Abstract
Aberrant DNA methylation is a characteristic feature of cancer including blood malignancies. Mutations in the DNA methylation regulators DNMT3A, TET1/2 and IDH1/2 are recurrent in leukemia and lymphoma. Specific and distinct DNA methylation patterns characterize subtypes of AML and lymphoma. Regulatory regions such as promoter CpG islands, CpG shores and enhancers show changes in methylation during transformation. However, the reported poor correlation between changes in methylation and gene expression in many mouse models and human studies reflects the complexity in the precise molecular mechanism for why aberrant DNA methylation promotes malignancies. This review will summarize current concepts regarding the mechanisms behind aberrant DNA methylation in hematopoietic malignancy and discuss its importance in cancer prognosis, tumor heterogeneity and relapse.
Keywords: DNA methylation, leukemia, lymphoma, epigenetics
DNA Methylation Patterning in Normal and Malignant Hematopoiesis
During differentiation, hematopoietic stem cells (HSCs) give rise to multi-potent progenitors, which subsequently undergo commitment to mature cell lineages of the blood. The maturation stages are characterized by dynamic changes in gene expression, governed by transcription factor networks and, at the epigenetic level, via distinct DNA methylation patterns [1] [2] [3]. Perturbation in DNA methylation status can be both a biomarker of malignancy or causative in disease progression [4]. Indeed, next-generation sequencing has revealed that a high percentage of leukemia and lymphoma patients harbor somatic mutations in enzymes regulating DNA methylation. DNA methylation is one of the oldest and best-studied epigenetic mechanisms. Originally, it was considered a relatively stable epigenetic mark associated with transcriptional repression. In the last years, the study of diverse and dynamic DNA methylation patterns at single base resolution has generated a great deal of interest, providing novel insights into the role of DNA methylation in hematopoietic homeostasis and transformation[1, 5–8]. Differentially methylated regions are found at promoters in the context of CpG islands (CGIs) (See Glossary), CGI shores [8] and active enhancers [9] (Box 1). In addition, large hypomethylated stretches in the genome known as “valleys” or “canyons” have been recently identified [10, 11] (Box 1 and Figure 1a). Together these studies underscore the complexity of DNA methylation in the genome. In this review, we will focus in the recent discoveries elucidating the impact of aberrant DNA in hematopoietic malignancy with an emphasis on the role of cytosine-modifying enzymes and their somatic mutations during transformation.
Box 1. DNA methylation dynamics and gene regulation.
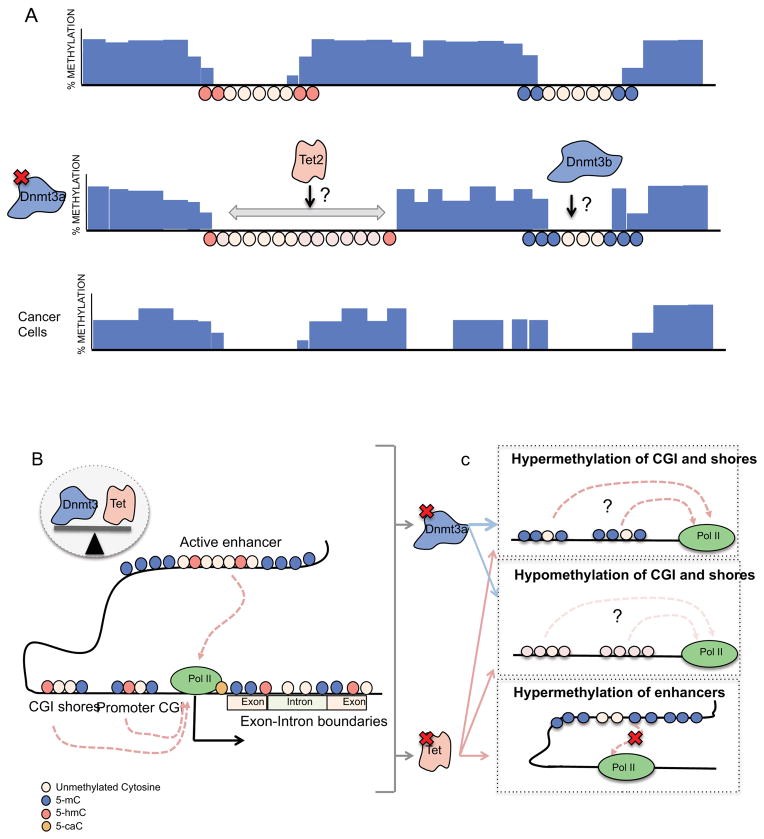
The development of large-scale DNA methylation mapping has uncovered the dynamic character of the methylome during cell differentiation and transformation. Approximately 60–80% of CpGs in mammalian cells are methylated and 20% of the CpG methylation is dynamic [128, 129]. Approximately 70% of gene promoters contain a CpG island [130]. Highly methylated CGIs are usually restricted to long term silenced genes whereas promoter regions of coding genes are usually hypomethylated supporting an open chromatin state and accessibility to transcription factors. In hematopoiesis, only modest global CGI methylation changes are observed throughout lineage commitment [2]. Focalized hypermethylation at the promoters of stem cell genes occurs during differentiation (Meis1, Smad6, and homeobox cluster [131]). Cell cycle genes, upregulated in progenitors cells become downregulated and hypermethylated in differentiated cells. Lymphoid cells exhibit increased promoter hypermethylation in key regulators of myeloid commitment. In contrast, hypomethylated CGIs are also maintained in other genes that are silenced during differentiation suggesting suppression of expression by mechanisms other than regulation of DNA methylation. Hypomethylated regions near CpG islands (CGI shores) (Figure 1b) show tissue and cancer specific methylation patterns [131]. During hematopoiesis, changes in DNA methylation patterns at CGI shores are inversely correlated with lineage gene expression more strongly than at CGIs [1]. 5-mC and 5-hmC in gene bodies have been positively correlated to gene expression and alternative splicing [8, 132, 133] (Figure 1b). Recently, 5-caC has been shown to regulate RNA Pol II elongation [134]. Consistent with the idea that DNA methylation abrogates transcription factor binding, several studies have also highlighted the correlation between DNA hypomethylation, transcription factor binding and enhancer activity in human cells [7, 135, 136]. Enrichment of 5hmC and TET1 occupancy is also associated with enhancers [7].
Large DNA hypomethylation regions in chromosomal domains have been identified (Figure1a). Conserved hypomethylated regions of more than 5kb (valleys) enriched in embryonic development genes have been identified in mammalian cells [137]. Jeon et al., have also characterized larger hypomethylated regions (canyons) that are also conserved through cell types [10]. Canyons show enrichment in H3K4me3 and/or H3K27me3 histone marks (Figure 1a). While an important percentage of the canyons expand upon DNMT3A depletion, some of them shrink or remain unchanged. Edges of those canyons enriched in H3K4me3 overlap with 5hmC marks. This finding suggests a model in which TET proteins and DNMT3A act concomitantly on canyon borders. The functional role in chromatin dynamics and epigenetics of the valleys and canyons is unknown.
