Abstract
Host-associated microbiomes influence host health. However, it is unclear whether genotypic variations in host organisms influence the microbiome in ways that have adaptive consequences for the host. Here, we show that wild accessions of Arabidopsis thaliana differ in their ability to associate with the root-associated bacterium Pseudomonas fluorescens, with consequences for plant fitness. In a screen of 196 naturally occurring Arabidopsis accessions we identified lines that actively suppress Pseudomonas growth under gnotobiotic conditions. We planted accessions that support disparate levels of fluorescent Pseudomonads in natural soils; 16S ribosomal RNA sequencing revealed that accession-specific differences in the microbial communities were largely limited to a subset of Pseudomonadaceae species. These accession-specific differences in Pseudomonas growth resulted in enhanced or impaired fitness that depended on the host’s ability to support Pseudomonas growth, the specific Pseudomonas strains present in the soil and the nature of the stress. We suggest that small host-mediated changes in a microbiome can have large effects on host health.
One of the most profound discoveries in biological research of the past decade is the overwhelming importance of host-associated microbial communities for the health of multicellular organisms1,2. Microbes growing in association with either an animal gut or plant roots (in the rhizosphere) can affect the health of their host. Because host genotype can shape the associated microbial communities3–6, it is possible that eukaryotic organisms have evolved the ability to cultivate specific beneficial microbiomes. In plants, the microbial composition of the surrounding soil is the largest determinant of the final rhizosphere community, and the effect of host genotype is minor by comparison3,4. As a result, it is unclear whether host genotype-mediated differences are important for host health, or if it is only random environmental encounters that matter. For a microbiome to be a host genotype-dependent adaptive trait (i.e. under natural selection), we reasoned that the following attributes of host-microbe interactions should be apparent in a single host species: first, populations of the host organism should possess genetic variation in traits that allow them to select environmentally adaptive microbes; and second, differences in microbial communities should differentially affect the fitness of the variant host organisms.
The plant rhizosphere is an ideal system to explore these two postulates. First, it is already known that plants shape their rhizosphere through exuded carbon and other metabolites7,8 and that there is genetic variation in these traits9. This variation presumably contributes to an observed variation in rhizosphere communities10–12. Second, microbes in the rhizosphere are known to affect plant health by assisting with host nutrient acquisition13, protecting against biotic and abiotic stress14,15, and altering plant growth and physiology16. Here, we sought to determine whether particular wild accessions of Arabidopsis thaliana differ in their ability to associate with particular soil microbes and thereby gain potential adaptive advantages.
To make this question experimentally tractable, we developed an assay (described below) to probe the interaction of Arabidopsis with a single-member rhizosphere community consisting of the beneficial root-associated bacterium Pseudomonas fluorescens. We used the wild plant Arabidopsis because of its small size and collections of naturally occurring inbred lines (accessions). P. fluorescens can confer benefits to members of the Brassicaceae including Arabidopsis by protecting them from biotic and abiotic stresses and promoting growth17–21, indicating that P. fluorescens is an important member of the indigenous rhizosphere community of Arabidopsis. We reasoned that enhanced or impaired ability of Arabidopsis to associate with P. fluorescens might confer an advantage under certain environmental conditions.
Results
To test whether natural variation in plants can contribute to an adaptive rhizosphere, we first developed a high-throughput assay that allowed us to screen a large number of plant genotypes for altered associations with rhizosphere bacteria. In this root–bacterium association assay, Arabidopsis plants are grown hydroponically in 48-well clear-bottom plates with the roots submerged and the shoots separated from the media by a floating mesh disk (Supplementary Fig. 1a). Plant roots are exposed to bacteria expressing green fluorescent protein (GFP), and bacterial fluorescence is measured with a plate reader. The plant growth media contains no added carbon, so bacteria are dependent on plant photosynthate for significant growth (Supplementary Fig. 1b). After testing a number of well-studied commensal P. fluorescens isolates in our assay (Supplementary Fig. 1b), we selected P. fluorescens WCS365. This strain was originally isolated from potato22, protects tomato plants from Fusarium oxysporum infection23,24, and has a broad host range25. WCS365 consistently grew to ~3 × 107 colony-forming units (CFU) per root of the Arabidopsis accession Col-0 in the hydroponic assay without causing any apparent disease symptoms or stress to the plant (Supplementary Fig. 1a,b).
To determine whether natural variation in Arabidopsis affects the levels of P. fluorescens WCS365 in the plant rhizosphere, we used the 48-well plate assay to screen a collection of 196 geographically diverse naturally occurring Arabidopsis accessions26. The 196 accessions supported an approximately 1.5 log range of P. fluorescens WCS365 growth, with the reference accession Col-0 supporting a slightly higher level than the average accession (Fig. 1a). We retested accessions that supported reduced levels of WCS365 in the primary screen (P < 0.05 by analysis of variance (ANOVA) and Tukey’s honest significant difference (HSD) test relative to Col-0) and identified five ‘incompatible’ accessions, RRS-10, Knox-10, Knox-18, Pna-10 and Pna-17, that consistently exhibited 0.5 to 1.5 logs less WCS365 growth than ‘compatible’ accessions Col-0, Sha and Ler-0 as measured by fluorescence, or by counting CFUs by plating the hydroponic media or the bacteria physically stuck to the plant roots (Fig. 1b).
Figure 1. Natural variation in Arabidopsis affects growth of Pseudomonas in the rhizosphere.
a, Using hydroponically grown Arabidopsis plants in 48-well plates, a collection of 196 naturally occurring accessions was screened for P. fluorescens growth in the rhizosphere. n = 6 plants per accession; P < 0.05 by ANOVA and Tukey’s HSD test (compared to Col-0) are shown in orange. b, Five accessions had consistently lower levels of P. fluorescens whether bacterial fluorescence, number of CFUs in the well, or number of CFUs attached to roots were measured; n ≥ 24 plants per treatment. c, Hydroponically grown plants were inoculated with B. subtilis 3610, E. coli OP50, previously sequenced Pseudomonas strains, or Pseudomonas isolates identified in this study. CFUs were either counted directly by plating bacteria in wells, or approximated by measuring bacterial fluorescence; n ≥ 12 plants per treatment. d, The culturable microbiome of Col-0 is enriched for fluorescent Pseudomonads relative to bulk soil and the microbiomes of RRS-10 and Knox-18. e, Fluorescent colonies per g of Cambridge or Carlisle soil or roots grown in these soils. b,c,e, Averages ± s.e.m. are shown; *P < 0.01 by t-test; letters designate P < 0.05 by ANOVA and Tukey’s HSD.
