Abstract
In the early mouse embryo, a specialized population of extraembryonic visceral endoderm (VE) cells called the anterior VE (AVE) establishes the anterior posterior (AP) axis by restricting gastrulation-inducing signals to the opposite pole. These cells arise at the distal tip of the egg cylinder stage embryo and then asymmetrically migrate to the prospective anterior following the path of an earlier arising and migrating population called the distal VE (DVE). The Nodal-signaling pathway has been shown to have a critical role in the generation of the DVE and AVE and in their migration. The Nodal gene is expressed in both the VE and in the pluripotent epiblast, which gives rise to the germ layers. Previous findings have provided conflicting evidence as to the relative importance of Nodal signaling from the epiblast vs. VE for AP patterning. Here we show that conditional mutagenesis of the Nodal gene specifically within the VE leads to reduced Nodal expression levels in the epiblast and incomplete or failed AVE migration. These results support a required role for VE Nodal to maintain normal levels of expression in the epiblast, and suggest signaling from both VE and epiblast is important for AVE migration.
Keywords: Nodal signaling, AVE, visceral endoderm, epiblast, Ttr-Cre
INTRODUCTION
The anterior-posterior (AP) axis of the mouse embryo is established through a complex set of mechanisms that include both inductive and inhibitory cell-cell signaling (Takaoka and Hamada, 2012; Zernicka-Goetz et al., 2009), as well as global cell movements (Takaoka et al., 2011) and, as recently shown, may also involve mechanical forces imposed by the uterus (Hiramatsu et al., 2013). Prior to implanting into the uterus, the blastocyst stage embryo contains two distinct cell populations, the outer trophectoderm (TE) and the pluripotent inner cell mass (ICM). The ICM contains precursors for the epiblast, the source of all fetal cell lineages, as well as for the extraembryonic primitive endoderm (PrE). Cells of these initially intermingled lineages segregate from each other, with the PrE forming a layer on the blastocoel side by the time of implantation at embryonic day (E) 4.5. Following implantation, the TE adjacent to the ICM differentiates into the extraembryonic ectoderm (ExE) and undergoes extensive proliferation, as does the epiblast. These two populations become aligned on a proximal-distal (PD) axis with respect to the uterine mesometrium, with the PrE derived visceral endoderm (VE) layer expanding to cover both the epiblast distally and the ExE proximally. At approximately E5.0, the epiblast undergoes epithelialization followed by the ExE, transforming the embryo into the egg cylinder, a process recently shown to involve embryo-intrinsic polarization cues (Bedzhov and Zernicka-Goetz, 2014). Other recent evidence suggests that mechanical forces from the uterus may also contribute to the elongated cylindrical shape by constraining growth to the PD axis (Hiramatsu et al., 2013). At E5.5, a molecularly and morphologically distinct population of VE cells arises at the distal end. These distal VE cells (DVE) subsequently migrate proximally, toward the junction of the epiblast and ExE (Rivera-Perez et al., 2003; Srinivas et al., 2004; Thomas and Beddington, 1996), guiding subsequently recruited distal cells as well (Morris et al., 2012; Takaoka et al., 2011). Upon migration, these cells are termed anterior VE (AVE), as they restrict gastrulation-inducing signals to the opposite side of the embryo by E6.5, thereby defining that side as the posterior and their own position as underlying the anterior head region (Stern and Downs, 2012).
Epiblast growth and egg cylinder elongation are essential for DVE formation, in order to displace the distal end of the embryo from inhibitory signals originating in the ExE (Mesnard et al., 2006; Rodriguez et al., 2005; Yamamoto et al., 2009). Recent evidence also supports a role for biomechanical tensile forces at the distal end, which by eliciting breaches in the basement membrane allows migration of distal epiblast cells into the forming DVE (Hiramatsu et al., 2013). The disproportionately high rate of proliferation of the epiblast compared to the VE is also critical to facilitate DVE migration (Stuckey et al., 2011). Several studies have shown that signaling by the TGF-beta related family member Nodal regulates epiblast proliferation (Hiramatsu et al., 2013; Mesnard et al., 2006; Stuckey et al., 2011), with Nodal null mutants being rounder and failing to organize an elongated egg cylinder epithelium (Mesnard et al., 2006). Beyond a role in epiblast proliferation, Nodal signaling has also been shown to be required for patterning the VE covering the epiblast and for DVE specific gene expression (Mesnard et al., 2006). Consistent with these findings, Nodal null mutants or embryos in which Nodal signaling has been blocked show no evidence of DVE or AVE (Brennan et al., 2001; Hiramatsu et al., 2013; Mesnard et al., 2006; Yamamoto et al., 2009), whereas embryos with reduced Nodal signaling form DVE and AVE, but exhibit defects in subsequent migration and AP patterning (Lowe et al., 2001; Norris et al., 2002; Takaoka and Hamada, 2012).
At the time of DVE formation, Nodal is expressed both in the epiblast and VE. An early study provided evidence for Nodal expression in the VE being critical for anterior patterning (Varlet et al., 1997). Anterior defects were found in chimeras made between Nodal mutant embryos and wild type ES cells. In that ES cells do not populate the extraembryonic compartment, the observed defects were ascribed to the lack of Nodal signaling from the completely mutant VE. However, subsequent studies found that absence of detectable Nodal mRNA in the VE due to deletion of a critical Nodal enhancer had no affect on AVE formation and AP patterning (Norris et al., 2002), whereas mosaic deletion of a Nodal conditional allele specifically from the epiblast did cause AP defects (Lu and Robertson, 2004). These later results have led to the proposal that Nodal signaling in the epiblast is paramount for AP patterning and the role of Nodal in the VE, while still unclear, is probably minor (Lu and Robertson, 2004).
The availability of the Tg(Ttr-cre)10-3Xyfu transgenic mouse line expressing Cre recombinase under the control of regulatory elements of the transthyretin (Ttr) gene (Moh et al., 2007), known to be expressed in the VE (Li et al., 2010), in conjunction with our previously validated conditional allele Nodaltm2Mku (Kumar et al., 2008), has allowed us to directly address the role of Nodal in the VE. We find that the absence of Nodal signaling originating from within the VE leads to substantially lower Nodal gene expression within the adjacent wild type epiblast. In addition, lowering or eliminating Nodal specifically from the VE prevents or limits the extent of DVE/AVE migration suggesting an intrinsic function for VE Nodal in AP patterning.
MATERIALS AND METHODS
Nodal-EYFP reporter strain
The Nodal-EYFP reporter was generated by microinjecting linearized recombinant Nodal-EYFP BAC DNA (Lyozin et al., 2014) into pronuclei of C57BL/6NCr zygotes, which were then transferred into pseudopregnant B6D2F1 females. Out of 9 founder males, embryos from four males fully recapitulated Nodal expression at different developmental stages. The line that gave the maximum fluorescent intensity at E5.5 was selected for further analysis. All experiments involving the use of animals were carried out on approved protocols in accordance with the policies and procedures set forth by the Animal Care and Use Committee of NCI-Frederick.
