Abstract
Genetic risk loci have been identified for a wide range of diseases through genome-wide association studies (GWAS), but the relevant functional mechanisms have been identified for only a small proportion of these GWAS-identified loci. By integrating results from the largest current GWAS of chronic obstructive disease (COPD) with expression quantitative trait locus (eQTL) analysis in whole blood and sputum from 121 subjects with COPD from the ECLIPSE Study, this analysis identifies loci that are simultaneously associated with COPD and the expression of nearby genes (COPD eQTLs). After integrative analysis, 19 COPD eQTLs were identified, including all four previously identified genome-wide significant loci near HHIP, FAM13A, and the 15q25 and 19q13 loci. For each COPD eQTL, fine mapping and colocalization analysis to identify causal shared eQTL and GWAS variants identified a subset of sites with moderate-to-strong evidence of harboring at least one shared variant responsible for both the eQTL and GWAS signals. Transcription factor binding site (TFBS) analysis confirms that multiple COPD eQTL lead SNPs disrupt TFBS, and enhancer enrichment analysis for loci with the strongest colocalization signals showed enrichment for blood-related cell types (CD3 and CD4+ T cells, lymphoblastoid cell lines). In summary, integrative eQTL and GWAS analysis confirms that genetic control of gene expression plays a key role in the genetic architecture of COPD and identifies specific blood-related cell types as likely participants in the functional pathway from GWAS-associated variant to disease phenotype.
INTRODUCTION
Genome-wide association studies (GWAS) have identified many disease-associated loci for common, genetically complex diseases (1), and these loci are enriched near promoters and DNaseI hypersensitive sites (2,3). Conversely, putative regulatory variants identified through expression quantitative trait (eQTL) mapping are significantly overrepresented in the catalog of GWAS-associated variants (4,5), and a number of well-established disease loci have been shown to regulate the transcription of genes in cis (6–9) and trans (10–12).
Chronic obstructive pulmonary disease (COPD) is a heritable disorder characterized by a dysregulated inflammatory response to inhaled toxins, such as cigarette smoke, resulting in loss of lung function. Recent COPD GWAS studies have identified four loci at genome-wide significance (near HHIP (13), FAM13A (14), the IREB2 and nicotinic acetylcholine receptor complex on 15q25 (13,15), and the 19q13 locus (16)), and there is evidence for genetic control of gene expression in three of these loci in either sputum samples from COPD subjects (6) or surgical lung tissue specimens (17). These studies demonstrate the existence of loci that are associated with COPD susceptibility and the expression of nearby genes (COPD eQTL). However, due to the high genomic prevalence of eQTL, simple overlap of significant GWAS and eQTL signals does not confirm a causal link between SNP, gene expression and phenotype.
We hypothesized that (1) integrative analysis of the most current, comprehensive COPD GWAS results with multi-tissue eQTL results from sputum and whole-blood samples in subjects with COPD would identify novel COPD eQTL, (2) detailed fine-mapping, colocalization and conditional analyses would better characterize the relationship between local GWAS and eQTL peaks and (3) integrating these results with regulatory functional annotation from ENCODE cell lines would provide further confirmation and refinement of functional pathways that may link these variants to disease phenotype.
RESULTS
Subject characteristics
The characteristics of the 121 COPD subjects analyzed in the eQTL analysis are shown in Supplementary Material, Table S1. The characteristics of the 9767 subjects (5812 cases and 3955 controls) in the GWAS analysis are listed in Supplementary Material, Table S2.
eQTL identification
In blood and sputum a total of 8745 and 5685 genes (representing 13 561 and 8071 unique probesets) were associated with at least one eQTL SNP at an FDR of 10%. Forty-eight percent of cis eQTL identified in blood were also observed in the sputum samples, and 74% of eQTL identified in sputum were also present in the blood.
Integrating eQTL and COPD GWAS
To test for enrichment of blood and sputum eQTL SNP in COPD GWAS results, we identified the set of ∼411 000 genotyped SNPs common to both the GWAS and eQTL datasets, and compared the proportion of observed cis eQTL P-values to that estimated in 1000 randomly acquired sets of the same number of SNPs (Supplementary Material, Figs S1 and S2). We observed significant enrichment of eQTL SNP for COPD-associated variants (P < 0.001 in both tissues). The magnitude of enrichment was similar in both tissues (∼3-fold enrichment over genomic baseline eQTL rate in top 250 GWAS hits), but the overall yield of eQTL from blood was higher than sputum.
Having observed significant enrichment of eQTL in COPD association data, we sought to identify specific COPD eQTL. SNPs associated with both local gene expression and COPD susceptibility at an FDR of 10% were observed for 36 unique probesets at 19 distinct genomic loci. To support the validity of these eQTL observations, we queried publically available databases of blood-derived eQTL results, and we identified SNP-gene level replication for 9 of the 19 identified COPD eQTL (Supplementary Material, Table S3) (18,19). The COPD eQTL most strongly associated with COPD in blood and/or sputum for each gene in each locus are listed in Table 1. Among these 19 loci were all four COPD susceptibility regions previously identified by GWAS—HHIP, FAM13A, IREB2/CHRNA3 and the 19q locus including ADCK4, CYP2A6 and EGLN2. All four loci exhibited regulatory genetic effects in whole blood. Only one—rs12914385 for IREB2 and PSMA4—was an eQTL in sputum.
Table 1.
