Abstract
Cardiac progenitor cells are multipotent and give rise to cardiac endothelium, smooth muscle, and cardiomyocytes. Here, we define and characterize the cardiomyoblast intermediate that is committed to the cardiomyocyte fate, and we characterize the niche signals that regulate commitment. Cardiomyoblasts express Hopx, which functions to coordinate local Bmp signals to inhibit the Wnt pathway, thus promoting cardiomyogenesis. Hopx integrates Bmp and Wnt signaling by physically interacting with activated Smads and repressing Wnt genes. The identification of the committed cardiomyoblast that retains proliferative potential will inform cardiac regenerative therapeutics. In addition, Bmp signals characterize adult stem cell niches in other tissues where Hopx-mediated inhibition of Wnt is likely to contribute to stem cell quiescence and to explain the role of Hopx as a tumor suppressor.
Lineage analyses during cardiac development in the chick and mouse over the last two decades have demonstrated that at least two pools of progenitor cells contribute to the heart (1). Cardiac progenitor cells (CPCs) derived from the cardiac crescent, or the first heart field (FHF), express Nkx2-5 and contribute to a primitive heart tube. After subsequent looping of the heart tube, additional progenitor cells are added to the arterial and venous poles of the heart from the second heart field (SHF). SHF cells arise just medial and posterior to the FHF in the cardiac crescent, and populate the pharyngeal arches and dorsomedial mesoderm prior to migration into the heart proper (2, 3).
Studies modeling cardiac development using differentiation of ES-derived embryoid bodies (EBs), have demonstrated that SHF progenitors, marked by Islet1 (Isl1) expression (4), are multipotent, with potential for differentiation into cardiomyocyte, endothelial, or smooth muscle lineages (5–7). Wnt signaling is necessary to promote and expand multipotent CPCs (8–10), and subsequent differentiation of cardiac myocytes is influenced by Bone morphogenetic protein (Bmp) signaling and Wnt inhibition (11, 12). However, the implications of these studies for in vivo cardiogenesis are unknown. The characteristics of an embryonic CPC niche are poorly described and the degree to which CPCs remain uncommitted during mid and late gestation has been unclear. For example, recent reports using inducible lineage tracing of early cardiac progenitors, marked by Mesp1 expression, suggest the existence of a relatively small pool of multipotent progenitors in vivo and at least some of these become committed at very early stages, perhaps soon after gastrulation (13, 14). Efforts to fully characterize the signaling pathways active during cell fate decisions in vivo have been hampered, at least in part, by the lack of specific markers of lineage commitment.
Here, we report that CPCs committed to the myocyte lineage can be prospectively identified prior to the expression of sarcomere genes based on Hopx expression, an atypical homeodomain protein expressed during early cardiac development and in multiple stem cell populations (15–19). Akin to an erythroblast in hematopoietic differentiation, we have termed these committed CPCs “cardiomyoblasts.” We show that SHF-derived cardiomyoblasts are specified in the distal outflow tract (OFT) within a zone of high Bmp and low Wnt signaling. Finally, we show that Hopx not only marks commitment, but that it also promotes myogenesis by interacting with an activated-Smad complex to repress Wnt.
Cardiomyoblasts are defined by Hopx expression
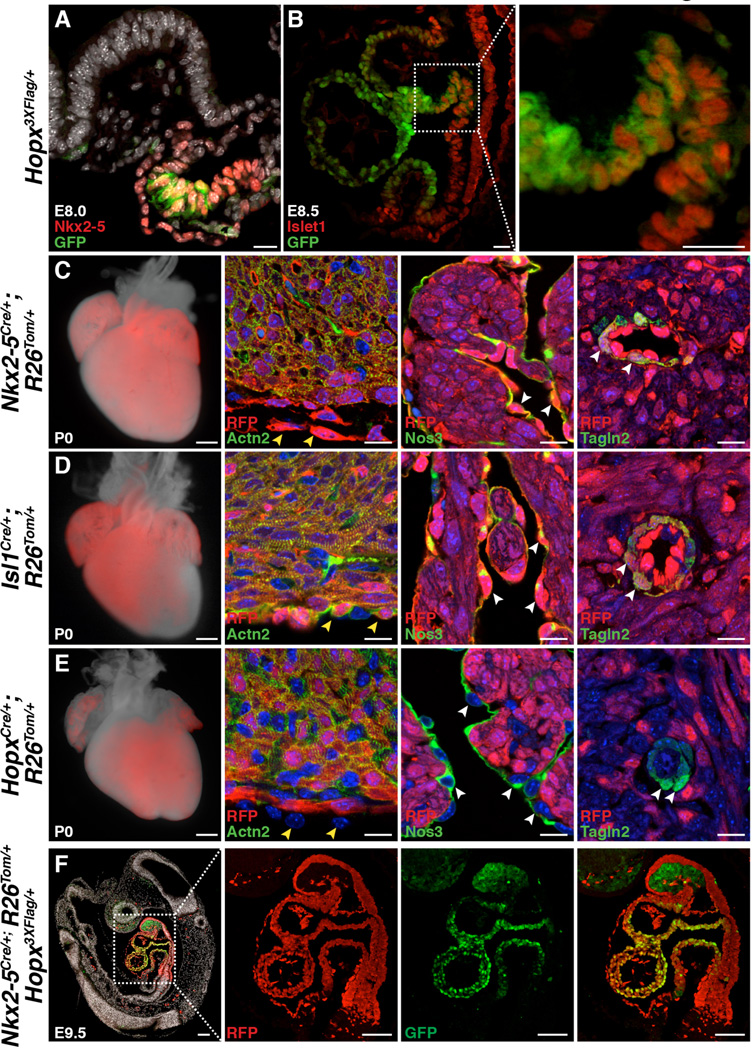
At early stages of cardiac development, Nkx2-5 and Isl1 mark populations of CPCs (2, 4), and Hopx expression initiates shortly after Nkx2-5 in precardiac mesoderm (15). We used a knock-in allele in which Hopx is epitope tagged and GFP is expressed in Hopx+ cells (18) to determine that Hopx is expressed in a subset of CPCs at E8.0 and E8.5 in the FHF and SHF, respectively (Fig. 1, A and B). As SHF progenitors enter the distal outflow tract (OFT), Isl1 expression is gradually extinguished and Hopx expression initiates providing a restricted region of co-expression in the distal OFT (Fig. 1B). Fate-mapping using Nkx2-5Cre/+ and cre-dependent reporter mice indicates that essentially the entire late-gestation heart derives from Nkx2-5+ precursors, including myocytes, smooth muscle, endothelium, and epicardium (Fig. 1C). Isl1+ cells also give rise to smooth muscle, endothelium, myocytes and epicardium, but some of the left ventricle myocardium and atria, derived from the FHF, is not labeled in Isl1 fate-mapping experiments (Fig. 1D). Lineage tracing experiments using a HopxCre/+ allele, in which we inserted cre following an internal ribosomal entry sequence (IRES) so as to avoid perturbing Hopx expression (20), demonstrate labeling in all four cardiac chambers (Fig. 1E and fig. S1, A and B). However, in contrast to Nkx2-5 and Isl1, Hopx-derivatives within the heart are entirely restricted to cardiac myocytes (Fig. 1E and fig. S2). Some cardiac fibroblasts derive from Nkx2-5 and Isl1-expressing precursors, but Hopx+ cells do not give rise to fibroblasts in the heart (fig. S2). Most cardiac myocytes derive from Hopx+ precursors, although some specialized myocytes surrounding the pulmonary veins and within the interatrial septum are not derived from Hopx+ cells (fig. S3). Analysis of E9.5 Nkx2-5Cre/+; R26Tom/+; Hopx3XFlag/+ embryos reveals that all Hopx+ cardiomyocytes at this time point derive from Nkx2-5+ precursors (Fig. 1F). Flow cytometry analysis of dissociated post-natal day 2 (P2) hearts, expressing a reporter allele, confirms the multi-lineage contribution of Nkx2-5 and Isl1, in contrast to Hopx+ cells (fig. S4). We do not detect Hopx in any non-myocyte cell types within the heart, consistent with our lineage tracing data (fig. S5).