In situ hybridization, PCR genotyping, X-gal staining and immunostaining
Whole mount in situ hybridization (WMISH) to detect the expression of various lineage specific mRNAs was done as described previously (Kumar et al., 2008). Following WMISH, embryos were imaged and lysed for genotyping as described (Kumar et al., 2014). Primers were as described previously for genotyping the Nodaltm2Mku and recombined alleles (Kumar et al., 2008), and Cre (Lowe et al., 2001). X-gal staining was done as previously described (Kumar et al., 2007). For whole mount immunostaining, embryos were dissected in ice-cold PBS with 0.02% BSA (PBS-BSA), fixed for 20 min in 2% PFA, washed in PBS-BSA and permeabilized in 0.1% Triton, 100mM glycine in PBS for 10 minutes at room temperature followed by 2 to 3 quick washes in PBS-BSA. Embryos were blocked overnight at 4° C in PBS containing 0.05% tween 20, 0.1% BSA, 10% goat serum. Embryos were incubated with anti-GFP (Abcam, ab290; diluted 1:1600 in PBS, 0.05% tween 20, 5% goat serum) for 2 hours at room temperature. After 5 washes in blocking solution, embryos were incubated with Alexa Fluor 488 goat anti-rabbit together with Alexa Fluor 633-conjugated Phalloidin (both 1:400 dilution; Molecular Probes). After washing, embryos were placed in anti-fade/glycerol for imaging.
Epifluorescence and immunofluorescence imaging
For epifluorescence imaging, embryos were dissected in PBS containing 10% FBS (PBS-Serum), placed in glass bottom MatTek dishes (No. 1.5, 10 mm diameter) in PBS-serum without fixative and imaged using a Zeiss SteREO Lumar V12 fluorescence stereomicroscope. Z-stack images of bright field and EYFP fluorescence were captured using NeoLumar S 1.5x objective, 46HeYFP reflector and AxiocamMR3 camera and analyzed using Zeiss AxioVision software. For confocal imaging, embryos were dissected in PBS-serum and fixed in 4% PFA, pH 7.4, for 30 minutes at room temperature. Embryos were washed in PBS and incubated with a 1:1,000 dilution of Alexa Fluor 633 Phalloidin in PBS overnight at 4° C. After washing five times with PBS, embryos were placed in MatTek dishes in Slow Fade anti-fade mounting media (S2828; Molecular probes) and covered with a glass coverslip. Images were acquired using a Zeiss LSM510 confocal microscope with Plan-Neofluor 40x/1.3 oil DIC objective using the following excitation and emission filters: for GFP excitation at 488, emission band pass 500–580; for EYFP excitation at 514 nm, emission at 535–590 nm band pass filter; for Alexa Fluor 633 Phalloidin excitation at 633 nm, and emission was collected using a 660–740 nm band pass filter. Following imaging, the coverslip was removed and the embryos were collected, washed in PBS-T and lysed and genotyped as described above.
Immunostained embryos were imaged with a cool snap HQ2 camera (Photometrics) using a wide field Nikon Eclipse Ti microscope with 20x optics (Plan Apo 20x DIC MN2, NA 0.8) and NIS element software. Differential interference contrast (DIC) images were captured simultaneously with GFP (FITC filter) and Alexa633 (Cy5 filter). The Alexa488 and Alexa633 images were deconvoluted using auto-quant software (Media cybernetics). The resulting images were exported as TIFF files and maximum intensity projection and composite images were created using ImageJ software.
RESULTS AND DISCUSSION
Evidence for lack of Nodal expression in the primitive endoderm
Nodal mRNA has been detected by whole mount in situ hybridization (WMISH) in the E4.5 blastocyst (Mesnard et al., 2006; Takaoka et al., 2006). These reports indicated expression occurs in both the epiblast and PrE. More recently, expression of a fluorescent protein transgene driven by the Nodal-signaling responsive ASE enhancer was found in ICM and individual PrE cells at E4.5, and Nodal mRNA expression was shown by WMISH to be absent from the PrE at E5.0 and present in the VE at E5.5 (Granier et al., 2011). To confirm this early pattern of expression in the PrE, we developed a fluorescence-based transgenic reporter to provide a sensitive and high-resolution analysis. We used a novel recombineering approach to generate a bacterial artificial chromosome (BAC) construct in which the first exon of Nodal was cleanly replaced with EYFP (Lyozin et al., 2014). This BAC construct includes 71 kb 5′ and 126 kb 3′ of the coding region, and thus contains all of the known upstream and intronic regulatory regions in their native orientation, and no extraneous introduced sequences other than EYFP (Fig. 1A). Preliminary results with transient transgenics showed EYFP epifluorescence faithfully replicating the Nodal mRNA expression pattern, as determined by WMISH at E6.0 (Fig. 1B) and at later stages (not shown). Further analysis was carried out with a stable transgenic line that was selected for maximal EYFP levels and appropriate post-implantation expression. To analyze peri-implantation stages, we first examined E3.5 Nodal-EYFP transgene positive embryos that were still within the zona pellucida and had not yet developed primitive endoderm (n=30), but none showed any specific EYFP epifluorescence. Expression was first detected at E4.5 using epifluorescence but we found that resolution was greatly improved by carrying out whole mount immunofluorescence using an anti-GFP antibody. At E4.5, 3 embryos out of a total of 7 analyzed showed immunofluorescence in a single ICM cell (Fig. 1C, left). In the remaining 4, either several cells or the entire ICM was positive (Fig. 1C, middle and right). As Nodal signaling regulates Nodal gene expression, there may be initially a single expressing ICM cell that then signals to adjacent cells, rapidly converting the entire ICM to Nodal expression.
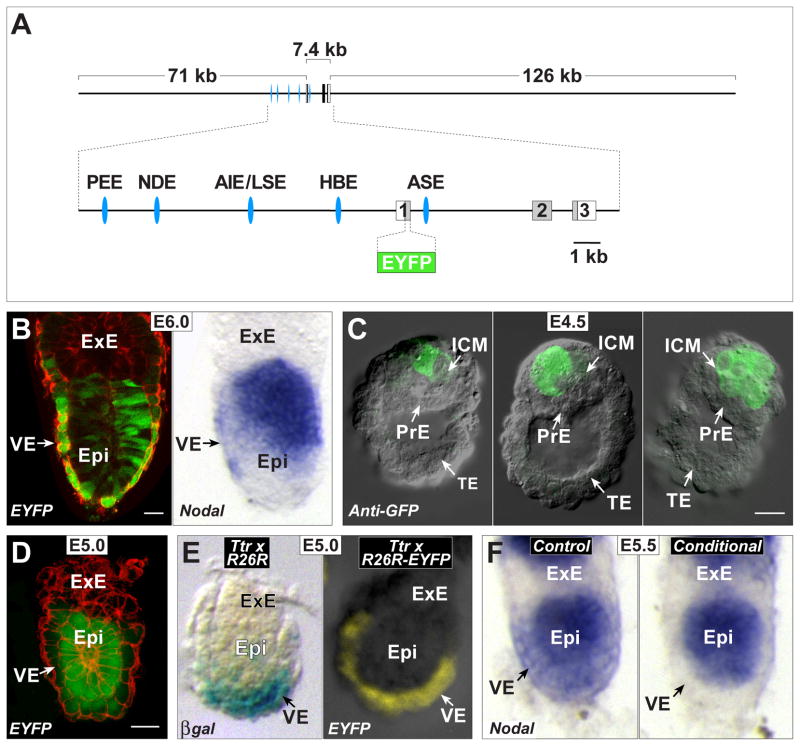
Fig. 1. Ttr-Cre activity coincides with first appearance of Nodal in VE.