Figure 1. DNA methylation dynamics in normal and malignant hematopoiesis.
A) Mammalian cells show 70–80% of global methylation in their genome. Large hypomethylated regions called “canyons” are enriched in H3K4me3 and/or H3K27me3. Edges of the canyons, especially those enriched in H3K4me3 show 5hmC marks. DNMT3A has been shown to play an important role in the maintenance of canyons. Dmnt3-null HSCs show normal global DNA methylated levels but alterations at canyon edges. Canyons occupying active chromatin (H3K4me3) usually expand and are enriched in hematopoietic stem cells genes (Meis1, Hox) that are known to be dysregulated in leukemia. Quiescent canyons (H3K27me3) do not expand with DNMT3A loss and often shrink suggesting that DNMT3B activity or other mechanisms drive the hypermethylation phenotype. DNMT3A mutant AML cells show global hypomethylation and newly formed hypomethylated valleys in their genome compared to normal cells. B) Hypomethylated areas of the genome are enriched in active regulatory elements such us enhancers and promoters. Hypomethylated CGIs and shores are enriched in active and poised genes, while hypermethylation of CGIs is related to gene silencing. Proper balance between DNMT3 and TET activity determine the DNA methylation status of CpGs. 5mC and 5hmC in genes bodies correlate to active transcription and exon splicing while 5caC interacts with RNA Pol II during elongation. C) DNMT3A mutant HSC and AML cells, display both differentially hypomethylated and hypermethylated CGIs. Hypomethylated CGIs are enriched in genes related to hematologic malignancies like Prdm16, Stat1, Ccnd1, Myc, Mn1, Msi2, Men1, Erg and Runx1. Changes in gene expression do not correlate with differentially methylated CGIs. TET2 mutant CMML patients and Tet2-null HSCs show a global increase in DNA methylation. In an AML mouse model (AML1-ETO) Tet2 depletion promotes hypermethylation of enhancers related to tumor suppressor genes. The activity of DNMT3 and TET proteins at the same genomic sites may suggest that these loci are tightly controlled by the balance of DNA methylation. The reported poor correlation between changes in methylation and gene expression in both mouse models and human samples may reflect non-coding and transcription independent roles for DNA methylation.
The Guardians of DNA Methylation
CpG methylation is mediated by a family of DNA methyltransferases (DMNT) that regulate both maintenance (DNMT1) and de novo (DNMT3A/B) methylation in the genome [12, 13]. During replication, DMNT1 adds methyl groups to the newly synthesized DNA strand ensuring the preservation of DNA methylation patterns during proliferation [14, 15]. De-novo methyltransferases, DMNT3A and DMNT3B, establish new DNA methylation patterns [16]. Antagonists of DNMT function have been identified in recent years that directly target 5mC for demethylation and have also emerged as key tumor suppressors of hematopoietic malignancy. The Ten-eleven-translocation enzymes (TET1, TET2 and TET3) progressively oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) [17, 18]. The oxidized forms of 5mC promote DNA demethylation through either active pathways involving base excision repair [19] or passively, by blocking re-methylation of newly synthesized DNA during replication [20].
Mutation of DNA Methylation Pathways in Leukemia
DNMT3A mutations are found in 30% of acute myeloid leukemia (AML), 7–15% myeloproliferative neoplasia (MPN) and 8% myelodysplastic syndrome (MDS) [21]. The majority of these mutations are heterozygous and cluster in the methyltransferase domain resulting in a truncated protein [22]. Approximately 60% of the missense mutations occur in the residue R882 causing decreased catalytic activity of the enzyme [23]. DNMT3A functions as an oligomer and the heterozygous dominant negative mutation R882H inhibits the wild type enzyme by disrupting its ability to homotetramerize resulting in 80% loss of methyltransferase activity [24]. Accordingly, the genomes of AML cases with DNMT3A mutation at R882 show an overall reduction in CpG methylation that was not found in cases with non-R882 mutation [24]. DNMT3A loss-of-function promotes a progressive expansion of long-term hematopoietic stem cells, probably due to an inability to properly repress self-renewal [25]. Moreover, conditional ablation of both Dnmt3a and Dnmt3b in mice result in a stem cell expansion and block in differentiation [26], indicating that de-novo DNA methylation is essential for normal HSC self-renewal and hematopoietic lineage commitment.
Loss-of-function mutations in TET family members are also prevalent in hematopoietic malignancy suggesting that cytosine demethylation imparts an important tumor suppressive role [27–29]. TET1 was first identified and cloned as a fusion partner with MLL in AML leading to the discovery of the gene family. However it is mutated in only 1% of de-novo AML patients [30]. TET3 mutations are very infrequent in myeloid malignancies and it has been found occasionally mutated in peripheral T-cell lymphomas and some chronic lymphoblastic leukemia [31, 32]. TET2 instead is one of the most frequently mutated genes in myeloid disease comprising AML (7–23%), Chronic Myelomonocytic Leukemia (CMML) (50%) and MDS (10–20%) [28, 29, 33]. Somatic deletion and loss-of-function mutations constitute 67% of the TET2 mutations while 33% of the mutations are missense mutations targeting the catalytic domain. Despite enrichment for heterozygous mutations, no evidence of a dominant-negative effect from mutation in TET2 has been reported.
TET catalytic activity can also be indirectly impaired by mutation of isocitrate dehydrogenase enzymes IDH1 and IDH2 [34]. In normal cells the IDH enzymatic activity converts isocitrate to α-ketoglutarate (α-KG), an essential cofactor for TET proteins and other epigenetic regulators such as the histone demethylase family of Jumonji proteins [35]. Initially, mutations in IDH genes were identified in glioblastomas. Subsequently, additional mutations have been identified in other blood tumors, namely 16–19% in AML, 2–5 % in MPN and 3 % in MDS [36, 37]. Missense mutations in the conserved residues R132 in IDH1 and R140 or R172 in IDH2 result in 2-hydroxyglutarate (2-HG) oncometabolite production instead of α-KG, thereby impeding TET protein activity. Indeed, IDH1/2-mutant AML is associated with more extensive promoter hypermethylation compared to other AML subtypes, indicating that aberrant cytosine methylation at specific loci may be causative in leukemogenesis [38].
Fingerprinting Malignant DNA Methylomes in Leukemia
Aberrant DNA methylation patterns are a characteristic feature of many cancers [39]. Hypermethylation of CpGs in the promoters of specific tumor suppressor genes was initially described as a hallmark feature of many cancer cells (refs) whereas global hypomethylation has been associated with genomic instability [40, 41]. Interestingly, large-scale genome-wide promoter DNA methylation profiling has revealed patterns that correlate with specific genetic and molecular subtypes of AML cases [4, 42]. In AML patients with DNMT3A R882 mutation there was no significant difference in global methylation levels compared to AML genomes without DNMT3A mutation when analyzed by mass spectrometry. In addition, promoter MeDIP-chip studies revealed 182 hypomethylated genomic loci [45]. Hypomethylation at CGI shores [24] and promoter CGIs of homeobox transcription factors have also been identified in AML R882 leukemia [46]. However these hypomethylated loci did not correlate with changes in expression of neighboring genes. Quantification of global 5hmC levels, by DNA dot blot or mass spectrometry, have shown that TET2 mutations correlate with a significant reduction in the levels of genomic 5hmC [47]. Figueroa et al. observed that TET2-mutated de-novo AMLs have 129 hypermethylated promoters compared with CD34+ bone marrow cells from healthy controls. [38]. Moreover, In TET2 mutant CMML samples, differentially hypermethylated CpG regions are enriched in enhancers, whereas promoter CGIs do not show significant changes compared to non-mutated samples [48]. Interestingly, IDH1/2 and TET2 mutations in patients tend to be mutually exclusive and both mutations confer an overlapping DNA hypermethylation signature [38, 49].