Fig. 1. Prospective identification of cardiomyoblasts.
(A) A subset of E8.0 Nkx2-5+ cells in the FHF precardiac mesoderm express Hopx. (B) A subset of E8.5 Isl1+ SHF cells in the outflow tract express Hopx (inset highlights distal outflow tract). (C,D) Nkx2-5+ (C) and Isl1+ (D) cells give rise to myocytes (Actn2+), epicardium (yellow arrowheads), endothelium (Nos3+, white arrowheads), and smooth muscle (Tagln2+, white arrowheads) at P0. (E) Hopx+ cells give rise to myocytes, not epicardium (yellow arrowheads), endothelium (white arrowheads), or smooth muscle (white arrowheads) at P0. (F) Hopx+ cells (GFP+) derive from Nkx2-5+ cells (RFP+) (E9.5, sagittal section). Scale bars: 500 µm (C–E, whole mount), 100 µm (F), 25 µm (A,B), 10 µm (C–E, histology).
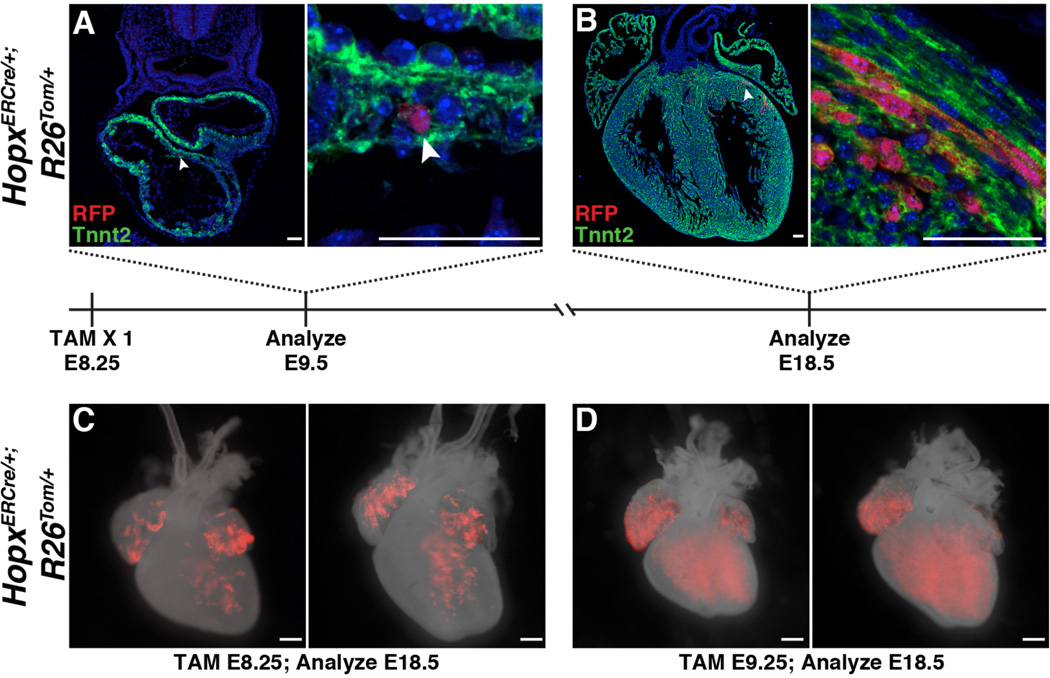
To confirm the fate of the earliest Hopx+ expressing cells we used a tamoxifen-inducible Hopx allele (HopxERCre/+) (19). Low doses of tamoxifen induction in HopxERCre/+; R26Tom/+ embryos reveal that single E8.25 Hopx+ cardiomyoblasts expand to form clusters of myocytes by E18.5 (Fig. 2, A and B) and Hopx+ cardiomyoblasts express markers of proliferation at early embryonic time points (fig. S6). In addition, inducing cre recombinase activity with a single dose of tamoxifen at E8.25, a time point correlating with Hopx expression within a subset of Nkx2-5+ progenitors, demonstrates lineage-labeled myocytes primarily in the left ventricle and both atria at E18.5 (Fig. 2C). However, similar lineage tracing of E9.25 Hopx+ cells demonstrate that cardiomyoblasts expressing Hopx at this later time point contribute to myocytes in both ventricles and both atria (Fig. 2D). Taken together, these results establish that cardiac Hopx-expression identifies a pool of progenitors committed entirely to the myocyte lineage (e.g. cardiomyoblasts) and that Hopx+ cardiomyoblasts expand during cardiogenesis. In addition, our results suggest that commitment of FHF and SHF CPCs occurs at distinct time points during cardiogenesis.
Fig. 2. Cardiomyoblasts expand during cardiogenesis.