Accessions RRS-10 and Knox-18 were chosen for further study because they had the greatest consistent reduction in growth of P. fluorescens WCS365 relative to Col-0. To determine whether the reduced growth of P. fluorescens WCS365 in the RRS-10 and Knox-18 rhizospheres was a general rule for the growth of any bacteria in the rhizosphere of these Arabidopsis genotypes, we tested RRS-10, Knox-18 and Col-0 for their association with several other bacterial species. In the rhizospheres of all three genotypes, we found equivalent growth of the human P. aeruginosa isolate PA14, the non-pathogenic strain Escherichia coli OP50, and the Arabidopsis-colonizing Bacillus subtilis 3610 (Fig. 1c). This indicates that the reduced level of P. fluorescens growth associated with RRS-10 and Knox-18 is not a general rule, but is specific to certain bacterial taxa.
To determine whether the reduced level of P. fluorescens WCS365 in the RRS-10 and Knox-18 rhizospheres is specific to WCS365 or whether other Pseudomonas strains behave similarly, we isolated fluorescent Pseudomonas strains from the roots of Arabidopsis plants growing naturally at several sites in eastern Massachusetts, USA (see Methods and Supplementary Table 1; accession codes for full-length 16S rRNA sequences from Pseudomonas strains WCS365, CH229, CH235, CH254, CH255, CH267 and CH271 are KP253039-45). We selected six fluorescent Pseudomonas strains from different plant roots representing three ecological sites and found that all showed less growth in the RRS-10 and Knox-18 rhizospheres than in the Col-0 rhizosphere (Fig. 1c). We also tested the sequenced Pseudomonas strains P. fluorescens Pf0-1 and P. brassicacearum NFM421 and the well-studied plant pathogen P. syringae pv. tomato DC3000 (Pto DC3000) and found that similarly to WCS365, RRS-10 and Knox-18 support lower rhizosphere levels of all three of these Pseudomonas strains relative to Col-0 (Fig. 1c). These data indicate that compared to Col-0, Knox-18 and RRS-10 support less growth of a diverse repertoire of P. brassicacearum, P. fluorescens and P. syringae strains.
To establish whether the Arabidopsis or the P. fluorescens genotype has a greater effect on P. fluorescens abundance in the rhizosphere, we tested WCS365, and the Arabidopsis rhizosphere isolates CH229 and CH267 on a set of nine Arabidopsis accessions spanning a range of compatibility with P. fluorescens WCS365. We found a strong correlation between the growth of P. fluorescens WCS365 with the growth of CH229 and CH267 (R2 = 0.95 for WCS365 vs. CH229 and R2 = 0.89 for WCS365 vs. CH267; Supplementary Fig. 2). Although the full-length 16S rRNAs from these strains are highly similar, geographical isolation combined with functional differences described below suggest that these are genotypically distinct bacterial strains. Collectively, these data suggest that Arabidopsis genotype rather than P. fluorescens genotype primarily determines the levels of certain Pseudomonas strains in the rhizosphere.
We also tested whether Col-0 supports higher levels of P. fluorescens than Knox-18 and RRS-10 in the context of an intact microbial community. We planted Knox-18, RRS-10 and Col-0 in two distinct soils collected from Cambridge and Carlisle, Massachusetts (soil chemistry and site locations are described in the Methods, Supplementary Table 2 and Supplementary Figs 3 and 4). The former is an unamended site with relatively low levels of endogenous fluorescent Pseudomonads (2.5 × 103 fluorescent CFU g−1), whereas the latter is a cultivated soil with a relatively high level of endogenous fluorescent Pseudomonads (6 × 104 fluorescent CFU g−1). In both soils, we found that relative to bulk soil, the culturable microbiome from Col-0 was enriched in fluorescent colonies from both soils when plated on King’s B (Fig. 1d,e). We also found enrichment in the number of fluorescent Pseudomonads in Knox-18 and RRS-10, but to a significantly reduced level relative to Col-0 (Fig. 1e). This indicates that the results obtained in our gnotobiotic hydroponic system are reflective of indigenous levels of fluorescent Pseudomonads in an intact microbiome.
To determine how the endogenous bacterial microbiomes differ between Col-0, RRS-10 and Knox-18, we planted these three genotypes in the Cambridge soil and Carlisle soils described above and sequenced partial 16S rRNAs to determine bacterial diversity in the rhizosphere and in bulk soil (Methods and Supplementary Methods). Using a 97% identity threshold, we identified a total of 15,891 species defined as operational taxonomic units (OTUs) that were represented by at least 25 reads in any five samples (Methods and Supplementary Methods; alpha diversity for each sample type is shown in Supplementary Fig. 5). Consistent with previous reports, we found that (1) the microbial community in the starting soil has the largest influence on the rhizosphere microbiome3,4,27—24.4% of the variation between samples (Supplementary Fig. 6); (2) that the rhizosphere is enriched for the phyla Proteobacteria and Firmicutes3,4,27 and depleted for Acidobacteria3,4 (Fig. 2a); and (3) that host genotype had a minor but significant effect on rhizosphere community3,4,28,29 (Fig. 2b–f).
Figure 2. The effect of Arabidopsis genotype is largely limited to OTUs in the family Pseudomonadaceae.
a, Abundance of 16S rRNA genes from major bacterial phyla in bulk soil and the rhizospheres of Col-0, RRS-10 and Knox-18. Arrows denote phyla that are significantly enriched or depleted in the rhizosphere of all three accessions in both soils. b,c, Genotype-constrained principal component analysis of bulk soil and rhizosphere samples from Cambridge (b) and Carlisle (c) soils. d, Venn diagrams showing the number of OTUs that are enriched or depleted in the rhizosphere of an individual plant genotype relative to the other two plant genotypes. In Cambridge soil, 60% (15/25) of genotype-dependent differences were Col-0-specific (P = 0.02 by chi-squared test); in Carlisle soil, 55.6% (10/18) of genotype-dependent differences were Col-0 specific (P = 0.03 by chi-squared test). e,f, OTUs in the Pseudomonadaceae that are significantly enriched in the rhizosphere of at least one plant genotype grown in soil from Cambridge (e), or Carlisle (f). *OTUs that are enriched in the Col-0 rhizosphere relative to RRS-10 and Knox-18; P < 0.01 by moderated t-test.
