ABSTRACT
Isolation of genomic DNA is one of the basic steps in many different molecular analyses. There are a few reports on methods of DNA isolation from milk, but many of them are time consuming and expensive, and require relatively large volumes of raw milk. In this study a rapid, sensitive, and efficient method of DNA extraction from milk somatic cells of various mammals (cattle, sheep, goats, horses) is presented. It was found that milk is a good source of genomic DNA, and to obtain a sufficient amount and quality of DNA, suitable for molecular analysis such as PCR, 10 mL of raw milk is sufficient. Thanks to this method, stress in animals can be reduced during collection of researched material. Therefore, this method could be widely used in molecular analyses.
KEYWORDS: DNA, isolation, mammals, milk somatic cells
Introduction
In recent years rapid growth of biotechnological sciences and wide practical application of their achievements has been observed. There are many molecular methods using genomic DNA as a basic research material. They include analyses of sequences of genes important in animal breeding; therefore, possible methods of quick, inexpensive, and efficient DNA isolation from different tissues are still sought after. Currently many techniques of isolation of nucleic acids from different biological materials, mainly from blood but also from meat, semen, hair follicles, and so forth, are described (1 2 3). Less frequently used materials are blood stains and bone marrow (4). There are also techniques that allow isolation of DNA from archival and archaeological materials, such as skulls, bones, and teeth (5, 6).
Despite the availability of many methods of extraction of nucleic acids from different bodily fluids and tissues, new ways of isolation allowing for increased yield and purity of DNA are sought after. This is especially important in livestock genetic research, where the most common tissue used for DNA isolation is blood, but very often breeders do not allow researchers to take it from their animals. This is related to the decreased productivity of animals exposed to stress and increased service costs. Therefore, milk appears to be a perfect material for the isolation of mammalian DNA, as obtaining milk samples is simple and noninvasive.
One of the natural components of raw milk are somatic cells, which include mostly polymorphonuclear leukocytes, macrophages, lymphocytes, and a small number of mammary epithelial cells (7). In milk the somatic cell count is affected by many factors such as species, breed, milk yield, stage of lactation, milking hygiene, stress, and individual predispositions (8). In healthy cows, the level of somatic cells in 1 mL of milk ranged from 2 × 104 to 2 × 105 (9). Cow’s milk in which the somatic cell count (SCC) exceeds 4 × 105 per 1 mL is considered to be unfit for human consumption (10). The increased level of somatic cells in milk can be associated with mastitis; therefore, SCC is regarded as an indicator of the technological quality of milk (11, 12).
Although the concentration of somatic cells in 1 mL of milk (usually 2 × 104 to 4 × 105) is much lower than the concentration of leukocytes in 1 mL of blood (usually 4 × 106 to 10 × 106), and despite the fact that milk contains inhibitors such as fat and protein, the isolation of DNA from milk is feasible (13 14 15 16). Hitherto known methods of DNA extraction from milk somatic cells are often time consuming, expensive, and require a relatively large volume of milk (15–50 mL) and use of toxic reagents (14 15 16 17 18 19 20). Bearing in mind the aforementioned, a new, fast, nontoxic, and inexpensive method of DNA isolation from small amounts of raw milk was developed at the Department of Cattle Breeding at the University of Agriculture in Krakow (patent application number: P.404 447 in Poland).
Material and metods
The research material consisted of 10 mL milk samples collected from cows (n = 250), sheep (n = 53), goats (n = 25), and mares (n = 10) by hand-milking after udder cleaning. First, contaminated milk streams were dismissed. Additionally, in order to test versatility of the studied method of DNA isolation, milk samples obtained from 2 female volunteers were included in the studies. Milk samples were stored at 4°C until the DNA extraction.
DNA isolation was performed according to Procedures 1 or 2 presented in Table 1. The choice of the procedure depends on the size of the somatic cells and milk proteins pellet (evaluated in step 1, which is the same in both procedures). It was assumed that in the case of the pellet diameter of <3.5 mm that indicates a low number of somatic cells in the milk sample (which occurs in milk samples taken from cows yielding more than 14 thousand liters of milk per lactation) or in case of the pellet diameter of >5.0 mm that is associated with the increased content of proteins in milk, Procedure 2 should be used. When the pellet diameter is within the range 3.5–5.0 mm, the shorter Procedure 1 is sufficient. If the strings of DNA clumps are invisible in the solution and to allow DNA to go into the mixture entirely, the time of cell lysis should be extended (see Step 3, Procedure 2). Then, the additional steps of protein and DNA precipitation should be performed (Steps 4–5, Procedure 2).