(A,B) E8.25 HopxERCre/+; R26Tom/+ embryos were induced with tamoxifen and harvested at E9.5 (A) and E18.5 (B). Individual Hopx-derived cells are labeled at E9.5 (A, B area highlighted by arrowhead is magnified on right). At E18.5, clusters of myocytes are identified (B). (C,D) HopxERCre/+; R26Tom/+ embryos were induced with tamoxifen at E8.25 (C) or E9.25 (D) and analyzed at E18.5 (n ≥ 2 litters per time point, 2 examples at each time point shown). Scale bars: 500 µm (C,D) and 50 µm (A,B).
Hopx promotes myogenesis by inhibiting Wnt signaling
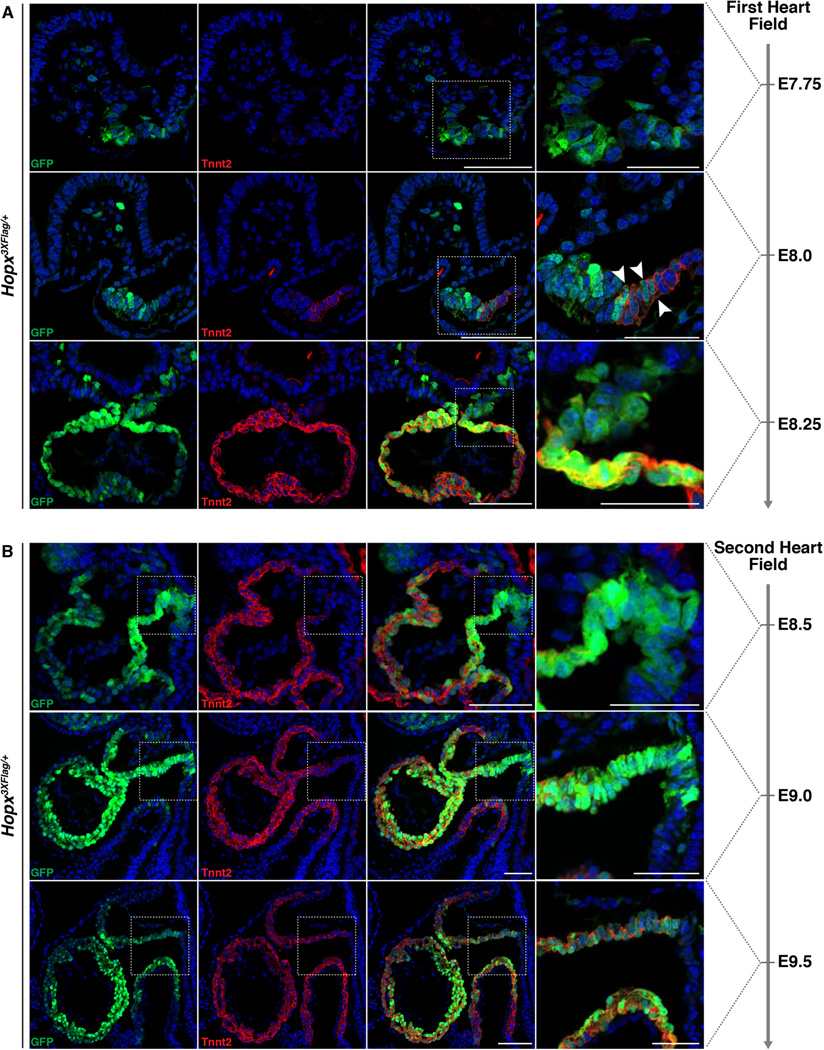
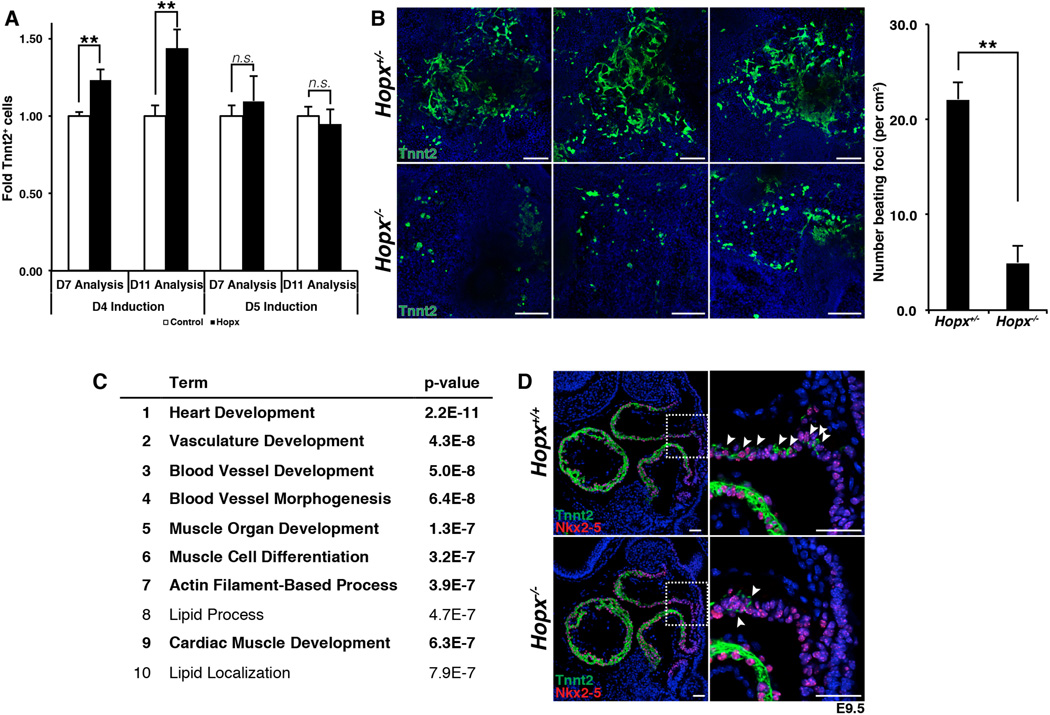
Hopx is expressed early during cardiomyocyte differentiation, and precedes that of troponin T (Tnnt2) in both the FHF and SHF (Fig. 3, A and B). In order to examine the function of Hopx during cardiomyogenesis, we overexpressed Hopx at various time points of embryonic stem (ES) cell differentiation. Hopx is normally detectable by day 5, before Tnnt2 and Mhy6 are expressed by maturing EBs (fig. S7A). Using established protocols (11, 21), we differentiated ES cells into EBs and expressed Hopx starting on day 4. Flow cytometry analyses of the resulting cultures at day 7 and day 11 indicate that precocious expression of Hopx induces a significant increase in the number of Tnnt2+ cells (Fig. 4A). When Hopx is induced only 1 day later, at day 5, no statistically significant increase in cardiomyogenesis is observed (Fig. 4A).
