Abstract
Human respiratory syncytial virus (RSV) is a major health challenge in the young and elderly owing to the lack of a safe and effective vaccine and proven antiviral drugs. Understanding the mechanisms by which viral genes and proteins modulate the host response to infection is critical for identifying novel disease intervention strategies. In this study, the RSV non-structural protein NS1 was shown to suppress miR-24 expression during infection. Lack of NS1 was linked to increased expression of miR-24, whilst NS1 overexpression suppressed miR-24 expression. NS1 was found to induce Kruppel-like factor 6 (KLF6), a transcription factor that positively regulates the transforming growth factor (TGF)-β pathway to induce cell cycle arrest. Silencing of KLF6 led to increased miR-24 expression via downregulation of TGF-β. Treatment with exogenous TGF-β suppressed miR-24 expression and induced KLF6. Confocal microscopy showed co-localization of KLF6 and RSV NS1. These findings indicated that RSV NS1 interacts with KLF6 and modulates miR-24 expression and TGF-β, which facilitates RSV replication.
Introduction
Human respiratory syncytial virus (RSV) is a ubiquitous negative-sense ssRNA virus that can cause severe lung disease following infection (CDC, 2013; Hall et al., 2009; Nair et al., 2010). RSV is a member of the genus Pneumovirus, family Paramyxoviridae with a non-segmented genome that encodes 10 genes and 11 proteins (NS1, NS2, N, P, M, SH, G, F, M2-1, M2-2 and L). The two non-structural proteins (NS1 and NS2), which are not part of the intact virion, are transcribed and translated during infection (Moore et al., 2008).
RSV infection rates are high and by age 2 years the majority of young children have experienced at least one infection (CDC, 2008, 2013; Hall et al., 2009; Mori et al., 2014; Nair et al., 2010; Stockman et al., 2012; Zhou et al., 2012). RSV infection in high-risk individuals, such as infants, young children, immunocompromised adults and the elderly, can manifest as serious pulmonary inflammatory disease including bronchiolitis and pneumonia (Hoffman et al., 2004; Moore et al., 2013; Openshaw & Chiu, 2013; Oshansky et al., 2009b; Psarras et al., 2004; Vicencio, 2010). There is also substantial evidence that early RSV infection can mediate airway remodelling, a feature that predisposes individuals to asthma development and exacerbation (Fong et al., 2000; Foronjy et al., 2014; Hirakawa et al., 2013; Hotard et al., 2015; Liesman et al., 2014; Meng et al., 2014; Piedimonte, 2002, 2003; Tan et al., 2008; Wu et al., 2011). Transforming growth factor (TGF)-β is expressed during RSV infection of lung epithelial cells (Gibbs et al., 2009; McCann & Imani, 2007; Mgbemena et al., 2011) and several studies indicate that it plays an important role in asthma development (de Faria et al., 2008; Fong et al., 2000; Gagliardo et al., 2013; Hoshino et al., 1998; Howell & McAnulty, 2006; Pelaia et al., 2007; Sharma et al., 2009). TGF-β also regulates aspects of the inflammatory cytokine response to RSV infection (Thornburg et al., 2010) and cell cycle arrest (Gibbs et al., 2009; McCann & Imani, 2007; Wu et al., 2011), although the mechanism(s) are not completely known. Kruppel-like factor 6 (KLF6), an evolutionarily conserved and ubiquitously expressed zinc-finger protein that belongs to the mammalian Sp1/KLF family of transcriptional regulators (Andreoli et al., 2010), is a critical transcription factor required for TGF-β gene expression during RSV infection of human lung epithelial cells (Mgbemena et al., 2011).
RSV has a tropism for respiratory epithelial cells, thus human type II lung epithelial (A549) cells are often used to model the host response to RSV infection (Gibbs et al., 2009; Martínez et al., 2007; Munday et al., 2010). RSV attaches to cells by its two major surface proteins, the attachment (G) and fusion (F) proteins, which bind to cellular glycosaminoglycans (Hallak et al., 2000). RSV F and G proteins also bind to several other cell constituents, such as CX3CR1 molecules (Tripp et al., 2001), Toll-like receptor 4 (TLR4) (Kurt-Jones et al., 2000), RhoA (Budge & Graham, 2004; Pastey et al., 1999), intercellular adhesion molecule 1 (Mastrangelo & Hegele, 2013; Mata et al., 2012) and nucleolin (Tayyari et al., 2011). These interactions may facilitate infection and contribute to tropism.
The virus–host interactions that follow RSV binding are modified and regulated by several RSV proteins, including non-structural and G proteins, which modulate pro-inflammatory and antiviral cytokine expression by host cells (Atreya et al., 1998; Bossert et al., 2003; Boyapalle et al., 2012; Elliott et al., 2007; Kotelkin et al., 2006; Liesman et al., 2014; Ling et al., 2009; Lo et al., 2005; Moore et al., 2008; Munir et al., 2011; Ren et al., 2011; Schlender et al., 2000; Spann et al., 2004, 2005; Wright et al., 2006; Wu et al., 2012; Xu et al., 2014). RSV non-structural proteins inhibit minigenome transcription and viral RNA replication (Atreya et al., 1998), and abrogate the innate host response to infection in part by controlling expression of host cell proteins involved in the cell cycle and replication (Atreya et al., 1998; Liesman et al., 2014; Wu et al., 2012). NS2 antagonizes IFN-β activation by binding to retinoic acid-inducible gene 1 (Ling et al., 2009), and NS1 and NS2 inhibit activation of IFN regulatory factor 3 (IRF3) (Bossert et al., 2003; Spann et al., 2005), promoting degradation of transcription factors for SOCS (suppressor of cytokine signalling) proteins (Moore et al., 2008; Xu et al., 2014). RSV lacking NS1 and NS2 genes are attenuated in vivo (Jin et al., 2000; Kong et al., 2007; Luongo et al., 2013; Straub et al., 2011; Teng & Collins, 1999; Teng et al., 2000; Whitehead et al., 1999). Normal human bronchial epithelial (NHBE) cells overexpressing RSV NS1 show reduced HLA-DR, CD80 and CD86 expression, and inhibition of T-helper Th1, Th2 and Th17 cell differentiation, whilst overexpression of RSV NS2 inhibits Th2 and Th17 cell differentiation (Qin et al., 2014). Codon deoptimized RSV is attenuated in vitro, but evokes a strong humoral response upon vaccination and challenge (Meng et al., 2014). These and other immune modulators inhibit the development of the antiviral state (Bossert et al., 2003; Elliott et al., 2007; Goswami et al., 2013; Lo et al., 2005; Moore et al., 2008; Oshansky et al., 2009a; Spann et al., 2005).