Table 1. Procedures of isolation of DNA from milk somatic cells.
| Step | Procedure 1 |
|---|---|
| 1 | Centrifuge 10 mL of raw milk at 7000 g for 10 minutes (4°C), remove the milk fat and most of the supernatant from above somatic cells and milk proteins pellet, and transfer the pellet with the remainder of the supernatant to a 2-mL sterile tube. Centrifuge the mixture at 5000 × g for 3 minutes (4°C) and remove the supernatant. |
| 2 | Wash the pellet with 1 mL of buffer (15 mM Tris-HCl (pH 7.4–7.6), 25 mM NaCl, 5 mM MgCl2,15 mM Na2HPO4, 2.5 mM EDTA, 1% sucrose). Centrifuge this mixture at 5000 × g for 3 minutes (4°C), remove the supernatant, and repeat this step until clear supernatant is obtained. |
| 3 | Add 1 mL of lysis buffer (pH = 8.8; 6% SDS, 3 mM MgCl2,15 mM Tris-HCl, 0.5% DMSO, 6% acetone) to the pellet and incubate this mixture at 65°C for approximately 20–30 minutes. After this time, the strings of DNA clumps will be visible in the liquid. |
| 4 | Attach the strings of DNA clumps to the wall of a new sterile 1.5-mL tube by pipette. Then, discard leftover supernatant that has dropped to the bottom and wash DNA twice with 100 µL of 70% ethanol. Centrifuge the mixture at 10000 × g for 1 minute at room temperature and discard the supernatant. |
| 5 | Dissolve the DNA pellet in 50–100 µL of deionized water or TE buffer (pH 8.0, 10 mM Tris,1 mM EDTA) |
| Step | Procedure 2 |
| 1–2 | The same as in Procedure 1. |
| 3 | Add 1 mL of lysis buffer (pH = 8.8; 6% SDS, 3 mM MgCl2,15 mM Tris-HCl, 0,5% DMSO, 6% acetone) to the pellet and incubate this mixture at 65°C for approximately 60–90 minutes. |
| 4 | Cool down the mixture to room temperature and add 450 µL of protein precipitation buffer (2.35 M NH4Cl, 1.15 M NaCl, 38% ethanol pH 5.0), vortex, and then centrifuge the mixture at 16000 × g for 8 minutes (10°C). |
| 5 | Transfer the supernatant to a new tube and add 600 µL of 100% isopropanol. Centrifuge the mixture at 10000 × g for 8 minutes and remove the supernatant. |
| 6 | Wash the DNA pellet twice with 70% ethanol and air dry. |
| 7 | Dissolve the DNA pellet in 50–100 µL of deionized water or TE buffer (pH 8.0, 10 mM Tris, 1 mM EDTA). |
The evaluation of the concentration and purity of the obtained genomic DNA was verified using a Nanodrop 2000 spectrophotometer (ThermoScientific) by measuring the UV absorption at wavelengths of 260 nm and 280 nm. The costs of DNA isolation, according to Procedures 1 and 2, were evaluated based on the prices of the Sigma Company reagents and compared with the costs of isolation used in other methods (Table 2). DNA size and quality were evaluated by electrophoresis in 1.0% agarose gel (Sigma) including SYBR Safe dye (Invitrogen). Electrophoresis was carried out in 1× TBE solution, at 80 V for 45 minutes and then the gel was observed under an LED light of DNR Bio-Imaging System (MicroBIS) (Fig. 1). The bovine SCD1 gene (302 bp), ovine DRB1 gene (296 bp), equine MSTN gene (204 bp), and caprine CAST gene (416 bp) were amplified by polymerase chain reaction (PCR) to verify the presence of amplifiable DNA in all isolates. Each amplification reaction of bovine gene was carried out in a mixture (25 uL) containing about 150 ng of genomic DNA, 2.5 mM of MgCl2, 0.3 uM of each primer (Forvard -5 'GCCACCTTATTCCGTTATGC 3' and reverse-5 'TGTTGCTTAACTTTCAAGGGTTT 3'), 200 M of dNTP mix, 1.75 U of Taq polymerase (ThermoScientific), and 1× Taq buffer. PCR reactions were run in a C-1000 thermal cycler (BioRad) under the following thermal conditions: initial denaturation at 95°C for 5 minutes, followed by 32 cycles of denaturation at 95°C for 40 seconds, annealing at 61°C for 35 seconds, elongation at 72°C for 35 seconds, and final elongation at 72°C for 7 minutes. PCR reactions of other genes were carried out according to the following procedures: ovine DRB1 gene, Shen at al (21).; equine MSTN gene, Li et al. (22); and caprine CAST gene, Sharma et al. (23). The PCR products were analyzed by electrophoresis in 2% agarose gel (Sigma) stained with SYBR Safe (Invitrogen) in a 1× TBE buffer (Fig. 2.).