Top COPD cis eQTLs in integrated eQTL–GWAS analysis
| Locus | SNP | COPD GWAS q-value | Sample | Best blood eQTL q-value | Best sputum eQTL q-value | eQTL genes |
|---|---|---|---|---|---|---|
| 4q22 | rs2869967 | <0.001 | Blood | 0.029 | >0.1 | FAM13A |
| rs2869663 | 0.011 | Blood | 0.003 | >0.1 | HERC3,FAM13A-AS1 | |
| rs6857847 | 0.019 | Blood | 0.082 | >0.1 | NAP1L5 | |
| 4q31 | rs720485 | <0.001 | Blood | 0.089 | >0.1 | HHIP |
| 15q25 | rs12914385 | <0.001 | Both | <0.001 | 0.020 | IREB2,CHRNA3,PSMA4 |
| rs1394371 | 0.003 | Blood | 0.060 | >0.1 | WDR61 | |
| rs578776 | 0.006 | Blood | 0.045 | >0.1 | PSMA4 | |
| 19q13 | rs7937 | <0.001 | Blood | 0.015 | >0.1 | ADCK4 |
| rs2604913 | 0.082 | Blood | 0.023 | >0.1 | C19orf54 | |
| rs3736329 | 0.082 | Blood | 0.030 | >0.1 | EGLN2 | |
| 3q21 | rs2811518 | 0.002 | Blood | 0.064 | >0.1 | SEC61A1,RUVBL1 |
| 15q26 | rs1993338 | 0.004 | Both | 0.004 | 0.003 | SNRPA1 |
| 2p16 | rs1177287 | 0.011 | Both | <0.001 | 0.052 | USP34,AHSA2,PEX13 |
| 16p11 | rs12446550 | 0.016 | Both | <0.001 | <0.001 | CCDC101,EIF3C,SBK1,TUFM |
| rs151228 | 0.030 | Sputum | >0.1 | 0.017 | EIF3C | |
| 14q32 | rs10134607 | 0.057 | Blood | 0.097 | >0.1 | CHGA |
| 20q11.22 | rs910397 | 0.057 | Blood | 0.073 | >0.1 | CBFA2T2 |
| 12q13 | rs2641530 | 0.063 | Both | <0.001 | 0.016 | ACVR1B |
| 20q13 | rs959581 | 0.063 | Blood | 0.069 | >0.1 | MRGBP |
| Sputum | >0.1 | 0.008 | OGFR | |||
| 11p13 | rs4756195 | 0.072 | Blood | 0.024 | >0.1 | CD44 |
| 17p11 | rs6502640 | 0.084 | Blood | 0.004 | >0.1 | LLGL1,SMCR8,TOP3A |
| 9q33 | rs10983326 | 0.085 | Blood | 0.085 | >0.1 | ASTN2 |
| 15q23 | rs8027920 | 0.085 | Blood | 0.052 | >0.1 | MYO9A |
| 8p23 | rs13282106 | 0.088 | Blood | 0.082 | >0.1 | MSRA |
| 20q11.21 | rs1205843 | 0.096 | Blood | 0.025 | >0.1 | PLAGL2,TPX2 |
| 12p11 | rs7960369 | 0.099 | Blood | 0.053 | >0.1 | CCDC91 |
Table entries represent the SNP most strongly associated with COPD for each probe harboring a significant cis eQTL locus that was also significantly associated with COPD susceptibility.
COPD GWAS FDR—q-value for association with COPD calculated from all SNP that were tested for association with COPD and found to be eQTL in blood or sputum at 10% FDR.
Best Blood eQTL FDR—smallest q-value for association with any blood probeset in cis window.
Best Sputum eQTL FDR—smallest q-value for association with any sputum probeset in cis window.
Sample—sample in which eQTL SNP-Probe pair attained FDR ≤ 0.1, i.e. whole blood or induced sputum.
Fine-mapping, colocalization and conditional analysis of COPD eQTLs
Coincidence of strong GWAS and eQTL signals at a specific locus is expected to occur for loci where the causal variant impacts the GWAS phenotype via gene expression; however, overlap of these signals may also occur by chance. To distinguish between these alternatives, we performed fine-mapping, colocalization and conditional analyses using 1000 Genomes imputed data to better characterize the relationship between GWAS and eQTL signals at each COPD eQTL. While fine-mapping alone is not always sufficient to identify the causal SNP at a given locus (20), it is likely to capture the causal variant and provide dense association information that can be used to infer the likelihood of a shared causal variant that explains the local pattern of GWAS and eQTL association (21). Using two methods to assess the local concordance between the GWAS and eQTL signal in whole blood, we identified five COPD eQTL that showed the strongest likelihood of harboring at least one shared causal variant (Table 2, results for all 19 COPD eQTL shown in Supplementary Material, Table S4). These five loci include three of the four previously identified COPD genome-wide significant loci (4q31, 15q25 and 19q13) and two novel COPD-associated loci at 2p16 near USP34 and 16p11 near CCDC101, SBK1 and TUFM.
Table 2.
Colocalization Between eQTL and COPD GWAS signals in fine-mapping analysis in whole blood
| Locus | Gene | Probeset | Lead eQTL SNP | Lead GWAS SNP | Distance (BP) | Colocalization | Local correlation |
|---|---|---|---|---|---|---|---|
| 2p16 | USP34 | 212980_at | rs2600671 | rs6545894 | 484425 | 0.91 | 0.63 |
| PEX13 | 235093_at | rs10181042 | rs1177292 | 123367 | 0.23 | 0.12 | |
| AHSA2 | 226334_s_at | rs1177306 | rs1177292 | 40210 | 0.11 | −0.14 | |
| 15q25 | CHRNA3 | 211587_x_at | rs138544659 | rs8042849 | 82772 | 0.80 | 0.71 |
| IREB2 | 214666_x_at | rs1394371 | rs8042849 | 93460 | 0.78 | 0.75 | |
| PSMA4 | 203396_at | rs72743158 | rs8042849 | 108516 | 0.34 | 0.44 | |
| WDR61 | 221531_at | rs72732595 | rs8042849 | 309014 | 0.20 | 0.12 | |
| 16p11 | CCDC101 | 221822_at | rs151231 | rs11074907 | 31116 | 0.80 | −0.19 |
| SBK1 | 226548_at | rs1074631 | rs40836 | 43571 | 0.79 | 0.31 | |
| TUFM | 238190_at | rs117382857 | rs11074907 | 386295 | 0.63 | 0.33 | |
| EIF3C | 236700_at | rs2650494 | rs11074907 | 295525 | 0.41 | 0.38 | |
| 19q13 | ADCK4 | 227324_at | rs7937 | rs7937 | 0 | 0.51 | 0.35 |
| EGLN2 | 227147_s_at | rs34950672 | rs7937 | 21519 | 0.23 | 0.45 | |
| C19orf54 | 222052_at | rs2545769 | rs7937 | 21473 | 0.06 | 0.23 | |
| 4q31 | HHIP | 237466_s_at | rs12504628 | rs138641402 | 9455 | 0.45 | 0.87 |
Distance—base-pair distance between lead GWAS SNP and lead blood eQTL SNP.
Lead eQTL SNP—SNP with the lowest P-value for association with target probeset in a 250 kb window.
Lead GWAS SNP—SNP with lowest P-value for association with COPD susceptibility in the corresponding eQTL 250 kb window.
Colocalization—posterior probability of the eQTL and GWAS signals sharing at least one causal variant.
Local correlation—correlation between eQTL and GWAS test statistics for SNPs in the top quartile for association with COPD susceptibility at a given locus.