Fig. 3. Hopx expression precedes troponin expression.
(A) Hopx expression precedes Tnnt2 expression in the precardiac mesoderm/FHF at early time points during cardiac development. Hopx+, Tnnt2+ cells are identified a few hours later (white arrowheads). (B). The distal outflow tract and SHF mesoderm harbor Hopx+, Tnnt2− cells at early time points during cardiac development. Scale bars: 100 µm except right most panels/insets which are 50 µm.
Fig. 4. Hopx promotes myogenesis.
(A) Precocious expression of Hopx in CPCs at day 4 of EB differentiation results in more Tnnt2+ cells measured by flow cytometry. (B) Differentiation of Hopx−/− EBs results in fewer Tnnt2+ cells and fewer beating foci compared to Hopx+/− EBs (day 10). Images from 3 different replicates are shown. (C) Gene ontology analysis from microarrays done with triplicate samples of Hopx−/− vs. Hopx+/− EBs (top 3000 genes down-regulated, ranked by fold change, FDR cutoff = 10%, day 8, GOTERM_BP_FAT). (D) Paucity of Tnnt2+ cells (arrowheads) in the distal outflow tract Hopx−/− compared to Hopx+/+ embryos (E9.5). ** p<0.05. Scale bars: 500 µm (B), 50 µm (D).
We performed reciprocal loss-of-function experiments by generating embryonic stem cells (ESCs) from Hopx−/− and littermate Hopx+/− blastocysts and differentiating them into cardiac cell types (20). Axin2 and Nkx2-5 are each expressed at comparable levels when we compared Hopx−/− and Hopx+/− EBs at days 3 and 4.75 (fig. S7B). However, there is a marked reduction of Tnnt2+ cells and beating foci upon differentiation of Hopx−/− EBs compared to Hopx+/− EBs (Fig. 4B). We performed microarrays from Hopx−/− and Hopx+/− EBs (day 8, n=3), and multiple myocyte related genes are down-regulated in Hopx−/− EBs compared to Hopx+/− control EBs [table S1, e.g. Mhy6 -30.3X, Myh7 -30.3X, Mybpc3 -13.0X, Ttn -7.0X, Tnnt2 -5.5X, Nkx2-5 -3.984X, false discovery rate (FDR) cutoff = 10%]. Gene ontology analysis of the top 3000 genes that are down-regulated confirmed enrichment for families of genes related to heart development and myogenesis (Fig. 4C). Although sarcomere genes are expressed in Hopx−/− hearts, the onset of Tnnt2 expression is delayed in SHF myoblasts as they enter the OFT, as evidenced by a lack of Tnnt2 expression in Nkx2-5+ precursors within the distal OFT of Hopx−/− versus Hopx+/+ hearts at E9.5 (Fig. 4D). The eventual expression of sarcomere genes in Hopx mutants suggests that redundant pathways exist for activation of the myogenic program.
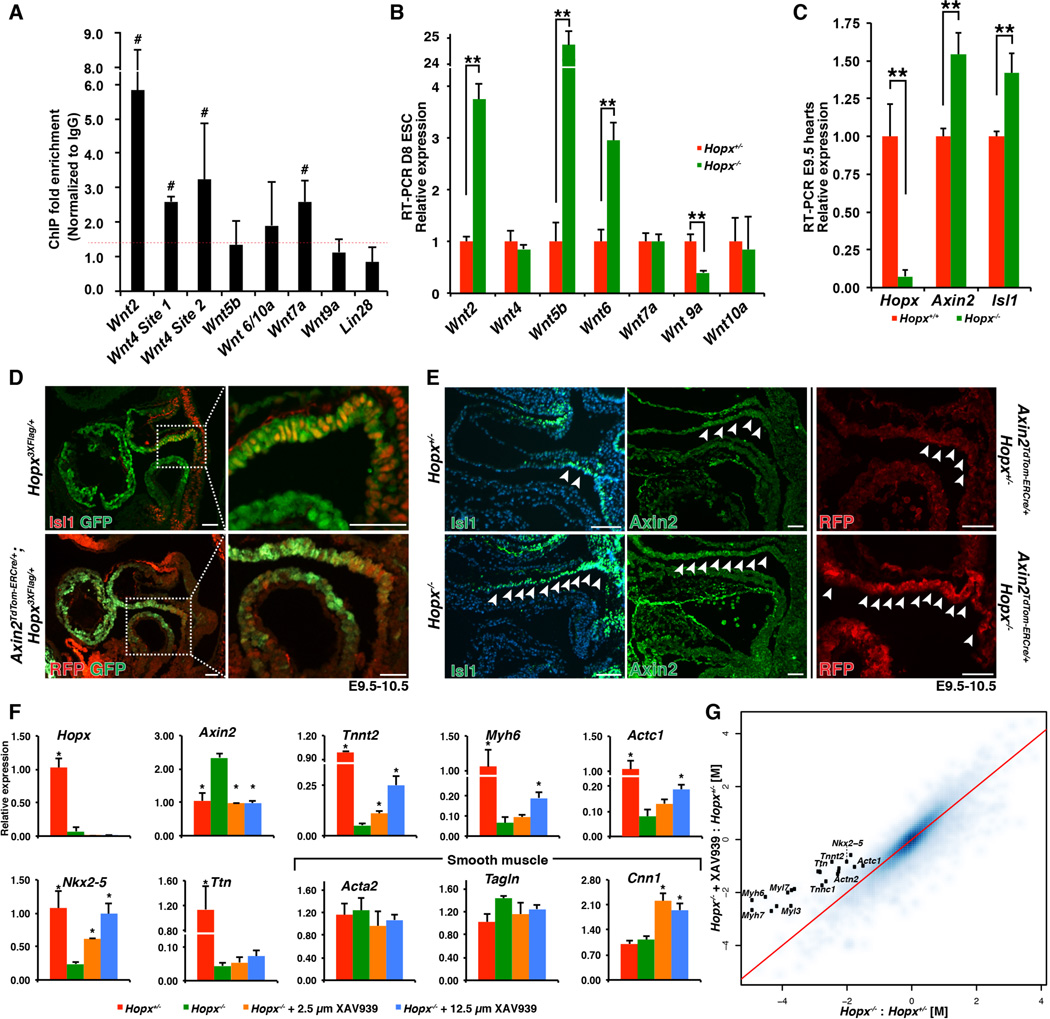
In order to further define the role of Hopx during myogenesis, we defined the genomic regions occupied by Hopx by performing chromatin-immunoprecipitation followed by massively paralleled sequencing (ChIP-seq). Although Hopx does not bind to DNA directly (22) it can interact with co-repressor complexes to inhibit gene expression. We performed ChIP-seq analysis of pooled chromatin derived from ~35 E9.5 microdissected murine embryonic hearts (table S2). KEGG analysis of the genes associated with the strongest 3000 peaks suggested that Hopx occupancy was enriched in genomic regions proximal to Wnt family member genes (p=3.9 × 10−4), and the Wnt signaling pathway was the top hit using PANTHER analysis (p=1.2 × 10−3).