The role of microRNAs (miRNAs) in post-transcriptional regulation of host genes responding to RSV infection is not fully understood. miRNA expression is known to be regulated by multiple processes including TLR4 signalling (Nahid et al., 2009, 2011a, b; O'Connell et al., 2007; Pauley et al., 2010; Schulte et al., 2013; Taganov et al., 2006), IRF3 activation (Liu et al., 2009), IFN-stimulated gene transcripts (Eis et al., 2005) and by viral proteins such as RSV G (Bakre et al., 2012). RSV infection of A549 cells affects a set of miRNAs (five that are induced and two which are repressed), particularly let-7f expression, which is induced primarily by RSV G (Bakre et al., 2012). Treatment of A549 cells with purified RSV G enhanced let-7f expression, a feature not observed following RSV F treatment. Importantly, modulation of the let-7 family of miRNAs with miRNA mimics and inhibitors affected RSV replication, indicating that RSV modulates host miRNA expression to affect the outcome of the antiviral host response and this is mediated to a considerable extent by RSV G protein expression (Bakre et al., 2012).
The data presented here, and that from related research (Thornburg et al., 2012), suggest that RSV NS1 suppresses host miRNA expression. In this study, it is shown that recombinant RSV lacking the NS1 gene induces miR-24 expression, and others have shown that ablation of NS1 and NS2 results in elevated let-7i and miR-30b expression (Thornburg et al., 2012). The molecular pathways by which RSV NS1 modulates miRNA expression have been fully elucidated, although the findings demonstrate a role for TGF-β in miR-24 suppression. Specifically, in this study it is shown that the NS1 induces transcription factor KLF6, a positive regulator of TGF-β (Mgbemena et al., 2011), which then suppresses miR-24 in a feed-forward pathway that further induces TGF-β. These findings demonstrate a novel mechanism in which RSV NS1 potentially interacts with KLF6 to modulate miR-24 and TGF-β, thus facilitating cell cycle arrest, reduced apoptosis and elevated RSV replication (Bakre et al., 2012; Gibbs et al., 2009; Mgbemena et al., 2011).
Results
RSV NS1 suppresses miR-24 expression
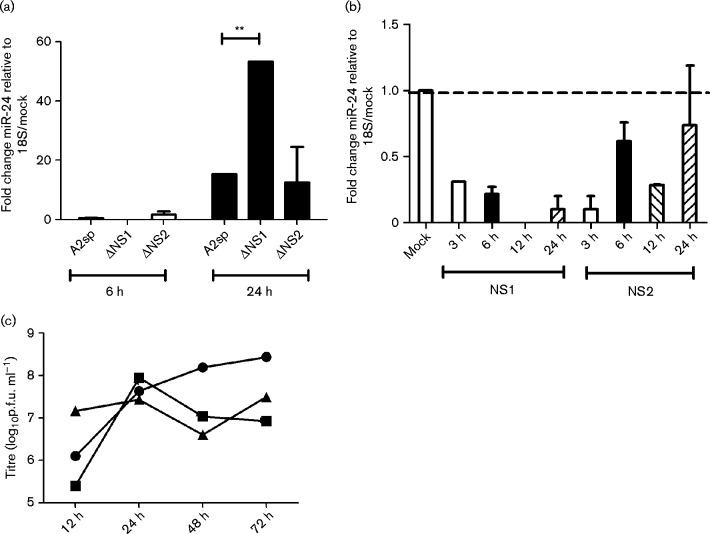
To investigate the role of RSV non-structural proteins in modulating miR-24 expression, A549 cells were infected (m.o.i. 1.0) with sucrose-purified (sp) or unpurified WT (RSV A2) or recombinant RSV A2 lacking NS1 [rA2ΔNS1EGFP (ΔNS1) or NS2 (rA2ΔNS2EGFP (ΔNS2)] genes. miR-24 expression was determined relative to 18S rRNA as described previously (Bakre et al., 2012). miR-24 expression was induced ∼20-fold at 24 h with RSV A2 infection and was further increased in ΔNS1 infection (Fig. 1a). Similar data were obtained for sucrose-purified and unpurified viruses. To confirm if this was modulated by NS1 and/or NS2, A549 cells were transfected with plasmids expressing RSV NS1/NS2 fused to EGFP or control plasmid expressing EGFP alone. NS1 and NS2 expression was validated by fluorescence microscopy beginning 6 h post-transfection (Fig. S1, available in the online Supplementary Material). miR-24 expression was suppressed by NS1 (but less so by NS2) overexpression at all time points tested, indicating that RSV NS1 suppressed miR-24 expression. Plaque titres showed equivalent replication of RSV A2, ΔNS1 and ΔNS2 to 24 h (Fig. 1c). RSV ΔNS1 and ΔNS2 viruses exhibited delayed growth 24 h post-infection (p.i.) due to an inability to counter the host innate immune response, as has been reported previously (Spann et al., 2005). As the opposite phenotypes occurred with NS1 deficiency versus overexpression, these data suggested that NS1 was the predominant regulator of miRNA expression and subsequent experiments were focused on elucidating this role.
