Abstract
Mitochondrial homoplasmy, which is maintained by strictly maternal inheritance and a series of bottlenecks, is thought to be an adaptive condition for metazoans. Doubly uniparental inheritance (DUI) is a unique mode of mitochondrial transmission found in bivalve species, in which two distinct mitochondrial genome (mtDNA) lines are present, one inherited through eggs (F) and one through sperm (M). During development, the two lines segregate in a sex- and tissue-specific manner: females lose M during embryogenesis, whereas males actively segregate it in the germ line. These two pivotal events are still poorly characterized. Here we investigated mtDNA replication dynamics during embryogenesis and pre-adulthood of the venerid Ruditapes philippinarum using real-time quantitative PCR. We found that both mtDNAs do not detectably replicate during early embryogenesis, and that the M line might be lost from females around 24 h of age. A rise in mtDNA copy number was observed before the first reproductive season in both sexes, with the M mitochondrial genome replicating more than the F in males, and we associate these boosts to the early phase of gonad production. As evidence indicates that DUI relies on the same molecular machine of mitochondrial maternal inheritance that is common in most animals, our data are relevant not only to DUI but also to shed light on how differential segregations of mtDNA variants, in the same nuclear background, may be controlled during development.
Introduction
In animals, the mitochondrial genome (mtDNA) is usually transmitted maternally by avoiding paternal inheritance through spermatozoa. This ‘strictly maternal inheritance' is partly responsible for offspring homoplasmy, namely, the presence of only one mitochondrial haplotype in an individual (as opposed to heteroplasmy). Homoplasmy is also thought to be maintained by a bottleneck during oogenesis that lowers the diversity of mtDNA variants in the mature egg (reviewed in Jokinen and Battersby, 2013, and Mishra and Chan, 2014). After fertilization, during early embryogenesis, another bottleneck is thought to occur due to an absence of mtDNA replication as cells rapidly divide, resulting into a drastic decline of mtDNA copy number per cell. The amplification of the reduced pool of mtDNA molecules inside cells takes place in a later developmental phase (Mishra and Chan, 2014). The maintenance of mitochondrial homoplasmy is commonly thought to be adaptive, because it avoids potentially harmful genomic conflicts that may arise between two different mtDNAs within a cell, and it promotes co-adaptation of interacting mitochondrial and nuclear genes (Hoekstra, 2011; Lane, 2011).
Heteroplasmy in an individual may result from maternal inheritance of multiple mtDNA variants that pass the bottleneck, from acquisition of de novo mutations during development and aging, or from paternal leakage. Heteroplasmy resulting from the coincidental presence of functional and deleterious mtDNAs is a cause of pathology in humans, when the harmful variant surpasses a certain copy-number threshold in a tissue or organ (Jokinen and Battersby, 2013; Wallace and Chalkia, 2013; Mishra and Chan, 2014). Stochastic segregation of heteroplasmic mtDNA variants during mitotic or meiotic cell division allows highly heterogeneous tissue distribution. Added to the stochastic segregation of mtDNA variants is the tissue-specific segregation of some mtDNA haplotypes that cannot be explained without invoking a selective process (Jokinen and Battersby, 2013; Wallace and Chalkia, 2013; Mishra and Chan, 2014). However, the mechanisms by which some mtDNA variants come to predominate in certain tissues or in the germ line remain poorly understood (see, for example, Burgstaller et al., 2014), and this may reflect a lack of suitable in vivo model systems in which to study these processes.
Only one exception to strictly maternal inheritance of mitochondria is known in animals, the ‘doubly uniparental inheritance' (DUI) of mtDNA (Skibinski et al., 1994a, 1994b; Zouros et al., 1994a, 1994b), a mode of mitochondrial transmission confirmed in eight families of bivalve mollusks (Theologidis et al., 2008). In the DUI system, two mtDNAs, named F and M, with high sequence divergence (Mytilus spp. nucleotide P-distance: 22–39% amino-acid P-distances in Ruditapes philippinarum and unionoids freshwater mussels: 34% and up to ~51%, respectively; reviewed in Zouros, 2013), co-exist in the same species and are inherited through separate routes. The F is transmitted by females through eggs to both sons and daughters, whereas the M is transmitted by males through spermatozoa to sons only (Breton et al., 2007; Passamonti and Ghiselli, 2009; Zouros, 2013; Breton et al., 2014). Females are essentially homoplasmic for the F mtDNA, whereas males are always heteroplasmic carrying both F and M mtDNAs. DUI females have been shown to produce progenies with variable sex ratios, that is, female biased, male biased, or balanced (females and males in similar proportions), a feature that appears to be dependent on the mother only (Saavedra et al., 1997; Kenchington et al., 2002; Ghiselli et al., 2012). Ghiselli et al. (2011) proposed the main steps of the DUI mechanism as a series of three consecutive checkpoints: in checkpoint #1, which takes place shortly after fertilization, sperm mitochondria enter the egg and are maintained as an aggregate in male embryos, whereas they disperse in females (observed in Mytilus by Cao et al., 2004; Obata and Komaru, 2005; Cogswell et al., 2006; and in R. philippinarum by Milani et al., 2011 and Milani et al., 2012); in checkpoint #2, the M mitochondrial genome disappears only from females, following the dilution and/or degradation of sperm mitochondria; in checkpoint #3, the M mtDNA is segregated in the male germ line and the F in the female one, becoming the dominant mtDNA in the gonad and the only mitochondrial line transmitted by sperms and eggs, respectively. The loss of M mtDNA in developing females, that is, checkpoint #2, has been studied in Mytilus (Sutherland et al., 1998; Sano et al., 2011) and appears to take place in the first 24 h of development. To our knowledge, the proposed checkpoint #3, which should occur before gamete production starts, has never been characterized so far.
The DUI system represents a promising model to study mechanisms controlling transmission, segregation, replication and expression of the mitochondrial genome. DUI males are naturally heteroplasmic for the two highly divergent F and M mtDNAs: gonads contain a high amount of M mtDNA and produce sperms carrying only the M genome, whereas levels of heteroplasmy in somatic tissues can be variable depending on species. In adult Mytilus males, the M mtDNA is usually at low concentrations or absent in somatic tissues (reviewed in Zouros, 2013), whereas in R. philippinarum, adult male soma can contain high quantities of M, even more than the F mtDNA (Passamonti and Scali, 2001; Ghiselli et al., 2011). The different relative quantities of the M genome in these two models could be explained by a different ‘degree' of aggregation of sperm mitochondria in male embryos: a weaker aggregation, as hypothesized in R. philippinarum, could let more copies of M to leak through the developing embryo, reaching blastomeres that give rise to somatic tissues (Milani et al., 2012). Males without M mtDNA, to our knowledge, have never been observed in natural conditions, but have only been produced artificially in Mytilus by chemically induced triploidization (Kenchington et al., 2009). Conversely, heteroplasmy is sometimes observed in female tissues, probably due to the failure of sperm mitochondria degradation (Ghiselli et al., 2011; Zouros, 2013), hybridization events (reviewed in Brannock et al., 2013) or being even typical of some populations where DUI disruption is frequent (Brannock et al., 2013). Cases of heteroplasmic females are mostly known for Mytilus, but in R. philippinarum they seem to be uncommon so far (Ghiselli et al., 2011).