Host genotype accounted for 1–2% of the difference in community structure depending on the soil and plant genotype (Fig. 2b,c). About 2% of the variation distinguished the Col-0 rhizosphere from both the Knox-18 and RRS-10 rhizospheres (constrained component 2 for both soils; Fig. 2b,c). The majority of the host genotype-mediated differences were observed at the OTU level, and of the OTU-specific differences, the majority distinguished Col-0 from RRS-10 and Knox-18 (Fig. 2d and Supplementary Figs 7 and 8). Of the OTUs that were specifically enriched in the Col-0 rhizosphere, a significant proportion was in the family Pseudomonadaceae. In the Cambridge soil, of the OTUs that were enriched to a greater degree in the Col-0 rhizosphere than in the Knox-18 and RRS-10 rhizospheres, 83.3% (5/6) were in the family Pseudomonadaceae; this is a significant enrichment relative to the 9.2% (8/87) Pseudomonadaceae OTUs that were enriched in association with all three plant genotypes (P = 8 × 10−7 by chi-squared test). Carlisle soil showed a similar trend where of the OTUs enriched to a greater degree in association with Col-0, 50% (3/6) were Pseudomonadaceae compared to 3.3% (3/91) of the total enriched OTUs that were Pseudomonadaceae (P = 0.02 by chi-squared test; Fig. 2e,f). The remaining OTUs that were more significantly enriched in association with Col-0 than RRS-10 and Knox-18 were all members of the Proteobacteria (Supplementary Table 4 and Figs 7 and 8). In Cambridge soil, there was a significant enrichment in the genus Pseudomonas and family Pseudomonadaceae in the rhizospheres of all three plant genotypes (Supplementary Fig. 9). Surprisingly, we did not find a significant enrichment of the genus Pseudomonas and family Pseudomonadaceae in rhizosphere samples from Carlisle soil (Supplementary Fig. 9). This is due to the presence of a single OTU in the family Pseudomonadaceae that was depleted in the Col-0 rhizosphere relative to bulk soil (Fig. 2f). Collectively, these data indicate that the mechanism by which Knox-18 and RRS-10 limit growth of Pseudomonas in the rhizosphere is largely limited to a subset of species (defined as OTUs with 97% identity) in the family Pseudomonadaceae.
To determine whether Knox-18 and RRS-10 fail to support bacterial growth, for instance by not providing a nutrient the bacteria require, or if these genotypes actively suppress bacterial growth we co-cultured Col-0 and RRS-10 or Col-0 and Knox-18 (by growing two plants in the same well) and measured P. fluorescens growth. We found that bacterial growth associated with the mixed genotype co-cultured plants was more similar to two RRS-10 or two Knox-18 plants than to two Col-0 plants (Fig. 3a). We also measured the growth of bacteria in root exudates and found that RRS-10 and Knox-18 exudates supported less growth of several P. fluorescens strains relative to Col-0 exudate after 1 day of bacterial growth; by 2 days, bacterial growth was equivalent in the exudates of all three accessions (Supplementary Fig. 10). These findings indicate that Knox-18 and RRS-10 actively inhibit bacterial growth by a mechanism that appears to be inducible.
Figure 3. Incompatible Arabidopsis accessions actively inhibit growth of Pseudomonas.
a, Knox-18 and RRS-10 plants grown in the same wells as Col-0 can inhibit growth of bacteria in trans; n = 18 plant pairs per treatment. b, Defence gene induction 24 h after treatment with P. fluorescens WCS365 measured by qRT-PCR. Averages ± s.d. of three biological replicates are shown; n = 24 plants per replicate. c, Bacteria grown in 48-well plates with Col-0, RRS-10 or Knox-18 were stained with DAPI (blue) and SYTOX (green); Scale bar: 5 µm. d, Quantification of SYTOX+ positive cells in the Arabidopsis rhizosphere.
a,d, *P < 0.001 compared to Col-0 by t-test; Averages ± s.e.m. are shown.
Next, we assessed whether the mechanism by which Knox-18 and RRS-10 inhibit Pseudomonas growth has signatures of characterized defence responses. We examined the expression of defence responsive genes including the flagellin-inducible genes MYB5130 and CYP71A1230 and the flagellin and salicylic acid-inducible genes GST631, WRKY3032 and WRKY3333 (we also tested expression of PR1 and PR2 but could not detect transcripts in roots under any conditions). We found that all defence genes tested had similar or higher induction levels in compatible interactions (in Col-0 and Sha) than in incompatible interactions (RRS-10 and Knox-18) (Fig. 3b). This indicates that the magnitude of defence responses do not correlate with bacterial growth inhibition but rather appear to be a signature of a compatible interaction.
Based on the observation that Knox-18 and RRS-10 actively inhibit bacterial growth, we hypothesized that these accessions are secreting an antimicrobial compound. A combination of bacterial vital dyes can be used to assess the mode of action of unknown antimicrobial compounds34. We found that Pseudomonas strains WCS365, CH229 and CH267 growing in the Knox-18 and RRS-10 rhizospheres exhibited an increase in membrane permeability as indicated by a 30- to 40-fold increase in the fraction of cells that took up the membrane-impermeable DNA-binding dye SYTOX (Fig. 3c,d). We also observed an increase in bacterial cell size and bacterial filament formation associated with Knox-18 and RRS-10 roots. No change in cell shape or the number of SYTOX-positive B. subtilis or P. aeruginosa cells was observed in association with RRS-10 and Knox-18 roots (Fig. 3c,d). The observed changes in P. fluorescens cell morphology are reminiscent of those induced by inhibitors of cell wall synthesis or inhibitors of cell division34. These data are consistent with Knox-18 and RRS-10 actively suppressing growth of some Pseudomonas strains through the production of an antimicrobial compound.