We independently validated Hopx occupancy close to several Wnt ligand transcriptional start sites by ChIP-qPCR (Fig. 5A). Consistent with the known function of Hopx as a transcriptional repressor (15), many of these ligands, including Wnt2, Wnt5b, and Wnt6, are expressed at higher levels at day 8 of differentiation in Hopx−/− EBs compared to Hopx+/− EBs (Fig. 5B). Axin2 and Isl1, both target genes of canonical Wnt signaling (23, 24), are expressed at significantly higher levels in E9.5 Hopx−/− hearts as compared to littermate controls (Fig. 5C). Immunohistochemistry of E9.5-10.5 control OFTs reveals that the distal OFT is a transition zone in which Axin2 and Isl1 expression diminish while Hopx expression is activated (Fig. 5D). In Hopx−/− embryos, Wnt signaling, as represented by Axin2 and Isl1 expression, is expanded into the proximal OFT compared to littermate controls (Fig. 5E). Overexpression of Hopx in EBs reduces Axin2 expression, whereas a mutant form of Hopx that does not effectively interact with Smad4 (discussed further below) does not have this effect (fig. S7C). These data suggest that Hopx represses Wnt signaling during cardiogenesis.
Fig. 5. Myogenesis requires inhibition of Wnt signaling.
(A) ChIP-qPCR from E9.5 hearts. Wnt2, Wnt4 sites 1 and 2, and Wnt7a demonstrate greater than 1.4X enrichment (red dashed line) over IgG in all replicates (denoted by #, n≥3 replicates). (B) Expression analysis of Hopx−/− vs. Hopx+/− EBs (day 8). (C) qRT-PCR from littermate E9.5 microdissected hearts. (D) Isl1 (upper panels) and Axin2 (lower panels, RFP expression reflects Axin2 in an Axin2TdTom-ERCre/+ embryo, sagittal sections) are each coexpressed with Hopx in the cardiac OFT. (E) Isl1 and Axin2 expression is expanded in Hopx−/− (arrowheads point to Isl1+ and Axin2+ cells; middle panels show Axin2 IHC (green), right panels show RFP IHC in Axin2TdTom-ERCre/+ embryos). (F) qRT-PCR analyses of Hopx+/− day 8 EBs (red), or Hopx−/− EBs (green) with either 2.5µM (orange) or 12.5µM (blue) XAV939 (*p<0.05 in comparison to Hopx−/−). (G) Comparison of the log2-transformed fold change (M) of genes differentially expressed in Hopx−/− vs. Hopx+/− EBs (x-axis) versus Hopx−/− + 12.5 µm XAV939 vs. Hopx+/− (y-axis) (n=3 samples). Sarcomere-related genes annotated with black dots. Genes above the red line are partially normalized by Wnt inhibition. ** p<0.05, Scale bars 50 µm.
During ES cell differentiation into cardiomyocytes, Hopx also functions to repress Wnt. KEGG analysis of the up-regulated genes in Hopx−/− versus Hopx+/− day 8 EB microarrays confirms overrepresentation of the Wnt signaling pathway in Hopx−/− cells (p=1.4 × 10−3). EBs lacking Hopx show elevation of Axin2 and significant impairment of sarcomere gene expression at day 8 of differentiation (Fig. 5F). Addition of XAV939, a potent inhibitor of Wnt signaling, at day 5 of differentiation to Hopx−/− EBs restores Axin2 to control levels and partially rescues sarcomere gene expression. Nkx2-5 is expressed by CPCs and its expression markedly increases over the course of cardiac differentiation of EBs (21). Differentiating Hopx−/− EBs, however, fail to up-regulate Nkx2-5 normally. Inhibition of Wnt in Hopx−/− EBs rescues Nkx2-5 to levels found in controls (Fig. 5F). Microarray analysis (table S3, n=3) confirms that expression of multiple sarcomere genes is rescued upon Wnt inhibition (Fig. 5G, e.g. Myh6, Myh7, Myl7, Myl3, Tnnc1, Actn2, Actc1). Taken together, these data suggest that Hopx repression of Wnt signaling promotes cardiomyogenesis.