The conclusions from these studies suggest that aberrant methylomes are associated with leukemic transformation. However, the impact of these epigenetic changes in the transcriptome is still unclear. Whole-genome methylation analysis to assess DNA methylation status in non-coding regions of the genome such us enhancers (Figure 1) and additional 5-hmC profiling studies may help elucidate the complex role of DNA methylation in leukemia.
DNMT, TET and IDH Mechanistic Studies In Vivo
In deletion and knock-in mouse models, Dnmt3a ablation [25], Tet inactivation [27, 51–54] or Idh mutant overexpression [55–57] have all been shown to cause progressive expansion of long-term hematopoietic stem cells. Dnmt3a-deficient mice do not develop spontaneous myeloid leukemia. However, mapping of DNA methylation patterns in Dnmt3a-null HSCs reveals differentially hypomethylated and hypermethylated gene patterns compared to wild-type cells. Although, a core set of genes crucial for HSC self-renewal (Gata3, Runx1, Homeobox transcription factors) and malignancy-promoting factors (Prdm16, Stat1, Ccnd1, Myc, Mn1, Msi2, Men1, Erg) show CGI hypomethylation and increased expression levels, their role in human AML has not been properly demonstrated. Whole genome bisulfite sequencing (WGBS) of Dnmt3a-null stem and progenitor cells revealed alterations in the edges of DNA methylation canyons, specially in those enriched in 5-hmC (Figure 1a) [10]. Expressed canyon-associated genes were significantly enriched for differentially expressed genes in humans with AML with and without DNMT3A mutation. This finding suggests a model in which TET proteins and DNMT3A act concomitantly on canyon borders. In addition, transgenic overexpression of the R882H mutation in mice causes a progressive MDS-like phenotype characterized by anemia, dysplastic features, and a block in erythroid differentiation [58]. Lethally irradiated mice transplanted with Dnmt3a-null HSCs develop a spectrum of leukemias (including MDS, AML, primary myelofibrosis, and T- and B-cell acute lymphoblastic leukemia within a year. Global hypomethylation was observed in all of the malignancies [59]. Taken together, these data support the notion that alteration in DNMT3A activity causes aberrant DNA methylation patterns contributing to an expansion of pre-leukemic stem cells.
Several murine mouse models have shown that Tet2 loss of function increases self-renewal and impairs normal hematopoietic differentiation resulting in a progressive expansion of the progenitor ckit+ compartment, increased GMP frequency, myeloid and erythroid expansion and splenomegaly from approximately 6 months of age [27, 51–54]. In addition, Tet2 loss in an AML1-ETO background originates widespread DNA hypermethylation affecting up to 25% of active enhancer elements associated with down-regulation of neighboring genes [54] (Figure 1b).
Conditional knock-in of the IDH1 (R132H) mutation in the mouse hematopoietic compartment causes 2-HG production and expands the hematopoietic progenitor pool. Idh1 mutant knock-in mice develop anemia, splenomegaly, and extra-medullary hematopoiesis (8–9 months of age) [55]. DNA methylation profiling of progenitor cells from the hematopoietic compartment confirmed the genome-wide trend towards hypermethylation seen in IDH1-mutant patients [38]. These functional, epigenetic, and genetic data suggest that TET2 and IDH mutations drive a similar pathway in pre-leukemic transformation
Prognostic Relevance of Aberrant DNA Methylation in Leukemia
Although focal DNA hypermethylation in the context of global hypomethylation is characteristic of AML, this deregulation is not uniform across the spectrum of these myeloid diseases. DNA methylation patterns strongly correlate with specific genetic and molecular subtypes of AML. It has been demonstrated that the methylation status of specific genes can predict the future survival of AML patients suggesting that DNA methylation is a biomarker for clinical outcome [4, 44, 61]. A microsphere-based multiplex technique called MELP (microsphere HpaII tiny fragment enrichment by ligation-mediated PCR) has been proposed as an inexpensive platform for simultaneous evaluation of DNA methylation at multiple loci [60]. Similarly, the analysis of DNA methylation of as few as 10 genes in patients with MDS has prognostic significance [62].
In contrast, the mutation status of the enzymes regulating DNA methylation has been associated to variable prognosis. DNMT3A-mutant AML has been linked to poor prognosis, anthracycline resistance and clonal tumor relapse in some studies [45, 63, 64]. Initially, the prognostic significance of TET2 mutations was unclear given contradictory findings [28, 33, 65]. However, in a study incorporating the largest number of patients, TET2 mutations in cytogenetically normal AML conferred a poor prognosis [63]. In patients with MDS and secondary AML, TET2 mutation predicted a high response rate to 5-azacytidine compared to TET2–wild-type [66], however, no overall effect on survival was reported. In contrast, another study determined that TET2 alterations in a similar cohort of patients might actually predict for decreased responsiveness to demethylating therapies [67]. Discrepancies in prognostic significance could be due to heterogeneity in the mutational landscape of patients [68]. Indeed, IDH mutations have been associated with a favorable prognosis in co-occurrence with NPM1, while variable prognoses have been associated with co-occurrence of IDH and FLT3 mutations [69].
DNA Methylation Heterogeneity in Clonal Pre-leukemic Transformation
Mouse models of mutations and loss of function in DNA methylation modifiers have shown that HSCs acquire an early onset self-renewal advantage and clonal expansion that resemble pre-leukemic lesions. Acquisition of a secondary mutation may promote transformation and expansion of a dominant clone generating distinct myeloid neoplasms, (AML, MDS and MPNs) [70]. It has been shown that human HSCs purified from patients with AML can harbor DNMT3A mutations in the absence of other common leukemia-associated mutations. DNMT3A-mutant HSCs were also found to be resistant to chemotherapy and tumor relapse correlated with the acquisition of secondary mutations [45, 72]. In this study blood cells of 5–6% of people older than 70 years contain mutations that may represent premalignant lesions that promote clonal hematopoietic expansion. The order in which mutations are acquired during AML transformation has been shown to influence the clonal evolution and clinical features of patients [73]. DNA methylation patterns characterize distinct forms of AML [4] and that progression from MDS to AML has been associated with increased aberrant DNA methylation [61]. Therefore, tumor clonal evolution could be explained by increased DNA methylation heterogeneity. Indeed, in chronic lymphocytic leukemia (CLL), disordered methylation plays a similar role to that of genetic instability in that it increases the likelihood of transformation [74].