Because we found evidence for active suppression of bacterial growth in the rhizosphere, we tested whether Knox-18 and RRS-10 can also suppress growth of the pathogenic Pseudomonas strain Pto DC3000 in a standard foliar pathogenicity assay. RRS-10 and Col-0 exhibited no significant difference in susceptibility to Pto DC3000, and Knox-18 was significantly more susceptible than Col-0 (~5-fold higher bacterial growth relative to Col-0, P < 0.05; Supplementary Fig. 11). This indicates that the differences in Pseudomonas growth associated with RRS-10 and Knox-18 are rhizosphere-specific.
The results described thus far establish that natural variation in Arabidopsis affects Pseudomonas abundance in the rhizosphere. Our next aim was to determine whether Arabidopsis genotype-mediated differences in Pseudomonas result in differences in plant health. P. fluorescens is generally known to promote plant growth and increase lateral root formation35–37. We observed growth promotion and increased lateral root formation on Col-0 grown vertically on solid agar plates (with no added carbon source) by P. fluorescens WCS365 and the majority of Massachusetts Pseudomonas isolates tested (four of six) (Supplementary Fig. 12). In contrast, RRS-10 and Knox-18 did not show increased biomass or numbers of lateral roots in the presence of P. fluorescens WCS365 or Pseudomonas isolates CH229 and CH267 (Fig. 4a–c) indicating that in our assays, a compatible host genotype is required for plant growth promotion by beneficial bacteria. We also observed growth inhibition of Knox-18 in the presence of strain CH229, indicating that some P. fluorescens strains might have a detrimental effect on incompatible accessions. This observation suggests a possible selection pressure for the evolution of a mechanism that limits Pseudomonas growth in the Arabidopsis rhizosphere.
Figure 4. Pseudomonas strains do not promote the growth of incompatible accessions.
a, Col-0, RRS-10 and Knox-18 grown on agar plates with no carbon source and inoculated with buffer or the indicated Pseudomonas strains. b, Quantification of fresh plant weight. c, Number of lateral roots on plants shown in a; n = 15 plants per treatment. d, Fresh shoot weight of plants grown in Cambridge soil and treated with 10 mM MgSO4, bacteria resuspended to an A600 of 0.01 in 10 mM MgSO4 or fertilizer. e, Fresh shoot weight of plants grown in a commercial soil/vermiculite mixture and treated as described in d; in b–d letters designate P < 0.05 by ANOVA and Tukey’s HSD tests; n ≥ 12 plants; averages ± s.e.m. are shown.
To determine whether host genotype confers a selective advantage when a more complex microbiome is present, we assessed the ability of Pseudomonas to promote Arabidopsis growth in both natural and commercial soils. For a natural soil, we chose the soil from Cambridge, Massachusetts, that has low levels of indigenous fluorescent Pseudomonads (2.5 ± 0.6 × 103 CFU g−1 soil; growth promotion and pathogen protection occur at P. fluorescens levels upwards of 105 CFU g−1 root38). We reasoned that this soil type would allow us to obtain a baseline plant weight that captures the plant genotype–environment and genotype–microbe interactions that are largely independent of fluorescent Pseudomonads. In this soil, the shoot weights of RRS-10, Knox-18 and Col-0 were not statistically significantly different (Fig. 4d). When the soil was inoculated with 105 CFU g−1 Pseudomonas strain CH267 (isolated from this soil), but not with equivalent amounts of WCS365 or CH229 (from different soils), we observed significant growth promotion only on Col-0 but not on RRS-10 or Knox-18 (Fig. 4d and Supplementary Fig. 13a). This indicates that both Arabidopsis and Pseudomonas genotype contribute to whether the Arabidopsis microbiome provides a benefit to its host.
In commercial soil, we were only able to observe P. fluorescens-dependent growth promotion on Col-0 under nutrient-limiting conditions (when plants were grown in very small cells and soil was diluted with vermiculite). Under these conditions, we did not observe growth promotion on RRS-10 or Knox-18 (Fig. 4e and Supplementary Fig. 13b). Growth promotion of all accessions in both natural and commercial soils was observed with fertilizer addition indicating that plants were indeed nutrient limited (Fig. 4e). We found that growth promotion in soil and on plates did not always correlate (Fig. 4). These results indicate that host genotype-mediated differences in microbiome community may not be important for growth under nutrient-rich soil conditions. However under conditions of stress (such as nutrient limitation), a host’s ability to cultivate beneficial microbes may provide an advantage.
