Abstract
Aim
To evaluate patients’ expectations regarding the perceived utility of whole-genome sequencing (WGS).
Materials & methods
We used latent class analysis to characterize individuals enrolled in the MedSeq Project based on their perceived utility of WGS. Multinomial logistic regression was used to evaluate associations between participant characteristics and latent classes.
Results
Findings characterized participants into one of three perceived utility groups: enthusiasts, who had a high probability of agreement with all utility items (23%); health conscious, who perceived utility in medically related areas (60%) or skeptics, who had a low probability of agreement with utility items (17%). Trust significantly predicted latent class.
Conclusion
Understanding differences in perceived utility of WGS may inform strategies for uptake of this technology.
Keywords: clinical utility, patient perspectives, perceived utility, personal utility, whole-genome sequencing
As whole-genome sequencing (WGS) is being integrated into clinical practice [1,2], there is a growing interest in the utility of this technology [3]. While testing for genetic conditions has been a part of routine clinical care for several decades, these tests were designed for specific genes and conditions, such as hereditary breast and ovarian cancer (BRCA1, BRCA2), Li-Fraumeni syndrome (TP53), familial hypertrophic cardiomyopathy (MYH7) and familial hypercholesterolemia (PCSK9) [4]. However, as the process of WGS is rapidly refined, the possibility for using one’s entire genome for a personalized approach to wellness appears attainable [3,5].
In the context of genetic testing, clinical utility has been defined as how likely the test is to significantly improve patient outcomes [5–7]. Given the broad scope of information that WGS will produce on an individual, in addition to the lack of consensus about how to accurately assess the clinical utility of genomic tests, some have argued for expanding the definition of utility to include personal utility [5]. Personal utility refers to using genomic information for decisions, actions or self-understanding that may be health-related, but may not have direct medical benefit [5]. While debate remains about the possibility of expanding the definition of utility to include information that may have no direct medical benefit [8], to date, little is known about patients’ perceived utility of WGS [5]. Understanding patients’ perceived utility of WGS is important because it will influence uptake of this new technology [9], as well as how individuals are likely to respond to WGS results [5,9].
In addition to understanding patients’ perceived utility of WGS, there have been few, if any, assessments exploring whether individual patient characteristics influence their perceptions about its utility. The objective of this study was to characterize perceived utility of WGS among participants enrolled in the MedSeq Project, a randomized controlled trial exploring the impact of integrating genomic medicine into the clinical care of primary care and cardiology patients [10]. Additionally, we evaluated whether patient attitudes, behaviors and demographic factors predict one’s perceived utility of WGS.
Materials & methods
Study overview
Details of the MedSeq Project have been described previously [10,11]. Briefly, cardiologists, primary care physicians and their patients were enrolled in the MedSeq Project. Both physicians and patients were evaluated with surveys and interviews at multiple time points, and patients were randomized to receive either standard of care (family history review or targeted genetic testing for patients with cardiomyopathies) or standard of care plus WGS. In this report, we focus on patients’ perceived utility of WGS at baseline, prior to receiving their WGS results or learning their randomization status. The Baylor College of Medicine Institutional Research Board and the Partners Human Research Committee approved this study.
Study design & participants
At the baseline visit, participants completed a baseline survey, an online family history assessment, underwent a blood draw and were randomized to either the WGS or standard of care arm (control arm). In total, 100 adults in the primary care cohort and 102 adults in the cardiology cohort completed the baseline visit. Primary care participants were between 40 and 70 years of age and generally healthy (individuals with cardiovascular disease or diabetes were specifically excluded), without an indication for genetic testing. Cardiology participants were >18 years of age, had a clinical diagnosis of hypertrophic or dilated cardiomyopathy and had previous or concurrent targeted genetic testing for their condition.
