Abstract
We explored the association between baseline gut microbiota (16S rRNA biomarker sequencing of stool samples) in 17 relapsing-remitting pediatric MS cases and risk of relapse over a mean 19.8 months follow-up. From the Kaplan-Meier curve, 25% relapsed within an estimated 166 days from baseline. A shorter time to relapse was associated with Fusobacteria depletion (p=0.001 log-rank test), expansion of the Firmicutes (p=0.003), and presence of the Archaea Euryarchaeota (p=0.037). After covariate adjustments for age and immunomodulatory drug exposure, only absence (vs presence) of Fusobacteria was associated with relapse risk (hazard ratio=3.2 (95%CI:1.2-9.0), p=0.024). Further investigation is warranted. Findings could offer new targets to alter the MS disease course.
Keywords: pediatric multiple sclerosis, gut microbiota, 16S rRNA, relapse risk, survival analyses, Kaplan-Meier, Cox regression
Background
Gut microbiota perturbations have been associated with disease activity in animal models of MS1,2,3 but the association with activity in MS subjects is unknown. In animal models representing relapsing-remitting MS, for instance, a germ-free environment has been associated with a milder disease course.1,2 In addition, oral administration of members of the Bacteroides phylum (Bacteroides Fragilis) have been associated with a lower ‘clinical’ score in relapsing models of MS.3,4 Currently, relatively little is known as to what might trigger or facilitate the onset of a new MS relapse. Environmental exposures such as stress, season, sunlight, vitamin D and recent viral infections have been linked to risk of relapse, with the presumed pathway(s) being through immune system modulation. Interestingly, these environmental factors also influence the gut microbiota which likewise modulates the immune system which is known to be affected in MS.1,2,3 Further, differences have been observed in the gut microbiota of individuals with and without MS, 3,5-8 including pediatric MS.5 Pediatric MS offers opportunity to study disease processes in the very early stages of MS, relatively close to the actual biological onset of disease, potentially limiting confounders. We explored the association between gut microbiota profiles in early pediatric MS and subsequent relapse risk.
Methods
Cohort selection
Children ≤18 years old with a first demyelinating event and at least 2 silent brain lesions or relapsing-remitting MS (McDonald criteria) attending a University of California, San Francisco (UCSF) pediatric MS clinic provided a baseline stool sample, as described previously.5 At baseline, all cases were within 2 years of symptom onset with no systemic antibiotic exposure in the previous 2 months.
Capture of clinical and demographic data
Baseline characteristics captured included demographic (e.g. age, sex), and clinical (e.g. disease duration, immunomodulatory drug (IMD) exposure). After stool collection, physician confirmed relapses were determined via structured forms and chart review by abstractors unaware of the child’s gut microbiota profile.
DNA extraction and 16S rRNA sequencing
DNA was extracted from stool using a cetyl trimethylammonium bromide method9 and the V4 region of the 16S rRNA gene was amplified in triplicate,5,10 combined, purified and pooled in equimolar concentrations prior to sequencing on the Illumina MiSeq platform. Reads were clustered at ≥97% similarity into operational taxonomic units and singly rarefied to 201,546 reads per sample. Taxonomy was assigned using the Greengenes database via QIIME (Quantitative Insights Into Microbial Ecology).11
Statistical analyses
The gut microbiota formed the exposure, expressed as phylum-level relative abundance and categorized according to the data distribution as either ‘absent vs. present,’ or ‘low vs high’ (≤ vs. >median) when detectable in >90% of cases. Phyla with sparse data were excluded (i.e. <20% of cases had detectable reads).
The outcome was the first on-study (post-baseline) relapse. Relapse-free cases were censored at their last clinic visit. Associations between each exposure and the outcome were explored using Kaplan-Meier curves, with the log-rank test to compare groups. After applying a conservative Bonferroni correction for multiple comparisons, those phyla remaining significant were assessed though multivariable Cox regression models, adjusting for potential confounders, including age and IMD drug exposure status (see Appendix, online).
In a sensitivity analysis, any child with an attack (either the onset attack or a relapse) within 30 days pre-stool sample was excluded and the log-rank tests were repeated.
Statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS Ver.22.0.NY:IBM Corp.2013). UCSF’s Institutional Review Board approved the study.
Results
Of the originally reported 18 MS cases,5 17 had up to 41.6 months (mean=19.8 months) post-baseline follow-up (one left the country and was excluded). Cohort characteristics are shown in the Table, additional characteristics (e.g. diet, body mass index) are available online (Supplementary Table). During follow-up, 7 relapsed (9 relapses were recorded in total from baseline). From Kaplan-Meier curves, 25% of cases relapsed within an estimated 166 days from baseline.
Table 1. Baseline characteristics of the pediatric multiple sclerosis (MS) casesϕ.
| Characteristic, n (%) unless stated otherwise | MS cases, n=17 |
|---|---|
| Sex: Girl | 10 (59%) |
| Boy | 7 (41%) |
| Age, years: mean (SD; range) | 12.5 years (SD=4.57; 4-17) |
| Age: ≤12 years old | 5 (29%) |
| >12 years old | 12 (61%) |
| Race: White | 8 (47%) |
| Non-white | 9 (53%) |
| Ethnicity: Hispanic | 8 (47%) |
| Non- Hispanic | 9 (53%) |
| Co-morbid condition [a]: present | 7 (41%) |
| Absent | 10 (59%) |
| MS-Specific clinical characteristics | |
| Age at MS symptom onset, years: mean (SD; range) | 12.1 years (SD=4.8; 4-17) |
| Disease duration [b], months: mean (SD; range) | 10.3 months (SD=6.6; 2.3-23.1) |
|
Time since last relapse or onset attack (onset attack considered): days: mean (SD; range) |
183 days (SD=140; 4 to 489 days) |
| Disability level - EDSS at enrolment, median (range) | 2.0 (0-4.0) |
| 0-<2.0 | 7 |
| 2.0-<3.0 | 7 |
| 3.0+ | 3 |
| Immunomodulatory drug exposure status [c]: IMD nai̇ve | 8 (47%) |
| IMD exposed | 9 (53%) |
| Corticosteroids – systemic [d]: No | 11 (65%) |
| Yes | 6 (35%) |
| Available prospective follow-up‡, months: mean (SD; range) | 19.8 months (SD=12.0; 1.8-41.6) |
Key: SD=standard deviation; EDSS=Expanded Disability Status Scale score; IMD=immunomodulatory drug
data shown are in relation to baseline (i.e. date of stool sample collection) unless otherwise stated. EDSS was assessed at the clinic visit nearest to the stool sample, i.e. at enrollment into the study
all prospective follow-up was expressed regardless if (or when) a relapse occurred, with the study end being the last clinic visit or contact for each child
the comorbid conditions for the 7 children were: headache, atopic dermatitis/eczema, long-term constipation, history of shingles, seizures, reactive airways disease and headache, scoliosis
disease duration: time from symptom onset to baseline (stool collection)
‘IMD naïve’ indicates never exposed pre-baseline. ‘IMD exposed’ indicates ever exposed pre-baseline. At baseline, all IMD exposed cases were still on an MS drug as follows: beta-interferon (n=3); glatiramer acetate (n=5); natalizumab (n=1). No child had switched or stopped an IMD (although one child had previously been exposed to plasma exchange before taking glatiramer acetate).