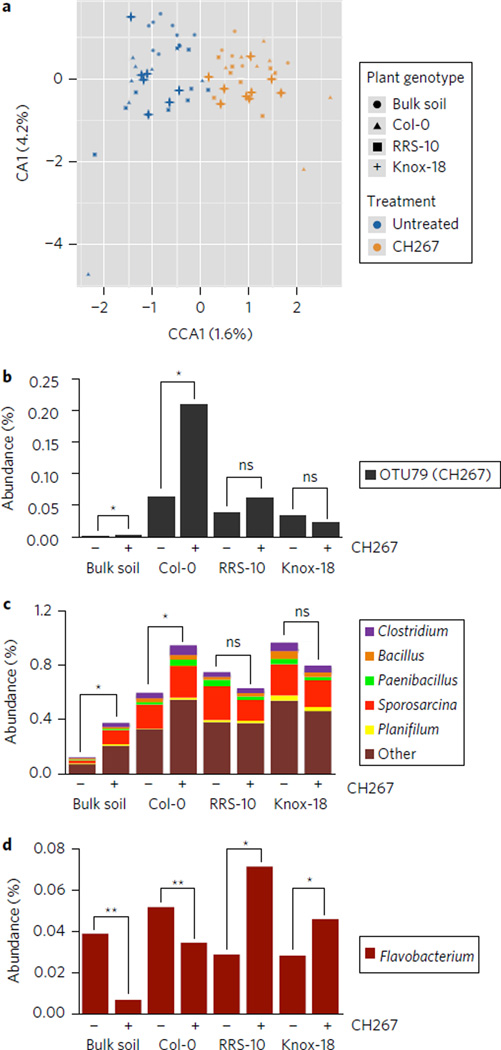
To determine how the plant rhizosphere changes upon artificial addition of Pseudomonas, we compared 16S rRNAs from the rhizosphere of plants grown in Cambridge soil with or without CH267 added. We found that addition of Pseudomonas CH267 had a minor but significant effect on the microbial community and explained 1.6% of the differences in microbiome composition (treatment-constrained CCA; Fig. 5). We found that upon addition of Pseudomonas CH267 (which likely corresponds to OTU79; Fig. 5b), both bulk soil and the Col-0 rhizosphere showed a significant enrichment of this OTU while the RRS-10 and Knox-18 rhizospheres did not (Fig. 5b). Addition of CH267 also resulted in a significant increase in the abundance of multiple genera within the Firmicutes in both bulk soil and in the Col-0 rhizosphere, but not in RRS-10 and Knox-18 rhizospheres (Fig. 5c).We found a reciprocal change in a single genus within the Bacteroidetes (Flavobacterium; Fig. 5d); a significant decrease in abundance of Flavobacterium was found in the Col-0 rhizosphere and in bulk soil, while an increase in abundance of Flavobacterium was observed in the rhizospheres of Knox-18 and RRS-10. We identified 46 OTUs that had distinct behaviours in the plant rhizospheres in untreated soil compared to CH267-treated soil (that is, enriched in the rhizosphere of plants grown in untreated soil and depleted in the rhizospheres of plants grown in soil treated with Pseudomonas CH267; Supplementary Table 4); the majority of these changes could be explained by differences in OTU abundance in treated versus untreated bulk soil (Supplementary Table 4). Because there were changes to the overall community composition, we cannot rule out the possibility that plant growth promotion in soil is an indirect result of the effect of Pseudomonas on the microbial community. These data indicate that addition of Pseudomonas CH267 to Cambridge soil at a level that is sufficient to confer plant growth promotion (1) results in significant changes to a small number of bacterial taxa and (2) has little effect on the overall rhizosphere microbiome community.
Figure 5. Addition of Pseudomonas has a minor but significant effect on soil and rhizosphere community composition.
a, Treatment-constrained principal component analysis of bulk soil and rhizosphere samples with or without the addition of Pseudomonas CH267. b, Relative abundance of OTU79 (which probably corresponds to Pseudomonas CH267) in rhizosphere and soil samples with and without CH267 added. c, Abundance (%) of genera in the Firmicutes in Cambridge soil with or without the addition of Pseudomonas CH267. Upon addition of CH267, all genera listed showed significant enrichment in bulk soil and the Col-0 rhizosphere and no significant difference in the rhizospheres of RRS-10 and Knox-18. d, Abundance (%) of the genus Flavobacterium with or without addition of CH267. In b–d *indicates a significant increase; **indicates a significant decrease by ANOVA and moderated t-test (P < 0.01); ns, not significant; averages ± s.e.m. are shown.
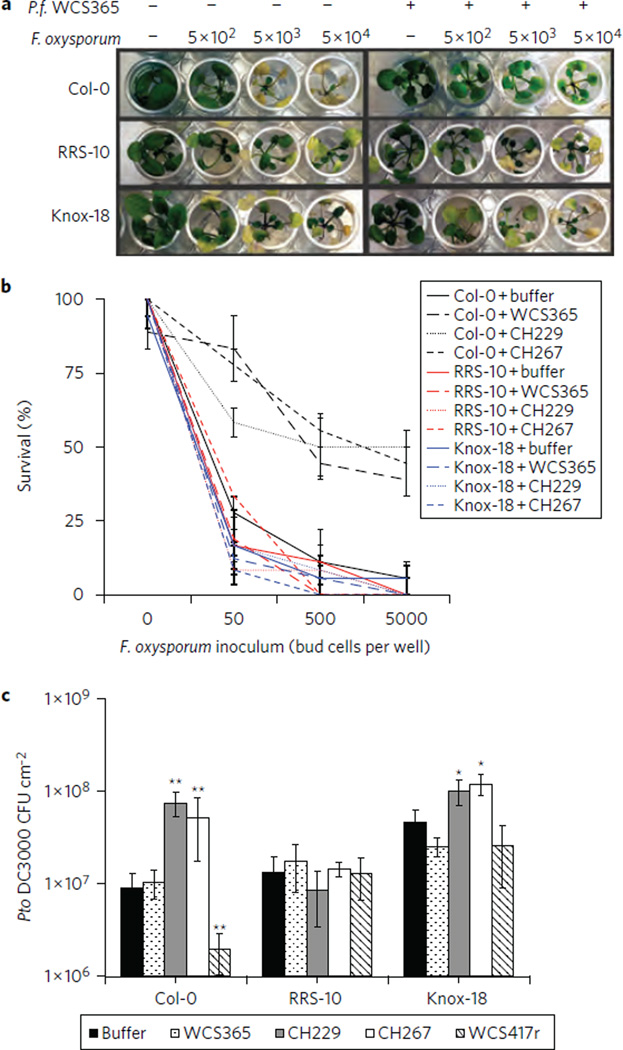
To study the effects of host genotype-mediated P. fluorescens levels on pathogen stress, we developed a hydroponic infection assay to study susceptibility to the fungal pathogen Fusarium oxysporum. We found that Col-0 had reduced susceptibility to F. oxysporum in the presence of P. fluorescens WCS365 and several Massachusetts Pseudomonas strains than in their absence (Fig. 6a,b and Supplementary Fig. 14a). In contrast to Col-0, incompatible accessions RRS-10 and Knox-18 did not exhibit P. fluorescens-dependent protection from F. oxysporum (Fig. 6a,b and Supplementary Fig. 14). This trend was consistent when measured as the percentage of plants that survived after 3 weeks (Fig. 6b), F. oxysporum levels in the rhizosphere (Supplementary Fig. 14b), or the number of infection events (Supplementary Fig. 14c,d). Not all rhizosphere-competent strains were able to protect Col-0, indicating that F. oxysporum growth is not simply being limited by depletion of nutrients from the medium (Supplementary Fig. 14a). These results indicate that Arabidopsis genotypes that curate rhizosphere colonization by Pseudomonas have a selective advantage in the presence of beneficial P. fluorescens and F. oxysporum.