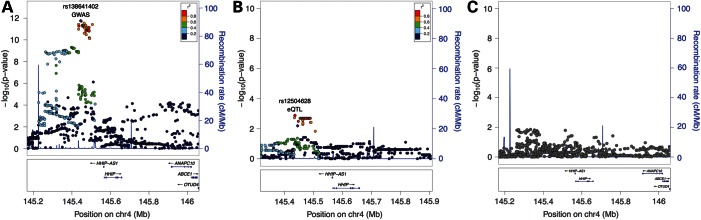
The 4q31 locus near HHIP demonstrates a strong local correlation between the eQTL and GWAS peak (Table 2), supporting the hypothesis that a disease-causing variant in this region plays a role in the regulation of the HHIP. Figure 1 illustrates the concordance between the COPD association and blood eQTL signal (Fig. 1A and B), and it demonstrates that conditioning on the lead eQTL SNP completely eliminates the GWAS signal in this region (Fig. 1C). These findings are consistent with functional data demonstrating allele-specific enhancer activity in this region likely mediated by altered binding of the transcription factor Sp3 (22).
Figure 1.
Locus Plots for COPD GWAS and whole-blood eQTL association near HHIP. COPD GWAS association results are shown in (A) whole-blood eQTL association is shown in (B) and COPD association conditioned on lead eQTL SNP rs12504628 demonstrates complete attenuation of the association signal (C).
Three eQTL were identified under the GWAS peak at the 19q13 locus for the genes ADCK4, EGLN2 and C19orf54 (Table 2), but colocalization analysis strongly prioritizes the ADCK4 eQTL association. Locus plots of the GWAS and eQTL signals in this region (Supplementary Material, Figs S3 and S4) demonstrate that rs7937 is both the lead GWAS and lead eQTL SNP for ADCK4 in this region. Conditioning on rs7937 eliminates the genetic association signal in the region, but this is not the case for the lead eQTL SNPs for EGLN2 and C19orf54 (Supplementary Material, Figs S5–S7).
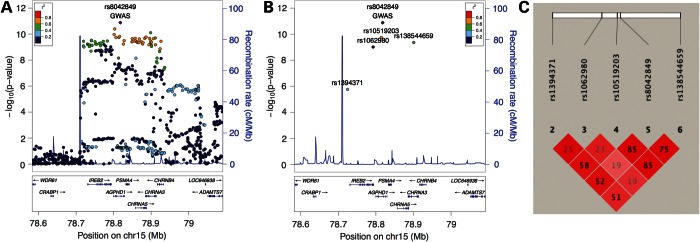
The 15q25 locus harbors six genes beneath the GWAS peak (Fig. 2). Three of these genes (CHRNA3, IREB2 and PSMA4—see Table 1) had associated eQTL and demonstrated moderate-to-strong correlation with the GWAS peak (Table 2). Table 3 shows the cis eQTL associations for the lead eQTL SNPs in this region, and the LD plot in Figure 2 indicates that rs10519203 and rs138544659 lie in a block of strong LD that includes the lead GWAS SNP rs8042849 (Fig. 2C). rs1394371 is in moderate LD with these SNPs, and rs1062980 is largely independent from this region. Three of the four SNPs regulate multiple genes in cis, and all SNPs are associated with expression levels of IREB2.
Figure 2.
Lead GWAS and eQTL SNPs in the 15q25 region. Locus plots for association with COPD susceptibility (A) and showing only the lead GWAS SNP (diamond) and lead eQTL SNPs (circles) for four probesets mapping to IREB2, CHRNA3 and PSMA4 (B). Linkage disequilibrium plot with R2 values for the lead GWAS and lead eQTLs SNPs (C).
Table 3.
COPD cis eQTL Lead SNPs for CHRNA3, IREB2 and PSMA4 in the 15q25 locus
| SNP | MAF | Position (bp) | eQTL Genes | Probeset | eQTL q-value | eQTL tissue |
|---|---|---|---|---|---|---|
| rs138544659 | 0.36 | 78900701 | PSMA4 | 203396_at | <0.001 | Sputum |
| CHRNA3 | 211587_x_at | <0.001 | Blood | |||
| IREB2 | 1555476_at | 0.003 | Blood | |||
| IREB2 | 1555476_at | 0.026 | Sputum | |||
| IREB2 | 214666_x_at | 0.017 | Blood | |||
| rs10519203 | 0.37 | 78814046 | PSMA4 | 203396_at | <0.001 | Sputum |
| CHRNA3 | 211587_x_at | <0.001 | Blood | |||
| IREB2 | 214666_x_at | <0.001 | Blood | |||
| IREB2 | 1555476_at | 0.002 | Blood | |||
| IREB2 | 1555476_at | 0.030 | Sputum | |||
| rs1062980 | 0.37 | 78792527 | IREB2 | 1555476_at | <0.001 | Blood |
| IREB2 | 1555476_at | <0.001 | Sputum | |||
| IREB2 | 214666_x_at | 0.063 | Blood | |||
| rs1394371 | 0.31 | 78724469 | IREB2 | 1555476_at | <0.001 | Blood |
| IREB2 | 214666_x_at | <0.001 | Blood | |||
| CHRNA3 | 211587_x_at | 0.001 | Blood | |||
| WDR61 | 221531_at | 0.039 | Blood | |||
| IREB2 | 1555476_at | 0.089 | Sputum |
Position (bp)—genomic base-pair position in hg19.
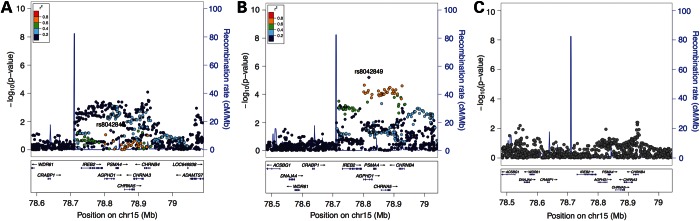
Conditioning on the lead eQTL, rs10519203, which is in strong LD with the lead GWAS SNP, markedly reduces the strength of the regional COPD association (Fig. 3A). However, an independent association signal is apparent after conditioning on this lead eQTL SNP (lead SNP for secondary signal, rs12440014, P = 8 × 10−5). Conditioning on rs1062980, which is associated only with IREB2 expression, eliminates this secondary signal (Fig. 3B), and simultaneously conditioning on rs10519203 and rs1062980 removes all significant association signals from this region (Fig. 3C).
Figure 3.
Locus plots for conditional association with COPD susceptibility in the 15q25 Locus. Association results conditioning on lead eQTL rs10519203 (A) and lead eQTL rs1062980 (B) attenuate different components of the overall association signal, and conditioning on both lead eQTL SNPs simultaneously completely attenuates the association (C).