Evidence indicates that DUI relies on the same molecular machine of strictly maternal inheritance typical of animals, with some modifications that let paternal inheritance of M mtDNA happen (Breton et al., 2014). The preferential segregation of M mtDNA in male germ line seems more linked to active processes than to replication rate only (Venetis et al., 2006; Ghiselli et al., 2011; Milani et al., 2011; Guerra et al., 2014), while the driving forces behind F and M distribution in somatic tissues are still unknown. Studying F and M behavior and replication dynamics during female and male developmental paths is of central importance not only to understand the mechanism of DUI but also to shed light on the factors that govern tissue-specific segregation of heteroplasmic variants during development, a topic of great importance, for example, in many human health issues (Lane, 2012; Jokinen and Battersby, 2013; Wallace and Chalkia, 2013; Mishra and Chan, 2014).
As a first step towards a better understanding of the mechanisms underlying mtDNA segregation and replication in a species with DUI, the present study explored the dynamics of F and M mtDNA amounts during development using real-time quantitative PCR (qPCR) in two groups of samples of R. philippinarum, respectively representing early (embryos and larvae) and advanced (sub-adults) developmental stages. Consistent with previous studies in mammals and other animal models (reviewed in Jokinen and Battersby, 2013, and Mishra and Chan, 2014), our results indicate no detectable F and M mtDNA replication during the first phases of development, resulting in a marked reduction of mtDNA copies per cell owing to an increase in cell number. This reduction is followed by tissue- and sex-specific mtDNA replication boosts, most probably related to massive cell proliferation associated with the start of gonad and/or gamete formation.
Materials and methods
Sample characteristics
Two groups of samples were analyzed in this work, provided by the ‘Centro Ricerche Molluschi (CRiM)' (Goro, Italy). The first group, named ‘embryo series', represents the early stages of R. philippinarum development and consists of individuals whose age was from 2 to 86 h post fertilization (hpf; a time interval that covers the developmental stages of eight-cell embryos, trochophore, D-larva, and veliger), divided into six different age subgroups (2, 6, 12, 24, 48, and 86 hpf; see Table 1). Spawning, fertilization, and embryo collection were performed in controlled lab conditions. We assume a balanced sex ratio of each stage in the embryo series, given that (1) the spawning involved dozens of different females taken from the field expected to produce progenies with varying sex bias, (2) the overall sex ratio of wild populations of DUI species is 1:1 (Kenchington et al., 2002; Ghiselli et al., 2012), and (3) we collected a high number of individuals per stage (see below). These samples were preserved in 100% ethanol and stored at 4 °C until use.
Table 1. Ratio statistics for each stage of the embryo series.
| hpf | N | mdn Nu | Nu MAD | avg Nu | Nu s.d. | mdn F | F MAD | avg F | F s.d. | mdn M | M MAD | avg M | M s.d. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 30 | 1.00 | 0.55 | 1.16 | 0.74 | 3 096.57 | 995.29 | 3 377.94 | 1 626.92 | 0.46 | 0.31 | 2.15 | 4.62 |
| 6 | 9 | 0.05 | 0.03 | 0.05 | 0.03 | 3 912.24 | 740.44 | 5 309.01 | 3 367.08 | 1.78 | 0.57 | 4.02 | 4.43 |
| 12 | 30 | 0.33 | 0.38 | 0.46 | 0.46 | 64.50 | 62.25 | 95.81 | 84.72 | 0.08 | 0.07 | 0.17 | 0.20 |
| 24 | 24 | 3.14 | 3.63 | 4.14 | 4.37 | 256.69 | 152.99 | 281.99 | 171.36 | 0.07 | 0.09 | 0.26 | 0.53 |
| 48 | 27 | 1.76 | 1.32 | 2.52 | 1.68 | 58.33 | 28.09 | 73.44 | 58.97 | 5.67E–03 | 5.03E–03 | 7.98E–03 | 6.40E–03 |
| 86 | 30 | 26.18 | 8.02 | 25.99 | 10.68 | 47.05 | 11.75 | 47.85 | 14.59 | 3.95E–05 | 4.10E–05 | 9.10E–05 | 1.87E–04 |
Abbreviations: avg, average; F, F mtDNA ratio; hpf, hours post fertilization; M, M mtDNA ratio; MAD, median absolute deviation; mdn, median; N, number of replicates considered for the analyses; Nu, nuclear ratio; s.d., standard deviation.
Total DNA was extracted from 10 pools of embryos/larvae for each of the six stages. Statistics were calculated on all suitable technical replicates available for each pool. Two hours post fertilization median Nu ratio is 1 because its median Cq in this stage was used as the calibrator to normalize the Nu ratios of the other stages (see Materials and methods).
The second group, named ‘young series', was collected in the field, and is composed of individuals from 1 to 20 mm of shell length: the largest ones (15–20 mm) were those nearest to their first reproductive season (Devauchelle, 1990). These samples were grouped into four distinct classes (named 1, 2, 3, and 4) based on their dimensions. Table 2 enlists dimension intervals of classes and the number of specimens for each class. Whole animals of the young series were put in 100% ethanol immediately after collection and conserved at −80 °C until use.
Table 2. Characteristics and sex ratio of the four classes of the young series.
| Class | Shell length interval (mm) | # of specimens | # of females | # of males |
|---|---|---|---|---|
| 1 | 1–3 | 10 | 5 | 5 |
| 2 | 5–6 | 13 | 8 | 5 |
| 3 | 9–15 | 5 | 3 | 2 |
| 4 | 18–20 | 16 | 8 | 8 |
Sex of the specimens was assigned after the qPCR experiments, based on the absence/presence of a M mtDNA signal (absence: female, presence: male; see text for details).
DNA extractions
For the embryo series, DNA was extracted using pools of individuals, because extractions from single individuals gave too little yield. The overall developmental stage of specimens was checked under a light microscope before extraction. Ten pools for each of the six stages were used for the extractions, for a total of 60 pools. We sampled ~100 individuals per pool, that corresponds to ~1000 individuals per stage, for a total of ~6000 among embryos and larvae. Each pool was identified with a unique name composed by the stage and a letter (Supplementary Table 1).
DNA extractions for the young series were performed from whole animals for the smallest specimens, or from bodies for the bigger specimens (the sample identified as ‘body' consists of the central visceral mass of the animal, with foot, gills, and digestive gland removed). To analyze the mtDNA content variation in other tissues, we also extracted DNA from adductor muscles and mantles from classes 2 and 4 specimens (see Table 3 for sample size of all tissues). To avoid contaminations, single animals were processed separately, a different scalpel was used to dissect each tissue, all reusable tools (glass plates and tweezers) were carefully washed and rinsed between dissections, and all disposable materials (gloves and table covers) were changed every time. After dissection, tissues were kept frozen in separate tubes at −80 °C until DNA extraction. A unique name composed of class number, a letter identifying the specimen, and a letter identifying the tissue was given to each juvenile sample (Supplementary Table 2).