Figure 6. Disease outcome depends on host genotype, the Pseudomonas strain present and the pathogen.
a, Accessions were grown hydroponically in 48-well plates and treated with buffer, P. fluorescens and/or F. oxysporum (For 815) as indicated. Representative images 14 days post inoculation with WCS365 are shown. b, Percentage survival of plants inoculated with P. fluorescens WCS365, CH229 or CH267 and For 815 after 21 days.
a,b, Average ± s.d. of three replicates with n = 6 plants per treatment per rep. c, Growth of Pto DC3000 in the leaves of adult soil-grown plants grown with P. fluorescens in the rhizosphere. n = 12 leaves from six plants; *P < 0.05; **P < 0.01 by Tukey’s HSD test; averages ± s.e.m. are shown.
Some strains of P. fluorescens in the rhizosphere can induce systemic resistance (ISR) to foliar pathogens including the leaf pathogen Pto DC300039. Consistent with previous studies, we found that P. fluorescens WCS417r could induce ISR and protect Col-0 from Pto DC300040 (Fig. 6c). We also tested two of the Massachusetts Pseudomonas isolates for their ability to induce ISR and found unexpectedly that they actually induced systemic susceptibility (ISS) to Pto DC3000 (Fig. 6c). Under conditions where we observed P. fluorescens-dependent ISR or ISS, we observed no effect on RRS-10 and a marginally significant effect on Knox-18 (Fig. 6c). This indicates that there may be a complex evolutionary trade-off to supporting P. fluorescens in the rhizosphere; depending on the P. fluorescens strain present in the soil, and the particular pathogen stresses, there may be advantages or disadvantages to generally supporting or inhibiting Pseudomonas growth.
Discussion
Our data indicate that accessions of Arabidopsis actively inhibit growth of some species within the Pseudomonadaceae while leaving the majority of the microbiome intact. While reminiscent of a defence response, this is distinct from the paradigm of well-characterized defence mechanisms in plants (that is, Resistance (R) genes) that achieve specificity at the perception level41, but where the inputs from perception of diverse pathogens converge on largely overlapping downstream pathways42. Known downstream defence mechanisms are largely blunt force antimicrobial instruments such as plant cell death, reactive oxygen species production, or production of broad-spectrum antimicrobials43,44 that would not be expected to limit the growth of a narrow group of bacterial taxa. We also did not detect increased expression of defence genes in incompatible accessions; in fact, defence gene induction was more closely correlated with compatibility (Fig. 2b). Together, these data indicate that the mechanism by which Knox-18 and RRS-10 limit growth of Pseudomonas strains in the rhizosphere may be distinct from well-characterized defence responses in plants.
We conclude that plant genotype is an essential determinant of whether a microbiome community provides benefit or harm to its host. Random encounters with beneficial microbes are not sufficient to explain the health benefits of the plant microbiome; if the plant genotype is incompatible with growth of the beneficial microbe, the plant will not benefit from its presence in the environment. We found that a particular microbiome will provide a benefit or detriment to the host depending on (1) the particular biotic stress and (2) the individual Pseudomonas strains present in the rhizosphere. This indicates that there may be an evolutionary trade-off to inhibiting growth of Pseudomonas. Our results indicate that plants within a single species have varied rhizosphere microbiomes, which result in divergent outcomes for plant health and physiology depending on precise biotic and abiotic stresses. Thus, multicellular organisms possess the machinery for ongoing microbiome-mediated adaptation in the face of changing nutrient, environmental and pathogen stresses.
Methods
Rhizosphere assay in 48-well plates
Eight-millimetre Teflon mesh disks (McMaster-Carr #1100t41) were sterilized by autoclaving and placed into sterile clear-bottom 48-well tissue culture plates (BD Falcon) with 250 µl Murashige and Skoog (MS) media containing 2% sucrose. Sterile imbibed seeds were placed individually at the centre of each disk. Plates were placed in the dark for 3 days so that the seedlings etiolated. Plates were subsequently transferred to a shelf with 16 h light/8 h dark (at a light fluence of 100 µE) at 22 °C. Ten days after plating, the media was removed and replaced with 270 µl 1/2X MS media with no sucrose. Two days after the media change, 30 µl of bacteria were added to a final absorbance A600 = 0.00002 (~3,000 CFU per well). Plates were read from the bottom using a SpectraMax M3 fluorescent plate reader (Molecular Devices) with 418/515(20) nm excitation/emission. Any plants with submerged leaves or showing symptoms of water stress were discarded. The collection of 196 accessions used in the primary screen was described previously26. For bacterial growth assays, a minimum of 24 plants was assayed per plant genotype per bacterial strain. For bacterial strains expressing GFP, CFUs per well were approximated by generating a standard curve for each strain. For strains not expressing GFP, media from the well was removed and serially diluted to determine CFUs. To measure bacterial growth in plant root exudate, plants were removed 2 days after the media change and bacteria were added to the wells as described above. Statistics were performed with StatPlus (AnalystSoft).
Natural soil collection, Arabidopsis population sites and isolation of new strains of P. fluorescens
‘Cambridge’ soil was collected near the Weld Boathouse in Cambridge, MA (42 °22′10″ N, 71 °7′18″ W). ‘Carlisle’ soil was isolated from a vegetable garden at GPS coordinates 42 °31′38″ N, 71 °21′4″ W in Carlisle, Massachusetts (Supplementary Fig. 4). Soil chemistry and texture was assayed by the University of Massachusetts soil and plant tissue testing laboratory and is summarized in Supplementary Table 2. Additional site information and soil collection methods are present in the Supplementary Methods. New strains of Pseudomonas were isolated from the roots of plants growing at these and other sights around Massachusetts (Supplementary Methods and Supplementary Table 1).