Epigenetic Dysregulation in Lymphoma
B and T lymphocytes of various developmental stages and immunophenotype can give rise to lymphoma, also known as non-Hodgkin lymphoma (NHL) [75–77]. Diffuse Large B-cell lymphoma (DLBCL), Follicular Lymphoma (FL) and Mantle cell lymphoma (MCL) are the most common B-cell lymphoma and together comprise over 60% of all Non-Hodgkin’s lymphoma (NHL). Peripheral T-cell lymphoma (PTCL) constitutes approximately 10% of NHL and can be further divided into at least 7 subtypes, with anaplastic large cell lymphoma (ALCL), peripheral T-cell lymphoma-not otherwise specified (PTCL-NOS), and angioimmunoblastic T-cell lymphoma (AITL) forming the majority of PTCL. Recent genomic studies in patients with B-NHL [78, 79] and PTCL [31, 80] have revealed that epigenetic modifiers, including histone modifying enzymes and regulators of DNA methylation such as TET2, DNMT3A and IDH2, are amongst the most frequently mutated genes.
DNA Methylation Status as a Biomarker of Lymphoma and Myeloma
B and T cell lymphoma with different histological subtypes are associated with distinct global DNA methylation states [81, 82]. DLBCL consists of two subtypes, germinal center B-cell like (GCB) and activated B-cell like (ABC) originally identified based on their gene expression patterns [83]. Subsequent studies have shown that GCB and ABC lymphomas can also be distinguished on the basis of differential methylation patterns affecting gene pathways related to cytokine signaling, germinal center B cell and NFκB signaling known to be dysregulated in these subtypes [84]. Hypermethylation of key genes involved in cell cycle regulation, survival signaling and DNA repair can also differentiate FL (germinal center tumors) from MCL (pre-germinal center) and B-CLL/SLL (pre- or post-germinal center tumors) [85]. WGBS analysis in Burkitt lymphoma, another germinal center derived B cell neoplasm, compared to FL showed that all tumor samples were globally hypomethylated when compared to normal germinal center B cells however the extent and variability of DNA methylation was distinct between the two tumor groups [86]. DNA methylation was found to associate specifically with down-regulation of nuclear factor (NF)-κB, hyper-methylation of JAK-STAT signaling pathway genes and the upregulation of cell cycle control genes in Burkitt lymphoma [86].
DNA methylation correlates with progression in Multiple Myeloma
Multiple myeloma (MM) is an incurable and aggressive clonal proliferation of plasma cells in the bone marrow that often arises from a premalignant state known as monoclonal gammopathy of undetermined significance (MGUS) [87]. MGUS and MM cells can also be distinguished from normal plasma cells on the basis of DNA methylation patterns. Most malignant cells display distinct regions of hypermethylation at promoters of tumor suppressor genes and B cell specific enhancers [88] embedded in global hypomethylation [89]. The progression from MGUS to MM is associated with increased hypomethylation in the absence of additional hypermethylation [88]. These data suggest that a disease-associated methylation signature can already exist in pre-malignant lymphoid cells.
Whether methylation signatures reflect the stage of normal lymphoid maturation from which B cell neoplasms arise or etiology of the disease is still under investigation. WGBS and high-density microarrays were recently used to analyze the DNA methylome of the entire human B cell differentiation program from stems cells to plasma cells [90]. Normal B cell differentiation leads to progressive DNA demethylation and is associated with massive reconfiguration of the DNA methylome during the germinal center reaction [90, 91]. Differentially methylated sites in B cell neoplasms overlapped with those undergoing dynamic methylation during normal differentiation. Comparing the DNA methylation profiles of normal B cells and their malignant cell counterparts, hypomethylation in DLBCL and MM predominantly affected CpGs in heterochromatin in contrast to hypomethylation in ALL that showed enrichment for CpGs in enhancers [90]. Hypermethyation of Polycomb repressed regions was also an epigenetic hallmark of most B cell neoplasms except for MM cells that already share this feature with their normal plasma cell counterpart and instead show hypermethylation of B cell specific enhancers [88, 90].
DNA Methylation Heterogeneity is a Risk Factor for Lymphoid Malignancies
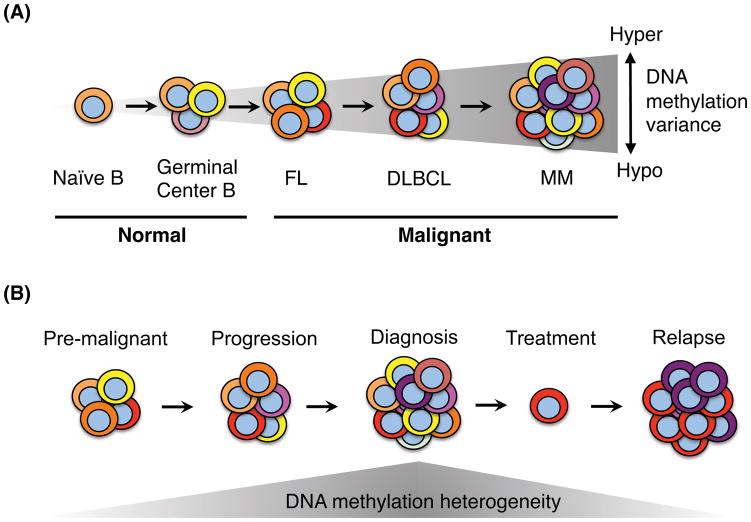
Epigenetic factors have been shown to contribute to the heterogeneity of lymphoma, both within (intra-tumor) and between individual (inter-tumor) lymphoma patient samples. DNA methylation heterogeneity is higher in FL and DLBCL compared to normal mature B cells, increases with disease severity and a higher degree of heterogeneity is associated with a poor prognosis [92–94] (Figure 2). In MM, methylation heterogeneity is even greater than in DLBCL, ranging from global hypo- to hypermethylation when compared to normal plasma cells or other lymphoid malignancies [88, 95] and MGUS are less heterogeneous than MM patients [88], again suggesting that an increase in methylation heterogeneity is associated with disease progression. DNA methylation patterns in DLBCL correlate with distinct lymphomagenic factors such as BCL6 or EZH2 mutation in a target-specific fashion and through spreading to neighboring promoters in the absence of insulator elements such as CTCF [92, 93].
Figure 2. DNA methylation heterogeneity in mature B cell lymphoma.
A) DNA methylation heterogeneity increases in germinal center B cells most likely as a result of sub-clonal expansion during an immune response. B-cell lymphomas display increased intra-tumor and inter-tumor methylation heterogeneity, ranging from hypo- to hyper-methylation when compared to normal B cells. MM shows the highest degree of DNA methylation heterogeneity compared to DLBCL and FL. B) Increased DNA methylation heterogeneity is associated with disease progression. A higher level of heterogeneity at diagnosis increases the risk of relapse. Relapsed tumors often display lower methylation heterogeneity than at diagnosis suggesting clonal evolution. A higher degree of DNA methylation heterogeneity at diagnosis may increase the probability that treatment-resistant clones can survive to promote relapse.