Fig. 1.
RSV NS1 modulates miR-24 expression. (a) Lack of RSV NS1 induces miR-24 expression. A549 cells were infected with sucrose-purified (sp) RSV A2 (A2sp), ΔNS1 or ΔNS2 virus (m.o.i. 1.0) for 6 or 24 h followed by RNA isolation and quantitative real-time (qRT)-PCR for miR-24 relative to A549 cells treated with 30 % sucrose as mock and 18S rRNA as a housekeeping control. (b) RSV NS1 overexpression suppresses miR-24. miR-24 expression was measured relative to 18S rRNA in A549 cells transfected with pEGFPC1-NS1 or pEGFPC1-NS2 (50 ng each) plasmids for varying times. Data shown are representative of three independent experiments. (c) WT and ΔNS1/ΔNS2 viruses grow at equivalent titres at early time points. Viral titres of RSV A2 (•), ΔNS1 (▪) or ΔNS2 (▴) viruses were determined by plating serial 10-fold dilutions of infected A549 lysates on Vero E6 cells as per the standard protocol and visualized by staining for RSV F protein using mAb-131-2A. Data represent mean ± sem of four biological replicates. ** P < 0.01.
RSV NS1 modulates TGF-β expression via KLF6
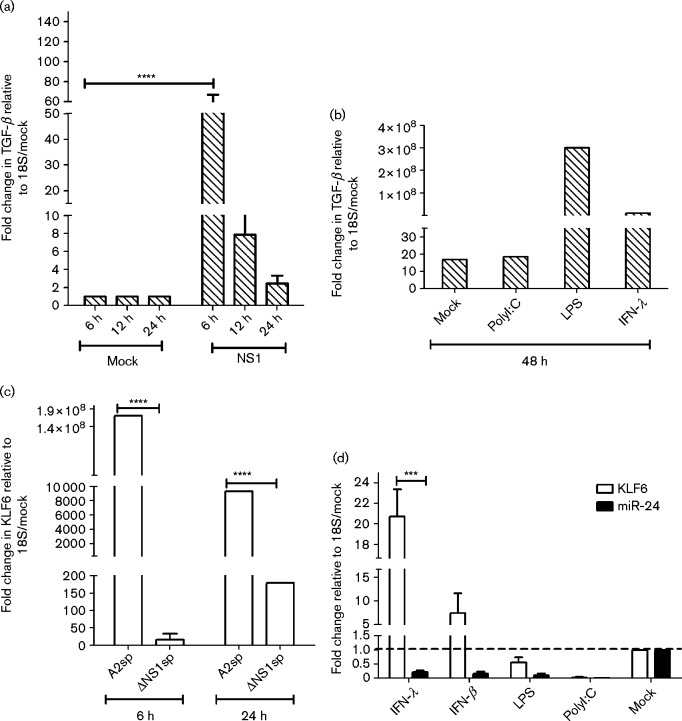
RSV non-structural proteins have been shown to interact with several host nuclear, cytosolic and mitochondrial factors, leading to suppression of the antiviral response, cell cycle regulation, DNA damage repair and culminating in G0/G1 phase cell cycle arrest (Atreya et al., 1998; Bossert et al., 2003; Boyapalle et al., 2012; Elliott et al., 2007; Lo et al., 2005; Munday et al., 2010; Munir et al., 2011; Ren et al., 2011; Schlender et al., 2000; Spann et al., 2004, 2005; Wu et al., 2012; Xu et al., 2014). TGF-β expression during RSV infection has been shown to be important for cell cycle inhibition in the G0/G1 phase (Gibbs et al., 2009; McCann & Imani, 2007; Wu et al., 2011) and miR-24 has been shown to be suppressed by TGF-β (Sun et al., 2008), as well as regulate TGF-β precursor processing (Luna et al., 2011). As preliminary data showed a role for NS1 in regulating miR-24 expression, the impact of NS1 overexpression on TGF-β expression was determined. The TGF-β transcript was significantly (P < 0.05) induced post-NS1 overexpression (Fig. 2a). This finding is in agreement with a previous study showing TGF-β was induced following RSV infection and caused cell cycle arrest (Gibbs et al., 2009). Additionally, as early as 6 h post-treatment, it was shown that treatment of NS1-transfected cells with recombinant IFN-λ and lipopolysaccharide (LPS) but not polyI : C or IFN-β, induced TGF-β expression (Fig. 2b). As type III IFNs constitute the majority of antiviral cytokines produced during RSV infection (both at early and late time points), these data suggest that type III IFNs expressed by RSV-infected cells may contribute to cell cycle arrest via induction of TGF-β in neighbouring uninfected and infected cells, facilitating viral replication and persistence.
Fig. 2.
RSV NS1 and IFN-λ induce TGF-β expression via KLF6. (a) Overexpression of RSV NS1 induces TGF-β. A549 cells were transfected with pEGFPC1-NS1 or pEGFPC1 alone for varying times and TGF-β expression was determined by qRT-PCR at different time points following NS1 overexpression. Data represent mean ± sem of three independent experiments. (b) Secondary triggers of TGF-β expression. NS1-transfected A549 cells were either mock treated, or treated with recombinant IFN-λ (10 ng ml− 1) or LPS (1 μg ml− 1), or transfected with high-molecular-mass polyI : C (10 ng ml− 1) for 24 h. TGF-β expression was determined by qRT-PCR. (c) RSV NS1 induces KLF6 expression. WT RSV A2 but not ΔNS1 infection induces KLF6 expression. A549 cells were infected with sucrose purified (sp) WT RSV A2 or recombinant ΔNS1 virus (m.o.i. 1.0) for 24 h followed by qRT-PCR analysis of KLF6 expression. Data represent two independent experiments. (d) Secondary triggers of KLF6 expression. pEGFPC1-NS1-transfected A549 cells were mock treated, or treated with IFN-λ (10 ng ml− 1), IFN-β (10 ng ml− 1) or LPS (1 μg ml), or transfected with high-molecular-mass polyI : C (10 ng ml− 1) for 24 h in two independent experiments. KLF6 or miR-24 expression was measured relative to 18S rRNA. *** P < 0.001; **** P < 0.0001.