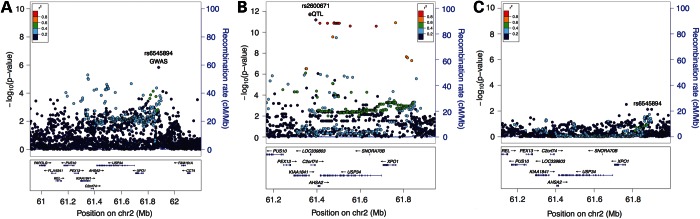
The 2p16 locus includes two genes with COPD eQTL signals, USP34 and PEX13, but the colocalization and local correlation signals are stronger for USP34. The GWAS signal near USP34 is diffuse, with association peaks in weak LD at both ends of the gene (Fig. 4A). The eQTL signal shows a similar broad-based pattern, and, interestingly, conditioning on the lead eQTL SNP at the 3′ end of the gene, rs2600671, attenuates the GWAS signal throughout the gene (Fig. 4B and C).
Figure 4.
Locus plots for COPD GWAS and whole-blood eQTL association near USP34. COPD GWAS association results are shown in (A) whole-blood eQTL Association is shown on (B) and COPD association conditioned on lead eQTL SNP rs2600671 demonstrates complete attenuation of the association signal (C).
The 16p11 locus demonstrates notable transcriptional complexity with four eQTL target genes within the GWAS locus (CCDC101, TUFM, SBK1 and EIF3C). The GWAS peak in this area is located downstream from CCDC101 (Supplementary Material, Fig. S8). To better capture the extent of eQTL signals in this region, the fine-mapping analysis was extended to include a 1 MB window around each gene. The eQTL signal for CCDC101 shows the strongest colocalization and mirrors the pattern of the GWAS peak (Supplementary Material, Fig. S9). Conditioning on rs151231 almost completely attenuates the GWAS signal in this region (Supplementary Material, Fig. S10). Of the other three eQTL genes in this locus, the SBK1 eQTL signal is very similar to that observed for CCDC101 and also shows strong colocalization.
Integration with ENCODE/Roadmap functional annotations
For nine lead eQTL SNPs from the five loci with the strongest colocalization between GWAS and eQTL signals, we examined ENCODE and Roadmap Epigenomics data for evidence of functional regulatory activity. Enhancer enrichment analysis demonstrates significant enrichment for enhancer regions within these loci primarily in T-cells and B-cell lymphoblastoid cell lines (Table 4). Of the nine lead eQTL SNPs from these loci, eight SNPs disrupt multiple transcription factor binding motifs as identified from ENCODE experiments (Supplementary Material, Table S5).
Table 4.
Enhancer enrichment analysis for 5 COPD eQTLs with strongest colocalization signal
| Cell type |
All enhancers |
Strongest enhancers |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | Description | Obs | Exp | Fold | P-value | Obs | Exp | Fold | P-value |
| CCCRA.NP | CD4+ CD25- CD45RA+ naive primary cells | 2 | 0.4 | 4.9 | 0.061 | 2 | 0.1 | 15.6 | 0.007 |
| CD4.MP | CD4 memory primary cells | 2 | 0.5 | 4.3 | 0.076 | 2 | 0.1 | 14.1 | 0.008 |
| R.MUC31 | Rectal Mucosa.Donor 31 | 2 | 0.4 | 5.2 | 0.054 | 2 | 0.2 | 12.1 | 0.011 |
| GM12878 | B-lymphocyte, lymphoblastoid | 2 | 0.5 | 3.8 | 0.096 | 2 | 0.2 | 10.4 | 0.015 |
| CD3.P | CD3 primary cells | 2 | 0.4 | 5.5 | 0.049 | 2 | 0.2 | 9.9 | 0.017 |
| CD4.NP | CD4 naive primary cells | 1 | 0.4 | 2.8 | 0.302 | 1 | 0 | 20.8 | 0.047 |
Enhancer enrichment analysis for five lead eQTL SNPs from COPD eQTLs with strongest colocalization signal (Table 2).
ID—ENCODE or Roadmap Epigenomic cell identification.
Obs, observed number of overlaps with enhancer regions; Exp, expected number of chance overlaps; Fold, fold enrichment over expected rate; P-value, calculated by binomial test compared with distance-matched (from transcription start site) controls (23).
Differential expression of COPD eQTL genes in COPD cases and smoking controls
For the probesets associated with COPD-associated SNPs, we performed differential expression analysis comparing the expression of these probesets in 144 subjects with COPD (of which the 121 subjects in the eQTL analysis comprise a subset) and 93 smoking controls from ECLIPSE. Of the 36 probesets analyzed, 9 were nominally associated with COPD status (P < 0.05, Supplementary Material, Table S6). Of these nine probesets, three lay within previously identified genome-wide significant loci for COPD susceptibility (FAM13A and FAM13A-AS1 at 4q22, ADCK4 at 19q13 and IREB2 and 15q25). For the two novel COPD eQTL prioritized through colocalization analysis, a probeset mapping to TUFM from the 16p11 locus met nominal significance, and a probeset mapping to USP34 showed borderline evidence of differential expression (P = 0.08).
Association of COPD eQTLs with smoking exposure
Two of the loci associated with COPD susceptibility via GWAS (15q25 and 19q13) have also been associated with measures of smoking behavior (24,25). To determine whether any of the loci identified in this analysis show a similar association with cumulative amount of pack-year smoking exposure, we related the lead eQTL SNPs from Table 2 to cumulative pack-year exposure in NHW smoking subjects in COPDGene, and we did not identify any additional significant associations (Supplementary Material, Table S7).
DISCUSSION
Using whole-genome expression arrays from whole blood and induced sputum in subjects with COPD, we have identified instances of genetic control of gene expression in 19 distinct loci that are also associated with COPD susceptibility. All four previously identified COPD GWAS loci harbor a cis eQTL SNP, suggesting that eQTLs play an important role in the genetic contribution to COPD susceptibility. Colocalization analyses prioritized five distinct loci with moderate to high likelihood of harboring a shared causal variant that explains the local GWAS and eQTL signals, and functional analyses of these loci using ENCODE and Roadmap Epigenomics data confirmed cell-type specific regulatory activity for all of these loci. Enhancer enrichment analysis shows significant enrichment of T and B-lymphocyte enhancer regions in these loci, and 8 of the 9 lead eQTL SNPs in these regions disrupt predicted transcription factor binding motifs. Four of these five loci show nominally significant differential expression between COPD subjects and smoking controls.
The enrichment of cis eQTL SNPs in complex genetic diseases has been well-demonstrated for GWAS-identified loci (1,6,9,11,26). The current study adds to our previous observations from sputum by demonstrating that eQTL analysis of whole blood from subjects with COPD shows similar enrichment of eQTL signals in top GWAS associations, but is more informative than sputum, perhaps due to the greater number of eQTL identified in whole blood. The utility of whole-blood eQTL data for integration with COPD GWAS results is further supported by enhancer enrichment analysis that identified significant enrichment for T and B-lymphocyte enhancer regions in the highest confidence set of COPD eQTL. In addition, we performed eQTL and GWAS fine mapping with densely imputed genotype data that improved our ability to localize causal variants and distinguish convergence of local eQTL and GWAS peaks due to shared causal variants versus chance overlap.