A relapse-associated methylation signature has also been identified in DLBCL. SMAD1, a regulator of TGF-β receptor activity, was found to be hypermethylated in chemoresistant DLBCL and its reactivation by exposure to low dose DNMT inhibitor, 5-azacitdine, was required for chemosensitization [96]. By characterizing the DNA methylome of DLBCL at diagnosis and relapse another study discovered a core set of genes that became aberrantly methylated at relapse, including SMAD6 and other regulators of the TGF-β pathway [93]. In addition, when intra-tumor methylation heterogeneity was compared at diagnosis and relapse in DLBCL, a model of evolutionary fitness emerged. Relapsed patients had a higher degree of methylation heterogeneity at diagnosis but lower heterogeneity at relapse compared to non-relapsed patients [93]. A greater degree of epigenetic heterogeneity at diagnosis may increase the probability that a resistant sub-clone will survive upon treatment. Importantly, intra-tumor methylation heterogeneity did not clearly correlate with genetic clonal heterogeneity in this study, suggesting there are as yet unknown mechanisms that could uncouple epigenetic and genetic clonality in tumors. Further characterization of DNA methylation patterns in various lymphoma subtypes, chemoresistant or relapsed patients and monitoring the epigenetic clonal evolution of this disease may help facilitate both diagnosis and clinical treatment.
TET2, DNMT3A and IDH2 Mutation in Lymphoma
In human lymphoid malignancies, several common mutations have been characterized linking B and T-cell lymphoma with myeloid cancer that converge on the deregulation of cytosine methylation. TET2 is mutated or deleted in 6–12% of DLBCL [52, 97], 19–51% of PTCL [31, 80] and up to 76% of AITL [52, 97–99] making it the most common genetic lesion associated with this disease. Methylation profiling of DNA from AML patients led to the first observation that TET2 mutant AMLs are characterized by a hypermethylation phenotype [38]. Genome-wide profiling of DLBCL patients has also identified a TET2 mutant hypermethylation signature compared to TET2 wild-type DLBCL [97]. The prognostic relevance of TET2 mutation in B cell lymphoma is as yet unknown.
Unlike TET2 mutations, which are found in both B and T-cell lymphoma, mutations in IDH and DNMT3A are strikingly absent in B cell malignancies yet prevalent in T-cell lymphoma. Mutations in IDH in myeloid malignancy also promote DNA hypermethylation [38, 55] by generating the oncometabolite 2-HG instead of α-KG, the substrate normally required for TET catalytic activity [100]. In AITL, IDH2 mutations exclusively at R172 have been found in approximately 20–45% of patients [98, 101]. DNMT3A mutation occurs in 11–33% of AITL patients [31, 80, 98, 99], making these epigenetic regulators some of the most frequently mutated genes along with TET2 in this disease. In T-cell lymphoma, TET2 mutations have been associated with advanced stage disease, and a shorter progression free survival [102]. However IDH2 mutation alone did not show prognostic value, possibly due to low numbers in the cohort or heterogeneous treatment regimens in the patients studied [101].
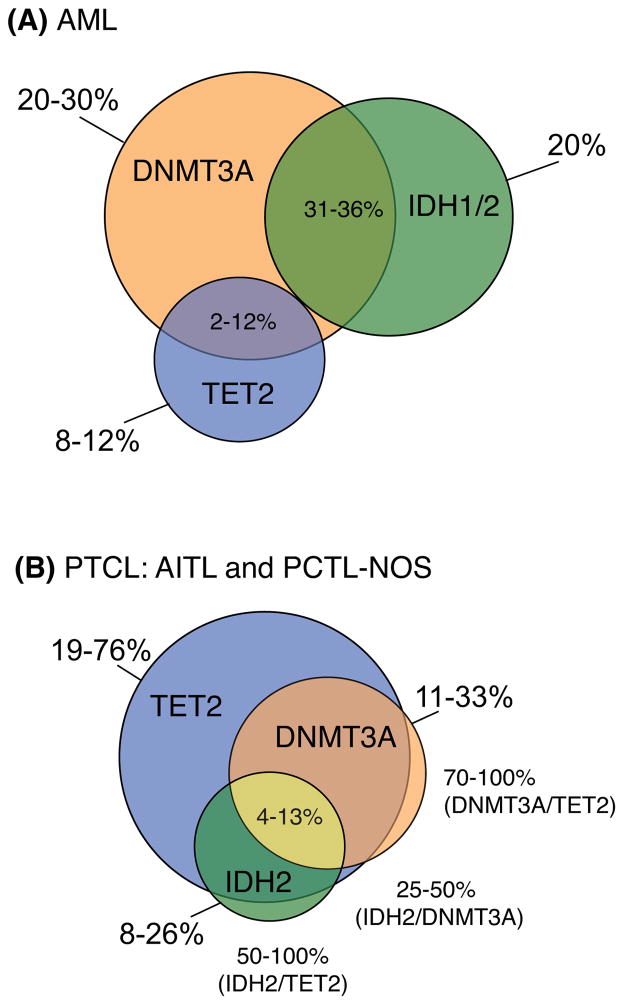
IDH mutations in AITL have the potential to indirectly inactivate TET enzymatic activity. However, unlike the mutual exclusivity often seen in AML between IDH and TET2 mutations, AITL patients can accumulate mutations in both genes [31, 98, 101] (Figure 3). In addition, AITL and PTCL-NOS harbor DNMT3A mutations that co-occur with TET2 mutations at a high frequency (73–100% of cases) [31, 98, 99]. In some cases, mutation in TET2 and DNMT3A were shown to arise in T-cell lymphoma patients with a previous history of MDS. In this study DNMT3A was found to be mutated prior to TET2 indicating that the two diseases may have a common ancestry [99]. Clonal evolution of biallelic mutations in TET2 has also been described in CD34+ cells of a patient that developed both B cell lymphoma and AML [52]. Together these studies provide further evidence of stem or progenitor cell contribution to lymphomagenesis.
Figure 3. Co-mutation in regulators of cytosine methylation in AML and PTCL.
A) In de-novo AML patients, mutation in DNMT3A is more frequent than IDH1/2 or TET2 and co-occurs with both enzymes independently, given that TET2 and IDH mutations are mutually exclusive. Mutation frequencies are shown per gene outside the venn diagram and in co-occurrence with DNMT3A within overlapping segments. B) Subsets of PTCL, in particular AITL and PCTL-NOS, have a high prevalence of TET2 mutation (>70%) that co-occurs 73–100% of the time with DNMT3A mutation. IDH1 is never seen to be mutated in these patients however IDH2 mutations readily co-occur with TET2 mutation and DNMT3A showing that co-mutation in all three genes is a distinct feature of T-cell lymphoma. Mutation frequencies are shown per gene and in co-occurrence with each other outside the venn diagram, or together in co-occurrence with TET2 mutations within the overlapping segment. Frequencies are representative of recent sequencing studies in AML [45, 63, 107, 138] and PTCL [31, 80, 98, 99, 102].