Hopx interacts with Smad4
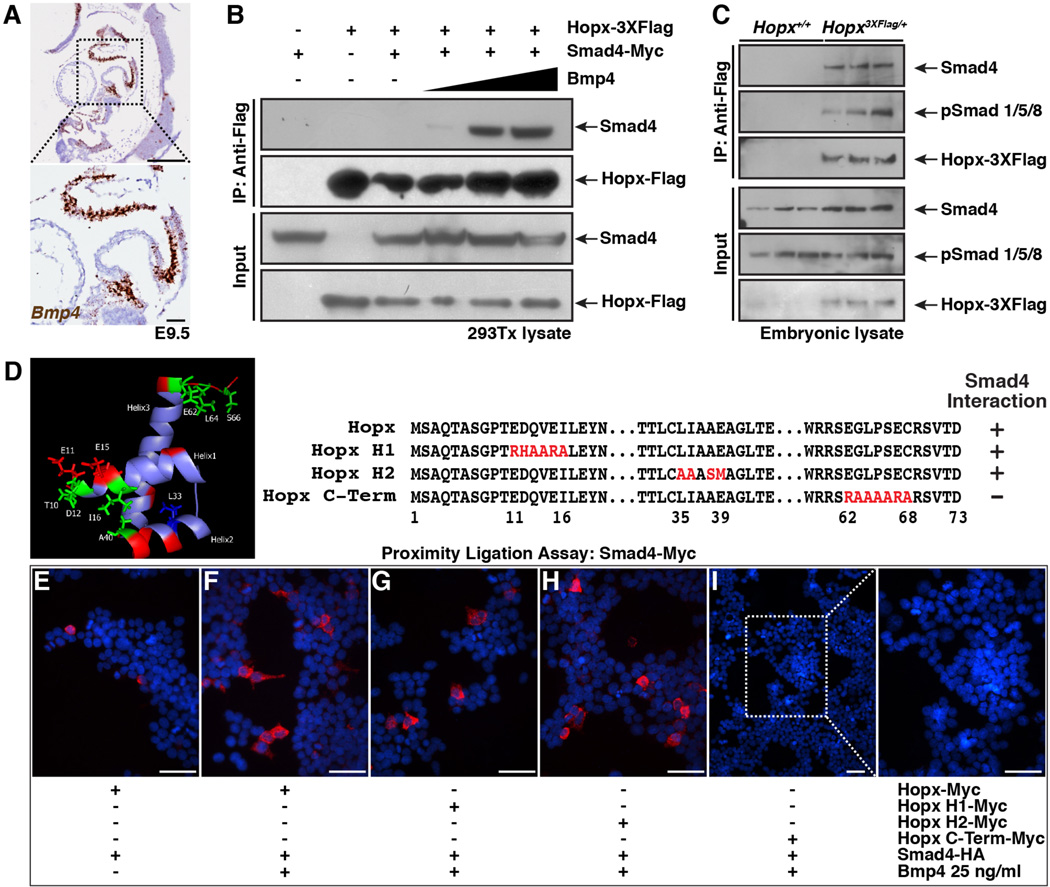
We purified Hopx-containing protein complexes from E9.5 Hopx3XFlag/+ murine hearts and identified protein components by mass spectrometry. Numerous members of the Mi-2/NuRD (nucleosome remodeling deacetylase) complex were identified (e.g. Hdac1, Hdac2, Rbbp4/7, MTA 1/2/3, and MBD3), consistent with the known association of Hopx with Hdac2 (22). In addition, Smad4 was identified as a Hopx-interacting protein. This finding is of particular interest because Bmp4, phospho-Smad1/5/8 and, to a lesser extent, Bmp2 are expressed in the OFT at E9.5, as we confirmed (Fig. 6A and fig. S8A). We confirmed the interaction between Hopx and Smad4 by co-immunoprecipitation of both factors overexpressed in 293Tx cells in the presence of increasing concentrations of recombinant Bmp4 and performing co-immunoprecipitation experiments. An interaction between Hopx and Smad4 that is dependent upon the presence of Bmp4 is detectable (Fig. 6B). Further, we confirmed that endogenous Hopx interacts with an activated Smad complex (Smad4 and phospho-Smad1/5/8) in co-immunoprecipitation experiments from E9.5-10 whole embryo lysates (Fig. 6C and fig. S8B).
Fig. 6. Hopx interacts with an activated Smad complex.
(A) Bmp4 expression in control E9.5 heart (RNAscope in situ hybridization, brown; sagittal section). Inset focuses on outflow and inflow tracts. (B) Hopx co-immunoprecipitates with Smad4 in 293Tx cells in a Bmp4 dependent manner (0, 2, 10, 25 ng/ml Bmp4, 7.5% input). (C). Hopx co-immunoprecipitates with Smad4 and phospho-Smad1/5/8 in vivo (E9.5-E10 tissue lysates 3 independent examples of each genotype shown, 7.5% input). (D) NMR structure of Hopx (22) and schema of mutant constructs. (E–I) Proximity ligation assay using transfected 293Tx cells. Presence of Bmp4 and transfected plasmids are indicated. Best representative images from n=3 experiments shown. Scale bars: 250 µm (A, top) and 50 µm (A bottom, E–I).
We have previously determined the structure of Hopx by NMR spectroscopy demonstrating a helix-turn-helix motif (22). Residues within the first and second alpha helix that are located close to one another were shown to be important for Hopx-mediated transcriptional repression, as were a distinct cluster of residues at the carboxyl terminus (shown in green, Fig. 6D). We mutated amino acids in the first and second helix and at the carboxyl terminus and assayed whether the mutants could interact with Smad4 using an in situ proximity ligation assay (Fig. 6E–I). We confirmed expression of Hopx constructs (fig. S8, C and D), and specificity of the proximity ligation assay (fig. S8E). Consistent with the co-immunoprecipitation experiments (Fig. 6B), the Smad4 interaction with Hopx is enhanced by Bmp4 (Fig. 6E and F). The Smad4-Hopx interaction is attenuated by mutation of residues at the carboxyl terminus of Hopx, but not by mutations in helix 1 or 2 (Fig. 6G–I). Consistent with a physical interaction between Hopx and Smad4, we detect enrichment of Smad4 occupancy by ChIP at Wnt ligand loci that are also occupied by Hopx (fig. S8F).
Hopx integrates Bmp and Wnt signaling
The data presented thus far indicate that Hopx expression defines a cardiomyoblast, and that Hopx modulates cardiomyogenesis by repressing Wnt signaling. Further, Hopx can interact with an activated Smad complex. In murine EBs, myogenesis requires inhibition of Wnt and is promoted by activation of Bmp (9, 11, 25–27). Hence, we sought to determine if Hopx functions to integrate Bmp signaling with Wnt repression.