Our findings confirm established associations for COPD GWAS loci and suggest novel associations. The eQTL SNP observed in blood from COPD subjects ∼100 kb upstream from HHIP has previously been shown to act as a long-range enhancer for HHIP in BEAS-2B cells, and it is in strong LD (0.93) with a functional SNP (rs1542725) that affects the binding of Sp3 (22). This eQTL was also observed to regulate HHIP in an eQTL study of resected lung tissue (17). In a study using liver tissue, it was involved in regulation of ANAPC10 but not HHIP (lead eQTL SNP rs1828591, ∼50 kb from the top eQTL SNP in our study) (12). This eQTL has not been observed in many other previous eQTL studies (27), suggesting that this eQTL may have tissue-specific or disease-specific activity. Further study will be needed to determine the disease relevant cell type(s) for this well-established COPD risk locus.
The chromosome 19q13 locus also harbors many likely COPD candidate genes, including CYP2A6, a member of the cytochrome p450 family of enzymes. However, analysis of the COPD GWAS and eQTL signals from this region clearly prioritizes ADCK4 as the gene in this region most likely to affect COPD susceptibility through genetic control of gene expression in blood, though it is possible that eQTL analysis from other tissues will identify additional candidate genes.
Genetic variants in the 15q25 locus have been associated through GWAS with multiple phenotypes including COPD, pulmonary function, lung cancer and smoking behavior (28). This locus contains a gene cluster of nicotinic acetylcholine receptor genes (CHRNA3, CHRNA5 and CHRNB4), the iron-responsive binding element IREB2, and the proteasome subunit PSMA4. Conditional analyses indicate the presence of two independent association signals in this region, and both signals can be completely attenuated by conditioning on neighboring lead eQTL SNPs. Previous studies have identified eQTL SNPs in this region for IREB2 and CHRNA5 (6,27), and eQTLs for CHRNA3 and PSMA4 have been observed in liver (12). Our data demonstrate the complexity of gene regulation in this locus, and implicate IREB2, CHRNA3 and PSMA4 as strong candidate genes for COPD susceptibility in this region.
Two novel COPD eQTL regions (2p15 and 16p11) showed strong likelihood of harboring a shared causal GWAS/eQTL variant. The lead eQTL SNP rs2600671 at 2p15 is located ∼20 kb downstream from its eQTL target USP34, and this variant lies within a region of enhancer activity in the immortalized lymphoblastoid cell line GM12878. This is also a DNase1 hypersensitive region in multiple cell lines, including human tracheal epithelium. However, the eQTL and GWAS signal in this region is broad, and precise localization of the causal variant will require additional investigation. USP34 is a ubiquitin-specific peptidase that is widely expressed across tissues, including lung, and has been implicated in regulation of Wnt/β-catenin signaling (29).
The 16p11 region demonstrates a complex pattern of genetic control of gene expression, with multiple variants affecting the expression of multiple nearby transcripts. While CCDC101 is the top-ranked gene in this region from colocalization analysis, it is not possible to clearly distinguish the eQTL/GWAS overlap of this gene from that of SBK1, which shows a nearly identical pattern of eQTL association. This region has been previously associated with Type I diabetes and inflammatory bowel disease through GWAS (30,31). Further work will be required to better define the causal variant or variants in this locus and their functional mechanisms.
These findings are consistent with the report from a recently published COPD GWAS meta-analysis combining the NHW GWAS data analyzed here with African-American subjects in COPDGene (32). Of the six genome-wide significant loci reported, three were found to have concordant eQTL signals in blood and sputum from COPD subjects. Two loci did not harbor eQTL signals in the studied tissues (TGFB2 and MMP12). One locus near RIN3 did have an eQTL signal in whole blood, though colocalization analysis did not provide strong support for a shared causal variant between the GWAS and eQTL signals. The identification of disease-related eQTL does not necessarily imply differential expression of the target genes between the disease and unaffected state, particularly because the magnitude of the difference in allele frequencies for a disease-associated SNP between COPD cases and control subjects is quite small. However, our analysis does demonstrate differential expression in whole blood between COPD cases and smoking controls for 9 of the 36 genes regulated by COPD-associated SNPs (and at 4 of the 5 most likely COPD eQTL loci).
This study has the following strengths and limitations. By performing eQTL analysis in subjects with COPD, we increased the likelihood that the eQTL signals identified would be relevant to the disease state. We were able to examine expression from two different sources in the same individuals, but future studies will be strengthened by the inclusion of additional tissue or cell-specific expression data. One of the pitfalls of integrating eQTL and GWAS data is over-interpreting overlap between the two association signals, and a particular strength of our study is the use of densely imputed genotype data to generate appropriate input for Bayesian colocalization analysis to better distinguish causal from coincidental GWAS/eQTL overlap. Also, because we had access to the primary GWAS genotype data, we were able to perform conditional COPD association analysis adjusting for the lead eQTL SNPs from each candidate region. Because the expression samples used in this study are a mixture of cell types, we are unable to draw strong conclusions regarding cell-type specificity of the identified COPD eQTL associations. However, cell-type specific information can be drawn from the enhancer enrichment analysis performed using ENCODE and Roadmap Epigenomics data.
In conclusion, these data demonstrate significant enrichment for whole blood and sputum eQTL in top signals from the largest GWAS meta-analysis to date for COPD. Of the 19 significant COPD eQTL identified in our study, the concordance of GWAS and eQTL signals at five loci show moderate-to-strong evidence of at least one shared causal variant. Functional analysis of the lead eQTL SNPs in these regions confirm a likely gene regulatory function for all of these loci, and in many loci there is experimental evidence of transcriptional activity in specific lung and blood-related cell lines, with enhancer enrichment strongest among T-cells and immortalized B-lymphocytes. In some of these regions, the gene regulatory signals are complex with evidence of multiple, independent causal variants.