Measurements
Our outcome of interest was perceived utility of WGS. Specifically, the following six items were used to measure perceived utility, Do you think your study results will: accurately identify your disease risk; influence what treatment you receive for your current or future medical problems; influence decisions you make about your medical care; influence your or your child’s reproductive decisions; influence what medications you take and influence your end-of-life planning. Response options to these questions were yes, probably yes, probably no and no. Predictors of perceived utility that were evaluated included: gender; age; marital status; education; income; study cohort (primary care vs cardiology); genomic knowledge (adapted with permission from Kaphingst et al. [12]); and intolerance of uncertainty [13]. Additionally, we assessed associations with trust, motivations for participation in the study and concerns about genetic test results. These attitude scales were constructed specifically for this analysis and were computed as summed totals of item responses based upon results from exploratory factor analyses. A listing of the items used to construct the trust, motivation and concern scales can be found in the Supplementary Materials. Trust in the context of our study was a composite of six novel items included on the baseline survey. Responses on the individual items ranged from strongly disagree (coded as 0) to strongly agree (coded as 4), and reflected trust in physicians (e.g., “My physician has enough knowledge to help me interpret my genetic information”); medical researchers (e.g., “I trust doctors who do medical research”) and the study investigators’ confidentiality of genetic information (e.g., “I trust that neither my identity nor my genetic information will be shared with third parties without my permission”). Finally, participants were evaluated on current versus future utility of WGS. Specifically, participants were asked, on a scale of 1–10, how useful do you think WGS will be for managing your health, both now and in the future.
Analysis
Summary statistics were generated for the following demographic characteristics: age (<45 years, 45–54 years, 55–64 years or ≥65 years); gender (male or female); race/ethnicity (non-Hispanic white or other); marital status (not married or living with partner); education level (college graduate, yes or no); annual household income (<US$100,000 or ≥US$100,000); and previous genetic testing (yes or no). Mean scores and standard deviations were determined for genetic knowledge, and intolerance of uncertainty as well as the scales measuring trust, motivation and concern. Differences in these characteristics by study cohort (primary care vs cardiology) were evaluated using chi-squared tests for categorical variables and t-tests for continuous variables at a significance level of 0.05.
Initial descriptive statistics for the six perceived utility items suggested that participants differed more in their pattern of responses to these items than in magnitude (low to high utility). To examine this possibility, a latent class analysis was conducted to identify salient patterns on this construct. Items were first dichotomized by collapsing ‘yes’ and ‘probably yes’ into ‘yes’ and ‘no’ and ‘probably no’ into ‘no’. Mplus 7.2 [14] was used to conduct both the latent class and subsequent prediction models. Solutions with two through five latent classes were inspected for fit and interpretability. Fit was assessed using information criteria (Akaike: AIC; Bayesian Information Criteria: BIC; and the sample size Adjusted BIC: aBIC) as well as the Vuong-Lo-Mendell-Rubin Likelihood Ratio Test (LMR-LRT) and the Bootstrapped LRT. For the information criteria, lower values indicate better fit. The likelihood ratio tests assess whether a model with k categories fits significantly better than one with k-1; therefore a significant LRT indicates that the model with 1 less category is more likely to adequately reproduce the data [15]. Because fit indices do not always provide a clear-cut answer on fit, interpretability of the results and size of the categories were also used to determine the optimal number of categories [16]. Once an acceptable latent class solution was obtained, a multinomial logistic regression model was fit to assess the relationship between the predictors and the classes defined by the latent class model. Odds ratios (OR) and 95% CI were calculated, along with p-values to evaluate the association between selected predictors and perceived utility classes. Differences between latent classes on current versus future utility of WGS were assessed using one-way analysis of variance (ANOVA).
Results
Primary care and cardiology participants’ sociodemographic characteristics are summarized in Table 1. Across the two cohorts, participants were predominantly white, middle-aged with college degrees and reported high annual household incomes. The only differences between cohorts were on age, gender, income and experience with genetic testing (p < 0.05).
Table 1.