Table 3. mtDNA ratio statistics in young series samples.
| Class | Sample type | ♀ | ♂ | mdn F | F MAD | avg F | F s.d. | mdn Fm | Fm MAD | avg Fm | Fm s.d. | mdn M | M MAD | avg M | M s.d. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | w | 5 | 5 | 7.77 | 2.93 | 7.34 | 2.57 | 10.60 | 1.65 | 10.40 | 4.83 | 5.50E–04 | 8.12E–04 | 2.33E–03 | 3.63E–03 |
| 2 | w | 2 | 3 | 7.58 | 4.29 | 8.05 | 3.55 | 9.26 | 9.89 | 18.72 | 19.50 | 1.46E–03 | 8.65E–04 | 1.78E–03 | 1.28E–03 |
| b | 6 | 2 | 5.57 | 3.85 | 7.44 | 5.86 | 9.36 | 7.08 | 10.60 | 6.37 | 1.02 | 1.52 | 1.71 | 2.04 | |
| w+b | 8 | 5 | 5.57 | 3.85 | 7.59 | 5.31 | 9.26 | 9.74 | 15.33 | 15.77 | 1.84E–03 | 1.43E–03 | 1.85E–03 | 1.54 | |
| a | 5 | 2 | 27.64 | 7.42 | 52.64 | 67.86 | 47.36 | 21.73 | 78.24 | 66.56 | 0 | NA | 0 | NA | |
| m | 6 | 2 | 13.56 | 9.94 | 16.13 | 12.41 | 27.01 | 30.99 | 33.23 | 30.85 | 0 | NA | 0 | NA | |
| 3 | b | 3 | 2 | 4 565.40 | 2 438.17 | 39 252.27 | 56 208.46 | 1 301.27 | 726.83 | 1 273.84 | 580.15 | 75.92 | 74.02 | 84.45 | 61.38 |
| 4 | b | 8 | 8 | 44 916.26 | 40 672.72 | 71 661.11 | 62 589.45 | 337.77 | 413.25 | 650.57 | 712.82 | 485.17 | 322.14 | 1 309.54 | 1 704.89 |
| a | 3 | 3 | 8 144.11 | 2 716.40 | 23 105.73 | 23 845.21 | 5 406.84 | 2 955.24 | 6 781.18 | 3 737.75 | 10.66 | 0.80 | 12.69 | 3.99 | |
| m | 4 | 4 | 2 952.54 | 957.90 | 3 019.82 | 846.25 | 1 754.79 | 897.45 | 1 683.86 | 898.44 | 71.98 | 3.76 | 66.23 | 12.23 |
Abbreviations: a, adductor muscle; avg, average; b, body; F, F mtDNA ratio in female samples; Fm, F mtDNA ratio in male samples; M, M mtDNA ratio in male samples; m, mantle; MAD, median absolute deviation; mdn, median; NA, calculation not possible; s.d., standard deviation; w, whole animal; ♀, number of female samples; ♂, number of male samples.
Statistics were calculated on all suitable technical replicates available for each sample. Three replicates were used for each sample (except for a female adductor, where two were used; Supplementary Table 2). No M mtDNA was detected in class 2 adductors and mantles.
Extractions of embryo pools were performed using MasterPure Complete DNA and RNA purification kit (Epicentre, Madison, WI, USA) or DNeasy Blood & Tissue kit (Qiagen, Mississauga, ON, Canada) (extraction modes for each pool are specified in Supplementary Table 1), whereas DNA from juvenile samples was extracted using MasterPure kit (reducing the quantities of reagents when needed to obtain a better yield). Concentration and quality of the extractions were determined with a NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA) or a Biodrop DUO (Biodrop, Cambridge, UK).
Real-time qPCR procedure
To quantify the amounts of F and M mtDNAs, real-time qPCR experiments were performed in multiplex using TaqMan chemistry, following the approach developed and utilized by Ghiselli et al. (2011) and Milani et al. (2012). Three targets were considered: the nuclear heat-shock protein 70 (hsp70) gene, the mitochondrial NADH dehydrogenase subunit 1 (nad1), and the small subunit of ribosomal RNA (12S) genes for the F and M mtDNA, respectively (refer to Ghiselli et al., 2011, for details on the choice of target sequences). Primers were provided by Alpha DNA (Montréal, Québec, Canada) and TaqMan probes by Sigma-Aldrich (The Woodlands, Texas, USA). See Supplementary Table 3 for the characteristics of primers and probes. All experiments were performed in triplicates on a PikoReal 96 Real-Time PCR System (Thermo Scientific, Burlington, ON, Canada) with the DyNAmo ColorFlash Probe qPCR Kit (Thermo Scientific), in a 10-μl total volume. The PCR cycle consisted of the following steps: uracil-N-glycosylase treatment for 2 min at 50 °C, an initial denaturation and polymerase activation for 7 min at 95 °C, and then 40 cycles of denaturation at 95 °C for 5 s and annealing plus extension at 60 °C for 30 s.
Following the method developed by Gallup and Ackermann (2008), we built a Stock1 dilution for all 60 embryo pools and three for young series samples, composed, respectively, of classes 1, 2, and 3 plus 4. All Stock1 dilutions were used to build four separate 1:10 dilution series (from 100 to 0.1 ng of DNA in reaction) to calculate the efficiencies of the targets in real time. For all four Stock1 dilution series, primers and probes concentrations were adjusted to obtain the best possible combination of efficiencies: these concentrations (enlisted in Supplementary Table 4) were then used for the quantifications of the respective samples. Efficiency of the three targets in each Stock1 dilution series (enlisted in Supplementary Table 4) was calculated automatically by PikoReal Software 2.2 (Thermo Scientific). Quantification experiments on all samples were performed using 100 ng of DNA in reaction, except for class 2 adductor muscles where 10 ng of DNA was used because of the low quantity of DNA obtained during extraction.
Statistical analyses
Ratios of F and M mtDNAs in samples from both series, normalized to the nuclear hsp70 quantity, were calculated using the efficiency-corrected ratio method (equation 3.5 from Pfaffl, 2004) for all reactions, whereas the procedure of Livak and Schmittgen (2001) for multiplex reactions was used to calculate average ratio values. Replicates in which the nuclear target failed to amplify, or showing amplification issues for the mitochondrial targets, were excluded from the subsequent calculations and analyses (see Supplementary Tables 1 and 2 for the excluded reactions). We avoided normalization to a calibrator sample for the mtDNA ratios in both series because: (1) we would have needed to choose it arbitrarily for each group, and (2) it is not technically correct to directly compare ratios from different experiments normalized to different calibrators. To calculate the growth in nuclear content in the embryo series, we normalized the content of each pool to the hsp70 median quantification cycle (Cq) of stage 2 hpf, the first of the series, using equation 3.3 from Pfaffl (2004). Statistical analyses (Wilcoxon rank-sum test, cluster analysis using the single-linkage method, linear models on the log10 transformed ratio values) and graphic elaborations were performed with R 3.1.0 (R Core Team, 2014) on RStudio 0.95 (RStudio, 2012).
Results
Three technical replicates were performed for each of the 133 samples, for a total of 399 triplex quantification reactions. Of these samples, 60 were from the embryo series and 73 from the young series. All Cq values and ratio calculations for each technical replicate in the embryo and young series, as well as the excluded replicates, are enlisted in Supplementary Tables 1 and 2, respectively. Summaries of the ratio distributions in both embryo and young series are enlisted in Tables 1 and 3, respectively.