MATERIALS AND METHODS
Ethics statement
Informed consent was obtained from all study subjects, and study approval was obtained from institutional review boards for all participating institutions.
eQTL analysis
eQTL analysis has been previously reported from 131 Caucasian subjects with COPD expression data from induced sputum samples in the ECLIPSE study (6). Here we report a novel integrative analysis of 121 of these 131 individuals for whom whole-blood gene expression data is also available. Gene expression profiling was performed using the Affymetrix Human U133 Plus2 array, and genome-wide genotyping was performed using the Illumina HumanHap 550 array with imputation performed to the 1000 Genomes v3 EUR reference panel using MaCH (33) and minimac (34). eQTL analysis was performed with the Bioconductor package GGtools for window sizes ranging from 50 to 500 kb around the gene transcription start and end site (35). Analyses were adjusted for age, gender, pack-years of smoking, principal components of genetic ancestry and the first 13 principal components of the expression data. Genome-wide eQTL analysis was performed using directly genotyped SNP data. For top loci, a fine-mapping eQTL analysis of genotypes imputed to the 1000 Genomes version 3 EUR reference panel was performed using a 250 kb cis window.
eQTL and GWAS integration
eQTL and GWAS results were merged by SNP. For the SNPs included in both the GWAS and eQTL results, FDR was calculated from the GWAS P-values using the method of Storey et al (36,37). Two methods were applied to quantify the degree of overlap between the GWAS and eQTL signals in a given locus—local correlation and Bayesian colocalization analysis (21).
Bioinformatic and functional assessment of transcription factor binding
For lead eQTL SNPs likely to be causal GWAS variants, we queried ENCODE and Roadmap Epigenomics data via the Haploreg interface to (1) perform enhancer enrichment analysis and (2) determine if these SNPs disrupted TFBS learned from ENCODE and Roadmap data. For both analyses, we included lead eQTL SNPs and any 1000 Genomes SNPs in perfect LD (i.e. r2 = 1) in our query. For enhancer enrichment analysis we queried all ENCODE and Roadmap cell lines included in Haploreg to determine if the specified SNP set overlapped with cell-type specific enhancer annotations at a rate beyond that expected by chance, and statistical significance was calculated via the binomial test implemented in Haploreg. For TFBS analysis, the impact was assessed for each SNP on predicted binding affinity of overlapping position weight matrices (PWMs) learned from ENCODE data (38).
Differential expression analysis between COPD cases and smoking controls
Differential gene expression was performed in whole blood from 144 subjects with COPD and 93 smoking controls from the ECLIPSE Study that underwent the same pre-processing procedure applied to the eQTL expression data. All of the 121 subjects from the eQTL analysis were included in the 144 COPD subjects. Differential expression analysis was performed using the limma R package without covariate adjustment (39). Nominal significance was defined as P ≤ 0.05.
COPD GWAS
COPD GWAS results from a combined analysis of non-Hispanic white subjects from the NETT-NAS, Norway GenKOLS, ECLIPSE and COPDGene studies (including African-American subjects in COPDGene) are reported separately (32). Conditional GWAS analyses were performed using logistic regression adjusting for the same covariates used in the primary GWAS analysis and the genotype or genotypes of interest. The false discovery rate was controlled using the method of Storey et al (36,37). Locus plots for GWAS and eQTL analyses were generated with LocusZoom (40).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by US National Institutes of Health (NIH) grants K08HL102265 and R01HL124233 (P.J.C.), K08HL097029 (M.H.C.), R01HL094635 (C.P.H.), R01NR013377 (C.P.H), R01 HL086601 (B.A.R.), P01HL105339 (E.K.S.), R01HL111759 (J.Q., E.K.S), R01HL089897 (J.D.C) and R01HL089856 (E.K.S). Additional support was provided by a grant from the Parker B Francis Foundation (M.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The COPDGene® project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens and Sunovion. The ECLIPSE study was supported by GlaxoSmithKline.
Supplementary Material
Acknowledgments
Conflict of Interest statement. R.T.-S. and J.H.R are current employees and shareholders of GlaxoSmithKline. D.S. has received sponsorship to attend international meetings, honoraria for lecturing or attending advisory boards and research grants from various pharmaceutical companies including Almirall, AstraZeneca, Boehringer Ingelheim, Chiesi, CIPLA, Forest, Genetech, GlaxoSmithKline, Merck, Novartis, Pfizer and Takeda. Since 2011, SR reports relationships with the following companies and institutions: AARC, American Board of Internal Medicine, Able Associates, Align2 Acton, Almirall, APT, AstraZeneca, American Thoracic Society, Beilenson, Boehringer Ingelheim, Chiesi, CIPLA, Clarus Acuity, CME Incite, COPDFoundation, Cory Paeth, CSA, CSL Behring, CTS Carmel, Dailchi Sankyo, Decision Resources, Dunn Group, Easton Associates, Elevation Pharma, FirstWord, Forest, GLG Research, Gilead, Globe Life Sciences, GlaxoSmithKline, Guidepoint, Health Advance, HealthStar, HSC Medical Education, Johnson and Johnson, Leerink Swan, LEK, McKinsey, Medical Knowledge, Medimmune, Merck, Navigant, Novartis, Nycomed, Osterman, Pearl, PeerVoice, Penn Technology, Pennside, Pfizer, Prescott, Pro Ed Communications, PriMed, Pulmatrix, Quadrant, Regeneron, Saatchi and Saatchi, Sankyo, Schering, Schlesinger Associates, Shaw Science, Strategic North, Summer Street Research, Synapse, Takeda, Telecon SC, ThinkEquity. In the past 3 years, EKS received honoraria and consulting fees from Merck and grant support and consulting fees from GlaxoSmithKline. D.A.L. is a consultant for and receives honoraria and grant support from GSK. He is the Chair of the GSK Respiratory Area Therapy Board.
Appendix
COPDGene® Investigators—Core Units
Administrative Core: James Crapo, MD (PI), Edwin Silverman, MD, PhD (PI), Barry Make, MD, Elizabeth Regan, MD, PhD, Rochelle Lantz, Lori Stepp, Sandra Melanson.
Genetic Analysis Core: Terri Beaty, PhD, Barbara Klanderman, PhD, Nan Laird, PhD, Christoph Lange, PhD, Michael Cho, MD, Stephanie Santorico, PhD, John Hokanson, MPH, PhD, Dawn DeMeo, MD, MPH, Nadia Hansel, MD, MPH, Craig Hersh, MD, MPH, Peter Castaldi, MD, MSc, Merry-Lynn McDonald, PhD, Jing Zhou, MD, PhD, Manuel Mattheissen, MD, PhD, Emily Wan, MD, Megan Hardin, MD, Jacqueline Hetmanski, MS, Margaret Parker, MS, Tanda Murray, MS.
Imaging Core: David Lynch, MB, Joyce Schroeder, MD, John Newell, Jr, MD, John Reilly, MD, Harvey Coxson, PhD, Philip Judy, PhD, Eric Hoffman, PhD, George Washko, MD, Raul San Jose Estepar, PhD, James Ross, MSc, Mustafa Al Qaisi, MD, Jordan Zach, Alex Kluiber, Jered Sieren, Tanya Mann, Deanna Richert, Alexander McKenzie, Jaleh Akhavan, Douglas Stinson.