Transcription factor KLF6 positively regulates the expression of TGF-β during RSV infection by binding to the TGF-β promoter and activating TGF-β transcription (Mgbemena et al., 2011). Silencing KLF6 abrogates TGF-β expression and reduces RSV replication (Mgbemena et al., 2011). KLF6 expression was found to be significantly (P < 0.05) increased in cells infected with RSV, but not with ΔNS1 RSV at 6 h p.i. At 24 h p.i., KLF6 was induced in ΔNS1-infected cells, but was significantly lower (P < 0.05) relative to WT infection, suggesting that NS1 is a major, but not the only, driver of KLF6 induction (Fig. 2c). To determine if KLF6 expression was IFN dependent or independent, cells were treated cells with LPS, polyI : C, or types I or III IFN, and it was found that treatment with exogenous IFN-λ and IFN-β, but not LPS or polyI : C, significantly (P < 0.05) induced KLF6, but not miR-24 expression (Fig. 2d). The substantial increase in TGF-β upon NS1 overexpression and increase in KLF6 expression in WT versus ΔNS1 infection supported the hypothesis that NS1 positively regulated TGF-β expression via induction of KLF6. Additionally, induction of KLF6 by IFN-λ/IFN-β but not LPS/polyI : C suggested that these cytokines could induce KLF6 expression via the Jak–Stat (Janus kinas–signal transducer and activator of transcription) pathway. Treatment with IFN-λ does cause cell cycle arrest at G0/G1 phase in A549 (Li et al., 2012) as well as other cell lines (Li et al., 2008, 2010).
Silencing KLF6 induces miR-24 and TGF-β treatment represses miR-24 expression
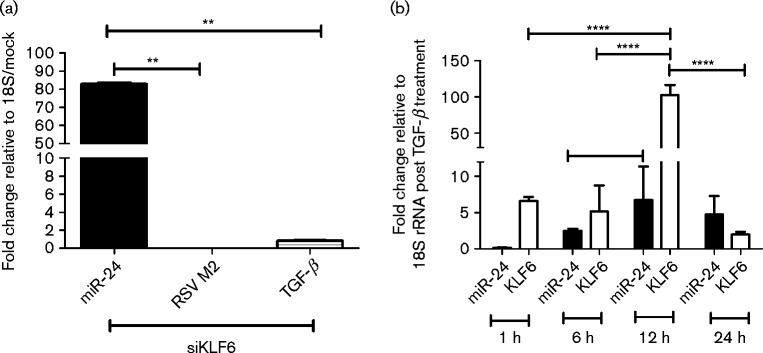
As KLF6 has been shown to induce TGF-β during RSV infection (Mgbemena et al., 2011) and TGF-β has been shown to suppress miR-24 expression, the effect of KLF6 gene silencing on miR-24 expression was examined. A549 cells were transfected with small interfering RNA (siRNA) SMARTpools (four different siRNAs per gene) targeting KLF6 (siKLF6) to knockdown KLF6 expression and subsequently infected with RSV. Expression levels of miR-24, RSV M2 and TGF-β were determined 24 h post-treatment using quantitative real-time qRT-PCR relative to mock-transfected cells. siKLF6 SMARTpools knocked down KLF6 mRNA (data not shown), led to decreased TGF-β and also reduced viral replication (Fig. 3a). In contrast, miR-24 expression was considerably upregulated, supporting the hypothesis that NS1-induced TGF-β suppresses miR-24 expression and removing KLF6, the positive inducer of TGF-β, relieves TGF-β-mediated miR-24 suppression (Fig. 3a).
Fig. 3.
KLF6 suppresses miR-24 via TGF-β. (a) KLF6 silencing induces miR-24, and represses TGF-β expression and viral replication. A549 cells were either mock treated or transfected with 25 nM siRNA SMARTpools against KLF6 in DharmaFECT 1 for 24 h. Plates with similar treatment were infected with RSV A2 (m.o.i. 1.0). Total RNA was isolated 24 h post-transfection using RNAzol RT and used to measure miR-24, RSV M2 and TGF-β expression relative to 18S rRNA. Data represent mean ± sem from two independent experiments. (b) Recombinant TGF-β suppresses miR-24 and induces KLF6. A549 cells were treated with recombinant human TGF-β (10 ng ml− 1) for 6, 12 and 24 h. Total RNA was isolated at the respective time points using RNAzol RT, and analysed for miR-24 and KLF6 expression relative to 18S rRNA. Data represent mean ± sem from three independent experiments. ** P < 0.01, **** P < 0.0001.