Microbiome assays
Microbiome samples included the plant root and soil that was firmly adhered to the root. Seven-day-old seedlings were transferred from plates (MS media with 2% sucrose) to pots. Plants were grown in 12 h days and 12 h nights at a fluence of 100 µE. Four seedlings were planted per 7.5 cm pot, ten pots were planted per genotype per soil type, and all four plants were pooled for microbiome analysis. An equivalent number of unplanted pots were used for ‘bulk soil’ samples. Rhizosphere and bulk soil samples (8 mm cores) were harvested 21 days after transplanting.
16S rRNA microbiome sample preparation, sequencing and analysis
DNA was extracted using the MoBio Powerlyser Power soil kit (MoBio). DNA from seedlings grown sterilely on MS plates (prior to transfer) was also included to confirm that there was no contamination and that the seeds did not harbour any entophytic microbes that survived sterilization. Samples were prepared for sequencing as described in the Earth Microbiome Project (http://www.earthmicrobiome.org/) using primers 515F and 806R and sequenced using MiSeq v3 600 cycles kit (Illumina).
The fastq files from sequencing reads were processed with the QIIME software package45 (Supplementary Methods). Taxa that were significantly enriched or depleted were determined using the R software package (http://www.r-project.org/) as described3 using ANOVA and a Bayesian model-based moderated t-test46 that takes into account the variance of all OTUs when determining significance.
Plant growth promotion plate assays
For plate assays, ten seeds were germinated on small square plates containing MS media with no sucrose. Five days after planting, seeds were thinned to five seedlings per plate and inoculated with 1 µl of 10 mM MgSO4 or bacteria diluted to A600 = 0.001 (~103 CFU per plant or ~105 CFU g−1 of root) in MgSO4. Ten days after inoculation plants were imaged on an Epson V750 Pro scanner and then removed from plates and fresh weight was determined.
For commercial soil experiments, seeds were sown directly in a 1:1 mix of Sunshine MVP (SunGro Horticulture) and vermiculite in 1.8 cm diameter cells. Eight days after sowing, plants were treated with 1 ml bacteria (A600 = 0.01 in 10 mM MgSO4; 105 CFU g−1 soil), 1 ml 10 mM MgSO4 or 1/4X Hoagland solution. Plants were imaged and weighed 2 weeks after inoculation.
For natural soil experiments, plants were grown as described for microbiome experiments (above) and treated with 5 ml bacteria (A600 = 0.01 in 10 mM MgSO4; 105 CFU g−1 soil), 5 ml 10 mM MgSO4 or 1/4X Hoagland solution. Plants were imaged and weighed 3 weeks after inoculation.
Infection assays
Pto DC3000 infection assays47 and Fusarium infection assays48 were performed as described with modifications (Supplementary Methods).
Data availability
Pseudomonas full-length 16S rRNA from new strains have been deposited in GenBank: WCS365, CH229, CH235, CH254, CH255, CH267 and CH271 (accession codes: KP253039, KP253040, KP253041, KP253042, KP253043, KP253044 and KP253045). Illumina HiSeq microbiome data are available on request from C.H.H. (haneymolbio.mgh.harvard.edu).
Supplementary Material
Acknowledgments
C.H.H. is funded by MGH Toteston and Fund for Medical Discovery Fellowship grant 2014A051303 and previously by the Gordon and Betty Moore Foundation through Grant GBMF 2550.01 to the Life Sciences Research Foundation. B.S.S. was funded by a Charles King Trust Sr. Postdoctoral Fellowship. This work was supported by NIH R37 grant GM48707 and NSF grants MCB-0519898 and IOS-0929226 awarded to F.M.A. We thank J. Meyer, D. McEwan, J. Griffitts and L. Shapiro for critical reading of the manuscript and A. Diener and members of the Ausubel Lab for helpful comments and discussion.
Footnotes
Author contributions
C.H.H. and F.M.A. conceived experiments and discussed results. C.H.H. and J.B. designed assays and performed experiments. B.S.S. and C.H.H. analysed data. C.H.H. wrote the manuscript with input from F.M.A., J.B. and B.S.S.
Supplementary information is available online.
Competing interests
The authors declare no competing financial interests.
References
- 1.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nature Rev. Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant. Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Bulgarelli D, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 4.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukherjee S, et al. Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature. 2014;505:103–107. doi: 10.1038/nature12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badri DV, Chaparro JM, Manter DK, Martinoia E, Vivanco JM. Influence of ATP-binding cassette transporters in root exudation of phytoalexins, signals, and in disease resistance. Front. Plant Sci. 2012;3:149. doi: 10.3389/fpls.2012.00149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Jones DL, Nguyen C, Finlay RD. Carbon flow in the rhizosphere: carbon trading at the soil-root interface. Plant Soil. 2009;321:5–33. [Google Scholar]
- 9.Rovira AD. Plant root exudates. Bot. Rev. 1969;35:35–57. [Google Scholar]
- 10.Lemanceau P, et al. Effect of two plant species, flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent Pseudomonads. Appl. Environ. Microbiol. 1995;61:1004–1012. doi: 10.1128/aem.61.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Germida JJ, Sicilliano SD, de Freitas JR, Arlette MS. Diversity of root-associated bacteria associated with field-grown canola (Brassica napus L.) and wheat (Triticum aestivum L.) FEMS Microbiol. Ecol. 1998;26:43–50. [Google Scholar]
- 12.Bouffaud ML, Poirier MA, Muller D, Moenne-Loccoz Y. Root microbiome relates to plant host evolution in maize and other Poaceae. Environ. Microbiol. 2014 doi: 10.1111/1462-2920.12442. [DOI] [PubMed] [Google Scholar]
- 13.Marschner H, Römheld V, Horst WJ, Martin P. Root-induced changes in the rhizosphere: Importance for the mineral nutrition of plants. Z. Pflanz. Bodenkunde. 1986;149:441–4456. [Google Scholar]
- 14.Couillerot O, Prigent-Combaret C, Caballero-Mellado J, Moenne-Loccoz Y. Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett. Appl. Microbiol. 2009;48:505–512. doi: 10.1111/j.1472-765X.2009.02566.x. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez RJ, et al. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008;2:404–416. doi: 10.1038/ismej.2007.106. [DOI] [PubMed] [Google Scholar]