(A) Structure of the approximately 204 kB BAC transgene containing the 7.4 kb Nodal coding region, and 71 kb upstream and 126 kb downstream. Shown enlarged below is the area containing the three exons, with the coding region displayed in gray. The approximate positions of the Proximal Epiblast Enhancer (PEE) (Ben-Haim et al., 2006; Vincent et al., 2003), Node Specific Enhancer (NDE) (Adachi et al., 1999; Norris and Robertson, 1999), Asymmetric/Left Side-Specific Enhancer (AIE/LSE) (Saijoh et al., 2005; Vincent et al., 2004), Highly Bound Element (HBE) (Papanayotou et al., 2014) and Asymmetric Enhancer (ASE) (Norris et al., 2002; Norris and Robertson, 1999) are displayed as blue ellipses. The coding region within the first exon was replaced with EYFP followed by a stop codon. (B) Left panel, a projection of 6 consecutive confocal sections from an E6.0 transient transgenic embryo showing EYFP fluorescence in green and cell outlines marked in red with Alexa-Fluor 633 phalloidin. The scale bar represents 25 microns. Right panel, WMISH for Nodal expression of a similarly staged embryo. Expression in posterior epiblast (Epi) is seen in both embryos. Strong EYFP signal in VE is likely due to protein perdurance, as mRNA expression in VE is diminishing at this stage. (C) DIC images of E4.5 transgenic embryos superimposed with anti-GFP immunofluorescence signal (green) showing expression in single or multiple cells of the ICM. No signal is detected in the PrE and trophectoderm (TE). The scale bar represents 25 microns. (D) Confocal section of an E5.0 stage transgenic embryo showing EYFP fluorescence (green) in epiblast and distal VE. Cell outlines marked in red with Alexa-Fluor 633 phalloidin. The scale bar represents 25 microns. (E) Left panel, E5.0 embryo derived from Ttr-Cre by R26R cross. The majority of the VE shows X-gal stained cells reflecting Cre activity. Right panel, a similarly staged embryo from a Ttr-Cre by R26R-EFYP cross showing epifluorescence signal throughout the VE. (F) WMISH for Nodal expression in two E5.5 littermates derived from a Ttr-Cre, Nodal +/− by homozygous Nodal f l cross. The control embryo on the left is Nodal f l/− but Cre negative, and shows Nodal expression throughout the epiblast and VE. The conditional mutant embryo on the right is Nodal f l/− and Cre positive, and shows loss of Nodal expression from the VE.
Surprisingly, we did not detect EYFP epifluorescence or anti-GFP immunofluorescence in the PrE at peri-implantation stages. At E5.0, however, EYFP epifluorescence was found in the VE of 7 out of 19 embryos, predominantly in the distal region (Fig. 1D), suggesting this is the stage when Nodal is first expressed in this lineage. The EYFP used in our construct is not the destabilized version. Thus, cells retain EYFP protein even after gene expression terminates, making it likely we would have detected even transient expression in the PrE. It is possible that WMISH analysis, used in the previously published studies, may not provide high enough resolution to distinguish between negative PrE cells and adjacent ICM cells positive for Nodal expression. Although the ASE-only reporter was found in the PrE, the EYFP Nodal reporter is likely a more accurate indicator of Nodal expression at these stages. The latter contains all of the Nodal regulatory elements in their normal configuration, whereas the former may be missing inhibitory elements that would normally prevent PrE expression. Indeed, the ASE enhancer has been shown to drive reporter expression at E8.5 in the floor plate of the neural tube (Norris and Robertson, 1999), a region that does not express Nodal, at least at levels detectable by WMISH.
Initial Ttr-Cre activity coincides with the onset of Nodal protein expression in the VE
We next determined the time of onset of Cre activity in the Tg(Ttr-cre)10-3Xyfu transgenic line, which expresses Cre recombinase under the control of regulatory elements from the transthyretin gene (Moh et al., 2007). A different Cre line using the same transthyretin regulatory domain was shown to specifically recombine floxed alleles in the VE of E5.5 embryos (Kwon and Hadjantonakis, 2009). To determine whether Cre activity is present at E5.0 or earlier in the VE, we mated mice carrying the Tg(Ttr-cre)10-3Xyfu transgene (hereafter referred to as Ttr-Cre) with the reporter strains R26R (Soriano, 1999) and R26R-EYFP (Srinivas et al., 2001). These reporter strains allow cells with Cre activity or descendants of these cells to be visualized by X-gal staining or EYFP fluorescence, respectively, in whole mount embryos. In 7 out of 23 (30%) E4.5 embryos that were genotyped as Ttr-Cre positive, we detected labeled cells within the PrE (not shown), suggesting this is the time of initial activation of Ttr-Cre. At E5.0, evidence of Ttr-Cre activity was found in 100% of transgene positive embryos, throughout the VE (Fig. 1E).
Based on the above results, Ttr-Cre should mediate recombination in the VE lineage prior to or at the onset of Nodal expression. To confirm this, we utilized the Nodaltm2Mku floxed allele (hereafter referred to as Nodal f l) and the Nodal null allele (Lowe et al., 2001). We bred the null allele onto the Ttr-Cre background and then crossed the resulting Ttr-Cre positive, Nodal+/− mice with homozygous Nodal f l mice (Fig. 2A, Scheme 1). Upon Ttr-Cre mediated deletion of Nodal f l, one quarter of the embryos from this cross will have two nonfunctional Nodal alleles within the VE (Fig. 2A, Scheme 1, iv). Analysis by WMISH showed the absence of Nodal expression in the VE of E5.5 conditional mutant embryos with this genotype (Fig. 1F, right), whereas in similarly staged control embryos lacking Ttr-Cre (Fig. 2A, Scheme 1, iii), Nodal expression was found in both the epiblast and VE as expected (Fig. 1F, left). These results confirm the temporal and spatial specificity of our experimental approach.