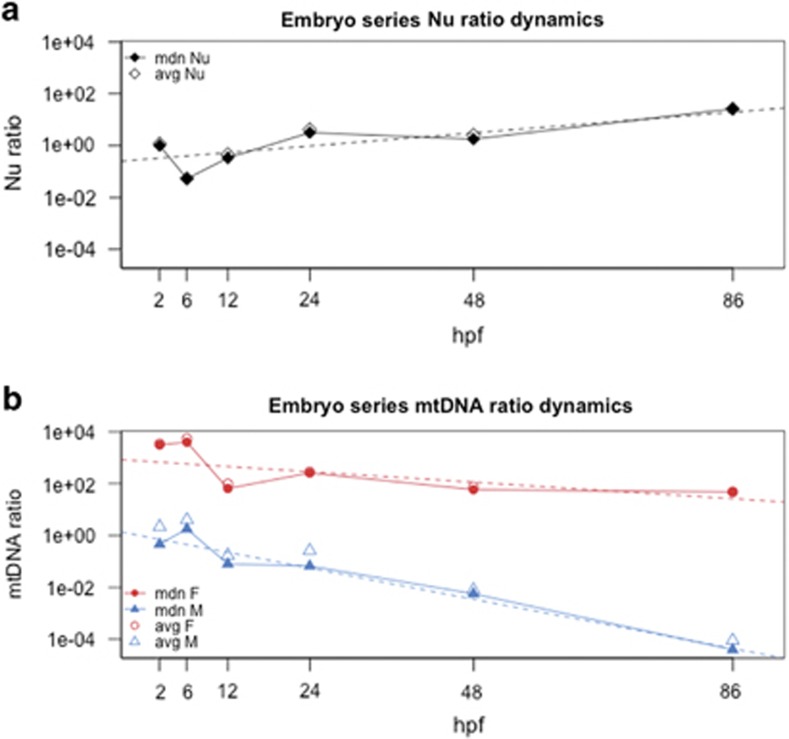
Trends in median ratios for the three targets of each stage in the embryo series are represented in Figure 1. Given the fluctuating ratios between stages 2 and 12 hpf, likely because of low nuclear target abundance, we cannot exclude erroneous estimations due to qPCR amplification issues in these stages. Nonetheless, the general trends of all targets are clear and supported by the linear models: the nuclear ratio grows one order of magnitude as expected from 2 to 86 hpf (linear model adjusted R2=0.56, P-value<2.2E–16; Figure 1a; Table 1), whereas both F and M mtDNAs content per cell decrease in time, from ~103 to ~101, and from ~10–1/101 to ~10–5, respectively (F mtDNA linear model adjusted R2=0.40, P-value<2.2E–16; M mtDNA linear model adjusted R2=0.86, P-value<2.2E–16; Figure 1b; Table 1). The M ratio trend becomes clearly distinct from the F one after 24 hpf, showing a steeper descending after this stage (Figure 1b; Table 1).
Figure 1.
Dynamics of the three targets in embryos and larvae from 2 to 86 hpf. Full lines, trends of median ratio; dotted lines, linear model trends calculated on all suitable technical replicates (see Results for statistics). Color code: black, nuclear hsp70; red, F mtDNA; blue, M mtDNA. Y axis is in log10 scale. See Table 1 for median absolute deviations and standard deviations of all ratios for each stage. (a) Nuclear target dynamics; stage 2 hpf median nuclear ratio is 1 because the median Cq of this stage has been used as the reference to calculate all nuclear ratios (see Materials and Methods). (b) F and M mtDNA dynamics. avg, average ratio; F, F mtDNA ratio; M, M mtDNA ratio; mdn, median ratio; Nu, nuclear hsp70 ratio.
Specimens of the young series have been considered ‘males' or ‘females' based on the presence or absence of M mtDNA signal in their bodies. This sexing method is not error free, but there is no other consistent way to sex clams at this developmental stage (see Discussion). On a total of 44 animals, 24 females and 20 males were determined this way (Table 2).
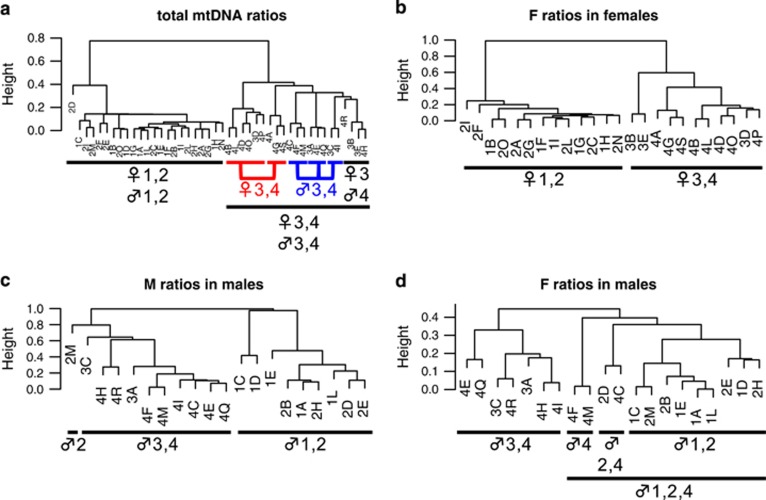
Cluster analysis separates whole animal and body samples of the young series in two main groups based on their total mtDNA ratio (average F ratio+average M ratio), as seen in Figure 2a. One cluster contains all specimens from classes 1 and 2 in a non-ordered manner, irrespective of the sample sex and/or class, whereas the other contains all specimens from classes 3 and 4, and can be divided in six sub-clusters, five of which are sex specific (Figure 2a). The total mtDNA content is not significantly different between females and males in classes 1 and 2, which all cluster together (Wilcoxon rank-sum test P-value=0.11). On the contrary, there is a significant difference between sexes in classes 3 and 4 (Wilcoxon rank-sum test P-value=3.97E–05), which supports the nearly perfect distinction between females and males in the cluster containing these two classes (clusters highlighted in colors in Figure 2a). The average ratios of F in females and M in males (Figures 2b and c) are each separated in two clusters, one containing specimens from the first two classes and the other those from the last two. Ratios of F mtDNA in males (Figure 2d) can be also divided into two major clusters containing either males of classes 3 and 4 or males from classes 1, 2, and 4.
Figure 2.