PFT QA Core, LDS Hospital, Salt Lake City, UT: Robert Jensen, PhD.
Biological Repository, Johns Hopkins University, Baltimore, MD: Homayoon Farzadegan, PhD, Stacey Meyerer, Shivam Chandan, Samantha Bragan.
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, PhD, Andre Williams, PhD, Carla Wilson, MS, Anna Forssen, MS, Amber Powell, Joe Piccoli.
Epidemiology Core, University of Colorado School of Public Health, Denver, CO: John Hokanson, MPH, PhD, Marci Sontag, PhD, Jennifer Black-Shinn, MPH, Gregory Kinney, MPH, PhDc, Sharon Lutz, MPH, PhD.
COPDGene® Investigators—Clinical Centers
Ann Arbor VA: Jeffrey Curtis, MD, Ella Kazerooni, MD.
Baylor College of Medicine, Houston, TX: Nicola Hanania, MD, MS, Philip Alapat, MD, Venkata Bandi, MD, Kalpalatha Guntupalli, MD, Elizabeth Guy, MD, Antara Mallampalli, MD, Charles Trinh, MD, Mustafa Atik, MD, Hasan Al-Azzawi, MD, Marc Willis, DO, Susan Pinero, MD, Linda Fahr, MD, Arun Nachiappan, MD, Collin Bray, MD, L. Alexander Frigini, MD, Carlos Farinas, MD, David Katz, MD, Jose Freytes, MD, Anne Marie Marciel, MD.
Brigham and Women's Hospital, Boston, MA: Dawn DeMeo, MD, MPH, Craig Hersh, MD, MPH, George Washko, MD, Francine Jacobson, MD, MPH, Hiroto Hatabu, MD, PhD, Peter Clarke, MD, Ritu Gill, MD, Andetta Hunsaker, MD, Beatrice Trotman-Dickenson, MBBS, Rachna Madan, MD.
Columbia University, New York, NY: R. Graham Barr, MD, DrPH, Byron Thomashow, MD, John Austin, MD, Belinda D'Souza, MD.
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr, MD, Lacey Washington, MD, H Page McAdams, MD.
Fallon Clinic, Worcester, MA: Richard Rosiello, MD, Timothy Bresnahan, MD, Joseph Bradley, MD, Sharon Kuong, MD, Steven Meller, MD, Suzanne Roland, MD.
Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, MD, MPH, Joseph Tashjian, MD.
Johns Hopkins University, Baltimore, MD: Robert Wise, MD, Nadia Hansel, MD, MPH, Robert Brown, MD, Gregory Diette, MD, Karen Horton, MD.
Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Los Angeles, CA: Richard Casaburi, MD, Janos Porszasz, MD, PhD, Hans Fischer, MD, PhD, Matt Budoff, MD, Mehdi Rambod, MD.
Michael E. DeBakey VAMC, Houston, TX: Amir Sharafkhaneh, MD, Charles Trinh, MD, Hirani Kamal, MD, Roham Darvishi, MD, Marc Willis, DO, Susan Pinero, MD, Linda Fahr, MD, Arun Nachiappan, MD, Collin Bray, MD, L. Alexander Frigini, MD, Carlos Farinas, MD, David Katz, MD, Jose Freytes, MD, Anne Marie Marciel, MD.
Minneapolis VA: Dennis Niewoehner, MD, Quentin Anderson, MD, Kathryn Rice, MD, Audrey Caine, MD.
Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, MD, MS, Gloria Westney, MD, MS, Eugene Berkowitz, MD, PhD.
National Jewish Health, Denver, CO: Russell Bowler, MD, PhD, David Lynch, MB, Joyce Schroeder, MD, Valerie Hale, MD, John Armstrong, II, MD, Debra Dyer, MD, Jonathan Chung, MD, Christian Cox, MD.
Temple University, Philadelphia, PA: Gerard Criner, MD, Victor Kim, MD, Nathaniel Marchetti, DO, Aditi Satti, MD, A. James Mamary, MD, Robert Steiner, MD, Chandra Dass, MD, Libby Cone, MD.
University of Alabama, Birmingham, AL: William Bailey, MD, Mark Dransfield, MD, Michael Wells, MD, Surya Bhatt, MD, Hrudaya Nath, MD, Satinder Singh, MD.
University of California, San Diego, CA: Joe Ramsdell, MD, Paul Friedman, MD.
University of Iowa, Iowa City, IA: Alejandro Cornellas, MD, John Newell, Jr, MD, Edwin JR van Beek, MD, PhD.
University of Michigan, Ann Arbor, MI: Fernando Martinez, MD, MeiLan Han, MD, Ella Kazerooni, MD.
University of Minnesota, Minneapolis, MN: Christine Wendt, MD, Tadashi Allen, MD.
University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, MD, Joel Weissfeld, MD, MPH, Carl Fuhrman, MD, Jessica Bon, MD, Danielle Hooper, MD.
University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, MD, Sandra Adams, MD, Carlos Orozco, MD, Mario Ruiz, MD, Amy Mumbower, MD, Ariel Kruger, MD, Carlos Restrepo, MD, Michael Lane, MD.
ECLIPSE Investigators
Bulgaria: Y. Ivanov, Pleven; K. Kostov, Sofia. Canada: J. Bourbeau, Montreal; M. Fitzgerald, Vancouver, BC; P. Hernandez, Halifax, NS; K. Killian, Hamilton, ON; R. Levy, Vancouver, BC; F. Maltais, Montreal; D. O′Donnell, Kingston, ON. Czech Republic: J. Krepelka, Prague. Denmark: J. Vestbo, Hvidovre. The Netherlands: E. Wouters, Horn-Maastricht. New Zealand: D. Quinn, Wellington. Norway: P. Bakke, Bergen. Slovenia: M. Kosnik, Golnik. Spain: A. Agusti, J. Sauleda, P. de Mallorca.Ukraine: Y. Feschenko, V. Gavrisyuk, L. Yashina, Kiev; N. Monogarova, Donetsk. United Kingdom: P. Calverley, Liverpool; D. Lomas, Cambridge; W. MacNee, Edinburgh; D. Singh, Manchester; J. Wedzicha, London. USA: A. Anzueto, San Antonio, TX; S. Braman, Providence, RI; R. Casaburi, Torrance CA; B. Celli, Boston; G. Giessel, Richmond, VA; M. Gotfried, Phoenix, AZ; G. Greenwald, Rancho Mirage, CA; N. Hanania, Houston; D. Mahler, Lebanon, NH; B. Make, Denver; S. Rennard, Omaha, NE; C. Rochester, New Haven, CT; P. Scanlon, Rochester, MN; D. Schuller, Omaha, NE; F. Sciurba, Pittsburgh; A. Sharafkhaneh, Houston; T. Siler, St. Charles, MO; E. Silverman, Boston; A. Wanner, Miami; R. Wise, Baltimore; R. ZuWallack, Hartford, CT.