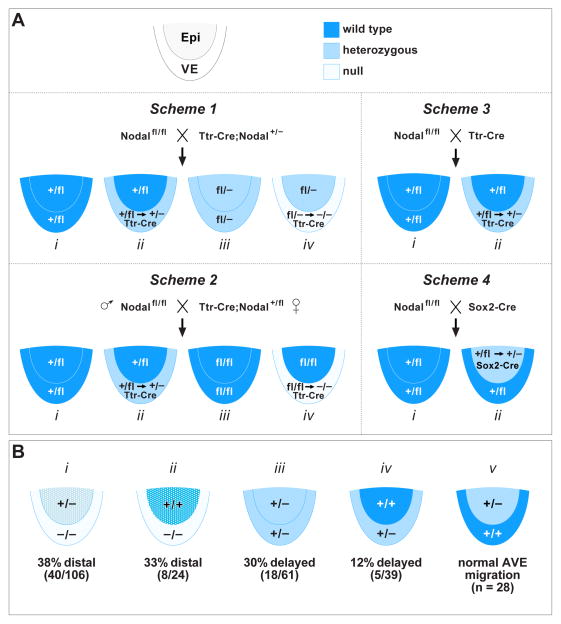
Fig. 2. Mating schemes and results for VE specific Nodal conditional mutants.
(A) Mating schemes utilized for generating conditional mutants. The embryonic portion of E5.0 embryos is shown schematically with the epiblast and VE labeled in different colors depending on the possible genotype. Dark blue, wild type; light blue, heterozygous; pale, homozygous null. Scheme 1: Nodal f l/fl crossed to (X) Ttr-Cre, Nodal +/−. There are four potential genotypes of which (iv) represents the VE specific conditional mutant. The epiblast is heterozygous (fl/−) for Nodal. Scheme 2: Nodal f l/fl male X Ttr-Cre, Nodal +/fl female. There are four potential genotypes of which (iv) represents the VE specific conditional mutant. The epiblast is wild type (fl/fl) for Nodal. Scheme 3: Nodal f l/fl X Ttr-Cre. There are two potential genotypes of which (ii) represents the VE specific conditional heterozygote. The epiblast is wild type (+/fl) for Nodal. Scheme 4: Nodal f l/fl X Sox2-Cre. There are two potential genotypes of which (ii) represents the epiblast specific conditional heterozygote. The VE is wild type (+/fl) for Nodal. (B) The four different epiblast and VE genotypes resulting from conditional mutagenesis, as well as the Nodal heterozygote, are shown with their associated frequency of delayed or incomplete/failed AVE migration, color coded as for (A) above. Dark blue and light blue stippling represents reduced Nodal expression in the epiblast due to absence of Nodal signaling from VE.
Ttr-Cre conditional mutagenesis of Nodal can broadly affect early patterning
To determine the importance of Nodal signaling from the VE, we examined the phenotype of Scheme 1 conditional mutant embryos at late E5.5 and later. At E6.0 – E6.5, embryos with the appropriate genotype (Fig. 2A, Scheme 1, iv), displayed a range of phenotypes (Fig. 3). Using confocal imaging, we found that 8 of 17 E6.0 – E6.5 conditional mutants were phenotypically normal, showing a fully migrated AVE (Fig. 3A, i). Of the other 9 conditional mutants, 4 showed elongated VE cells more distal than normal (Fig. 3A, ii), and 3 had elongated cells restricted to the distal end (Fig. 3A, iii). The 2 remaining conditional mutants were more severely affected, showing a phenotype similar to Nodal null mutants including underdeveloped epiblast (Fig. 3A, iv), and detachment of the VE at the distal end (Fig. 3A, v).
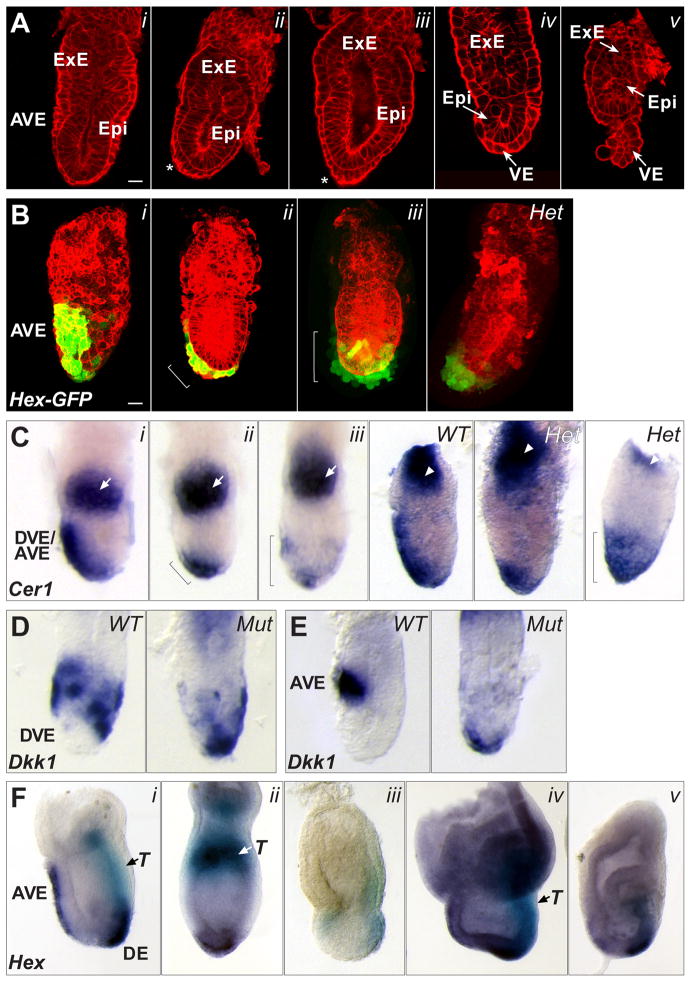
Fig. 3. AVE migration and other patterning defects in Scheme 1 conditional mutants.