within the previous 2 months
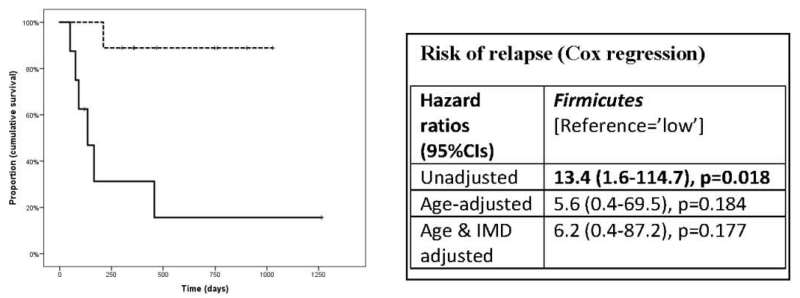
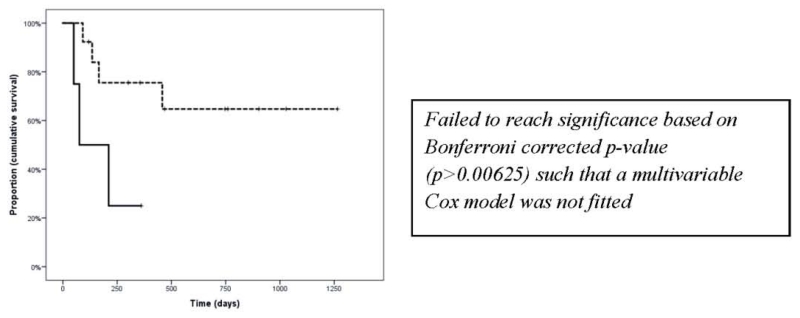
Eight of 13 phyla identified had sufficient data to be examined (i.e. detected in >20% cases). A shorter time to relapse was associated with absence of Fusobacteria (p=0.001, log-rank test), a higher abundance of Firmicutes (p=0.003) and presence of the Archaea Euryarchaeota (p=0.037), see Figure. No remarkable associations were observed with the remaining phyla (Bacteroidetes, Actinobacteria, Verrucomicrobia, Proteobacteria and Tenericutes) and time to relapse (all p>0.05, data not shown). Only two phyla (Firmicutes and Fusobacteria) reached significance after a Bonferroni correction (p<0.00625) and Cox regression models were built for these only (see Figure). After covariate adjustments, the Fusobacteria phylum remained significant; its absence (vs presence) being associated with a 76% (95%CI:55%-90%) chance of an earlier relapse (HR=3.2 (95%CI:1.2-9.0), p=0.024 age and IMD exposure adjusted).
Figure 1. Association between gut microbiota (phylum-level) and relapse for: Fusobacteria [panel A, top], Firmicutes [panel B: middle] and Euryarchaeota [panel C, bottom].
Panel A: Fusobacteria: Kaplan-Meier curves (left), absent (dashed line) vs. present (solid line). p=0.001, log-rank test.
Panel B: Firmicutes: Kaplan-Meier curves (left), lower relative abundance (dashed line) vs. higher (solid line), p=0.003. log-rank test.
Panel C Euryarchaeota: Kaplan-Meier curves (left), absent (dashed) vs present (solid linegreen), p=0.037, log-rank test.
Key: IMD=immunomodulatory drug.
Hazard ratios indicate risk of relapse from baseline and are derived from Cox regression hazards models. Age at baseline (stool collection) and IMD exposure at baseline (exposed vs. naïve) were used to adjust models as shown. Bold indicates p<0.05.
Binomial categories for each phylum were created based on the data distribution as either absent versus present or high versus low (≤ vs. > median relative abundance). Of the Fusobacterium phyla identified, the genera were either Fusobacterium or Leptotrichia (genus was the lowest taxonomic level available).
In the sensitivity analyses, exclusion of the participant with a relapse within 30 days prior to the stool sample did not change the direction of findings; the differences were more significant for Firmicutes (p=0.002, log-rank test) and Fusobacteria (p=0.00033) but not Euryarchaeota (p=0.041).
Discussion
Findings suggest that gut microbiota composition may be associated with subsequent relapse risk. Absence of Fusobacteria was associated with over three times the hazard of an earlier relapse relative to a child with measurable levels of this phylum (see Figure). This remained significant after adjustment for potential confounders.
While data were suggestive that presence or higher abundance of Firmicutes and Euryarchaeota might also be associated with relapse, neither remained significant after either adjustment for multiple testing (Euryarchaeota) or potential confounders (Firmicutes). Nonetheless, as both these phylum have been associated with inflammatory conditions such as inflammatory bowel disease (Crohns) and obesity,12 it may be of value to consider these in larger studies. Our findings may help in the optimal design of future studies.