Table 2. Comparison of different methods of DNA isolation from milk somatic cells (time of isolation, volume of milk, cost per sample).
| Author | Method | Time of isolation | Volume of milk (mL) | Cost (USD)/Sample |
|---|---|---|---|---|
| Our method Procedure 1 | method with acetone | 0.5 h | 10 | 0.16 |
| Procedure 2 | salting-out method | 1–1.5 h | 10 | 0.30 |
| Amills et al. (1997) (24) | chelex method | 0.7 h | 0.01 | 0.22 |
| d'Angelo et al. (2007) (15) | salting-out method | 2.5 h | 40 | 4.30 |
| Yang et al. (2013) (26) | organic extraction method | 2 days | 1 | 3.17 |
| Liu et al. (2014) (16) | organic extraction method | 2 days | 13 | 2.05 |
Figure 1.
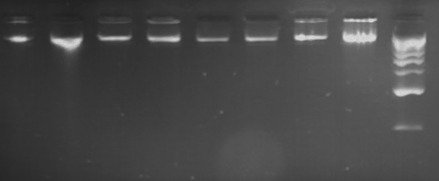
Electrophoresis of DNA in 1% agarose gel next to a 1 kb ladder; lanes 1, 2 - ovine DNA; lanes 3,4- bovine DNA; lanes 5, 6 - caprine DNA; lanes 7, 8 - equine DNA, lane 9- marker PerfectTM 1 kb DNA Ladder, EURx .
Figure 2.
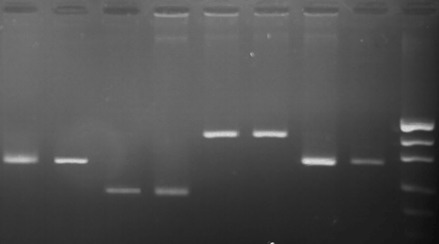
Agarose gel electrophoresis of PCR products; lanes 1,2- ovine DRB1 gene (296 bp); lanes 3,4- equine MSTN gene (204 bp) lanes 5, 6 - caprine CAST gene (416 bp) lanes 7, 8- bovine SCD1 gene (302 bp); lane 9- marker pUC19 DNA/MspI (Thermo Scientific).
Results
The results of the study proved that all DNA samples isolated from raw milk were of high purity and quality (Table 3, Fig. 1). Concentrations of the genomic DNA ranged from 10 ng/µL (sheep) to 2809.6 ng/µL (goats). The high standard deviations from the average amounts of the obtained DNA resulted from the fact that in several milk samples increased somatic cell counts occurred (observed as 3–4 times bigger cell pellets: step 2 in the isolation procedures), not detected before milking (by using reagents for the mastitis diagnosis: MASTIRAPID (Biowet).
Table 3. Concentration and purity of DNA isolated from the milk of different animal species, using the method of Pokorska et al. (2015).
| Species | Number of samples | Concentration of DNA |
Purity of DNA |
||||
|---|---|---|---|---|---|---|---|
| Average concentration of DNA (ng/µL)* | Minimum concentration of DNA (ng/µL)* | Maximum concentration of DNA (ng/µL)* | Standard deviation | 260/280 (nm) | Standard deviation | ||
| Cattle | 250 | 684.64 | 50.0 | 2090.0 | 471.88 | 1.79 | 0.06 |
| Sheep | 53 | 198.63 | 10.0 | 1322.4 | 318.94 | 1.60 | 0.30 |
| Goat | 25 | 867.86 | 53.4 | 2809.6 | 824.30 | 1.84 | 0.11 |
| Horse | 10 | 33.00 | 13.0 | 53.0 | 28.28 | 1.81 | 0.21 |
| Human | 2 | Woman 1: concentration, 148 ng/µL; purity, 1.83 Woman 2: concentration, 300 ng/µL; purity, 1.85 | |||||
*All DNA samples were dissolved in 100 µL TE buffer.
The PCR amplification products are shown in Fig. 1. The bands were clear and single; no nonspecific products or dimmer fragments were found. Therefore, the results of the amplification reactions indicate that the described method can be used to extract high quality template DNA required for molecular analysis.
While comparing different methods of DNA isolation from milk somatic cells, it was found that the presented method not only requires a relatively small amount of milk but is less expensive and less time-consuming (especially Procedure 1) than those described by other authors (Table 2).