(A) Confocal sections of five E6.0 – E6.5 conditional mutants, stained with Alexa-Fluor 633 phalloidin to outline cells, showing no defects (i); a partially moved AVE (ii, asterisk); distal elongated VE cells (iii, asterisk); reduced epiblast growth (iv); overall reduced size and detached VE (v). The scale bar in i represents 25 microns; all embryos are at the same scale. (B) Confocal projections of an E6.5 wild type embryo (i), showing Hex-GFP marked AVE cells along the anterior; an E6.5 conditional mutant (ii), showing Hex-GFP marked cells partially moved; an E6.5 conditional mutant (iii), showing Hex-GFP marked cells all around the distal region; an E6.5 Nodal heterozygous embryo (Het), showing distal Hex-GFP marked cells. Cell outlines marked with Alexa-Fluor 633 phalloidin. The scale bar in i represents 25 microns; all embryos are at the same scale. (C) WMISH for Cerl mRNA expression in a late E5.5 wild type embryo (i), showing anterior location of DVE/AVE cells; a late E5.5 conditional mutant (ii), showing Cerl expressing cells only partially moved toward the anterior (bracket); a late E5.5 conditional mutant (iii), showing Cerl expressing cells all around the distal region (bracket); an E6.0 Nodal WT embryo showing Cerl expressing cells along the anterior; a heterozygous (Het) littermate showing distal Cerl expressing cells; another Het embryo showing Cerl expressing cells all around the distal region (bracket). White arrows in panels i – iii mark Esrrβ expression in the ExE. White arrowheads in the right panels mark Mash2 expression in the ExE. (D) Left, WMISH for Dkk1 mRNA expression in an early E6.0 wild type embryo. The distal region lacks expressing cells. Right, Dkk1 mRNA expression in an E6.0 conditional mutant embryo showing abnormal distal location of Dkk1 expressing cells. (E) Left panel, WMISH for Dkk1 mRNA expression in an E6.5 wild type embryo. Dkk1 expressing cells are found at the AVE. Right panel, Dkk1 mRNA expression in an E6.5 conditional mutant embryo showing abnormal distal location of Dkk1 expressing cells. (F) WMISH for Hex and T (Brachyury) mRNA expression in an E7.5 wild type embryo (i). Hex expressing cells (dark purple) are found in the AVE and definitive endoderm (DE). T expressing cells (light blue) mark mesoderm forming at the primitive streak. E7.5 conditional mutants are misshapen and can show distal Hex and proximal T expression (ii); lower Hex and T expression levels (iii); an expanded Hex expression domain with normal T levels (iv); reduced Hex and T expression domains (v).
The presence of elongated VE cells other than at the anterior in the majority of affected conditional mutants (7/9) suggested that in the absence of Nodal signaling from within the VE, the AVE can arise but may fail to migrate fully or at all. To investigate this further, we introduced onto the Nodal f l background the Hex-GFP transgene, which marks Hex-expressing DVE and AVE cells with GFP (Rodriguez et al., 2001). We then followed the Scheme 1 breeding strategy (Fig. 2A) to again generate conditional mutant embryos. In wild type control embryos at E6.0 – E6.5, Hex-GFP marked cells are found along the prospective anterior (Fig. 3B, i). In 8 of 18 conditional mutants at a similar stage, Hex-GFP was found either only partially moved from the distal end (Fig. 3B, ii) or completely distal, with one embryo showing GFP-marked cells around the entire distal circumference of the embryo (Fig. 3B, iii). To extend these findings, we carried out WMISH for Cer1, which marks a subset of DVE/AVE cells overlapping with those expressing Hex (Yamamoto et al., 2004). In late E5.5 control embryos, Cer1 marks the DVE migrating along the prospective anterior (Fig. 3C, i). All similarly staged conditional mutant embryos showed Cer1 expression, confirming DVE formation in mutants. However, in 18 of 56 we found more distal expression (Fig. 3C, ii), which in one case involved an expansion of the Cer1 expression domain, such that it encompassed the distal one-third of the embryo (Fig. 3C, iii). We also examined the gene for the Wnt antagonist Dkk1, which provides guidance cues for DVE/AVE migration (Kimura-Yoshida et al., 2005). Normally, Dkk1 mRNA expressing cells are found just in front and transiently just behind AVE cells as they migrate (Fig. 3D, left panel), progressing to a completely anterior position at E6.5 (Fig. 3E, left panel). In 7 of 15 conditional mutant embryos, we found Dkk1 in the distal-most cells, an area where expression is not normally found (Fig. 3D, E; right panels). The abnormal distal localization of Dkk1, along with the expanded distal expression of Hex-GFP and Cer1 in conditional mutants, suggests that removing Nodal completely from the VE potentially affects not only DVE/AVE migration, but may also lead to changes in cell fate specification in the distal VE. This is consistent with the finding that Nodal null mutants do not properly specify the embryonic portion of the VE (Mesnard et al., 2006).
In the experiments described above, we found 40 out of 106 (38%) E5.5 and E6.5 Nodal VE conditional mutant embryos to be phenotypically abnormal with respect to DVE/AVE migration (Fig. 2B, i). Migration occurs normally in the remaining mutants, suggesting that levels of Nodal signaling in these embryos are above a critical threshold. It is possible that recombination was not complete by the time Nodal gene expression commenced in these individuals. Alternatively, there may be a variable degree of rescue by Nodal signaling originating in the epiblast. To further analyze the phenotypic abnormalities, we examined later stage embryos using two-color WMISH for T (Brachyury) and Hex expression. At E7.5, control embryos show Hex expression in the anteriorly migrated AVE and a second domain in the nascent definitive endoderm (DE) at the anterior end of the primitive streak (Fig. 3F, i; dark purple). T is found in the posterior primitive streak mesoderm (Fig. 3F, i; light blue). Of 21 conditional mutants examined, 11 (52%) showed an abnormal phenotype. Three were oddly shaped with only a single domain of Hex in the distal VE, and T expression in the proximal epiblast (Fig. 3F, ii). While the presence of T indicates gastrulation, the altered location of mesoderm formation suggests a failure to convert the PD axis into the AP axis. The remaining 8 conditional mutants were also distorted in shape, some severely, and had reduced or expanded expression of T and/or Hex (Fig. 3G, iii–v). The increased frequency of severe defects in E7.5 conditional mutants compared to E6.5 (52% vs. 38%) suggests that continued development in the absence of Nodal signaling from the VE has a cumulative negative effect.
Loss of Nodal solely from the VE affects DVE/AVE migration
As part of this analysis, we were surprised to find that 18 of 61 (30%) E6.5 Nodal heterozygous embryos (Fig. 2B, iii) showed Hex-GFP marked cells at a more distal position than wild type (Fig. 3B, Het), and a more distal Cer1 expression domain (Fig. 3C, Het, left), including one with an expanded distal domain (Fig. 3C, Het, right). Perhaps not unexpectedly, no abnormalities or altered expression domains were seen in Nodal heterozygous embryos at E7.5 (not shown), suggesting that the more distal expression domains found in E6.5 heterozygotes represent a delay in AVE movement due to Nodal levels being halved in both the epiblast and VE. This transient phenotype in the Nodal heterozygote suggests that the range in expressivity and penetrance found in Scheme 1 conditional mutants might be due to reduced signaling from the heterozygous epiblast, in addition to the complete loss of Nodal signaling from the VE. To explore the relative contribution of epiblast and VE expression further, we modified the conditional strategy such that the epiblast would be completely wild type and loss of the Nodal gene would occur solely within the VE. This involved generating Ttr-Cre animals carrying the Nodal f l allele, rather than the Nodal null allele, for crossing with Nodal f l animals (Fig. 2A, Scheme 2). Complicating this strategy was the discovery that Ttr-Cre mediates recombination of the Nodal f l allele in the paternal germline. There is no report detailing Ttr gene expression in the mouse germline, suggesting this represents ectopic activity. Therefore for all timed mating experiments, we bred Ttr-Cre; Nodal +/f l females, which showed no evidence for germline recombination, with Nodal f l homozygous males.