TGF-β treatment has been shown to increase RSV replication in A549 as well as primary NHBE cells (Gibbs et al., 2009). TGF-β has also been shown to inhibit miR-24 expression in other cell types (Cao et al., 2012; Sun et al., 2008) via Smad3 transcription factor. To validate the KLF6 silencing observations, cells were treated with recombinant TGF-β for various times (1, 6, 12 and 24 h), and expression of KLF6 and miR-24 determined by qRT-PCR. Treatment with recombinant TGF-β1 led to a rapid induction of KLF6 expression which peaked at 12 h, whilst miR-24 levels increased by twofold and remained unchanged at 24 h (Fig. 3b).
miR-24 repression during infection modulates multiple cellular pathways
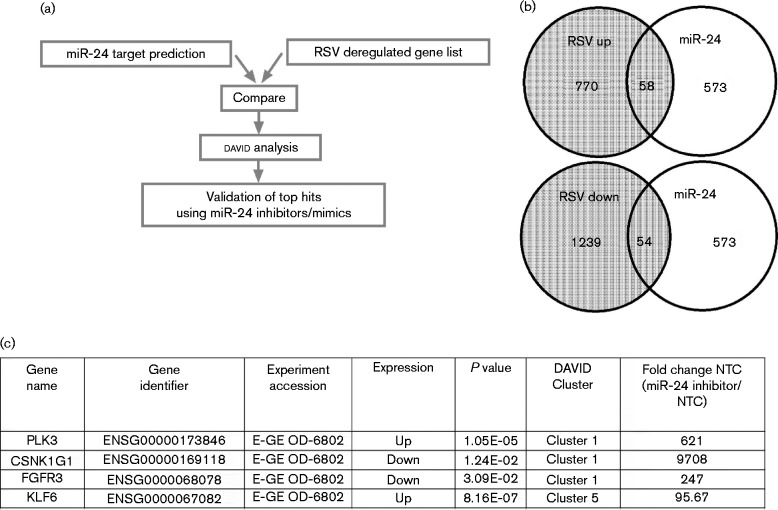
Inhibition of miR-24 in A549 cells has been shown to prevent cell cycle progression (Cheng et al., 2005). To determine the broader impact of miR-24 repression during RSV infection, potential miR-24 targets were short-listed by comparing the top 500 predicted miR-24 targets based on their PCT (probability of conserved targeting) scores (Friedman et al., 2009; Garcia et al., 2011; Grimson et al., 2007; Lewis et al., 2005) with those deregulated during RSV infection (Fig. 4a, b). An overlapping set of genes was generated which was further categorized into functional clusters using david (Huang et al., 2009). Clusters with enrichment scores ≥ 1.3 (corresponding to non-log P ≤ 0.05) were considered significant and 10 target genes [Pim-1 proto-oncogene-1 (PIM1), polo-like kinase 3 (PLK3), dual-specificity tyrosine phosphorylation-regulated kinase 2 (DYRK2), serine threonine kinase 10 (STK10), cyclin-dependent kinase inhibitor 1 (CDKN1A), Nemo-like kinase (NLK), casein kinase γ1 (CSNK1G1), fibroblast growth factor 3 (FGFR3), platelet-derived growth factor β (PDGFRB) and KLF6 that met the above criteria were chosen for validation for miR-24 regulation using a miR-24 inhibition assay (Fig. 4c). It was hypothesized that miR-24 inhibitor would relieve miR-24 repression of genuine target genes relative to mock-transfected or negative controls as measured by qRT-PCR. A549 cells were transfected with miR-24 inhibitors, or negative controls, for 24 h as described previously (Bakre et al., 2012) and expression levels of the 10 target genes chosen above were measured by qRT-PCR using gene-specific primers relative to 18S rRNA. miR-24 inhibitor transfection increased expression of PLK3, CSNK1G1, FGFR3 and KLF6 (Fig. 4c). Expression of these genes was significantly (P < 0.05) repressed in cells infected with ΔNS1 virus (data not shown). Sequence analysis showed that the 3′ UTRs of CSNK1G1 harboured two miR-24-binding sites, whilst PLK3, FGFR3 and KLF6 each had a single miR-24-binding site (Fig. S2). These data suggest that NS1-mediated miR-24 suppression induced the expression of PLK3, CSNK1G1, FGFR3 and KLF6 via a miR-24 interaction with the cognate 3′ UTR.
Fig. 4.
miR-24 regulates multiple cellular pathways. (a) Work flow used to shortlist miR-24 target genes. Top target predictions were compared to public microarray data on RSV infection. (b, c) Subsets of genes (b) identified were validated using miR-24 inhibition experiments (c). A549 cells (2 × 105) were transfected with miR-24 inhibitor (25 nM) or mock or non-targeting control (NTC) for 24 h followed by analysis of gene expression using target-specific primers. Expression was calculated relative to 18S rRNA and non-targeting control from two or three independent experiments.
