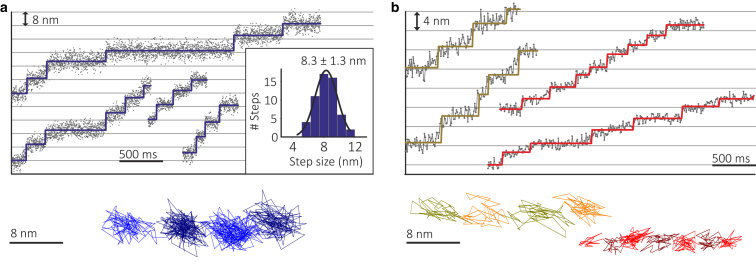
Next, we tracked MTs bound to kinesin motors attached to the glass surface in the presence of 10 μM ATP. We reduced the kinesin concentration until MT swivelling occurred, indicating that single kinesin motors are interacting with the MT (19, 20). At this point we could clearly resolve sequential 8 nm steps (Fig. 2 a; Movie S1 in the Supporting Material) in agreement with previous reports. The data were recorded at 1000 frames/s, two orders of magnitude faster than the current state of the art (12). A histogram of step sizes agrees with the previously reported behavior of single kinesin-1 molecules moving along an MT (17, 18).
Figure 2.
Analysis of MT motion driven by surface-bound kinesin. (a) Representative time traces showing 8 nm steps for MTs bound to single kinesins, including a representative xy trajectory acquired at 1000 frames/s. (b) Tracking results for higher motor densities exhibiting smaller but distinct fractional steps, including representative xy trajectories acquired at 100 frames/s. To see this figure in color, go online.
At higher motor densities, we frequently observed fractional steps on the order of 4 and even 2 nm, most likely resulting from several motors acting on a single MT (Fig. 2 b). To resolve these steps, we averaged the original 1000 frames/s movie to 100 frames/s, which improved the localization precision from 1.4 to 0.4 nm (Fig. 1 b). Although such small steps are difficult to discern in the time traces alone, they are clearly visible in the corresponding xy trajectories. We emphasize that we could not precisely control the number of motors interacting with a single MT in these experiments, a shortcoming that could be addressed in the future by using fluorescently labeled kinesin together with correlative iSCAT and imaging as demonstrated by tracking individual, dual-labeled myosin 5a molecules and quantum-dot-labeled virions (21, 22).
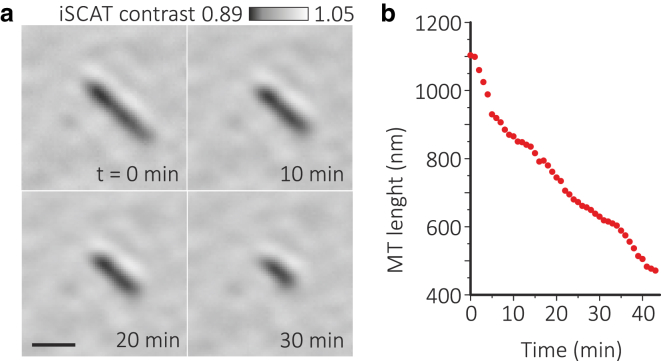
Finally, we present label-free imaging of a single unlabeled MT shrinking after removal of free tubulin and taxol from solution (Fig. 3; Movie S2). This measurement was taken over 40 min; we recorded 1 s movies every minute, even though the sample was constantly illuminated. Since iSCAT detection relies on light scattering instead of fluorescence emission, the illumination time can be arbitrarily long without suffering from photobleaching or blinking. In addition, nonresonant iSCAT illumination is less likely to cause photodamage to MTs due to sample heating and free radical formation, as is often encountered in the presence of fluorescent dyes.
Figure 3.
Shrinking of a microtubule upon the washout of taxol. (a) Time-lapse images acquired over 40 min. Scale bar, 500 nm. (b) MT length as a function of time. To see this figure in color, go online.
Conclusion
Our results demonstrate that light scattering from MTs when detected coherently in an interferometric geometry generates images of sufficient quality to enable high-speed center-of-mass tracking and length measurements with subnanometer precision without the need for any labels. These capabilities should enable detailed studies of how multiple motors interact with single MTs, how they coordinate and how their function scales with ATP concentration. Observation times and speeds are only limited by data storage and camera speed, respectively, enabling access to previously unattainable short (submillisecond) and long (hours) timescales while maintaining nanometer precision. In principle, nanometer-precise MT tracking could have been achieved at up to 1 MHz with an appropriate camera, given that the iSCAT contrast of individual MTs is comparable to 20 nm gold particles (23). Combination with single-molecule fluorescence and superresolution imaging could enable simultaneous localization of MTs and attached motors. Similarly, the recently demonstrated single protein sensitivity of iSCAT (15, 24) could be used for real-time monitoring of MT assembly and disassembly down to the single protein level.
Author Contributions
J.A. and K.L. performed research; J.O.A contributed analytic tools; J.A. analyzed the data; R.A.C. provided essential reagents; and J.A. and P.K. wrote the article with feedback from R.A.C.
Acknowledgments
The authors thank Adam Wollman for introducing them to the MT gliding assay. The data underpinning the results presented here can be accessed free of charge at http://ora.ox.ac.uk.