Characteristics of the MedSeq Project participants by study cohort.
| Characteristics | Total (n = 202) | Study cohort | p-value | |
|---|---|---|---|---|
| Primary care (n = 100) | Cardiology (n = 102) | |||
| Socio-demographic | ||||
| Age (years), n (%): | <0.001 | |||
| – <45 | 33 (17.9) | 11 (11.0) | 22 (21.6) | |
| – 45–54 | 51 (25.4) | 32 (32.0) | 19 (18.6) | |
| – 55–64 | 79 (39.1) | 51 (51.0) | 28 (27.5) | |
| – ≥65 | 39 (19.3) | 6 (6.0) | 33 (32.4) | |
| – Mean (SD) | 55.3 (11.3) | 54.8 (7.3) | 55.9 (14.2) | 0.495 |
| Gender, n (%): | 0.035 | |||
| – Male | 101 (50.0) | 42 (42.0) | 59 (57.8) | |
| – Female | 101 (50.0) | 58 (58.0) | 43 (42.2) | |
| Race/ethnicity, n (%): | 1.000 | |||
| – Non-Hispanic white | 177 (87.6) | 87 (87.0) | 90 (88.2) | |
| – Other | 25 (12.4) | 13 (13.0) | 12 (11.8) | |
| Marital status, n (%): | 1.000 | |||
| – Not married | 52 (25.7) | 26 (26.0) | 26 (25.5) | |
| – Married or living with partner | 150 (74.3) | 74 (74.0) | 76 (74.5) | |
| Education level, n (%): | 0.104 | |||
| – Did not graduate from college | 38 (18.8) | 14 (14.0) | 24 (23.5) | |
| – College graduate | 164 (81.2) | 86 (86.0) | 78 (76.5) | |
| Annual household income, n (%): | 0.003 | |||
| – <US$100,000 | 71 (36.4) | 26 (26.5) | 45 (46.4) | |
| – ≥US$100,000 | 124 (63.6) | 72 (73.5) | 52 (53.6) | |
| Other characteristics | ||||
| Previous genetic testing, n (%): | <0.001 | |||
| – No | 136 (67.3) | 89 (89.0) | 47 (46.1) | |
| – Yes | 59 (29.2) | 7 (7.0) | 52 (51.0) | |
| – Unsure | 7 (3.5) | 4 (4.0) | 3 (2.9) | |
| Genetic knowledge, mean (SD) | 10.0 (1.1) | 10.0 (1.2) | 10.0 (1.1) | 0.962 |
| Intolerance of uncertainty, mean (SD) | 27.9 (8.2) | 28.0 (8.4) | 27.8 (8.0) | 0.844 |
| Motivation, mean (SD) | 10.7 (2.6) | 10.7 (2.6) | 10.6 (2.5) | 0.718 |
| Concerns, mean (SD) | 8.2 (5.1) | 8.7 (4.8) | 7.6 (5.4) | 0.129 |
| Trust, mean (SD) | 17.4 (3.2) | 16.9 (3.5) | 17.8 (2.8) | 0.068 |
| WGS report useful now, mean (SD) | 7.2 (2.2) | 7.1 (2.3) | 7.4 (2.1) | 0.315 |
| WGS report useful future, mean (SD) | 8.3 (1.8) | 8.3 (1.9) | 8.3 (1.7) | 0.864 |
Motivation, concerns and trust are each six item scales; concerns and trust have a potential range of 0–24, where 0 = no concern/no trust and 24 = high concern/high trust; motivation has a potential range of 0–18, where 0 = no motivation and 18 = high motivation. Bold p-values represent statistically significant results.
SD: Standard deviation; WGS: Whole-genome sequencing.