Modeling TET Loss of Function in Lymphoma
The impact of TET2 mutation on DNA methylation status in T cell lymphoma has not been tested rigorously. However, a recent study has reported that loss of Tet2 function in mice leads to a phenotype resembling T lymphoma after a long period of latency (>1 year) [103]. The Tet2-deficient T lymphoma cells in mice arise from CD4 T cells that display follicular T helper cell (TFH)-like features (CD4+, PD1+, CXCR5+) [103]. This finding is consistent with the prevalence for TET2 mutation in PTCL of TFH cell origin such as AITL and a subset of PTCL-NOS with T-FH-like features [102]. Methylation and hydroxymethylation analysis comparing normal T cells and Tet2-deficient lymphoma CD4 T cells in mice revealed greater increases in 5mC across TSS, gene body and CpG islands than losses in 5hmC [103]. An increase in 5mC specifically at an intronic silencer region of Bcl-6 was associated with upregulation in expression [103]. Bcl-6 is a master regulator of TFH differentiation [104], is oncogenic when overexpressed and drives DLBCL [105]. How hypermethylation can influence the activity of both enhancer or silencer regions of the genome has been overlooked in many studies that focus on promoter methylation analysis and may help shed light on what is often a perplexing lack of correlation between locus specific DNA methylation changes and gene expression in the literature.
Using genetic models of TET1 deletion, our laboratory has shown that loss of TET1 function promotes non-Hodgkin-like B cell lymphoma in mice [106]. TET1 mutations have been reported at low frequency in AML (~1%) [107] compared to T-ALL (~14%) [108] and translocations involving MLL and TET1 have been detected in AML [109], T cell lymphoma [110] and B-ALL [111] however TET1 was not found to be mutated in several whole genome and exome sequencing studies in B-NHL [78, 79]. Instead, it was shown that the TET1 gene is silenced and its expression is significantly down-regulated in B-cell lymphoma patients compared to normal mature B cells [106]. Multiple studies in solid tumors have described a correlation between decreased expression of the TET proteins and loss of 5hmC in the absence of mutation in these enzymes [112, 113]. Future studies that combine gene expression and methylation status, specifically of epigenetic regulators, not just mutational analysis, may help shed light on how epigenetic heterogeneity can become amplified in lymphoma.
Targeting Aberrant DNA Methylation
Epigenetic treatments that target aberrant DNA methylation and silenced chromatin approved by the FDA include histone deacetylase inhibitors (HDACis), such as vorinostat [114], and DNMT inhibitors (DNMTis), azacitidine and decitabine [115]. Multiple trials are currently testing the efficacy of using DNA hypomethylating agents alone and in combination with standard chemotherapy to treat patients with PTCL, MM, DLBCL, MDS and AML [116–118]. Re-expression of tumor suppressors including apoptotic genes, differentiation factors and cell cycle regulators, has been the main focus of epigenetic therapies with the goal of removing aberrant silencing at these loci. However, given the recent findings that link DNA methylation heterogeneity with a poor prognosis in lymphoma [92, 94], pre-treating patients with HDACi or DNMTi may help level the epigenetic playing field for standard treatment regimens. Recently it was shown that treatment of DLBCL with low-dose DNMTi reprogrammed chemoresistant cells to become sensitive to standard chemotherapy [96]. Removing epigenetic heterogeneity may provide a means to decrease clonal diversity thus limiting the possibility that epigenetically resistant sub-clones can survive treatment and drive relapse. Response rate to DNMTi in TET2 mutant MDS patients is greater than TET2 wild-type [119] suggesting a dependency of these tumors on aberrant methylation for survival. However, it is not clear whether DNMT3A, IDH or TET2 mutation status and prognosis in AML, MDS or PTCL correlates specifically with increased epigenetic heterogeneity.
Concluding Remarks
Single-base resolution mapping of DNA methylation has greatly advanced the field of epigenetics in the last few years. These studies have revealed how patterns in DNA methylation are dynamic in both coding and non-coding genomic loci, can define leukemia and lymphoma subtypes and predict disease aggression or therapeutic outcome. Previous high-throughput studies in patients utilized bisulfite conversion or methylation-sensitive digestion assays coupled with array technologies to determine DNA methylation changes in the genome. These studies however were focused primarily on measuring DNA changes at promoters and CpG islands and did not differentiate between 5mC or 5hmC, as both residues are protected during the bisulfite-conversion reaction. Recent studies have shown that enzymatic and chemical treatment of DNA (oxidation or reduction) alone or coupled with bisulfite conversion can allow for single-base resolution mapping of 5mC, 5hmC, 5fC and 5caC [120–125], paving the way for future sequencing studies to map these DNA methylation intermediates in the genome. Given the accumulating evidence of a role for TET proteins as tumor suppressors in many human malignancies along with DNMT3A and IDH, these studies will provide essential mechanistic insight into the regulation of cytosine-modification patterning in normal or malignant hematopoiesis. In addition, DNA methylation sequencing can now be successfully carried out at the single-cell level [126, 127]. The imperative of single-cell studies should be to track DNA methylation clonal heterogeneity in leukemia, lymphoma or myeloma at diagnosis and relapse. The ability to identify epigenetic signatures associated with sub-clonal populations that can harbor cell-intrinsic treatment resistance will be the goal of these studies (See Outstanding Questions Box). The collective information from both genetic and epigenetic sequencing studies should greatly improve our understanding of how to combat these hematopoietic diseases and provide a rationale for targeting aberrant DNA methylation in combination with conventional chemotherapy.
Outstanding Questions.
What is the molecular mechanism behind DNA methylation and chromatin dynamics?
Do 5hmC, 5fC and 5caC have unique roles in DNA methylation patterning, chromatin dynamics and gene transcription?
How does DNA methylation heterogeneity influence treatment resistance?
Can we predict tumor relapse based on the DNA methylation heterogeneity at diagnosis?
Should aberrant DNA methylation be targeted in combination with conventional chemotherapy?
Trends Box.
Mutations in DNA methylation enzymes (DNMT3A, TET1/2 and IDH1/2) are frequent in leukemia and lymphoma.
Dynamic DNA methylation patterns in coding and non-coding regions are found during hematopoietic transformation.
Aberrant DNA methylation can define leukemia and lymphoma subtypes, predict disease aggression and therapeutic outcome.
Acknowledgments
This research was supported by the US National Institutes of Health 5RO1CA194923, 1R01CA169784, 1R01CA133379, R01CA149655 and 5R01CA173636), the Leukemia & Lymphoma Society (TRP#6340-11 and LLS#6373-13), The Chemotherapy Foundation, The V Foundation for Cancer Research, Alex’s Lemonade Stand Foundation for Childhood Cancer, St. Baldrick’s Cancer Research Foundation and the Howard Hughes Medical Institute (I.A.). The work was also supported by the New York State Department of Health (#CO030132).