P.K. is supported by an European Research Council starting grant (NanoScope), J.A. by a Marie Curie Fellowship (330215), J.O.A. by a scholarship from CONACyT (scholar: 213546), and R.A.C. by the Wellcome Trust (103895/Z/14/Z).
Editor: E. Michael Ostap.
Footnotes
Two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)04695-0.
Supporting Material
The image size is 1.9 × 1.9 μm, 60 × 60 pixels (31.8 nm/pixel). The movie was acquired at a frame rate of 1000 frames/s, with every frame shown.
The image size is 2.2 × 2.2 μm, 70 × 70 pixels (31.8 nm/pixel). The movie was acquired for 1 second every minute over 40 minutes with the sample continuously illuminated. The original frame rate was 1000 frames/s but has been averaged to 1 frame/s.
References
- 1.Mitchison T., Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 2.Vale R.D. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 3.Kirschner M., Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 4.Howard J., Hyman A.A. Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Horio T., Hotani H. Visualization of the dynamic instability of individual microtubules by dark-field microscopy. Nature. 1986;321:605–607. doi: 10.1038/321605a0. [DOI] [PubMed] [Google Scholar]
- 6.Walker R.A., O’Brien E.T., Salmon E.D. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howard J., Hudspeth A.J., Vale R.D. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 8.Amos L.A., Amos W.B. The bending of sliding microtubules imaged by confocal light microscopy and negative stain electron microscopy. J. Cell Sci. Suppl. 1991;14:95–101. doi: 10.1242/jcs.1991.supplement_14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez-Medina B., Block S.M. Visualizing individual microtubules by bright field microscopy. Am. J. Phys. 2010;78:1152–1159. [Google Scholar]
- 10.Gell C., Bormuth V., Howard J. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 2010;95:221–245. doi: 10.1016/S0091-679X(10)95013-9. [DOI] [PubMed] [Google Scholar]
- 11.Nitzsche B., Bormuth V., Diez S. Studying kinesin motors by optical 3D-nanometry in gliding motility assays. Methods Cell Biol. 2010;95:247–271. doi: 10.1016/S0091-679X(10)95014-0. [DOI] [PubMed] [Google Scholar]
- 12.Leduc C., Ruhnow F., Diez S. Detection of fractional steps in cargo movement by the collective operation of kinesin-1 motors. Proc. Natl. Acad. Sci. USA. 2007;104:10847–10852. doi: 10.1073/pnas.0701864104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobsen V., Stoller P., Sandoghdar V. Interferometric optical detection and tracking of very small gold nanoparticles at a water-glass interface. Opt. Express. 2006;14:405–414. doi: 10.1364/opex.14.000405. [DOI] [PubMed] [Google Scholar]
- 14.Ortega-Arroyo J., Kukura P. Interferometric scattering microscopy (iSCAT): new frontiers in ultrafast and ultrasensitive optical microscopy. Phys. Chem. Chem. Phys. 2012;14:15625–15636. doi: 10.1039/c2cp41013c. [DOI] [PubMed] [Google Scholar]
- 15.Ortega Arroyo J., Andrecka J., Kukura P. Label-free, all-optical detection, imaging, and tracking of a single protein. Nano Lett. 2014;14:2065–2070. doi: 10.1021/nl500234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalafut B., Visscher K. An objective, model-independent method for detection of non-uniform steps in noisy signals. Comput. Phys. Commun. 2008;179:716–723. [Google Scholar]
- 17.Svoboda K., Schmidt C.F., Block S.M. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 18.Yildiz A., Tomishige M., Selvin P.R. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 19.Hunt A.J., Howard J. Kinesin swivels to permit microtubule movement in any direction. Proc. Natl. Acad. Sci. USA. 1993;90:11653–11657. doi: 10.1073/pnas.90.24.11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua W., Chung J., Gelles J. Distinguishing inchworm and hand-over-hand processive kinesin movement by neck rotation measurements. Science. 2002;295:844–848. doi: 10.1126/science.1063089. [DOI] [PubMed] [Google Scholar]
- 21.Kukura P., Ewers H., Sandoghdar V. High-speed nanoscopic tracking of the position and orientation of a single virus. Nat. Methods. 2009;6:923–927. doi: 10.1038/nmeth.1395. [DOI] [PubMed] [Google Scholar]
- 22.Andrecka J., Ortega Arroyo J., Kukura P. Structural dynamics of myosin 5 during processive motion revealed by interferometric scattering microscopy. eLife. 2015;4:e05413. doi: 10.7554/eLife.05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y.H., Chang W.L., Hsieh C.L. Shot-noise limited localization of single 20 nm gold particles with nanometer spatial precision within microseconds. Opt. Express. 2014;22:9159–9170. doi: 10.1364/OE.22.009159. [DOI] [PubMed] [Google Scholar]
- 24.Piliarik M., Sandoghdar V. Direct optical sensing of single unlabelled proteins and super-resolution imaging of their binding sites. Nat. Commun. 2014;5:4495. doi: 10.1038/ncomms5495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The image size is 1.9 × 1.9 μm, 60 × 60 pixels (31.8 nm/pixel). The movie was acquired at a frame rate of 1000 frames/s, with every frame shown.
The image size is 2.2 × 2.2 μm, 70 × 70 pixels (31.8 nm/pixel). The movie was acquired for 1 second every minute over 40 minutes with the sample continuously illuminated. The original frame rate was 1000 frames/s but has been averaged to 1 frame/s.