Cluster analysis results for the average mtDNA ratios in whole animals and bodies of the young series (specimens from 1 to 20 mm). Sample names are composed of a number, indicating their class, and a letter, to distinguish the samples. Symbols under the clusters indicate the sex of the specimens inside them (♀, females; ♂, males), whereas numbers indicate their class. See Supplementary Table 2 for average ratio values and standard deviations of each specimen. (a) Dendrogram of average total mtDNA ratios (average F ratio+average M ratio). Classes 1+2 are clearly separated from classes 3+4. Females and males from classes 3 and 4 tend to group in sex-specific clusters (highlighted in red and blue, respectively). A cluster contains two class 3 females (3B and 3E) plus two class 4 males (4H and 4R): the two females have the lowest total mtDNA ratios among class 3 and 4 females, whereas the two males have the highest among males of the same classes. There is no difference in total mtDNA ratio between females and males of classes 1 and 2, whereas females and males of classes 3 and 4 are significantly different (see also Results for Wilcoxon rank-sum test P-value of these comparisons). (b) Dendrogram of average F ratio in females. Specimens from classes 1 and 2 are well separated from classes 3 and 4. (c) Dendrogram of average M ratio in males. Males from classes 1 and 2 cluster separately from those of classes 3 and 4; specimen 2M is an outlier of class 2 with high quantities of M mtDNA (Supplementary Table 2); therefore, it is more closely related to the cluster containing classes 3 and 4. (d) Dendrogram of average F ratio in males. Animals from classes 3 and 4 tend to cluster together, but three specimens from class 4 (4C, 4F, and 4M) cluster closer to males from classes 1 and 2: these animals have low F mtDNA ratios, more similar to those of classes 1 and 2 (Supplementary Table 2).
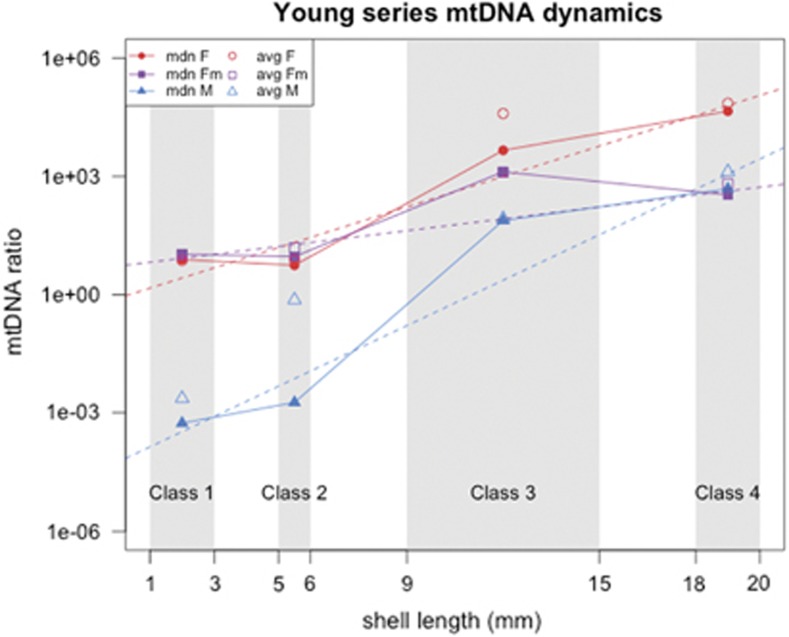
Table 4 reports the significance of Wilcoxon tests performed on mtDNA ratio comparisons in whole animal and body samples, within class and between classes (see also Table 3 for ratio statistics). There is little difference in F ratios between females and males in classes 1 and 2, whereas in classes 3 and 4 females have more F than males. F levels in males are always higher than M, except in class 4, where the two ratio distributions are comparable. F ratios in females are higher than M ratios in males in all classes. All ratios remain similar between classes 1 and 2, but they rise significantly between classes 2 and 3. From class 3 to class 4, F ratios in females and M ratios in males continue to grow, whereas F ratios in males show a slightly significant decrease (Table 3). The dynamics of median mtDNA ratios in whole animal and body samples, and the respective linear model trends, are represented in Figure 3: linear models support the growing trend of all targets, with the M showing the strongest rise (F ratio in females linear model adjusted R2=0.88, P-value<2.2E–16; F ratio in males linear model adjusted R2=0.59, P-value=3.70E–13; M ratio linear model adjusted R2=0.86, P-value<2.2E–16).
Table 4. Wilcoxon rank-sum test P-values of the mtDNA ratio comparisons in the young series.
| Sample type | Classes | Ratios | P-value | Significance | Proportions |
|---|---|---|---|---|---|
| Whole animal+body | 1 | F vs Fm | 0.011 | * | F<Fm |
| 2 | F vs Fm | 0.146 | NS | F~Fm | |
| 3 | F vs Fm | 3.996E–04 | *** | F>Fm | |
| 4 | F vs Fm | 6.202E–14 | *** | F>Fm | |
| 1 | Fm vs M | 1.289E–08 | *** | Fm>M | |
| 2 | Fm vs M | 1.728E–06 | *** | Fm>M | |
| 3 | Fm vs M | 2.165E–03 | ** | Fm>M | |
| 4 | Fm vs M | 0.071 | NS | Fm~M | |
| 1 | F vs M | 1.289E–08 | *** | F>M | |
| 2 | F vs M | 4.033E–08 | *** | F>M | |
| 3 | F vs M | 3.996E–04 | *** | F>M | |
| 4 | F vs M | 6.202E–14 | *** | F>M | |
| 1 vs 2 | F | 0.484 | NS | 1~2 | |
| 2 vs 3 | F | 5.186E–08 | *** | 2<3 | |
| 3 vs 4 | F | 0.020 | * | 3<4 | |
| 1 vs 2 | Fm | 0.806 | NS | 1~2 | |
| 2 vs 3 | Fm | 3.69E–05 | *** | 2<3 | |
| 3 vs 4 | Fm | 0.029 | * | 3>4 | |
| 1 vs 2 | M | 0.057 | NS | 1~2 | |
| 2 vs 3 | M | 5.16E–05 | *** | 2<3 | |
| 3 vs 4 | M | 3.368E–06 | *** | 3<4 | |
| Mantle | 2 | F vs Fm | 0.494 | NS | F~Fm |
| 4 | F vs Fma | 1.115E–03 | ** | F>Fm | |
| 4 | Fm vs M | 4.396E–03 | ** | Fm>M | |
| 4 | F vs M | 4.396E–03 | ** | F>M | |
| 2 vs 4 | F | 2.312E–08 | *** | 2<4 | |
| 2 vs 4 | Fm | 1.077E–04 | *** | 2<4 | |
| Adductor | 2 | F vs Fm | 0.029 | * | F<Fm |
| 4 | F vs Fmb | 0.062 | NS | F~Fm | |
| 4 | Fm vs M | 9.091E–03 | ** | Fm>M | |
| 4 | F vs M | 9.091E–03 | ** | F>M | |
| 2 vs 4 | F | 1.53E–06 | *** | 2<4 | |
| 2 vs 4 | Fm | 3.996E–04 | *** | 2<4 | |
| Mantle vs adductor | 2 | F | 2.37E–03 | ** | Mantle<adductor |
| 2 | Fm | 0.309 | NS | Mantle~adductor | |
| 4 | F | 6.804E–06 | *** | Mantle<adductor | |
| 4 | Fm | 6.804E–06 | *** | Mantle<adductor | |
| 4 | M | 0.100 | NS | Mantle~adductor |
Abbreviations: F, F mtDNA ratio in female samples; Fm, F mtDNA ratio in male samples; M, M mtDNA ratio in male samples; NS, not significant.
This test significance remained the same (P-value=9.288E–03, significance=**) when the male mantle showing an M mtDNA signal was removed from the comparison.