Glossary Box
- Canyons
conserved large (at least 5kb) low-methylated regions of the genome
- CpG islands (CGIs)
DNA regions enriched in cytosine nucleotides followed by guanine nucleotide. Cytosines in CpG context can be methylated to form 5-methylcytosine. In mammal genomes around 20% of CpGs are dynamically methylated
- CGI shores
CpG-enriched regions located up to 2kb away from the promoter CGIs
- Epigenetic inter-tumor heterogeneity
epigenetic variations found in different patient samples of the same type of tumor
- Epigenetic intra-tumor heterogeneity
distinct cellular populations showing specific epigenetic patterns within a tumor
- Promoter CGIs
short CpG-enriched regions in the promoter of a gene. 70% of the mammalian genes harbor CGIs in their respective promoters, most of them unmethylated. Unmethylated promoter CGIs correspond to nucleosome free regions at the transcription start site of the gene. Hypermethylation of promoter CGIs is associated with repression of gene transcription
- Valleys
large genomic domains that are generally devoid of DNA methylation that are enriched in developmental genes (see also “Canyons”)
- Whole-genome bisulfite sequencing (WGBS)
Bisulfite treatment converts cytosine residues to uracil, but leaves 5mC and 5hmC residues unaffected. Bisulfite-treatment of genomic DNA coupled with high-throughput sequencing techniques allows for single-base resolution mapping of methylation patterns
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ji H, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467(7313):338–42. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bock C, et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47(4):633–47. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nat Rev Immunol. 2011;11(7):478–88. doi: 10.1038/nri2991. [DOI] [PubMed] [Google Scholar]
- 4.Figueroa ME, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borgel J, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42(12):1093–100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 6.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500(7463):477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27(4):361–8. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 10.Jeong M, et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet. 2014;46(1):17–23. doi: 10.1038/ng.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bestor T, et al. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203(4):971–83. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 13.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19(3):219–20. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 14.Schermelleh L, et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35(13):4301–12. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67(3):946–50. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 16.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122(10):3195–205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 17.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–33. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo JU, et al. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–34. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cimmino L, et al. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell. 2011;9(3):193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15(3):152–65. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gowher H, et al. Mutational analysis of the catalytic domain of the murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J Mol Biol. 2006;357(3):928–41. doi: 10.1016/j.jmb.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita Y, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29(25):3723–31. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 24.Russler-Germain DA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25(4):442–54. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Challen GA, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 2014;15(3):350–64. doi: 10.1016/j.stem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko M, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 2011;108(35):14566–71. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdel-Wahab O, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144–7. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langemeijer SM, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41(7):838–42. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 30.Lorsbach RB, et al. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17(3):637–41. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 31.Palomero T, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46(2):166–70. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ko M, et al. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev. 2015;263(1):6–21. doi: 10.1111/imr.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tefferi A, et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23(7):1343–5. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chowdhury R, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcucci G, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(14):2348–55. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18(6):553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 40.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galm O, Herman JG, Baylin SB. The fundamental role of epigenetics in hematopoietic malignancies. Blood Rev. 2006;20(1):1–13. doi: 10.1016/j.blre.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 42.Akalin A, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8(6):e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li N, et al. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010;52(3):203–12. doi: 10.1016/j.ymeth.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Bullinger L, et al. Quantitative DNA methylation predicts survival in adult acute myeloid leukemia. Blood. 2010;115(3):636–42. doi: 10.1182/blood-2009-03-211003. [DOI] [PubMed] [Google Scholar]
- 45.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qu Y, et al. Differential methylation in CN-AML preferentially targets non-CGI regions and is dictated by DNMT3A mutational status and associated with predominant hypomethylation of HOX genes. Epigenetics. 2014;9(8):1108–19. doi: 10.4161/epi.29315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–43. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez C, et al. TET2 mutations are associated with specific 5-methylcytosine and 5-hydroxymethylcytosine profiles in patients with chronic myelomonocytic leukemia. PLoS One. 2012;7(2):e31605. doi: 10.1371/journal.pone.0031605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rampal R, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;9(5):1841–55. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamazaki J, et al. TET2 Mutations Affect Non-CpG Island DNA Methylation at Enhancers and Transcription Factor-Binding Sites in Chronic Myelomonocytic Leukemia. Cancer Res. 2015;75(14):2833–43. doi: 10.1158/0008-5472.CAN-14-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang H, et al. STAT3 Inhibition Enhances the Therapeutic Efficacy of Immunogenic Chemotherapy by Stimulating Type 1 Interferon Production by Cancer Cells. Cancer Res. 2015;75(18):3812–22. doi: 10.1158/0008-5472.CAN-15-1122. [DOI] [PubMed] [Google Scholar]
- 52.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rasmussen KD, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29(9):910–22. doi: 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sasaki M, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–9. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaturvedi A, et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013;122(16):2877–87. doi: 10.1182/blood-2013-03-491571. [DOI] [PubMed] [Google Scholar]
- 57.Chen C, et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 2013;27(18):1974–85. doi: 10.1101/gad.226613.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu J, et al. DNMT3A Arg882 mutation drives chronic myelomonocytic leukemia through disturbing gene expression/DNA methylation in hematopoietic cells. Proc Natl Acad Sci U S A. 2014;111(7):2620–5. doi: 10.1073/pnas.1400150111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayle A, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125(4):629–38. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wertheim GB, et al. Microsphere-based multiplex analysis of DNA methylation in acute myeloid leukemia. J Mol Diagn. 2014;16(2):207–15. doi: 10.1016/j.jmoldx.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang Y, et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009;113(6):1315–25. doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen L, et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28(4):605–13. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–89. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fernandez HF, et al. Anthracycline dose intensification in acute myeloid leukemia. N Engl J Med. 2009;361(13):1249–59. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 66.Itzykson R, et al. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25(7):1147–52. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 67.Pollyea DA, et al. Impact of TET2 mutations on mRNA expression and clinical outcomes in MDS patients treated with DNA methyltransferase inhibitors. Hematol Oncol. 2011;29(3):157–60. doi: 10.1002/hon.976. [DOI] [PubMed] [Google Scholar]
- 68.Taskesen E, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117(8):2469–75. doi: 10.1182/blood-2010-09-307280. [DOI] [PubMed] [Google Scholar]
- 69.Yamaguchi S, et al. IDH1 and IDH2 mutations confer an adverse effect in patients with acute myeloid leukemia lacking the NPM1 mutation. Eur J Haematol. 2014;92(6):471–7. doi: 10.1111/ejh.12271. [DOI] [PubMed] [Google Scholar]
- 70.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–78. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gu H, et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7(2):133–6. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–33. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kent DG, Ortmann CA, Green AR. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med. 2015;372(19):1865–6. doi: 10.1056/NEJMc1503143. [DOI] [PubMed] [Google Scholar]