Discussion
Collection of milk samples, compared to taking blood samples, is much easier and less stressful for animals because it does not require venipuncture. In the literature several different DNA extraction methods from milk cellular elements can be found. Classical Phenol-Chloroform and quick and reliable Chellex resin methods have been used for DNA extraction from bovine and caprine milk (18, 24). DNA of mammary gram-positive bacteria have been isolated from whole milk using a specific, sensitive, and rapid method based on the lysing and nuclease-inactivating properties of the chaotropic agent (guanidinium thiocyanate) as well as on the properties of nucleic acid to bind with the silica particles (25). This method is especially important in the detection and identification of bacterial strains that are mainly responsible for the occurrence of udder inflammation and thereby cause large dairy industry losses.
D’Angelo et al. (15) used a simple salting out procedure for extracting DNA from milk somatic cells. This method is relatively sensitive and free from toxic organic solvents but to obtain a sufficient amount of DNA (2.12 to 610.12 ug per milk sample) a large volume of milk (40 mL) is necessary. The advantage of this method is that it does not require organic extraction, overnight incubation or expensive reagents. Several years later, Liu et al. (16) proposed a new and efficient method of DNA isolation from a small amount of milk (13 mL) to amplificate the long-fragments of DNA. Similarly to the method presented in this paper, this method allows for obtaining large quantities of highly pure DNA; it is, however, time-consuming, requires overnight incubation with proteinase K, and uses toxic reagents. Another method for the isolation of DNA from a small volume of milk (1 mL) was proposed by Yang and Li (26). This method is also efficient but takes a lot of time and requires expensive reagents. Other methods of DNA isolation from macrophages, neutrophils, and lymphocytes found in milk are described in many extraction protocols (17, 18, 27) but these methods are also very time consuming.
Psifidi et al. (19) and Usman et al. (20), while using commercial kits for the isolation of DNA from blood and tissue to isolate DNA from milk somatic cells, obtained similar amounts and quality of genomic DNA as in the present study; however, their approach requires much larger volumes of raw milk (50 mL).
To summarize, milk somatic cells are a good source of genomic DNA. The method presented in this paper allows for quick isolation of DNA from relatively small volumes of raw milk; therefore, the cost and time of analysis are reduced. The use of raw milk as a source of DNA is less stressful for animals than collecting other tissues intravitally. Another advantage is no need to use toxic reagents. Thus, this method can be practically applied to the fast, safe, and economical extraction of DNA.
References
- Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual. 4th Ed. Vol. 1. New York: Cold Spring Harbor Laboratory Press; 2012. pp. 2–80. [Google Scholar]
- Suenaga E, Nakamura H. Evaluation of three methods for effective extraction of DNA from human hair. J Chrom B. 2005;820(1):137–141. doi: 10.1016/j.jchromb.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Griffin J. Methods of sperm DNA extraction for genetic and epigenetic studies. Methods Mol Biol. 2013;927:379–384. doi: 10.1007/978-1-62703-038-0_32. [DOI] [PubMed] [Google Scholar]
- Deggerdal A, Larsen F. Rapid isolation of PCR-ready DNA from blood, bone marrow and cultured cells, based on paramagnetic beads. Bio Techn. 1997;22:554–557. doi: 10.2144/97223pf02. [DOI] [PubMed] [Google Scholar]
- Geigl EM. On the circumstances surrounding the preservation and analysis of very old DNA. Archaeometry. 2002;44:337–342. [Google Scholar]
- Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat Protoc. 2007;2(7):1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- Boutinaud M, Jammes H. Potential uses of milk epithelial cells: a review. Reprod Nutr Dev. 2002;42:133–147. doi: 10.1051/rnd:2002013. [DOI] [PubMed] [Google Scholar]
- Rupp R, Boichard D, Bertrand C, Bazin S. Overview of milk somatic cell counts in the French dairy cattle breeds. Prod Anim. 2000;13:257–267. [Google Scholar]