Embryos derived from this mating scheme were phenotypically examined at E6.0 – E6.5, and analyzed for markers of the DVE/AVE by WMISH. Genotyping identified 24 conditional mutants (Fig. 2A, Scheme 2, iv) and of these, 8 had a more symmetric shape than normal and showed distal expression of Hex and Cer1 whereas all wild type embryos showed anterior expression at these stages (Fig. 4A, B). This frequency of 33% (Fig. 2B, ii) is only slightly less than the 38% found for Scheme 1 (Fig. 2B, i). Thus, increasing the Nodal gene dosage in the epiblast from heterozygous to wild type leads to only minimal improvement in DVE/AVE migration, suggesting that Nodal signaling from the VE may be critical for this process. At E7.0, 4 out of 16 (25%) Scheme 2 conditional mutants also showed other overt phenotypic defects (Fig. 4C). However, the frequency of these defects was lower and the severity milder than observed in Scheme 1 conditional mutants suggesting some degree of rescue due to increased Nodal signaling from the wild type epiblast.
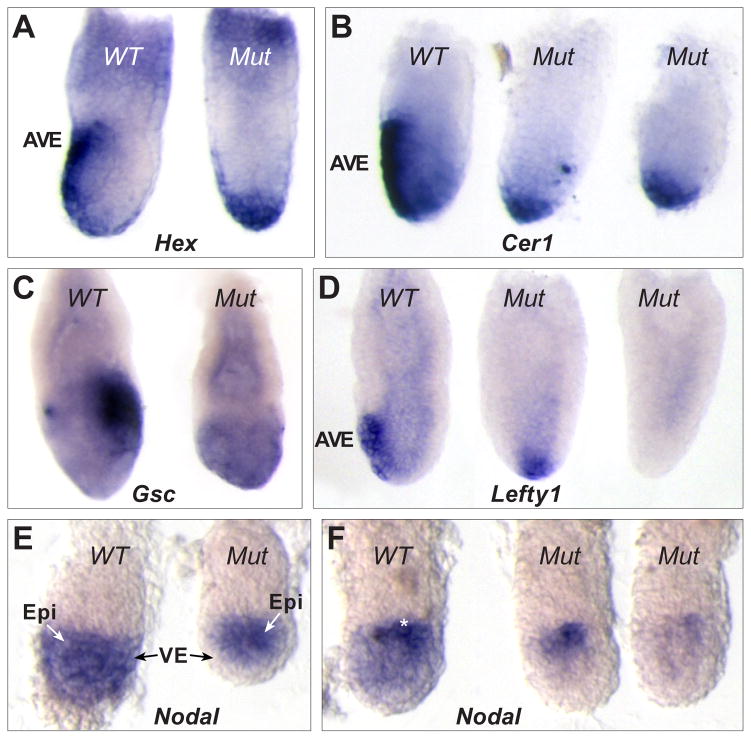
Fig. 4. AVE migration defects and reduced epiblast Nodal in Scheme 2 conditional mutants.
(A) Two E6.5 littermates after WMISH for Hex mRNA. The wild type embryo (left) shows Hex in AVE cells while the conditional mutant shows distal Hex expression. (B) WMISH for Cerl mRNA in three E6.5 littermates. The wild type embryo (left) shows Cerl expression in the AVE while the conditional mutants (center, right) show distal expression. (C) WMISH for Goosecoid (Gsc) mRNA in two E6.5 littermates. The wild type embryo (left) shows Gsc in primitive streak cells at the posterior. The conditional mutant (right) shows only background levels and is smaller and misshapen. (D) Three E6.5 littermates following WMISH for Lefty1 mRNA expression. The wild type embryo (left) shows Lefty1 in the AVE while the conditional mutants show either distal (center) or no expression (right). (E) WMISH for Nodal mRNA in two early E5.5 littermates. The wild type embryo (left) shows Nodal throughout the epiblast and VE. The conditional mutant (right) lacks Nodal expression in the VE. (F) WMISH for Nodal mRNA expression in three E5.5 littermates. The wild type embryo (left) shows reduced Nodal in the distal epiblast and VE and strong proximal expression (white asterisk). The conditional mutants (right, center) completely lack Nodal expression in the VE and distal epiblast and show reduced proximal expression compared to wild type.
Loss of Nodal from the VE leads to reduced Nodal expression in the epiblast
We also examined the expression of Lefty1, which is a target of Nodal signaling and a DVE/AVE marker (Fig. 4D, left). Out of 10 E6.5 conditional mutants, 5 showed normal anterior Lefty1 expression. However, in 4 mutants expression was distal (Fig. 4D, center), and one conditional embryo lacked Lefty1 expression altogether (Fig. 4D, right). This differential effect on Lefty1 prompted us to confirm that Scheme 2 conditional mutant embryos indeed completely lack Nodal expression in the VE from earlier stages. At early E5.5, wild type embryos show Nodal expression throughout the epiblast and VE (Fig. 4E, left). Examination of 10 similarly staged Scheme 2 conditional mutants showed that all 10 had complete absence of Nodal from the VE (Fig. 4E, right). In slightly older wild type embryos, Nodal expression becomes reduced distally in both the VE and epiblast and is upregulated in the proximal epiblast (Fig. 4F, left). When we analyzed 10 similarly staged mutants we found 4 with significantly reduced levels of Nodal expression in the epiblast, with upregulation in the proximal epiblast also affected (Fig. 4F, center and right). Along with previous findings showing reduced Nodal reporter expression in embryo explants cultured without VE or ExE (Beck et al., 2002; Guzman-Ayala et al., 2004), our results indicate that Nodal expression in the VE has an important role in sustaining proper levels of Nodal in the adjacent epiblast. Nodal signaling from the epiblast was previously shown to trigger Nodal expression in the VE (Brennan et al., 2001). Our results suggest that there is a reciprocal interaction, with VE Nodal signals either directly influencing epiblast Nodal gene expression or acting indirectly. Nodal ligand expressed in the VE may act as a sink for the Nodal signaling inhibitors Lefty1 and Cer1 expressed in the DVE/AVE. The absence of Nodal from the VE may result in an expansion of the functional domain of these inhibitors in the epiblast, contributing to the loss of Nodal gene expression observed in the proximal epiblast.