REFERENCES
- 1.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Nat. Acad. Sci. U.S.A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez-Arcelus M., Sekowska M., et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Degner J.F., Pritchard J.K., Pai A.A., Pique-Regi R., Veyrieras J.-B., Gaffney D.J., Pickrell J.K., De Leon S., Michelini K., Lewellen N., et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raby B.A., Murphy A., Chu J.-H., Xu M., Carey V.J., Lazarus R., Liu A., Szefler S.J., Strunk R., Demuth K., et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum. Mol. Genet. 2010;19:4745–4757. doi: 10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverman E., Cho M.H., Hersh C.P., Rennard S., Qiu W., Riley J.H., Anderson W.H., Singh D., Bakke P., Gulsvik A., et al. Genetics of sputum gene expression in chronic obstructive pulmonary disease. PLoS ONE. 2011;6:e24395. doi: 10.1371/journal.pone.0024395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui J., Stahl E.A., Saevarsdottir S., Miceli C., Diogo D., Trynka G., Raj T., Mirkov M.U., Canhao H., Ikari K., et al. Genome-wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLoS Genet. 2013;9:e1003394. doi: 10.1371/journal.pgen.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G., Diogo D., Wu D., Spoonamore J., Dancik V., Franke L., Kurreeman F., Rossin E.J., Duclos G., Hartland C., et al. Human genetics in rheumatoid arthritis guides a high-throughput drug screen of the CD40 signaling pathway. PLoS Genet. 2013;9:e1003487. doi: 10.1371/journal.pgen.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E., et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 10.Elbein S.C., Gamazon E.R., Das S.K., Rasouli N., Kern P.A., Cox N.J. Genetic risk factors for type 2 diabetes: a trans-regulatory genetic architecture? Am. J. Hum. Genet. 2012;91:466–477. doi: 10.1016/j.ajhg.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehrmann R.S.N., Jansen R.C., Veldink J.H., Westra H.-J., Arends D., Bonder M.J., Fu J., Deelen P., Groen H.J.M., Smolonska A., et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schadt E.E., Storey J.D., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillai S.G., Ge D., Zhu G., Kong X., Shianna K.V., Need A.C., Feng S., Hersh C.P., Bakke P., Gulsvik A., et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho M.H., Pillai S.G., Crapo J.D., Silverman E., Hersh C.P., Boutaoui N., Klanderman B., Sylvia J., Ziniti J., DeMeo D., et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho M.H., Wilk J.B., Silverman E., Artigas M.S., Crapo J.D., Shrine N., Loehr L., Zhao J., Manichaikul A., Lopez L., et al. Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am. J. Resp. Crit. Care. 2012;186:622–632. doi: 10.1164/rccm.201202-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverman E., Cho M.H., Pillai S.G., Hersh C.P., Rennard S., Regan E.A., Castaldi P.J., Wan E.S., Siedlinski M., Demeo D.L., et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum. Mol. Genet. 2012;21:947–957. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sin D., Lamontagne M., Couture C., Postma D.S., Timens W., Paré P.D., Hogg J.C., Nickle D., Laviolette M., Bossé Y. Refining susceptibility loci of chronic obstructive pulmonary disease with lung eqtls. PLoS ONE. 2013;8:e70220. doi: 10.1371/journal.pone.0070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westra H.-J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryu D., Cho S., Kim H., Lee S., Kim W. GEPdb: a database for investigating the ternary association of genotype, gene expression and phenotype. Bioinformatics. 2014;30:2540–2542. doi: 10.1093/bioinformatics/btu240. [DOI] [PubMed] [Google Scholar]
- 20.Battle A., Montgomery S.B. Determining causality and consequence of expression quantitative trait loci. Hum Genet. 2014;133:727–735. doi: 10.1007/s00439-014-1446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X., Hersh C.P., Silverman E., Cho M.H., Baron R.M., Hardin M., Zielinski J., Hawrylkiewicz I., Sliwinski P., Mancini J.D., et al. Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Hum. Mol. Genet. 2012;21:1325–1335. doi: 10.1093/hmg/ddr569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M., et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloom A.J., Baker T.B., Chen L.-S., Breslau N., Hatsukami D., Bierut L.J., Goate A. Variants in two adjacent genes, EGLN2 and CYP2A6, influence smoking behavior related to disease risk via different mechanisms. Hum. Mol. Genet. 2014;23:555–561. doi: 10.1093/hmg/ddt432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong H., Yang X., Kaplan L.M., Molony C., Schadt E.E. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am. J. Hum. Genet. 2010;86:581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Degner J.F., Pickrell J.K., Bell J. eQTL Browser. Available at http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl .
- 28.Hindorff L.A., MacArthur J., Morales J., Junkins H.A., Hall P.N., Klemm A.K., Manolio T.A. A Catalog of Published Genome-Wide Association Studies. Available at www.genome.gov/gwasstudies .
- 29.Lui T.T.H., Lacroix C., Ahmed S.M., Goldenberg S.J., Leach C.A., Daulat A.M., Angers S. The ubiquitin-specific protease USP34 regulates axin stability and Wnt/β-catenin signaling. Mol. Cell. Biol. 2011;31:2053–2065. doi: 10.1128/MCB.01094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett J.C., Clayton D.G., Concannon P., Akolkar B., Cooper J.D., Erlich H.A., Julier C., Morahan G., Nerup J., Nierras C., et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franke A., McGovern D.P.B., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R., et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho M.H., McDonald M.-L.N., Zhou X., Mattheisen M., Castaldi P.J., Hersh C.P., Demeo D.L., Sylvia J.S., Ziniti J., et al. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir. Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carey V.J., Davis A.R., Lawrence M.F., Gentleman R., Raby B.A. Data structures and algorithms for analysis of genetics of gene expression with Bioconductor: GGtools 3.x. Bioinformatics. 2009;25:1447–1448. doi: 10.1093/bioinformatics/btp169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storey J.D. A direct approach to false discovery rates. J. R. Statist. Soc. B. 2002;64:479–498. [Google Scholar]
- 37.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kheradpour P., Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth G.K. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer-Verlag; 2005. limma: Linear Models for Microarray Data; pp. 397–420. Statistics for Biology and Health. [Google Scholar]
- 40.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.