RSV NS1 co-localizes with KLF6
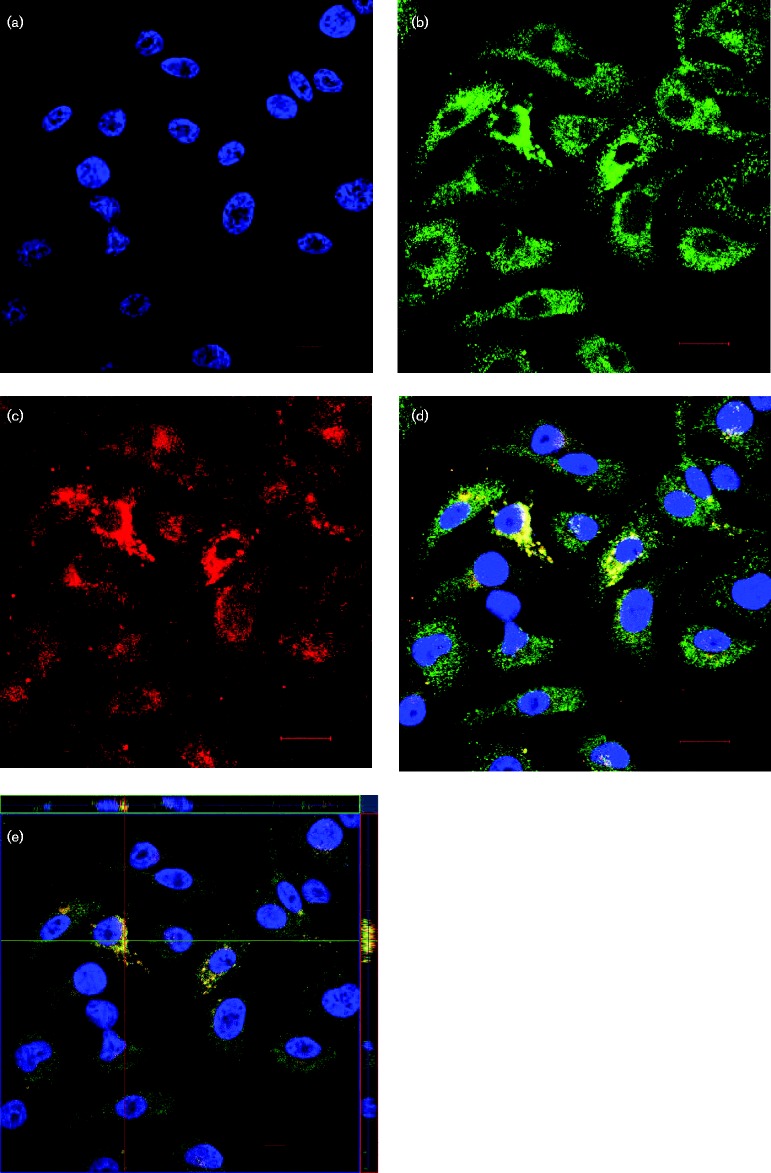
Given the finding that RSV NS1 induces KLF6 expression, which induces TGF-β to suppress miR-24 activity, the possibility that RSV NS1 interacted with KLF6 was examined in A549 cells. A549 cells expressing EGFPC1-NS1 stained for EGFP and KLF6 expression, and examined by confocal microscopy, showed that the fluorescence signals for NS1 overlapped with KLF6, suggesting that these proteins co-localize (Fig. 5d). To determine if this was the case, orthogonal optical sections were analysed in the xy, xz and yz planes to determine the localization of the fluorescence signals for NS1 and KLF6. The fluorescence signals for NS1 and KLF6 localized to the same positions, suggesting that these proteins co-localize and potentially interact (Fig. 5e). To validate if KLF6 and NS1 interact, co-immunoprecipitation assays were performed using an anti-GFP antibody to pull down KLF6 in EGFPC1-NS1-transfected A549 cells followed by a Western blot for KLF6. Co-immunoprecipitation failed to pull down detectable KLF6 by GFP pull-downs. This could have been due to either a genuine lack of direct KLF6 and NS1 interaction, or KLF6 and NS1 may have interacted transiently or indirectly through an accessory protein. Analysis of the KLF6 interactome using string database (Jensen et al., 2009) identified NOP56 ribonucleoprotein (NOP56), cadherin 1 (CDH1) and histone deacetylase 3 (HDAC3), three proteins that have been identified previously as NS1-interacting partners (Wu et al., 2012) (Fig. S3). These data suggested that NS1 may modulate the expression of KLF6 via interacting with these proteins.
Fig. 5.
RSV NS1 interacts with KLF6. A549 cells were transfected with 1.5 μg pEGFPC1-NS1 plasmid for 24 h in a 24-well plate and then transferred to chamber slides. Cells were allowed to adhere overnight, fixed, and stained for GFP expression using an anti-GFP antibody (green) and KLF6 (red). Nuclei were stained with DAPI (1 μg ml− 1). Confocal images were captured using a Zeiss LSM 710 inverted confocal microscope using an oil immersion lens at × 63. Images are representative of three fields. (a) DAPI alone, (b) GFP alone, (c) KLF6 alone, (d) merged, and (e) orthogonal projection of the optical section showing co-localization of NS1 and KLF6. Bar, 10 μm.
Discussion
Understanding how viral proteins modify host immune responses for viral survival and persistence is crucial to the development of effective countermeasures. Previous reports suggested that RSV non-structural proteins could modulate the expression of miRNAs during infection (Bakre et al., 2012; Thornburg et al., 2012). Here, it is demonstrated that a lack of NS1 during RSV infection significantly (P ≤ 0.05) induced miR-24 expression in type II lung epithelial cells (Fig. 1a) and that overexpression of NS1 substantively suppressed miR-24 expression (Fig. 1b). Furthermore, it is shown that NS1 induces expression of KLF6, a positive regulator of TGF-β, and silencing KLF6 represses TGF-β production and viral replication, but induces miR-24 expression. Confocal microscopy identified co-localization of NS1 and KLF6 proteins, suggesting potential interaction. Although co-immunoprecipitation experiments did not adequately demonstrate NS1–KLF6 interaction, analysis of KLF6-interacting proteins identified three proteins (CDH1, HDAC3 and NOP56) that were reported previously to interact with NS1. These data support our proposed model where RSV NS1 suppresses miR-24 via a KLF6/TGF-β-meditated pathway to affect multiple cellular pathways. It was shown previously that inhibiting miR-24 reduces RSV replication (Bakre et al., 2012), and this may be a mechanism to ensure viral persistence and delay clearance. Two different genomic loci (19p13 as a miR-23b–27b–24 and 9q22 as part of miR-23a–27a–24-2) encode miR-24, but express a single mature miR-24 (Chhabra et al., 2010). A549 cells lack the chromosome 19 locus (Xie et al., 2013) and hence express miR-24 from the chromosome 9 locus only. Irrespective of origin, miR-24-1 and miR-24-2 transcripts have the same mature sequence, and thus the same target gene repertoires.