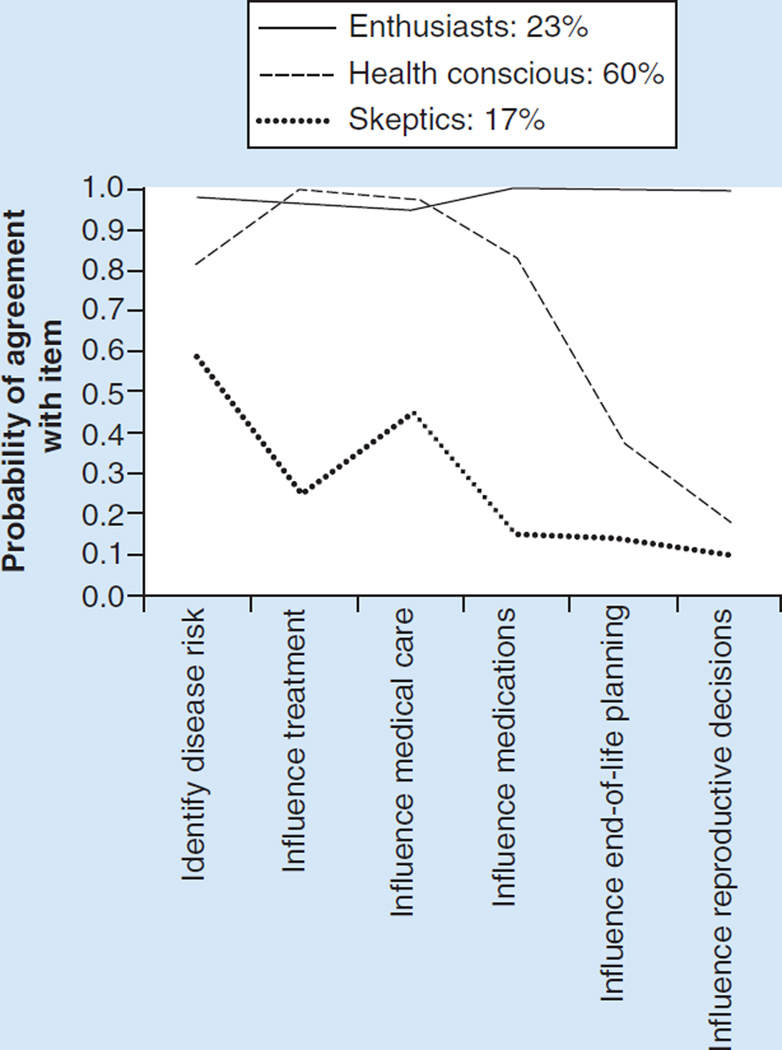
Table 2 provides the fit statistics for the latent class analysis on the perceived utility items. Specifically, we explored two, three, four and five class solutions for categorizing participants based on their perceived utility of WGS. A three class solution was determined to be optimal based on both fit indicators and interpretability. Figure 1 shows the three classes that were identified. Class 1, which we call ‘enthusiasts’ (n = 47; 23%), is defined by a high probability of agreement with all of the perceived utility items. The second class we labeled ‘health conscious’ (n = 121; 60%) as they perceived WGS to have utility in medically focused items but not for reproductive or end-of-life planning, which are related to personal and not clinical utility. The third class are ‘skeptics’ (n = 34; 17%) as they had the lowest probability of agreement with any utility items.
Table 2.
Indices of fit for the latent class analysis.
| Model Parameters | Two class solution | Three class solution | Four class solution | Five class solution |
|---|---|---|---|---|
| LL | −558.276 | −540.205 | −533.365 | −527.758 |
| AIC | 1142.551 | 1120.41 | 1120.73 | 1123.517 |
| BIC | 1185.299 | 1186.176 | 1209.51 | 1235.318 |
| aBIC | 1144.115 | 1122.816 | 1123.977 | 1127.606 |
| LMR-LRT (2* LL diff) | 141.262 (p <0.0001) | 36.141 (p <0.0001) | 13.681 (p = 0.02) | 11.213 (p = 0.041) |
| Boostrap LRT | 141.262 (p <0.0001) | 36.141 (p <0.0001) | 13.320 (p = 0.02) | 10.918 (p = 0.045) |
| Entropy | 0.803 | 0.832 | 0.907 | 0.974 |
The LL provides an indication of how well the model parameters are able to reproduce the data; in general higher values (closer to 0) indicate better fitting models. The AIC, BIC, and aBIC are information criteria used to assess comparative model fit; in general smaller values indicate a better fit, however, in latent class analysis the BIC has been found to underestimate the number of categories according to Nylund et al. [15] and often the optimal solution has one more class than indicated by BIC. The LMR-LRT and Bootstrap LRT test the null hypothesis that the model with one less class (k-1) fits adequately; thus a rejection of the null for a k class solution indicates that the k-1 class is preferred. A significance level of 0.01 was chosen when comparing solutions using the LMR-LRT and Bootstrap LRT. Entropy is an indicator of classification uncertainty and values closer to 1.0 indicate less uncertainty.
aBIC: Sample size Adjusted Bayesian Information Criteria; AIC: Akaike; BIC: Bayesian Information Criteria; LL: Log Likelihood; LMR-LRT: Vuong-Lo-Mendell-Rubin Likelihood Ratio Test; LRT: Likelihood Ratio Test.