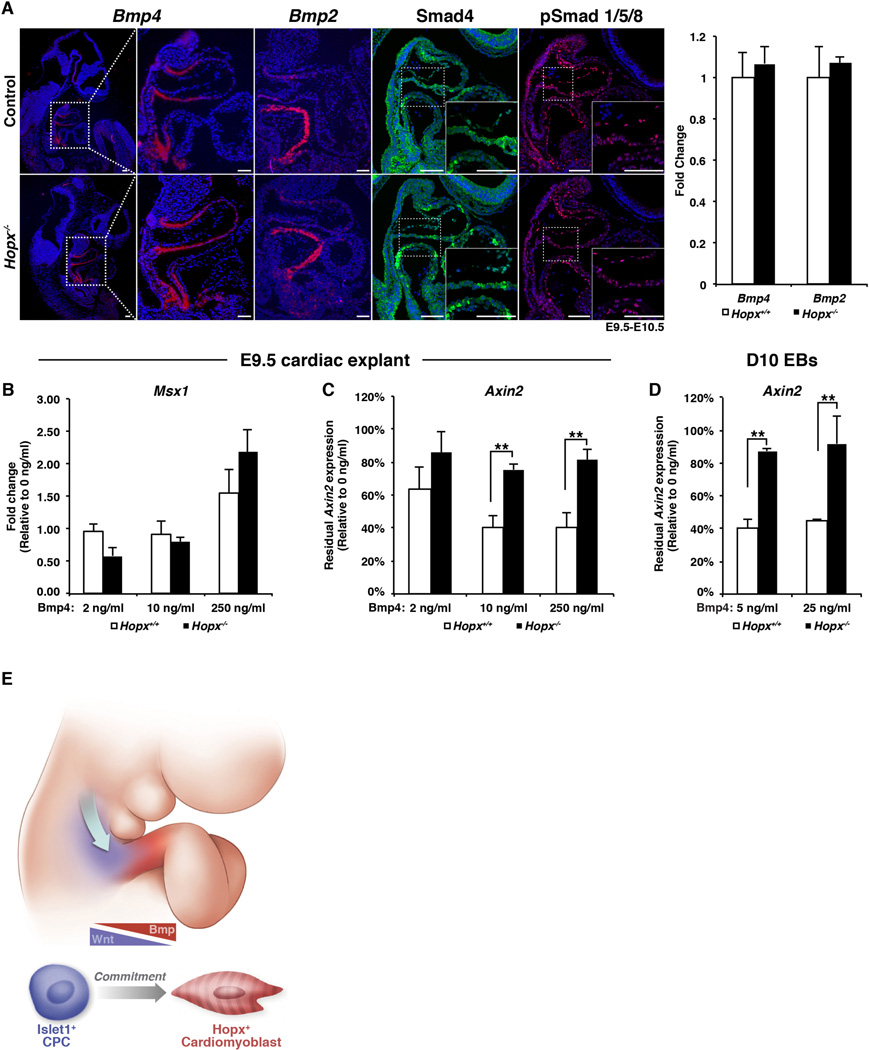
First, we confirmed that Bmp4 and Bmp2 levels are unchanged in Hopx−/− embryonic hearts by in situ hybridization and qRT-PCR (Fig. 7A). Protein expression and nuclear localization of Smad4 and phosphorylated Smad1/5/8 is also unchanged (Fig. 7A). Once in the nucleus, the active Smad complex functions to enhance transcription of Bmp target genes such as Msx1. Control E9.5 cardiac explants respond to exogenous Bmp4 by up-regulating Msx1 in a dose-dependent fashion (Fig. 7B). A similar response is seen in Hopx−/− explants, indicating an intact Bmp response system (Fig. 7B).
Fig. 7. Bmp signaling represses Wnt.
(A) Expression of Bmp4, Bmp2 (in situ hybridization), Smad4, phospho-Smad1/5/8 (IHC) in control and Hopx−/− E9.5-10.5 embryonic hearts, and qRT-PCR of E9.5 hearts (right panel). (B,C) qRT-PCR of E9.5 hearts after culture in increasing concentrations of Bmp4. (D) qRT-PCR of day 10 Hopx+/− and Hopx−/− EBs after differentiation with Bmp4. Hopx−/− explants and EBs failed to repress Axin2 as effectively as controls in the presence of Bmp4 (n≥3 for each experiment in B–D). (E) Model: A “zone-of-commitment” in the developing OFT. Wnt-activated, Isl1+ CPCs stream into the OFT and are exposed to Bmp signaling. Hopx+ cardiomyoblasts are committed to the myocyte lineage. ** p<0.05, Scale bars: 100 µm.
Bmp signaling can result in repression of Wnt activity in various tissues, although the mechanism has been unclear (28–31). Addition of recombinant Bmp4 decreases Axin2 expression in Hopx+/+ cardiac explants compared to vehicle-treated controls (Fig. 7C, white bars). However, Axin2 expression in Hopx−/− explants is relatively unresponsive to Bmp4 (Fig. 7C, black bars). We also tested whether Hopx participates in Bmp-mediated Wnt inhibition during cardiac differentiation of EBs (Fig. 7D). Bmp4 treatment of Hopx+/− EBs starting at day 5 results in a 60% decrease in Axin2 by day 10 of differentiation compared to vehicle treated EBs (white bars, Fig. 7D). However, Bmp4 minimally affects Axin2 expression in Hopx−/− EBs (black bars, Fig. 7D). These data suggest that Hopx is required for Bmp-mediated repression of Wnt signaling during cardiogenesis.
Discussion
Here, we have shown that cardiac precursor cells that express Hopx are irreversibly committed to the myocyte lineage, thereby defining progenitor cells that we call cardiomyoblasts. Hopx expression not only marks cardiomyoblasts, but it also functions to enhance cardiomyogenesis by linking Bmp signaling with repression of Wnt. A subset of committed myoblasts derived from the FHF express Hcn4, which is also expressed by endothelial cells during cardiac development (32, 33). However, the lack of a definitive marker of a cardiomyoblast has hampered a detailed analysis of cardiomyocyte commitment. Our studies provide such a marker, and reveal that commitment of FHF and SHF CPCs occurs at distinct time points and in different locations during cardiogenesis. In the SHF, Isl1+, Wnt-activated CPCs stream into the OFT from the surrounding mesoderm, where they encounter local Bmp4 signals. CPCs then express Hopx, down-regulate Wnt, and become committed to the myocyte fate. Thus, the distal OFT is a “zone of commitment” in the developing heart (Fig. 7E).
Markers of lineage commitment, and the signals that modulate lineage decisions, are likely to inform regenerative and stem cell approaches for cardiac disease. In the hematopoietic system, detailed understanding of these processes has allowed for definitive identification of various progenitor cells of the blood lineages, leading to the development of important therapies for human diseases (e.g. erythropoietin and granulocyte-macrophage stimulating factor). The ability to identify committed, but undifferentiated cardiomyocyte precursors may facilitate development of cardiac regenerative therapies including those using ES and iPS cells (34).
Reciprocal signaling between Bmp and Wnt has been recognized in multiple progenitor populations (28–31, 35). However, the mechanisms that coordinate these pathways in progenitor cell niches have remained elusive. Our current work raises the possibility that Hopx-mediated integration of Bmp signaling to repress Wnt may be active in other progenitor populations. For example, Hopx is expressed by +4 stem cells in the intestine (19) where niche Bmp signals repress Wnt (28, 36). Recent work by the Fuchs laboratory also suggests that balance between Bmp and Wnt signaling influences the fate of hair follicle cells as they differentiate into various progeny lineages (35) and Hopx may play a role in this process (18). HOPX is a tumor suppressor gene implicated in colorectal and other cancers (37, 38). Hijacking of developmental pathways is emerging as a potent mechanism of carcinogenesis (39). Loss of Hopx in cancer stem cells could result in uncoupling of niche-mediated Bmp signaling and quiescence through loss of Wnt repression.