- 74.Landau DA, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26(6):813–25. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annual review of immunology. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin’s lymphoma. The Non-Hodgkin’s Lymphoma Classification Project. Blood. 1997;89(11):3909–18. [PubMed] [Google Scholar]
- 77.Foss FM, et al. Peripheral T-cell lymphoma. Blood. 2011;117(25):6756–67. doi: 10.1182/blood-2010-05-231548. [DOI] [PubMed] [Google Scholar]
- 78.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nature genetics. 2011;43(9):830–7. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sakata-Yanagimoto M, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(2):171–5. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 81.Martin-Subero JI, et al. A comprehensive microarray-based DNA methylation study of 367 hematological neoplasms. PloS one. 2009;4(9):e6986. doi: 10.1371/journal.pone.0006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matsuda I, Imai Y, Hirota S. Distinct global DNA methylation status in B-cell lymphomas: immunohistochemical study of 5-methylcytosine and 5-hydroxymethylcytosine. Journal of clinical and experimental hematopathology : JCEH. 2014;54(1):67–73. doi: 10.3960/jslrt.54.67. [DOI] [PubMed] [Google Scholar]
- 83.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 84.Shaknovich R, et al. DNA methylation signatures define molecular subtypes of diffuse large B-cell lymphoma. Blood. 2010;116(20):e81–9. doi: 10.1182/blood-2010-05-285320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo J, et al. Differential DNA methylation of gene promoters in small B-cell lymphomas. American journal of clinical pathology. 2005;124(3):430–9. doi: 10.1309/LCGN-V77J-464L-NFD6. [DOI] [PubMed] [Google Scholar]
- 86.Kretzmer H, et al. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat Genet. 2015;47(11):1316–25. doi: 10.1038/ng.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364(11):1046–60. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 88.Agirre X, et al. Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers. Genome Res. 2015;25(4):478–87. doi: 10.1101/gr.180240.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salhia B, et al. DNA methylation analysis determines the high frequency of genic hypomethylation and low frequency of hypermethylation events in plasma cell tumors. Cancer Res. 2010;70(17):6934–44. doi: 10.1158/0008-5472.CAN-10-0282. [DOI] [PubMed] [Google Scholar]
- 90.Kulis M, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47(7):746–56. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shaknovich R, et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood. 2011;118(13):3559–69. doi: 10.1182/blood-2011-06-357996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De S, et al. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PLoS genetics. 2013;9(1):e1003137. doi: 10.1371/journal.pgen.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pan H, et al. Epigenomic evolution in diffuse large B-cell lymphomas. Nat Commun. 2015;6:6921. doi: 10.1038/ncomms7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chambwe N, et al. Variability in DNA methylation defines novel epigenetic subgroups of DLBCL associated with different clinical outcomes. Blood. 2014;123(11):1699–708. doi: 10.1182/blood-2013-07-509885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hopp L, Loffler-Wirth H, Binder H. Epigenetic Heterogeneity of B-Cell Lymphoma: DNA Methylation, Gene Expression and Chromatin States. Genes (Basel) 2015;6(3):812–40. doi: 10.3390/genes6030812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Clozel T, et al. Mechanism-based epigenetic chemosensitization therapy of diffuse large B-cell lymphoma. Cancer Discov. 2013;3(9):1002–19. doi: 10.1158/2159-8290.CD-13-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Asmar F, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica. 2013;98(12):1912–20. doi: 10.3324/haematol.2013.088740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Odejide O, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123(9):1293–6. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. The New England journal of medicine. 2012;366(1):95–6. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 100.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer cell. 2010;17(3):225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cairns RA, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119(8):1901–3. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lemonnier F, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120(7):1466–9. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 103.Muto H, et al. Reduced TET2 function leads to T-cell lymphoma with follicular helper T-cell-like features in mice. Blood Cancer J. 2014;4:e264. doi: 10.1038/bcj.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31(3):457–68. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 105.Cattoretti G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7(5):445–55. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 106.Cimmino L, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16(6):653–62. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.De Keersmaecker K, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45(2):186–90. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lorsbach RB, et al. Leukemia : official journal of the Leukemia Society of America. 3. Vol. 17. Leukemia Research Fund; U.K: 2003. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) pp. 637–41. [DOI] [PubMed] [Google Scholar]
- 110.Ittel A, et al. First description of the t(10;11)(q22;q23)/MLL-TET1 translocation in a T-cell lymphoblastic lymphoma, with subsequent lineage switch to acute myelomonocytic myeloid leukemia. Haematologica. 2013;98(12):e166–8. doi: 10.3324/haematol.2013.096750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Burmeister T, et al. The MLL recombinome of adult CD10-negative B-cell precursor acute lymphoblastic leukemia: results from the GMALL study group. Blood. 2009;113(17):4011–5. doi: 10.1182/blood-2008-10-183483. [DOI] [PubMed] [Google Scholar]
- 112.Yang H, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32(5):663–9. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kudo Y, et al. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 2012;103(4):670–6. doi: 10.1111/j.1349-7006.2012.02213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Grant S, Easley C, Kirkpatrick P. Vorinostat. Nature reviews Drug discovery. 2007;6(1):21–2. doi: 10.1038/nrd2227. [DOI] [PubMed] [Google Scholar]
- 115.Derissen EJ, Beijnen JH, Schellens JH. Concise drug review: azacitidine and decitabine. Oncologist. 2013;18(5):619–24. doi: 10.1634/theoncologist.2012-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tan J, et al. Novel histone deacetylase inhibitors in clinical trials as anti-cancer agents. J Hematol Oncol. 2010;3:5. doi: 10.1186/1756-8722-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saunthararajah Y. Key clinical observations after 5-azacytidine and decitabine treatment of myelodysplastic syndromes suggest practical solutions for better outcomes. Hematology Am Soc Hematol Educ Program. 2013:511–21. doi: 10.1182/asheducation-2013.1.511. [DOI] [PubMed] [Google Scholar]
- 118.Falini B, et al. Perspectives for therapeutic targeting of gene mutations in acute myeloid leukaemia with normal cytogenetics. Br J Haematol. 2015;170(3):305–22. doi: 10.1111/bjh.13409. [DOI] [PubMed] [Google Scholar]
- 119.Bejar R, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124(17):2705–12. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336(6083):934–7. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 121.Booth MJ, et al. Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nat Chem. 2014;6(5):435–40. doi: 10.1038/nchem.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Booth MJ, et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc. 2013;8(10):1841–51. doi: 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu H, et al. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat Biotechnol. 2014;32(12):1231–40. doi: 10.1038/nbt.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xia B, et al. Bisulfite-free, base-resolution analysis of 5-formylcytosine at the genome scale. Nat Methods. 2015;12(11):1047–50. doi: 10.1038/nmeth.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–80. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guo H, et al. Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nat Protoc. 2015;10(5):645–59. doi: 10.1038/nprot.2015.039. [DOI] [PubMed] [Google Scholar]
- 127.Smallwood SA, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11(8):817–20. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rollins RA, et al. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16(2):157–63. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sun D, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14(5):673–88. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103(5):1412–7. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41(12):1350–3. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479(7371):74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jones PA. The DNA methylation paradox. Trends Genet. 1999;15(1):34–7. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 134.Wang L, et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 2015;523(7562):621–5. doi: 10.1038/nature14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wiench M, et al. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30(15):3028–39. doi: 10.1038/emboj.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xie W, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153(5):1134–48. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ribeiro AF, et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119(24):5824–31. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]