This test significance remained the same (P-value=0.388, not significant) when the male adductor showing an M mtDNA signal was removed from the comparison.
Significance of P-values: *<0.05; **<0.01; ***<0.001.
The Wilcoxon rank-sum test has been applied to mtDNA ratio comparisons: for couples of targets in the same class and sample type; for single targets between sexes in the same sample type and class; for mantle and adductor muscle only, for single targets between tissues. Comparisons involving the M ratio of class 2 adductors and mantles have not been performed, as no M mtDNA was detected in these tissues.
Figure 3.
Dynamics of mtDNA ratios in whole animal and body samples from the young series (specimens from 1 to 20 mm). Full lines, trends of median ratio; dotted lines, ratio linear models. Lines color code: red, F mtDNA in females; purple, F mtDNA in males; blue, M mtDNA in males. Gray squares in the background define the dimensional interval of each class (Table 2), and points are positioned on the average shell length of the respective class (class 1, 2 mm; class 2, 5.5 mm; class 3, 12 mm; class 4, 19 mm) for graphical purposes only. Y axis is in log10 scale. Class 2 values comprise both bodies and whole animals. See Table 3 for ratio statistics, and Results for linear model statistics. avg, average ratio; F, F mtDNA ratio in females; Fm, F mtDNA ratio in males; M, M mtDNA ratio in males; mdn, median ratio.
No M mtDNA was detected in class 2 adductor muscles and mantles (Table 3), whereas it was detected in traces in only one mantle and one adductor sample from two different males of class 4 (Table 3; Supplementary Table 2). Table 4 shows the significance of the Wilcoxon test comparisons for mtDNA ratios in these tissues. In the mantle, F ratios are similar between females and males in class 2, and in class 4 are higher in females; in the adductor muscle, this difference is slightly significant between females and males in class 2, and not significant in class 4 (the significance of these tests in class 4 remains the same even when removing the heteroplasmic mantle and adductor samples; see Table 4 description). M levels of mantle and adductor samples in males from class 4 are always lower than F levels (Table 3). Finally, F ratios in heteroplasmic tissues are lower than those in all other male and female F-homoplasmic samples (Wilcoxon rank-sum tests: mantles P-value=2.45E–03, adductors P-value=9.88E–04); the same goes for the total mtDNA ratios, as the traces of M in the heteroplasmic tissues do not significantly affect the total (data not shown). F levels rise significantly between classes 2 and 4 in both sexes and in both tissue types. The mantle always has lower F ratios than the adductor muscle, except in class 2 male tissues where the distributions are comparable. No significant difference is found in the M content of the heteroplasmic male mantle and adductor samples.
Discussion
MtDNA replication is dormant during early embryogenesis
The observed decrease of F mtDNA content per cell in our pools of embryos and larvae (Figure 1b) is evidently an outcome of the denominator of the ratio, that is, the amount of nuclear target, which increases from 2 to 86 hpf (Figure 1a). At the same time, this indicates a weak, undetectable or absent mtDNA replication until 86 hpf (broadly corresponding to the veliger stage), which results in a significant decrease of mtDNA copies per cell. The absence of detectable replication of the maternally transmitted F genome during R. philippinarum embryogenesis is consistent with previous reports of reduction in mtDNA content per cell during early development in other animal groups (Milani et al., 2012). Interestingly, in R. philippinarum, the paternally transmitted M mtDNA seems to behave like the F at least until 24 hpf. Compared with the F ratio, the M mtDNA ratio shows a stronger drop, with a well distinguishable decrease after 24 hpf (Figure 1b; Table 1). As above, this drop can be explained by the absence of detectable replication of the M genome during early embryogenesis, coupled with the loss of M mtDNA in embryos that are going to develop into females (according to the DUI routes of mtDNA transmission).
Early embryogenesis of DUI species is thought to be the time frame in which sperm mitochondria are eliminated/diluted in females and retained in male offspring, that is, checkpoint #2 described by Ghiselli et al. (2011). Several works on Mytilus suggest that in this system checkpoint #2 is not a sudden event. In the pioneering work of Sutherland et al. (1998) on Mytilus edulis, the presence/absence of M mtDNA was tested in larvae at 18, 24, and 48 hpf using PCR. Eighteen hours post fertilization larvae from both female-biased and balanced progenies showed an identical M mtDNA signal, whereas starting from 24 hpf the signal was absent in the female-biased progenies and in some larvae from the balanced progenies (see Saavedra et al., 1997, and Kenchington et al., 2002, for discussions on the topic). Sano et al. (2011) used real-time qPCR to assess the F/M mtDNA ratio variation up to 24 hpf in Mytilus galloprovincialis larvae of female- and male-biased offsprings. In male-biased larvae, the F/M ratio remained the same up to 24 hpf, meaning that no sperm mitochondria elimination occurred (and that the relative quantities of F and M did not change), whereas in female-biased larvae this ratio increased after 3 hpf, that is, M mtDNA has started to diminish earlier in this group. From these studies it appears that in Mytilus checkpoint #2 begins before 24 hpf, and that after that time M mtDNA is no more detectable in females. In R. philippinarum, Milani et al. (2012) found no changes in both F and M mtDNA copy number up to 2 hpf, using a multiplex real-time qPCR approach on pools of embryos from different crosses, hence the authors concluded that checkpoint #2 happens after 2 hpf in this species.
The pools of R. philippinarum embryos used in this study were composed of specimens coming from a mass spawning fertilization event that involved dozens of males and females, so no prior information about offspring sex bias was available. Nevertheless, considering the large number of animals per pool, the fact that they were born from many different females producing differently sex-biased progenies, and that the overall sex ratio of wild populations of DUI species does not deviate significantly from 1:1 (Kenchington et al., 2002; Ghiselli et al., 2012), we can assume a balanced sex ratio for each stage. Therefore, the drop of M mtDNA ratio starting from 24 hpf can be ascribable to checkpoint #2, if we assume that the M mtDNA becomes undetectable from a certain point on from half of the embryos (the females; Sutherland et al., 1998), while it persists without amplification in the other half (the males). We also have to consider a stronger effect of dilution for the M mitochondrial genome in males compared with the F, given the much smaller initial quantity of M, as seen in Mytilus (Sutherland et al., 1998; Sano et al., 2011). Even if the exact timing of checkpoint #2 could not be precisely determined given the chosen experimental approach, and considering that the M mtDNA observed after 24 hpf is most likely carried only by males, the loss of M from females probably ends around 24 hpf in R. philippinarum, a situation similar to that of M. edulis observed by Sutherland et al. (1998).
Approaching adulthood: sex-specific mtDNA dynamics in young clams
One way to ascertain the sex of a bivalve is to check for the type of gametes produced by an individual: this system however requires reproductively mature adults. In pre-adulthood DUI bivalves, whose dimensions and age might not be suitable for a microscopic examination of the gonads, the only approach to predict the sex of a specimen is to check for the presence or absence of the M mtDNA, as no nuclear sex markers are known yet for bivalves. Indeed, before their first reproductive season, females are expected to have already become homoplasmic for F, whereas males should be heteroplasmic (both are consequences of checkpoint #2; Ghiselli et al., 2011). This molecular approach, however, can be the subject to two potential sources of error: heteroplasmic females scored as males, and males lacking M mtDNA sexed as females.