- 16.Vacheron J, et al. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013;4:356. doi: 10.3389/fpls.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weller DM, et al. Induced systemic resistance in Arabidopsis thaliana against Pseudomonas syringae pv. tomato by 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology. 2012;102:403–412. doi: 10.1094/PHYTO-08-11-0222. [DOI] [PubMed] [Google Scholar]
- 18.Pieterse CM, et al. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, et al. The use of Pseudomonas fluorescens P13 to control sclerotinia stem rot (Sclerotinia sclerotiorum) of oilseed rape. J. Microbiol. 2011;49:884–889. doi: 10.1007/s12275-011-1261-4. [DOI] [PubMed] [Google Scholar]
- 20.Jalili F, et al. Isolation and characterization of ACC deaminase-producing fluorescent pseudomonads, to alleviate salinity stress on canola (Brassica napus L.) growth. J. Plant Physiol. 2009;166:667–674. doi: 10.1016/j.jplph.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Pallai R, Hynes RK, Verma B, Nelson LM. Phytohormone production and colonization of canola (Brassica napus L.) roots by Pseudomonas fluorescens 6–8 under gnotobiotic conditions. Can. J. Microbiol. 2012;58:170–178. doi: 10.1139/w11-120. [DOI] [PubMed] [Google Scholar]
- 22.Brand I, et al. Isolation and characterization of a superior potato root-colonizing Pseudomonas strain. In: Keel C, Knoller B, De ´fago G, editors. Proceedings of the International Workshop on Plant Growth Promoting Rhizobacteria. IOBC/WPRS Bulletin XIV/8. 1991. pp. 350–354. [Google Scholar]
- 23.Kamilova F, Lamers G, Lugtenberg B. Biocontrol strain Pseudomonas fluorescens WCS365 inhibits germination of Fusarium oxysporum spores in tomato root exudate as well as subsequent formation of new spores. Environ. Microbiol. 2008;10:2455–2461. doi: 10.1111/j.1462-2920.2008.01638.x. [DOI] [PubMed] [Google Scholar]
- 24.Bolwerk A, et al. Interactions in the tomato rhizosphere of two Pseudomonas biocontrol strains with the phytopathogenic fungus Fusarium oxysporum f. sp. radicis-lycopersici. Mol. Plant Microbe Interact. 2003;16:983–993. doi: 10.1094/MPMI.2003.16.11.983. [DOI] [PubMed] [Google Scholar]
- 25.Simons M, et al. Gnotobiotic system for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol. Plant Microbe Interact. 1996;9:600–607. doi: 10.1094/mpmi-9-0600. [DOI] [PubMed] [Google Scholar]
- 26.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeAngelis KM, et al. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3:168–178. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- 28.Weinert N, et al. PhyloChip hybridization uncovered an enormous bacterial diversity in the rhizosphere of different potato cultivars: many common and few cultivar-dependent taxa. FEMS Microbiol. Ecol. 2011;75:497–506. doi: 10.1111/j.1574-6941.2010.01025.x. [DOI] [PubMed] [Google Scholar]
- 29.Bouffaud ML, et al. Is diversification history of maize influencing selection of soil bacteria by roots? Mol. Ecol. 2012;21:195–206. doi: 10.1111/j.1365-294X.2011.05359.x. [DOI] [PubMed] [Google Scholar]
- 30.Millet YA, et al. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell. 2010;22:973–990. doi: 10.1105/tpc.109.069658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mammarella ND, et al. Apoplastic peroxidases are required for salicylic acid-mediated defense against Pseudomonas syringae. Phytochemistry. 2014;112:110–121. doi: 10.1016/j.phytochem.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng X, et al. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta. 2012;236:1485–1498. doi: 10.1007/s00425-012-1698-7. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Z, Qamar SA, Chen Z, Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 34.Nonejuie P, Burkart M, Pogliano K, Pogliano J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl Acad. Sci. USA. 2013;110:16169–16174. doi: 10.1073/pnas.1311066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 36.Zamioudis C, Mastranesti P, Dhonukshe P, Blilou I, Pieterse CM. Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiol. 2013;162:304–318. doi: 10.1104/pp.112.212597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srivastava S, et al. Gene expression profiling through microarray analysis in Arabidopsis thaliana colonized by Pseudomonas putida MTCC5279, a plant growth promoting rhizobacterium. Plant Signal. Behav. 2012;7:235–245. doi: 10.4161/psb.18957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raaijmakers JM, et al. Dose-response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathology. 1995;85:1075–1081. [Google Scholar]
- 39.Pieterse CM, et al. Induced systemic resistance by beneficial microbes. Ann. Rev. Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 40.Pieterse CM, van Wees SC, Hoffland E, van Pelt JA, van Loon LC. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell. 1996;8:1225–1237. doi: 10.1105/tpc.8.8.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eitas TK, Dangl JL. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi D, Innes RW. Recent advances in plant NLR structure, function, localization, and signaling. Front. Immunol. 2013;4:3483. doi: 10.3389/fimmu.2013.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Ann. Rev. Phytopathol. 2013;51:245–266. doi: 10.1146/annurev-phyto-082712-102314. [DOI] [PubMed] [Google Scholar]
- 44.Wu L, Chen H, Curtis C, Fu ZQ. Go in for the kill: How plants deploy effector-triggered immunity to combat pathogens. Virulence. 2014;5:710–721. doi: 10.4161/viru.29755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerr MK. Linear models for microarray data analysis: hidden similarities and differences. J. Comput. Biol. 2003;10:891–901. doi: 10.1089/106652703322756131. [DOI] [PubMed] [Google Scholar]
- 47.Daudi A, et al. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell. 2012;24:275–287. doi: 10.1105/tpc.111.093039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diener AC, Ausubel FM. RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics. 2005;171:305–321. doi: 10.1534/genetics.105.042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Pseudomonas full-length 16S rRNA from new strains have been deposited in GenBank: WCS365, CH229, CH235, CH254, CH255, CH267 and CH271 (accession codes: KP253039, KP253040, KP253041, KP253042, KP253043, KP253044 and KP253045). Illumina HiSeq microbiome data are available on request from C.H.H. (haneymolbio.mgh.harvard.edu).