Finally, although Hopx-deficiency leads to thinned myocardium and cardiac rupture in a portion of embryos, cardiomyogenesis is not altogether blocked. Inhibition of Wnt signaling in SHF cardiomyoblasts is delayed, but not completely prevented and some Hopx−/− mice live to adulthood. This suggests that, not surprisingly, alternative mechanisms reinforce Wnt repression independent of Hopx (40). Further insights into the mechanisms that coordinate signaling in the niche are likely to inform our ability to harness the potential of regenerative medicine.
Supplementary Material
Acknowledgments
We thank the Epstein laboratory, S. Evans, R. Schwartz, N. Palpant, and C. Murry for helpful discussions and sharing of reagents, H. Aghajanian for critical reading of the manuscript and J. Schug and the Next Generation Sequencing Core for sequencing, ChIP-seq analysis, and microarray analysis. Data associated with this manuscript are available in GEO (GSE67254). We thank J. LeLay for help with initial ChIP-seq experiments, J. Jeong and S. Eun Kwon for help with bioinformatic analyses, the Wistar Proteomics Core for mass spectrometry analysis, and L. Guo for artistic help with the model figure. The work was supported by NIH K08 HL119553-02 to R.J., NIH 5-T32-GM-007170 to M.G., NIH U01 HL100405, R01 HL071546, the Cotswold Foundation and the Spain Fund for Regenerative Cardiology to J.A.E.
Footnotes
Supplementary Materials
Materials and Methods
Figs. S1–S8 and Tables S1–S3
References (41–51)
References
- 1.Epstein JA, Franklin H. Epstein Lecture. Cardiac development and implications for heart disease. N Engl J Med. 2010;363:1638–1647. doi: 10.1056/NEJMra1003941. [DOI] [PubMed] [Google Scholar]
- 2.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 3.Waldo KL, et al. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- 4.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Moretti A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 7.Wu SM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 8.Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naito AT, et al. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci U S A. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paige SL, et al. Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 2010;5:e11134. doi: 10.1371/journal.pone.0011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Nostro MC, Cheng X, Keller GM, Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008;2:60–71. doi: 10.1016/j.stem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lescroart F, et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nature cell biology. 2014;16:829–840. doi: 10.1038/ncb3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devine WP, Wythe JD, George M, Koshiba-Takeuchi K, Bruneau BG. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife. 2014;3 doi: 10.7554/eLife.03848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F, et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 16.De Toni A, et al. Regulation of survival in adult hippocampal and glioblastoma stem cell lineages by the homeodomain-only protein HOP. Neural Dev. 2008;3:13. doi: 10.1186/1749-8104-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin CH, et al. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- 18.Takeda N, et al. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development. 2013;140:1655–1664. doi: 10.1242/dev.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeda N, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Information on materials and methods is available at the Science Web site.
- 21.Christoforou N, et al. Mouse ES cell-derived cardiac precursor cells are multipotent and facilitate identification of novel cardiac genes. J Clin Invest. 2008;118:894–903. doi: 10.1172/JCI33942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kook H, et al. Analysis of the structure and function of the transcriptional coregulator HOP. Biochemistry. 2006;45:10584–10590. doi: 10.1021/bi060641s. [DOI] [PubMed] [Google Scholar]
- 23.Lin L, et al. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci U S A. 2007;104:9313–9318. doi: 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen ED, et al. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cagavi E, et al. Functional Cardiomyocytes Derived from Isl1 Cardiac Progenitors via Bmp4 Stimulation. PLoS One. 2014;9:e110752. doi: 10.1371/journal.pone.0110752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim SY, et al. Enhancing Human Cardiomyocyte Differentiation from Induced Pluripotent Stem Cells with Trichostatin A. Methods in molecular biology. 2014 doi: 10.1007/7651_2014_160. [DOI] [PubMed] [Google Scholar]
- 27.Ueno S, et al. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He XC, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nature genetics. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- 29.Kandyba E, et al. Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Proc Natl Acad Sci U S A. 2013;110:1351–1356. doi: 10.1073/pnas.1121312110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plikus MV, et al. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature. 2008;451:340–344. doi: 10.1038/nature06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song J, et al. Smad1 transcription factor integrates BMP2 and Wnt3a signals in migrating cardiac progenitor cells. Proc Natl Acad Sci U S A. 2014;111:7337–7342. doi: 10.1073/pnas.1321764111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang X, et al. HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res. 2013;113:399–407. doi: 10.1161/CIRCRESAHA.113.301588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spater D, et al. A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nature cell biology. 2013;15:1098–1106. doi: 10.1038/ncb2824. [DOI] [PubMed] [Google Scholar]
- 34.Chong JJ, et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–277. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genander M, et al. BMP Signaling and Its pSMAD1/5 Target Genes Differentially Regulate Hair Follicle Stem Cell Lineages. Cell Stem Cell. 2014;15:619–633. doi: 10.1016/j.stem.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li N, et al. Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem cell reports. 2014;3:876–891. doi: 10.1016/j.stemcr.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung WK, et al. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell. 2013;23:725–738. doi: 10.1016/j.ccr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita K, Katoh H, Watanabe M. The homeobox only protein homeobox (HOPX) and colorectal cancer. International journal of molecular sciences. 2013;14:23231–23243. doi: 10.3390/ijms141223231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karamboulas C, Ailles L. Developmental signaling pathways in cancer stem cells of solid tumors. Biochim Biophys Acta. 2013;1830:2481–2495. doi: 10.1016/j.bbagen.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 40.O'Connell DJ, et al. A Wnt-bmp feedback circuit controls intertissue signaling dynamics in tooth organogenesis. Sci Signal. 2012;5:ra4. doi: 10.1126/scisignal.2002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 42.Yang L, et al. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 2006;133:1575–1585. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi YS, et al. Distinct functions for Wnt/beta-catenin in hair follicle stem cell proliferation and survival and interfollicular epidermal homeostasis. Cell Stem Cell. 2013;13:720–733. doi: 10.1016/j.stem.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nature genetics. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 46.Kaartinen V, et al. Cardiac outflow tract defects in mice lacking ALK2 in neural crest cells. Development. 2004;131:3481–3490. doi: 10.1242/dev.01214. [DOI] [PubMed] [Google Scholar]
- 47.Nagy A. Manipulating the mouse embryo : a laboratory manual. 3rd. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2003. p. 764. [Google Scholar]
- 48.Addis RC, et al. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 2013;60:97–106. doi: 10.1016/j.yjmcc.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Addis RC, et al. Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS One. 2011;6:e28719. doi: 10.1371/journal.pone.0028719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trivedi CM, et al. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev Cell. 2010;19:450–459. doi: 10.1016/j.devcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kook H, et al. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest. 2003;112:863–871. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.