Interestingly, the frequency of E5.5 embryos with reduced Nodal expression in epiblast (4 out of 10; 40%) was only slightly higher than the frequency of DVE/AVE migration defects (8 out of 24; 33%) found in Scheme 2 conditional mutants at E6.0 and later. This correspondence is consistent with specific threshold Nodal levels being required within the epiblast, below which DVE/AVE migration is affected. Thus, it is possible that the main role of Nodal signaling specifically from the VE is to maintain optimal Nodal gene expression levels in the adjacent epiblast, rather than having an intrinsic function in migration. To explore further the relative contribution from each tissue layer, we generated embryos heterozygous for Nodal in either the VE or the epiblast. Timed matings between Nodal f l homozygotes and Ttr-Cre carrier animals, wild type for Nodal, were used for generating VE-specific heterozygotes (Fig. 2A, Scheme 3, ii). Timed matings between Nodal f l homozygotes and animals carrying Sox2-Cre (Hayashi et al., 2002) were used for generating epiblast-specific heterozygotes (Fig. 2A, Scheme 4, ii). Interestingly, we found that 5 out of 39 (12%) of VE-specific heterozygotes had distal expression of DVE/AVE markers at E6.5 (Fig. 2B, iv), whereas none of the 28 epiblast-specific heterozygotes examined showed any evidence for DVE/AVE delay (Fig. 2B, v). These results are consistent with a requirement for optimal Nodal expression levels intrinsic to the VE to allow proper DVE/AVE migration.
In conclusion, the findings presented here help to clarify the early pattern of Nodal expression as well as its function from within the VE for DVE/AVE migration. In contrast to previously published results, we find no evidence for Nodal expression in the PrE using our Nodal reporter, which is under the same regulation as the endogenous Nodal gene. Rather, the earliest expression detected is in the VE at E5.0. VE-specific conditional mutagenesis reveals a critical function for Nodal signals arising in the VE for DVE/AVE migration. This is likely due in no small part to the role VE Nodal plays in maintaining normal levels of Nodal expression in the epiblast. However, our results showing that lowering Nodal levels specifically within the VE delays DVE/AVE migration suggests a function intrinsic to Nodal signals originating in the VE, consistent with the original contention of a critical role for VE Nodal in AP patterning (Varlet et al., 1997). An intrinsic function in the VE has been questioned given the conflicting finding that normal AP development occurs in the absence of detectable Nodal in the VE of ASE enhancer knockout embryos (Norris et al., 2002). However, analysis of the very earliest stage of Nodal expression in the VE was not reported in that study. It is possible that sufficient initial VE Nodal expression does occur, perhaps mediated by the newly identified early acting HBE enhancer (Papanayotou et al., 2014), to allow normal DVE/AVE migration. However, due to the absence of the ASE, expression may not be maintained leading to the lack of detectable mRNA in the VE at later stages. It should also be noted that a previous analysis of embryos mutated for both Lefty1 and Cer1 also provided evidence for an intrinsic function, as these mutants were shown to have increased levels of Nodal signaling throughout the VE and a delay in DVE/AVE migration (Perea-Gomez et al., 2002; Yamamoto et al., 2004). The similar delay in DVE/AVE migration obtained with lowering Nodal signaling shown here indicate that this process must be extremely sensitive to VE Nodal levels, and suggest that there is a very narrow optimal window.
Highlights.
BAC transgenic reporter indicates no Nodal expression in primitive endoderm
Visceral endoderm specific mutation of Nodal affects DVE/AVE migration
Visceral endoderm Nodal signals regulate epiblast Nodal gene expression levels
Acknowledgments
We thank the Transgenic Mouse Model Laboratory for generation of Nodal EYFP transgenic mice; and Dr. Terry Yamaguchi for comments on the manuscript. This work was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services and nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi H, Saijoh Y, Mochida K, Ohishi S, Hashiguchi H, Hirao A, Hamada H. Determination of left/right asymmetric expression of nodal by a left side-specific enhancer with sequence similarity to a lefty-2 enhancer. Genes Dev. 1999;13:1589–1600. doi: 10.1101/gad.13.12.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S, Le Good JA, Guzman M, Ben Haim N, Roy K, Beermann F, Constam DB. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat Cell Biol. 2002;4:981–985. doi: 10.1038/ncb890. [DOI] [PubMed] [Google Scholar]
- Bedzhov I, Zernicka-Goetz M. Self-Organizing Properties of Mouse Pluripotent Cells Initiate Morphogenesis upon Implantation. Cell. 2014;156:1032–1044. doi: 10.1016/j.cell.2014.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim N, Lu C, Guzman-Ayala M, Pescatore L, Mesnard D, Bischofberger M, Naef F, Robertson EJ, Constam DB. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell. 2006;11:313–323. doi: 10.1016/j.devcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- Granier C, Gurchenkov V, Perea-Gomez A, Camus A, Ott S, Papanayotou C, Iranzo J, Moreau A, Reid J, Koentges G, Saberan-Djoneidi D, Collignon J. Nodal cis-regulatory elements reveal epiblast and primitive endoderm heterogeneity in the peri-implantation mouse embryo. Dev Biol. 2011;349:350–362. doi: 10.1016/j.ydbio.2010.10.036. [DOI] [PubMed] [Google Scholar]
- Guzman-Ayala M, Ben-Haim N, Beck S, Constam DB. Nodal protein processing and fibroblast growth factor 4 synergize to maintain a trophoblast stem cell microenvironment. Proc Natl Acad Sci USA. 2004;101:15656–15660. doi: 10.1073/pnas.0405429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev. 2002;119(Suppl 1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Hiramatsu R, Matsuoka T, Kimura-Yoshida C, Han SW, Mochida K, Adachi T, Takayama S, Matsuo I. External mechanical cues trigger the establishment of the anterior-posterior axis in early mouse embryos. Dev Cell. 2013;27:131–144. doi: 10.1016/j.devcel.2013.09.026. [DOI] [PubMed] [Google Scholar]