KLF6 belongs to the KLF family and encodes three isoforms ranging from 237 to 283 aa in length. KLF6 is expressed in the lung, and has been shown to be induced during infection by RSV and hepatitis C virus (Papic et al., 2012), and by bacterial pathogens (Kidane et al., 2013). In macrophages, KLF6 is induced by LPS and IFN-γ, and regulates macrophage polarization by suppressing peroxisome proliferator-activated receptor-γ (Date et al., 2014). KLF6 also acts as a co-activator for NFκB-regulated expression of p65-dependent pathways (Zhang et al., 2014). In this study, silencing KLF6 significantly induced miR-24 expression, suggesting that KLF6 and miR-24 can interact in a bidirectional manner. RSV NS1 can modulate this interaction, as confocal microscopy indicated co-localization of KLF6 with RSV NS1 in cells transfected with plasmids expressing EGFP-tagged NS1. Furthermore, deletion of NS1 (rA2ΔNS1EGFP) resulted in reduced KLF6 expression relative to WT RSV infection at 6 and 24 h p.i. (Fig. 2c).
In A549 cells infected with RSV, KLF6 regulates the induction of both TGF-β (Mgbemena et al., 2011) and inducible nitric oxide synthase (Mgbemena et al., 2013), inhibiting the cells in G0/G1 phase and preventing apoptosis during RSV infection (Mgbemena et al., 2012). miR-24 inhibition is also known to block the cell cycle in A549 cells (Cheng et al., 2005). Based on this evidence, it was hypothesized that RSV NS1 induces cell cycle arrest by promoting KLF6 transcription of TGF-β and inhibiting miR-24 expression. Indeed, the overexpression of NS1 induced elevated TGF-β expression. Treatment of NS1-expressing cells with either LPS or IFN-λ further elevated TGF-β expression. IFN-λ and also IFN-β treatment induced the expression of KLF6, which would also lead to elevated TGF-β. Treatment with IFN-λ causes cell cycle arrest in A549 (Li et al., 2012) and other cell lines (Li et al., 2008, 2010), and may do so via elevated TGF-β. The release of IFN-λ during infection may also induce TGF-β expression and cell cycle arrest in neighbouring uninfected cells during RSV infection. In this study, the link between TGF-β, KLF6 and miR-24 was demonstrated in A549 cells by the reduction in both TGF-β expression and RSV replication, and by the significant induction (P < 0.05) of miR-24 expression when KLF6 was silenced. Furthermore, treatment of A549 cells with TGF-β suppressed miR-24 and induced KLF6, indicating a feed-forward loop to induce further TGF-β expression. TGF-β has been shown previously to regulate miR-24 expression (Sun et al., 2008).
These data demonstrate a mechanism by which RSV induces cell cycle arrest in infected cells via TGF-β expression, which is promoted by NS1-mediated inhibition of miR-24 and elevation of KLF6 expression. Inhibition of miR-24 has also been shown to deregulate the cell cycle in A549 cells (Cao et al., 2012; Cheng et al., 2005) by inhibiting the anti-apoptotic factor XIAP (X-linked inhibitor of apoptosis), a protein induced during RSV persistence (Nakamura-López et al., 2011). However, cellular processes other than cell cycle arrest are also affected by inhibition of miR-24. Recently, miR-24 was shown to modulate the replication of highly pathogenic influenza H5N1. Inhibition of miR-24 induced furin activity and increased viral replication for H5N1, but not H1N1 viruses (Loveday et al., 2015). As furin is also involved in cleavage of RSV F (Basak et al., 2001; Bolt et al., 2000; Sugrue et al., 2001), it is possible that downregulation of miR-24 may induce furin activity for efficient conversion of the RSV F0 to F1/F2 forms required for adhesion and formation of RSV filaments (Krzyzaniak et al., 2013). miR-24 also regulates the processing of the pre-TGF-β1 isoform via furin (Luna et al., 2011), such that reduced miR-24 expression would lead to elevated furin-mediated TGF-β1 processing. TGF-β1 induces miR-23 but not miR-24 expression in A549 cells and suppresses miR-24 in myocytes, both in a Smad3-dependent manner (Cao et al., 2012).
In summary, this study identifies for the first time, to the best of our knowledge, a mechanism by which RSV NS1 suppresses the expression of miR-24 and induces KLF6 expression to promote TGF-β-mediated cell cycle arrest and RSV replication. miR-24 may prove to be a viable antiviral target, as the induction of miR-24 may overcome RSV-induced cell cycle arrest and inhibition of apoptosis, thus improving viral clearance from infected cells.
Methods
Cell culture, virus propagation and recombinant TGF-β treatment
A549 cells (ATCC CCL-185) were cultured in Dulbecco's modified Eagle's medium (DMEM) (HyClone) containing 5 % heat-inactivated FBS (Hyclone) (Bakre et al., 2012; Oshansky et al., 2009a). Mycoplasma-free stocks of RSV strain A2 and those lacking the NS1 or NS2 genes (rA2ΔNS1EGFP or rA2ΔNS2EGFP, respectively) (Webster Marketon et al., 2014) were expanded in Vero E6 cells (ATCC CCL-81) and maintained in DMEM (Hyclone) supplemented with 5 % heat-inactivated FBS (Hyclone) as described previously (Bakre et al., 2012; Oshansky et al., 2009a). Recombinant TGF-β (Peprotech) was reconstituted in 10 mM citric acid buffer as per the manufacturer's instructions. Plasmids pEGFPC1-NS1 and pEGFPC1-NS2 were grown in Escherichia coli TOP10 and isolated using standard protocols.
miRNAs, cell transfection and cell cytotoxicity
All transfection procedures were performed in triplicate for at least three independent experiments. A549 cells (2 × 104 cells per well) were plated in 96-well Costar flat-bottom tissue culture plates (Corning) for 12 h at 37 °C and subsequently transfected for 18 h with 25 nM miRNA-specific inhibitor or mimic, or non-targeting miRNA mimics or inhibitor to Caenorhabditis elegans miR-67 (inhibitor negative control; Dharmacon) using DharmaFECT 1 (ThermoFisher) as per the manufacturer's instructions. Cell cultures were evaluated post-transfection for cell cytotoxicity using Alamar blue dye (Serotec). Alamar blue fluorescence reduction was measured at the end of 1.5 h incubation using a Tecan Safire X2 at excitation 530 nm/emission 590 nm as per the manufacturer's recommendations.