Examples for such exceptions are mostly known in the Mytilus species complex (Brannock et al., 2013; Zouros, 2013). In Mytilus, males without M mtDNA have been obtained artificially by inducing triploidy and/or through interspecific crosses (Kenchington et al., 2009). Natural hybridization among different Mytilus species also appears to induce the retention of the M mtDNA in females (Brannock et al., 2013). However, these two ‘unordinary' situations, triploidy and hybridization, can interfere with sex determination and/or fate of paternal mitochondria in the two sexes. In contrast to what has been observed in Mytilus, female mitochondrial heteroplasmy appears to be uncommon in R. philippinarum: in the study of Ghiselli et al. (2011), only 2 of 33 females (6.06%) were found carrying the M mtDNA in their soma, and until now they are the only known ascertained examples of heteroplasmic females in this species. Moreover, males lacking the M mtDNA have never been observed in R. philippinarum. To our knowledge, no experiment has been done yet in this species to induce triploidy, and to check if this phenomenon can decouple maleness and presence of M mtDNA as in Mytilus.
Under normal conditions, it appears that the fate of paternal mitochondria in developing individuals of R. philippinarum is quite stable and not subject to important variations: males always carry the M mtDNA, and females have low probabilities of being heteroplasmic. Our samples are born under near-natural conditions: by doing this, we avoided hybridization and minimized other factors that could alter the behavior of M mitochondria in females or males and/or produce exceptional individuals. Under these circumstances, and given the results from our embryo series (that is, M mtDNA signal in females may be lost after 24 hpf), the utmost care in handling samples to avoid contamination among males and females, and the sensitivity of our experimental approach, we are confident that categorizing as females all the animals in whose bodies no trace of M mtDNA was detected, and as males all heteroplasmic animals, is the best possible approximation of the real sex of the utilized juvenile clams.
Our results for these juvenile clams suggest a general positive correlation between the dimension of an individual and its mtDNA content (Table 3; Figure 3). For example, regarding mantle and adductor muscle tissues, we found that class 2 specimens have always less mtDNA than those from class 4 (Table 4). Our results also show that class 4 females tend to have more F than males in their mantles, but not in their adductor muscles (Table 4), and that the difference remains the same even when comparing female tissues versus male homoplasmic tissues only. The reason for this difference in F content in mantle but not in adductor muscle between sexes remains obscure, and can be putatively ascribed to a sampling effect. It is however interesting to notice that the two heteroplasmic male tissues have significantly less F, and total, mtDNA than homoplasmic ones: again, the number of heteroplasmic tissues is low (one mantle and one adductor) and a larger sample size would be needed to confirm this observation, but some kind of conflict during mtDNA replication in these tissues that lowers the F mitochondrial genome ratio cannot be excluded a priori. Interestingly, in both sexes of class 4, the adductor muscle has higher mtDNA ratios than the mantle. The presence of a higher quantity of mtDNA suggests that the former tissue has substantially higher energy needs than the latter. This is expected, as the adductor, that is, the muscle that opens and closes the valves, is a much more active organ than the mantle.
In whole animals and bodies of juveniles, between classes 1 and 2, the total and single (F in females, M in males, F in males) mtDNA contents do not change significantly (Figures 2 and 3; Table 4). The similarity between sexes in total mtDNA content (Figure 2a) indicates that the low ratios of M in males (Table 3) do not appreciably contribute to the total in these two classes. There is, on the contrary, a disparity between sexes in total mtDNA content in classes 3 and 4 (Figure 2a), which seems to be due to the rapid increase of M content in males, which starts at very low levels in classes 1 and 2, and becomes dominant over the F in most males of class 4 (Table 4; Figure 3; Supplementary Table 2). Specifically, in males from classes 1–4, the F ratio shows a slow growth (Figure 3): some class 4 males even have levels of F similar to class 1 and 2 individuals (Figure 2d). The M mtDNA increase is sudden, and very similar to that of the F mtDNA in females, maybe even more accelerated (Figure 3). Thus, both F mtDNA in female bodies and M mtDNA in male bodies are subject to a strong boost in replication between classes 2 and 3, which goes on up to class 4 (Figure 3; Tables 3 and 4; females and males of classes 1+2 and 3+4 are indeed well distinguishable using these two ratios; Figures 2b and c).
What is the cause of this mtDNA boost? It may be due to the cellular proliferation and/or growing energetic demands of the organs located in the bodies during development, compared with the mantle and adductor muscle, for example. On the other hand, organs in the bodies include the gonads, and the occurrence of a replication boost just before the first reproductive season, together with the observation that in males only the M mtDNA, that is going to be transmitted by sperm, is showing this increase, may hint to another non-mutually excluding explanation. We know that in DUI species, the male gonad holds high quantities of M mtDNA (both in R. philippinarum and Mytilus; Ghiselli et al., 2011; Zouros, 2013), and also that in R. philippinarum adults of both sexes, the gonad has the highest mtDNA content (Ghiselli et al., 2011; Milani et al., 2014). Knowing that the largest analyzed animals are those closest to the first reproductive season (Devauchelle, 1990), the observed mtDNA boosts in both females and males of classes 3 and 4 are most probably the outcome of the first gonad formation.
Conclusions
In this study, we characterized the sex-specific replication trends of F and M mtDNAs during embryogenesis and early development in the DUI species R. philippinarum. As seen in other animal species, we observed that mtDNA replication does not happen during the first cellular divisions, and that tissue- and sex-specific mtDNA contents are present in juvenile individuals. We also pinpointed two events of pivotal importance to study how DUI can occur in place of strictly maternal inheritance: the loss of M mtDNA in females but not in males (around 24 hpf), and the start of gonad production (in young clams with a shell length >6 mm, probably). Studying these two temporal windows in the male path of development will allow us to understand what mechanisms can maintain a paternally transmitted mtDNA through the generations.
Data archiving
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.q3hs1.