Figure 1.
Probability of agreement with utility items by perceived utility class.
Predictors of perceived utility class (i.e., enthusiasts, health conscious or skeptics) are presented in Table 3. Trust was the only significant predictor across perceived utility classes. Specifically, enthusiasts were more likely to be trusting compared with skeptics (OR: 1.23; 95% CI: 1.02–1.48; p = 0.033), and the health conscious were also more likely to be trusting compared with skeptics (OR: 1.20; 95% CI: 1.02–1.40; p = 0.027). While enthusiasts had more motivations for participation in the study compared with skeptics (OR: 1.31; 95% CI: 1.01–1.70; p = 0.044), the same was not true for the health conscious compared with skeptics (OR: 1.16; 95% CI: 0.93–1.44; p = 0.184). Factors including gender, age, income, education, study cohort and genomic knowledge were not significant predictors of participants’ perceived utility class.
Table 3.
Multinomial logistic regression analysis predicting perceived utility class.
| Characteristic | Enthusiasts vs skeptics (reference) | The health conscious vs skeptics (reference) |
||||
|---|---|---|---|---|---|---|
| OR | 95% CI | p-value | OR | 95% CI | p-value | |
| Gender | 1.92 | 0.56–6.59 | 0.301 | 2.18 | 0.75–6.32 | 0.151 |
| Age | 1.00 | 0.95–1.05 | 0.917 | 0.99 | 0.95–1.04 | 0.690 |
| Income | 1.09 | 0.77–1.54 | 0.622 | 1.32 | 0.98–1.78 | 0.070 |
| Education | 0.84 | 0.32–2.16 | 0.711 | 1.28 | 0.57–2.87 | 0.547 |
| Marital status | 0.74 | 0.18–3.07 | 0.674 | 0.89 | 0.26–3.00 | 0.847 |
| Study cohort | 0.88 | 0.27–2.84 | 0.829 | 0.84 | 0.32–2.25 | 0.732 |
| Genetic knowledge | 0.78 | 0.42–1.45 | 0.430 | 0.62 | 0.35–1.08 | 0.092 |
| Intolerance of uncertainty | 1.01 | 0.93–1.09 | 0.877 | 0.98 | 0.92–1.05 | 0.516 |
| Motivations | 1.31 | 1.01–1.70 | 0.044 | 1.16 | 0.93–1.44 | 0.184 |
| Concerns | 1.12 | 0.98–1.28 | 0.092 | 1.08 | 0.96–1.21 | 0.193 |
| Trust | 1.23 | 1.02–1.48 | 0.033 | 1.20 | 1.02–1.40 | 0.027 |
Bold p-values represent statistically significant results.
OR: Odds ratio.
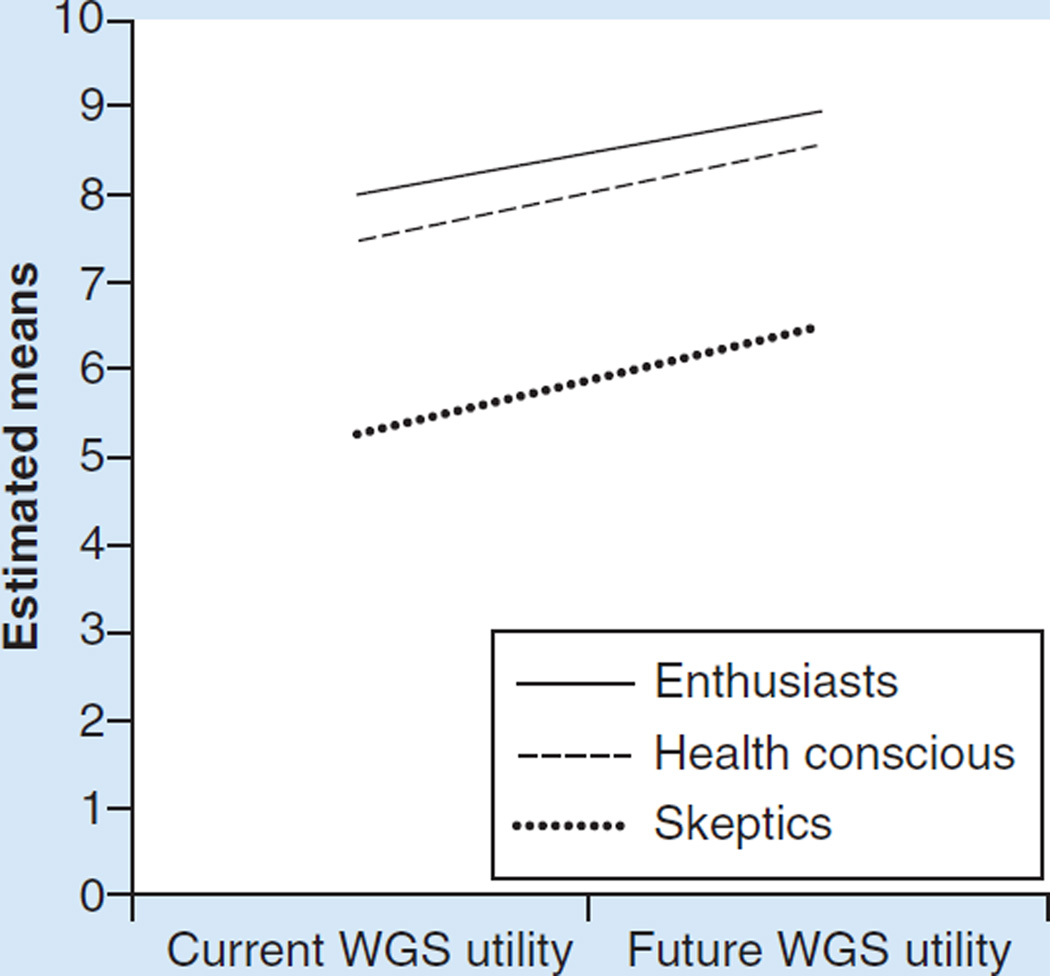
There were significant differences by perceived utility class both on current (p < 0.001) and future utility (p < 0.001). In pairwise comparisons, enthusiasts did not differ from the health conscious. However, both groups differed significantly (p < 0.001) from skeptics. While enthusiasts had the highest responses for both current and future utility, each group believed the utility would increase over time (Figure 2).
Figure 2.
Current and future utility of whole-genome sequencing by perceived utility class.
WGS: Whole-genome sequencing.
Discussion