The literature for Fusobacteria, which houses a large number a distinct species, is mixed; its presence being associated with health (in animals13) and disease, including colorectal cancer,14 which interestingly, individuals with MS appear at lower risk of developing (relative to matched general population controls15). This suggests that additional analyses examining the specific Fusobacteria species associated with reduced risk of relapse is warranted.
Of the Fusobacteria identified, the genera were either Fusobacterium or Leptotrichia (genus was the lowest taxonomic level available). Fusobacteria comprise of anaerobic, gram negative bacteria, as does the major phylum Bacteroidetes. Members of the Bacteroidetes phylum, such as Bacteroides fragilis have been shown to ameliorate the animal model experimental autoimmune encephalomyelitis (EAE) via polysaccharide A expression.3,4 It is possible that a similar pathway explains the observed association with the Fusobacteria.
Our study included children very close to disease onset, i.e. the onset attack. It remains possible that there were residual effects on the gut microbiota from previous (pre-baseline) attacks (including the onset attack). Longitudinal stool sampling, combined with clinical data, are required to understand the impact of sequential relapses. It is possible that those with frequent relapses are caught in a self-perpetuating cycle of inflammation in which the gut microbiota represents a propagating factor and reservoir of pro-inflammatory signaling. Nonetheless, removing those with a relapse within 30 days pre-stool did not change the direction of findings. It is not possible to determine whether the relative absence of Fusobacteria resulted from outgrowth of another competing microbe or direct demise of multiple members of this phylum. A better understanding of the gut microbiota’s role in modifying MS relapse risk may identify novel drug targets and improved outcomes in MS.
Supplementary Material
Highlights.
Gut microbiota was associated with subsequent relapse risk in pediatric multiple sclerosis
Fusobacteria depletion increased relapse risk (hazard ratio=3.2;95%CI:1.2-9.0)
Findings could offer new targets to alter the MS disease course
Acknowledgements
Gratitude is extended to the families and children for participation in this study. Special thanks to Dr Yinshan Zhao for helpful comments and critical review of the manuscript
Funding Statement: This work was supported by the National MS Society RG4861A3/1 (PI Waubant), National Institutes of Health NS071463 (PI Waubant), The Race to Erase MS (PI Waubant) and the Canada Research Chair program (PI Tremlett). The funding source(s) had no role in the study design, collection, analysis or interpretation of the data, or in the decision to submit the article for publication.
Footnotes
Statistical analyses were performed by Helen Tremlett (University of British Columbia)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributorship statement: The corresponding author (HT) takes responsibility for the integrity of the data and the accuracy of the data analysis and had full access to the data. DF performed the gut microbiota analyses. HT performed the statistical analyses. EW, JH and JG facilitated sample and data collection. SR facilitated electronic linkage and quality assurance of data from environmental risk factors parent study. All authors were involved in the current study design, contributed to interpretation of data. SL and EW designed the original study and obtained funding. DF drafted the microbiota methods. HT drafted the remaining manuscript. All authors revised the manuscript and approved of the final version to be published.
Competing Interests Statements:
Helen Tremlett is funded by the Canada Research Chair program. She has received research support from the National Multiple Sclerosis Society, the Canadian Institutes of Health Research, and the UK MS Trust; speaker honoraria and/or travel expenses to attend conferences from the Consortium of MS Centres (2013), the National MS Society (2012, 2014), Bayer Pharmaceuticals (2010), Teva Pharmaceuticals (2011), ECTRIMS (2011, 2012, 2013, 2014), UK MS Trust (2011), the Chesapeake Health Education Program, US Veterans Affairs (2012), Novartis Canada (2012), Biogen Idec (2014), American Academy of Neurology (2013, 2014, 2015). Unless otherwise stated, all speaker honoraria were either donated to an MS charity or to an unrestricted grant for use by her research group.