- Pantoja JCF, Hulland C, Ruegg PL. Somatic cell counts status across the dry period as a risk factor for the development of clinical mastitis in the subsequent lactation. J Dairy Sci. 2009;92:139–148. doi: 10.3168/jds.2008-1477. [DOI] [PubMed] [Google Scholar]
- Council Directive 92/46/EEC. European Union: Health rules for the production and placing on the market of raw milk, heat-treated milk and milk based products regulations. CELEX-EUR Off J 1992; L 268:1–31. [Google Scholar]
- Hunt KM, Williams JE, Shafii B, et al. Mastitis is associated with increased free fatty acids, somatic cell count, and interleukin-8 concentrations in human milk. Breastfeed Med. 2013;8:105–110. doi: 10.1089/bfm.2011.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Singh NK, Bhadwal MS. Relationship of somatic cell count and mastitis: an overview. Asian Austral J Anim. 2011;24:429–438. [Google Scholar]
- Murphy MA, Shariflou MR, Moran C. High quality genomic DNA extraction from large milk samples. J Dairy Res. 2002;69:645–649. doi: 10.1017/s0022029902005848. [DOI] [PubMed] [Google Scholar]
- Feligini M, Frati S, Curik VC, Brambilla A. Caprine αs1-casein polymorphism: Characterization of A, B, E and F variants by means of various biochemical and molecular techniques. Food Tech Biotechol. 2005;43:123–132. [Google Scholar]
- D’Angelo F, Santillo A, Sevi A, Albenzio M. Technical note: a simple salting-out method for DNA extraction from milk somatic cells: investigation into the goat CSN1S1 gene. J Dairy Sci. 2007;90:3550–3552. doi: 10.3168/jds.2007-0149. [DOI] [PubMed] [Google Scholar]
- Liu YF, Gao JL, Yang YF, Ku T, Zan LS. Novel extraction method of genomic DNA suitable for long-fragment amplification from small amounts of milk. J Dairy Sci. 2014;97:6804–6809. doi: 10.3168/jds.2014-8066. [DOI] [PubMed] [Google Scholar]
- Ernst CA, Dentine MR. Genotyping of dairy cattle using direct amplification from somatic cells in small volumes of milk. Anim Genet 1992; 23(Suppl. 1):56. [Google Scholar]
- Lipkin E, Shalom A, Khatib H, Soller M, Friedmann A. Milk as a source of deoxyribonucleic acid and as a substrate for the polymerase chain reaction. J Dairy Sci. 1993;76:2025–2032. doi: 10.3168/jds.S0022-0302(93)77536-0. [DOI] [PubMed] [Google Scholar]
- Psifidi A, Dovas CI, Banos G. A comparison of six methods for genomic DNA extraction suitable for PCR-based genotyping applications using ovine milk samples. Mol Cell Probes. 2010;24:93–98. doi: 10.1016/j.mcp.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Usman T, Yu Y, Liu C, Fan Z, Wang Y. Comparison of methods for high quantity and quality genomic DNA extraction from raw cow milk. Genet Mol Res. 2014;13(2):3319–3328. doi: 10.4238/2014.April.29.10. [DOI] [PubMed] [Google Scholar]
- Shen H, Han G, Jia B, Jiang S, Du Y. MHC-DRB1/DQB1 Gene polymorphism and its association with resistance/susceptibility to cystic echinococcosis in Chinese Merino sheep. J Parasitol Res. 2014;2014:272601. doi: 10.1155/2014/272601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liu D-H, Cao Ch-N, et al. Single nucleotide polymorphisms of myostatin gene in Chinese domestic horses. Gene. 2014;538(2014):150–154. doi: 10.1016/j.gene.2013.12.027. [DOI] [PubMed] [Google Scholar]
- Sharma R, Maitra A, Pandey AK, Singh LV, Mishra BP. Single nucleotide polymorphisms in Caprine Calpastatin. Russ J Genet. 2013;49(4):441–447. doi: 10.7868/s0016675813040139. [DOI] [PubMed] [Google Scholar]
- Amills M, Francino O, Jansa M, Sanchez A. Isolation of genomic DNA from milk samples by using Chelex resin. J Dairy Res. 1997;64:231–238. doi: 10.1017/s0022029997002161. [DOI] [PubMed] [Google Scholar]
- Cremonesi P, Castiglioni B, Malferrari G, et al. Technical note: improved method for rapid DNA extraction of mastitis pathogens directly from milk. J Dairy Sci. 2006;89:163–169. doi: 10.3168/jds.S0022-0302(06)72080-X. [DOI] [PubMed] [Google Scholar]
- Yang FL, Li XS. Somatic cell counts positive effects on the DNA yield extracted directly from Murrah Buffalo milk. J Curr Res Sci. 2013;1(5):392–395. [Google Scholar]
- Amills M, Francino O, Sanchez A. Nested PCR allows the characterization of TaqI and PstI RFLPs in the second exon of the caprine MHC class II DRB gene. Vet Imm Immunopat. 1995;48:313–321. doi: 10.1016/0165-2427(95)05442-9. [DOI] [PubMed] [Google Scholar]