Overall, most participants in the MedSeq Project perceived some utility in using WGS for health-related decision making. However, our results suggested that there were nuances to participants’ perceived utility as we found participants varied in their pattern of responses and fell into one of three groups. Specifically, enthusiasts were likely to agree with all perceived utility items. The health conscious, who made up the largest group of participants, perceived WGS would have utility in medical care, treatment and the identification of disease risk, but that it would not likely play a role in end-of-life or reproductive planning. Finally, skeptics were the smallest group and did not perceive much utility in any WGS items. While there were differences between enthusiasts, the health conscious and skeptics in terms of their perceived utility of WGS, each group indicated that WGS information would be more useful in the future.
A notable difference between enthusiasts and the health conscious is that enthusiasts perceived utility in WGS for decisions related to reproduction and end-of-life planning. As these items are related to personal utility rather than clinical utility [5], it may be that enthusiasts perceive a broader definition of utility of WGS, even beyond their physicians’ perceived utility of this technology [11]. If confirmed, it will be important for physicians to understand that some patients may expect to make more personal decisions based on WGS, which may necessitate a broader discussion of the nonclinical implications of this information.
The only factor that was significantly different across classes was trust. While trust did not differ based on study cohort (primary care vs cardiology), it was significantly higher among enthusiasts and health conscious when compared with skeptics. As trust items included one’s trust in the interpretation and dissemination of genetic information by health professionals, our findings that skeptics, who were the least likely to perceive utility in WGS, appear to be consistent with the notion that those least enthusiastic about the potential of WGS are those who are least likely to have trust in the system for translating WGS in terms of health-related decisions. Therefore, it is possible that as clinicians become more familiar with WGS, and the contextualization of these data with their patients, individuals who previously perceived little utility in this information may improve their outlook. As the importance of trust has been suggested as being related to one’s attitudes about receiving individual genomic research results [17,18], future research will need to examine perceived utility of WGS among patients with historical distrust of medicine, doctors and biomedical research.