Douglas Fadrosh has no disclosures
Ali Faruqi has no disclosures
Janace Hart has no disclosures
Shelly Roalstad has no disclosures
Jennifer Graves is funded by the Race to Erase MS and the National MS Society
Susan Lynch is funded by the NIH, Sloan Foundation, Cystic Fibrosis Foundation, Broad Foundation, Jannsen Pharmaceuticals, Gilead and Pfizer. She has recently or currently acts as an ad hoc consultant for Janssen Pharmaceuticals, Regeneron, Boston Consulting Group, Theravance and Novartis. She volunteers as a members of the Scientific Advisory Board of Second Genome and has received honoraria for lectures from American Thoracic Society, American Academy of Allergy Asthma and Immunology, Georgia Regents University, Alta Bates and Kaiser Permanente. She holds four patents and has received royalties for IP licensed by KaloBIos Inc.
Emmanuelle Waubant is funded by the NIH, the NMSS, and the Race to Erase MS. She has received honorarium for one educational lecture from Genentech. She volunteers on an advisory board for a Novartis trial. She has received honorarium or travel support from ACTRIMS, ECTRIMS, and AAN.
The US Network of Pediatric MS Centers (authors listed in alphabetical order): Greg Aaen1, Anita Belman2, Leslie Benson3,, Charlie Casper4, Tanuja Chitnis3, Mark Gorman3, Yolanda Harris7, Lauren Krupp2, Tim E Lotze6, Sabina Lulu7, Jayne Ness5, Cody Olsen4, Erik Roan4, Moses Rodriguez5, John Rose4, Timothy C Simmons4, Jan-Mendelt Tillema5, Wendy Weber4, Bianca Weinstock-Guttman9
1. Loma Linda University, Loma Linda, CA, United States; 2. Stony Brook University, Stony Brook, NY, United States; 3. Harvard University, Cambridge, MA, United States; 4. University of Utah, Salt Lake City, UT, United States; 5. Mayo Clinic, Rochester, MN, United States; 6. Baylor College of Medicine, Houston, TX, United States; 7. University of California, San Francisco, San Francisco, CA, United States; 8. University of Alabama, Birmingham, AL, United States; 9. State University of New York at Buffalo, Buffalo, NY, United States
References
- 1.Berer K, Mues M, Koutrolos M, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–41. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 2.Lee YK, Menezes JS, Umesaki Y, Sarkis K, Mazmaniana SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mielcarz DW, Kasper LH. The Gut Microbiome in Multiple Sclerosis. Curr Treat Options Neurol. 2015 Apr;17(4):344. doi: 10.1007/s11940-015-0344-7. doi: 10.1007/s11940-015-0344-7. [DOI] [PubMed] [Google Scholar]
- 4.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S, et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- 5.Tremlett H, Fadrosh DW, Faruqi AA, et al. Gut microbiota in early pediatric multiple sclerosis: a case-control study. 2016 doi: 10.1111/ene.13026. Under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T, et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS One. 2015;10(9):e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantarel BL, Waubant E, Chehoud C, Kuczynski J, DeSantis TZ, Warrington J, et al. Gut microbiota in multiple sclerosis: possible influence of immunomodulators. J Invest Med. 2015;63(5):729–34. doi: 10.1097/JIM.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rumah KR, Linden J, Fischetti VA, Vartanian T. Isolation of Clostridium perfringens type B in an individual at first clinical presentation of multiple sclerosis provides clues for environmental triggers of the disease. PLoS One. 2013;8(10):e76359. doi: 10.1371/journal.pone.0076359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeAngelis KM, et al. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3:168–78. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- 10.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.López-Cepero AA, Palacios C. Association of the Intestinal Microbiota and Obesity. P R Health Sci J. 2015;34:60–4. [PubMed] [Google Scholar]
- 13.Suchodolski JS, Dowd SE, Wilke V, Steiner JM, Jergens AE. 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS ONE. 2012;7:e39333. doi: 10.1371/journal.pone.0039333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2015 doi: 10.1136/gutjnl-2015-310101. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingwell E, Bajdik C, Phillips N, et al. Cancer risk in multiple sclerosis: findings from British Columbia, Canada. Brain. 2012;135:2973–2979. doi: 10.1093/brain/aws148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.