- Kimura-Yoshida C, Nakano H, Okamura D, Nakao K, Yonemura S, Belo JA, Aizawa S, Matsui Y, Matsuo I. Canonical Wnt signaling and its antagonist regulate anterior-posterior axis polarization by guiding cell migration in mouse visceral endoderm. Dev Cell. 2005;9:639–650. doi: 10.1016/j.devcel.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Kumar A, Lualdi M, Lewandoski M, Kuehn MR. Broad mesodermal and endodermal deletion of Nodal at postgastrulation stages results solely in left/right axial defects. Dev Dyn. 2008;237:3591–3601. doi: 10.1002/dvdy.21665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Lualdi M, Loncarek J, Cho YW, Lee JE, Ge K, Kuehn MR. Loss of function of mouse Pax-Interacting Protein 1-associated glutamate rich protein 1a (Pagr1a) leads to reduced Bmp2 expression and defects in chorion and amnion development. Dev Dyn. 2014;243:937–947. doi: 10.1002/dvdy.24125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Yamaguchi T, Sharma P, Kuehn MR. Transgenic mouse lines expressing Cre recombinase specifically in posterior notochord and notochord. Genesis. 2007;45:729–736. doi: 10.1002/dvg.20346. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Hadjantonakis AK. Transthyretin mouse transgenes direct RFP expression or Cre-mediated recombination throughout the visceral endoderm. Genesis. 2009;47:447–455. doi: 10.1002/dvg.20522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li YP, Fu XY, Deng CX. Anterior visceral endoderm SMAD4 signaling specifies anterior embryonic patterning and head induction in mice. International journal of biological sciences. 2010;6:569–583. doi: 10.7150/ijbs.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe LA, Yamada S, Kuehn MR. Genetic dissection of nodal function in patterning the mouse embryo. Development. 2001;128:1831–1843. doi: 10.1242/dev.128.10.1831. [DOI] [PubMed] [Google Scholar]
- Lu CC, Robertson EJ. Multiple roles for Nodal in the epiblast of the mouse embryo in the establishment of anterior-posterior patterning. Dev Biol. 2004;273:149–159. doi: 10.1016/j.ydbio.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Lyozin GT, Bressloff PC, Kumar A, Kosaka Y, Demarest BL, Yost HJ, Kuehn MR, Brunelli L. Isolation of rare recombinants without using selectable markers for one-step seamless BAC mutagenesis. Nat Methods. 2014 doi: 10.1038/nmeth.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnard D, Guzman-Ayala M, Constam DB. Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development. 2006;133:2497–2505. doi: 10.1242/dev.02413. [DOI] [PubMed] [Google Scholar]
- Moh A, Iwamoto Y, Chai GX, Zhang SS, Kano A, Yang DD, Zhang W, Wang J, Jacoby JJ, Gao B, Flavell RA, Fu XY. Role of STAT3 in liver regeneration: survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab Invest. 2007;87:1018–1028. doi: 10.1038/labinvest.3700630. [DOI] [PubMed] [Google Scholar]
- Morris SA, Grewal S, Barrios F, Patankar SN, Strauss B, Buttery L, Alexander M, Shakesheff KM, Zernicka-Goetz M. Dynamics of anterior-posterior axis formation in the developing mouse embryo. Nat Commun. 2012;3:673. doi: 10.1038/ncomms1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris DP, Brennan J, Bikoff EK, Robertson EJ. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 2002;129:3455–3468. doi: 10.1242/dev.129.14.3455. [DOI] [PubMed] [Google Scholar]
- Norris DP, Robertson EJ. Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements. Genes Dev. 1999;13:1575–1588. doi: 10.1101/gad.13.12.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanayotou C, Benhaddou A, Camus A, Perea-Gomez A, Jouneau A, Mezger V, Langa F, Ott S, Saberan-Djoneidi D, Collignon J. A novel nodal enhancer dependent on pluripotency factors and smad2/3 signaling conditions a regulatory switch during epiblast maturation. PLoS Biol. 2014;12:e1001890. doi: 10.1371/journal.pbio.1001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Gomez A, Vella FD, Shawlot W, Oulad-Abdelghani M, Chazaud C, Meno C, Pfister V, Chen L, Robertson E, Hamada H, Behringer RR, Ang SL. Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell. 2002;3:745–756. doi: 10.1016/s1534-5807(02)00321-0. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Mager J, Magnuson T. Dynamic morphogenetic events characterize the mouse visceral endoderm. Dev Biol. 2003;261:470–487. doi: 10.1016/s0012-1606(03)00302-6. [DOI] [PubMed] [Google Scholar]
- Rodriguez TA, Casey ES, Harland RM, Smith JC, Beddington RS. Distinct enhancer elements control Hex expression during gastrulation and early organogenesis. Dev Biol. 2001;234:304–316. doi: 10.1006/dbio.2001.0265. [DOI] [PubMed] [Google Scholar]
- Rodriguez TA, Srinivas S, Clements MP, Smith JC, Beddington RS. Induction and migration of the anterior visceral endoderm is regulated by the extra-embryonic ectoderm. Development. 2005;132:2513–2520. doi: 10.1242/dev.01847. [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Oki S, Tanaka C, Nakamura T, Adachi H, Yan YT, Shen MM, Hamada H. Two nodal-responsive enhancers control left-right asymmetric expression of Nodal. Dev Dyn. 2005;232:1031–1036. doi: 10.1002/dvdy.20192. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Rodriguez T, Clements M, Smith JC, Beddington RS. Active cell migration drives the unilateral movements of the anterior visceral endoderm. Development. 2004;131:1157–1164. doi: 10.1242/dev.01005. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern CD, Downs KM. The hypoblast (visceral endoderm): an evo-devo perspective. Development. 2012;139:1059–1069. doi: 10.1242/dev.070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey DW, Clements M, Di-Gregorio A, Senner CE, Le Tissier P, Srinivas S, Rodriguez TA. Coordination of cell proliferation and anterior-posterior axis establishment in the mouse embryo. Development. 2011;138:1521–1530. doi: 10.1242/dev.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka K, Hamada H. Cell fate decisions and axis determination in the early mouse embryo. Development. 2012;139:3–14. doi: 10.1242/dev.060095. [DOI] [PubMed] [Google Scholar]
- Takaoka K, Yamamoto M, Hamada H. Origin and role of distal visceral endoderm, a group of cells that determines anterior-posterior polarity of the mouse embryo. Nat Cell Biol. 2011;13:743–752. doi: 10.1038/ncb2251. [DOI] [PubMed] [Google Scholar]
- Takaoka K, Yamamoto M, Shiratori H, Meno C, Rossant J, Saijoh Y, Hamada H. The mouse embryo autonomously acquires anterior-posterior polarity at implantation. Dev Cell. 2006;10:451–459. doi: 10.1016/j.devcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Thomas P, Beddington R. Anterior primitive endoderm may be responsible for patterning the anterior neural plate in the mouse embryo. Curr Biol. 1996;6:1487–1496. doi: 10.1016/s0960-9822(96)00753-1. [DOI] [PubMed] [Google Scholar]
- Varlet I, Collignon J, Robertson EJ. nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development. 1997;124:1033–1044. doi: 10.1242/dev.124.5.1033. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 2003;17:1646–1662. doi: 10.1101/gad.1100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Norris DP, Le Good JA, Constam DB, Robertson EJ. Asymmetric Nodal expression in the mouse is governed by the combinatorial activities of two distinct regulatory elements. Mech Dev. 2004;121:1403–1415. doi: 10.1016/j.mod.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Beppu H, Takaoka K, Meno C, Li E, Miyazono K, Hamada H. Antagonism between Smad1 and Smad2 signaling determines the site of distal visceral endoderm formation in the mouse embryo. J Cell Biol. 2009;184:323–334. doi: 10.1083/jcb.200808044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Saijoh Y, Perea-Gomez A, Shawlot W, Behringer RR, Ang SL, Hamada H, Meno C. Nodal antagonists regulate formation of the anteroposterior axis of the mouse embryo. Nature. 2004;428:387–392. doi: 10.1038/nature02418. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Morris SA, Bruce AW. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet. 2009;10:467–477. doi: 10.1038/nrg2564. [DOI] [PubMed] [Google Scholar]