To our knowledge, this is the first study examining the views of patients on the use of WGS in the context of routine clinical practice. Physician perspectives on the utility of WGS have been previously evaluated using data from the MedSeq Project [11]. Physicians noted that WGS would likely improve risk stratification of patients, lead to better prevention strategies and inform pharmacogenomics strategies. Additionally, like participants evaluated in this analysis, physicians did expect the utility of WGS to increase in the future [11]. Therefore, both patients and physicians in this study largely see the promise and limitations of WGS in its current state.
Our study must be considered in light of certain limitations. One potential limitation is that our population is not representative of the general population in terms of sociodemographic factors. The MedSeq Project population is highly knowledgeable about genomic concepts, well-educated and relatively affluent. This may have influenced their overall sense of perceived utility of WGS. Additional research in more diverse populations will be necessary to inform the future of genomics practice. Another potential weakness is the sample size. For instance, we were limited in our ability to conduct subgroup analyses and to make conclusions on moderate differences between groups. However, our assessment is an important first step in characterizing how individuals perceive the utility of WGS.
Conclusion
WGS and other genome sequencing technologies are increasingly being integrated into clinical care as diagnostic and screening tools. While clinical utility can be clearly defined in relationship to improved patient outcomes, some have argued that genomic sequencing may have other types of utility, such as personal utility [5], and that uptake of these approaches will partially depend on patients’ perceived utility of these technologies [9]. There were differences in the MedSeq Project population in terms of perceived utility, which suggests that understanding these expectations based on patient characteristics may inform how to address misconceptions about WGS and how to target specific groups for pre-WGS counseling. Ultimately, a valuable aspect of the MedSeq Project will be to evaluate how different categories of individuals (enthusiasts, health conscious and skeptics) eventually incorporate WGS in their health-related decisions.
Future perspective
Rapid advances in genomic technology will likely change the way patients utilize this information in their approach to healthcare and wellness. However, how patients’ perceive the utility of this technology remains to be defined. Understanding how individuals incorporate information from WGS into their daily lives remains a next frontier in genomic medicine.
Supplementary Material
Executive summary.
Background
While whole-genome sequencing (WGS) is becoming more common in clinical practice, little is known about patients’ perspectives on the perceived utility of this technology.
Methods
We evaluated expectations regarding the perceived utility of WGS among patients enrolled in the MedSeq Project.
Surveys administered at enrollment assessed multiple domains, including perceived utility and other patient characteristics.
Latent class analysis was used to categorize individuals based on their perceived utility of WGS.
Multinomial logistic regression was used to evaluate associations between participant characteristics and latent classes.
Results
Participants were categorized into one of three classes: enthusiasts, who had a high probability of agreement with all perceived utility items (23%); health conscious, who perceived utility primarily in medically related, but not all, areas (60%); or skeptics, who had a low probability of agreement with all utility items (17%).
The only variable that was significantly different across classes was trust (odds ratio: enthusiasts vs skeptics = 1.23; 95% CI: 1.02–1.48; odds ratio: health conscious vs skeptics = 1.20; 95% CI: 1.02–1.40).
Discussion
Overall, most participants in the MedSeq Project perceived some utility in using WGS for health-related decision making.
Our results suggested that there were nuances to participants’ perceived utility as we found participants varied in their pattern of responses and fell into one of three groups.
Conclusion
Ultimately, a valuable aspect of the MedSeq Project will be to evaluate how different categories of individuals (enthusiasts, health conscious and skeptics) eventually incorporate WGS in their health-related decisions.
Acknowledgments
The authors thank the members and participants of the MedSeq Project for their important contributions. The authors thank 5AM Solutions, Inc. (Rockville, MD, USA), for their help in customizing the workflow of the “My Family Health Portrait” web tool for this study.
This work was supported by the US NIH (U01HG006500). RC Green’s research is supported by grants from NIH, DOD and Illumina. RC Green has received compensation for advisory services or speaking from Invitae, Prudential, Arivale, Illumina, AIA, Helix and Roche.