Acknowledgments
We are grateful to Edoardo Turolla from the ‘Centro Ricerche Molluschi' (Goro, Italy) for providing the samples. We want to thank Beatrice Fabbri and Marco Masironi for their invaluable help in the laboratory, the Thermo Scientific technical support staff for their assistance, and the anonymous reviewers that helped improving the paper. This work was supported by the Italian PRIN09 grant and the ‘Canziani bequest' (MP), and by the National Sciences and Engineering Research Council (SB).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Brannock PM, Roberts MA, Hilbish T. (2013). Ubiquitous heteroplasmy in Mytilus spp. resulting from disruption in doubly uniparental inheritance regulation. Mar Ecol Prog Ser 480: 131–143. [Google Scholar]
- Breton S, Doucet Beaupré H, Stewart DT, Hoeh WR, Blier PU. (2007). The unusual system of doubly uniparental inheritance of mtDNA: isn't one enough? Trends Genet 23: 465–474. [DOI] [PubMed] [Google Scholar]
- Breton S, Milani L, Ghiselli F, Guerra D, Stewart DT, Passamonti M. (2014). A resourceful genome: updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends Genet 30: 555–564. [DOI] [PubMed] [Google Scholar]
- Burgstaller JP, Johnston IG, Jones NS, Albrechtová J, Kolbe T, Vogl C et al. (2014). MtDNA segregation in heteroplasmic tissues is common in vivo and modulated by haplotype differences and developmental stage. Cell Rep 7: 2031–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Kenchington E, Zouros E. (2004). Differential segregation patterns of sperm mitochondria in embryos of the blue mussel (Mytilus edulis. Genetics 166: 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell AT, Kenchington ELR, Zouros E. (2006). Segregation of sperm mitochondria in two- and four-cell embryos of the blue mussel Mytilus edulis: implications for the mechanism of doubly uniparental inheritance of mitochondrial DNA. Genome 49: 799–807. [DOI] [PubMed] [Google Scholar]
- Devauchelle N. (1990) Sexual development and maturity of Tapes philippinarum. In: Tapes philippinarum Biologia e Sperimentazione. Ente Sviluppo Agricolo, Veneto, Italy. pp 48–62. [Google Scholar]
- Gallup JM, Ackermann MR. (2008). The ‘PREXCEL-Q Method' for qPCR. Int J Biomed Sci 4: 273–293. [PMC free article] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Passamonti M. (2011). Strict sex-specific mtDNA segregation in the germ line of the DUI species Venerupis philippinarum (Bivalvia: Veneridae). Mol Biol Evol 28: 949–961. [DOI] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Chang PL, Hedgecock D, Davis JP, Nuzhdin SV et al. (2012). De Novo assembly of the Manila Clam Ruditapes philippinarum transcriptome provides new insights into expression bias, mitochondrial doubly uniparental inheritance and sex determination. Mol Biol Evol 29: 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra D, Ghiselli F, Passamonti M. (2014). The largest unassigned regions of the male- and female-transmitted mitochondrial DNAs in Musculista senhousia (Bivalvia Mytilidae). Gene 536: 316–325. [DOI] [PubMed] [Google Scholar]
- Hoekstra RF. (2011) Nucleo-cytoplasmic conflict and the evolution of gamete dimorphism In: Togashi T, Cox PA (eds). The Evolution of Anisogamy. Cambridge University Press: Cambridge, UK. pp 111–130. [Google Scholar]
- Jokinen R, Battersby BJ. (2013). Insight into mammalian mitochondrial DNA segregation. Ann Med 45: 149–155. [DOI] [PubMed] [Google Scholar]
- Kenchington E, MacDonald B, Cao L, Tsagarakis D, Zouros E. (2002). Genetics of mother-dependent sex ratio in blue mussels (Mytilus spp.) and implications for doubly uniparental inheritance of mitochondrial DNA. Genetics 161: 1579–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenchington EL, Hamilton L, Cogswell A, Zouros E. (2009). Paternal mtDNA and maleness are co-inherited but not causally linked in mytilid mussels. PLoS ONE 4: e6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N. (2011). Mitonuclear match: optimizing fitness and fertility over generations drives ageing within generations. Bioessays 33: 860–869. [DOI] [PubMed] [Google Scholar]
- Lane N. (2012). The problem with mixing mitochondria. Cell 151: 246–248. [DOI] [PubMed] [Google Scholar]
- Livak KL, Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Maurizii MG, Passamonti M. (2011). Doubly uniparental inheritance of mitochondria as a model system for studying germ line formation. PLoS ONE 6: e28194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Passamonti M. (2012). Sex-linked mitochondrial behavior during early embryo development in Ruditapes philippinarum (Bivalvia Veneridae) a species with the doubly uniparental inheritance (DUI) of mitochondria. J Exp Zool B Mol Dev Evol 318: 182–189. [DOI] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Iannello M, Passamonti M. (2014). Evidence for somatic transcription of male-transmitted mitochondrial genome in the DUI species Ruditapes philippinarum (Bivalvia: Veneridae). Curr Genet 60: 163–173. [DOI] [PubMed] [Google Scholar]
- Mishra P, Chan DC. (2014). Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol 15: 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata M, Komaru A. (2005). Specific location of sperm mitochondria in mussel Mytilus galloprovincialis zygotes stained by MitoTracker. Dev Growth Differ 47: 255–263. [DOI] [PubMed] [Google Scholar]
- Passamonti M, Scali V. (2001). Gender-associated mitochondrial DNA heteroplasmy in the venerid clam Tapes philippinarum (Mollusca Bivalvia). Curr Genet 39: 117–124. [DOI] [PubMed] [Google Scholar]
- Passamonti M, Ghiselli F. (2009). Doubly uniparental inheritance: two mitochondrial genomes, one precious model for organelle dna inheritance and evolution. DNA Cell Biol 28: 79–89. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2004) Quantification strategies in real-time PCR. In: Bustin SA (ed). A-Z of Quantitative PCR. International University Line: La Jolla, CA, USA. pp 87–112. [Google Scholar]
- R Core Team. (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- RStudio. (2012) RStudio: Integrated Development Rnvironment for R. Boston, MA, USA.
- Saavedra C, Reyero MI, Zouros E. (1997). Male-dependent doubly uniparental inheritance of mitochondrial DNA and female-dependent sex ratio in the mussel Mytilus galloprovincialis. Genetics 145: 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano N, Obata M, Ooie Y, Komaru A. (2011). Mitochondrial DNA copy number is maintained during spermatogenesis and in the development of male larvae to sustain the doubly uniparental inheritance of mitochondrial DNA system in the blue mussel Mytilus galloprovincialis. Dev Growth Differ 53: 816–821. [DOI] [PubMed] [Google Scholar]
- Skibinski DO, Gallagher C, Beynon CM. (1994. a). Mitochondrial DNA inheritance. Nature 368: 817–818. [DOI] [PubMed] [Google Scholar]
- Skibinski DOF, Gallagher C, Beynon CM. (1994. b). Sex-limited mitochondrial DNA transmission in the marine mussel Mytilus edulis. Genetics 138: 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland B, Stewart D, Kenchington ER, Zouros E. (1998). The Fate of Paternal Mitochondrial DNA in Developing Female Mussels, Mytilus edulis: Implications for the Mechanism of Doubly Uniparental Inheritance of Mitochondrial DNA. Genetics 148: 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologidis I, Fodelianakis S, Gaspar MB, Zouros E. (2008). Doubly uniparental inheritance (DUI) of mitochondrial DNA in Donax trunculus (Bivalvia: Donacidae) and the problem of its sporadic detection in Bivalvia. Evolution 62: 959–970. [DOI] [PubMed] [Google Scholar]
- Venetis C, Theologidis I, Zouros E, Rodakis GC. (2006). No evidence for presence of maternal mitochondrial DNA in the sperm of Mytilus galloprovincialis males. Proc Biol Sci 273: 2483–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Chalkia D. (2013). Mitochondrial DNA Genetics and the Heteroplasmy Conundrum in Evolution and Disease. Cold Spring Harb Perspect Biol 5: a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR. (1994. a). An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proc Natl Acad Sci USA 91: 7463–7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR. (1994. b). Mitochondrial DNA inheritance. Nature 368: 818. [DOI] [PubMed] [Google Scholar]
- Zouros E. (2013). Biparental inheritance through uniparental transmission: the doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol Biol 40: 1–31. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.