Footnotes
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/full/10.2217/pme.15.45
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Clinical Trials Registration: ClinicalTrials.gov #NCT01736566.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
- 1.Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet. Med. 2013;15(4):258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayer AN, Dimmock DP, Arca MJ, et al. A timely arrival for genomic medicine. Genet. Med. 2011;13(3):195–196. doi: 10.1097/GIM.0b013e3182095089. [DOI] [PubMed] [Google Scholar]
- 3.Green ED, Guyer MS. National Human Genome Research I. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470(7333):204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 4.Caskey CT, Gonzalez-Garay ML, Pereira S, McGuire AL. Adult genetic risk screening. Annu. Rev. Med. 2014;65:1–17. doi: 10.1146/annurev-med-111212-144716. [DOI] [PubMed] [Google Scholar]
- 5.Bunnik EM, Janssens AC, Schermer MH. Personal utility in genomic testing: is there such a thing? J. Med. Ethics. 2014;41(4):322–326. doi: 10.1136/medethics-2013-101887. [DOI] [PubMed] [Google Scholar]
- 6.Foster MW, Mulvihill JJ, Sharp RR. Evaluating the utility of personal genomic information. Genet. Med. 2009;11(8):570–574. doi: 10.1097/GIM.0b013e3181a2743e. [DOI] [PubMed] [Google Scholar]
- 7.Sanderson S, Zimmern R, Kroese M, Higgins J, Patch C, Emery J. How can the evaluation of genetic tests be enhanced? Lessons learned from the ACCE framework and evaluating genetic tests in the United Kingdom. Genet. Med. 2005;7(7):495–500. doi: 10.1097/01.gim.0000179941.44494.73. [DOI] [PubMed] [Google Scholar]
- 8.Eckstein L, Garrett JR, Berkman BE. A framework for analyzing the ethics of disclosing genetic research findings. J. Law. Med. Ethics. 2014;42(2):190–207. doi: 10.1111/jlme.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopits IM, Chen C, Roberts JS, Uhlmann W, Green RC. Willingness to pay for genetic testing for Alzheimer’s disease: a measure of personal utility. Genet. Test. Mol. Biomarkers. 2011;15(12):871–875. doi: 10.1089/gtmb.2011.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vassy JL, Lautenbach DM, McLaughlin HM, et al. The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials. 2014;15:85. doi: 10.1186/1745-6215-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vassy JL, Christensen KD, Slashinski MJ, et al. ‘Someday it will be the norm’: physician perspectives on the utility of genome sequencing for patient care in the MedSeq Project. Per. Med. 2015;12(1):23–32. doi: 10.2217/PME.14.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaphingst KA, McBride CM, Wade C, et al. Patients’ understanding of and responses to multiplex genetic susceptibility test results. Genet. Med. 2012;14(7):681–687. doi: 10.1038/gim.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buhr K, Dugas MJ. The Intolerance of Uncertainty Scale: psychometric properties of the English version. Behav. Res. Ther. 2002;40(8):931–945. doi: 10.1016/s0005-7967(01)00092-4. [DOI] [PubMed] [Google Scholar]
- 14.Muthen BO. Mplus. 2012 www.statmodel.com. [Google Scholar]
- 15.Nylund KL, Asparouhov T, Muthen BO. Deciding on the number of classes in latent class analysis and growth mixture modeling: a Monte Carlo simulation study. Structural Equation Modeling. 2007;14(4):535–569. [Google Scholar]
- 16.Masyn KE. Statistical analysis. In: Little TD, editor. The Oxford Handbook of Quantitative Methods. Oxford, UK: Oxford University Press; 2013. [Google Scholar]
- 17.Hobbs A, Starkbaum J, Gottweis U, Wichmann HE, Gottweis H. The privacy-reciprocity connection in biobanking: comparing German with UK strategies. Pub. Health Gen. 2012;15(5):272–284. doi: 10.1159/000336671. [DOI] [PubMed] [Google Scholar]
- 18.Lemke AA, Halverson C, Ross LF. Biobank participation and returning research results: perspectives from a deliberative engagement in south side Chicago. Am. J. Med. Genet. A. 2012;158A(5):1029–1037. doi: 10.1002/ajmg.a.34414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.