RNA isolation and qRT-PCR
Total RNA was isolated using a RNAzol RT kit (MRC Gene) as per the manufacturer's protocol. Briefly, following transfection, infection or treatment, cells were harvested in RNAzol RT reagent and lysed by repeated pipetting. DNA, proteins and carbohydrates were precipitated, and purified RNA quantified and treated with RQ1 RNase-free DNase I (Promega). Then, 100 ng DNase-treated RNA was used for cDNA synthesis using a miRNA cDNA synthesis kit (Agilent) as per the manufacturer's protocol, diluted as required, and used as template for qRT-PCRs with gene-specific primers (Table S1) and SYBR Green-based Maxima Thermoscript qPCR master mix (ThermoFisher) in an Agilent Mx3000P or Mx3005P instrument with 5 μM final concentration for both forward and reverse primers. Oligonucleotides were obtained desalted from IDT Biosciences and reconstituted to 10 μM working concentrations. Cycling conditions were: initial denaturation 95 °C/10 min, 40 cycles of 95 °C/15 s, 60 °C/30 s and 72 °C/30 s, followed by denaturation curve analysis. Annealing temperatures were pre-optimized using gradient PCRs. Data shown are mean ± sem from three biological replicates with three replicates each.
Confocal microscopy
A549 cells were transfected with 100 ng pEGFPC1-NS1 plasmid using Lipofectamine 2000 (Invitrogen) as per the manufacturer's suggested protocol. Transfected cells were trypsinized, transferred to Lab-Tek II chamber slides (Nunc) and incubated overnight at 37 °C to allow for cell attachment. Cells were fixed with 4 % formaldehyde (ThermoFisher) in PBS for 10 min and permeabilized using 0.1 % NP-40 (Sigma) in PBS for 10 min. Fixed and permeabilized cells were blocked using 5 % BSA (Sigma) and subsequently stained for GFP using mouse anti-GFP antibody (Abcam) coupled to Alexa Fluor 488 (Life Technologies). KLF6 was detected using rabbit anti-KLF6 polyclonal antibody (Abcam) coupled to Alexa Fluor 546 (Life Technologies). Nuclei were stained using DAPI (Life Technologies) (1 μg ml− 1) for 10 min. Slides were mounted in Prolong Gold Anti-Fade (Life Technologies) and stored in the dark at room temperature for drying, followed by storage at 4 °C. Confocal laser scanning microscopy was performed using a Zeiss LSM 710 instrument. Z-stacks, and images were captured and analysed using Zen Black 2012 software (Zeiss). Scale bar = 10 μm.
Statistics
Statistical analysis was performed using GraphPad Prism version 5.0 using one/two-way ANOVA. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, **** P < 0.0001.
Acknowledgements
This work was supported by National Institutes of Health grant 1021RR211342 to R. A. T and through the Georgia Research Alliance support to R. A. T.
Supplementary Data
Supplementary Data
References
- Andreoli V., Gehrau R.C., Bocco J.L. (2010). Biology of Krüppel-like factor 6 transcriptional regulator in cell life and death IUBMB Life 62 896–905 10.1002/iub.396 . [DOI] [PubMed] [Google Scholar]
- Atreya P.L., Peeples M.E., Collins P.L. (1998). The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication J Virol 72 1452–1461 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakre A., Mitchell P., Coleman J.K., Jones L.P., Saavedra G., Teng M., Tompkins S.M., Tripp R.A. (2012). Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication J Gen Virol 93 2346–2356 10.1099/vir.0.044255-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak A., Zhong M., Munzer J.S., Chrétien M., Seidah N.G. (2001). Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses: a comparative analysis with fluorogenic peptides Biochem J 353 537–545 10.1042/0264-6021:3530537 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt G., Pedersen L.O., Birkeslund H.H. (2000). Cleavage of the respiratory syncytial virus fusion protein is required for its surface expression: role of furin Virus Res 68 25–33 10.1016/S0168-1702(00)00149-0 . [DOI] [PubMed] [Google Scholar]
- Bossert B., Marozin S., Conzelmann K.K. (2003). Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3 J Virol 77 8661–8668 10.1128/JVI.77.16.8661-8668.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyapalle S., Wong T., Garay J., Teng M., San Juan-Vergara H., Mohapatra S., Mohapatra S. (2012). Respiratory syncytial virus NS1 protein colocalizes with mitochondrial antiviral signaling protein MAVS following infection PLoS One 7 e29386 10.1371/journal.pone.0029386 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budge P.J., Graham B.S. (2004). Inhibition of respiratory syncytial virus by RhoA-derived peptides: implications for the development of improved antiviral agents targeting heparin-binding viruses J Antimicrob Chemother 54 299–302 10.1093/jac/dkh355 . [DOI] [PubMed] [Google Scholar]
- Cao M., Seike M., Soeno C., Mizutani H., Kitamura K., Minegishi Y., Noro R., Yoshimura A., Cai L., Gemma A. (2012). MiR-23a regulates TGF-β-induced epithelial–mesenchymal transition by targeting E-cadherin in lung cancer cells Int J Oncol 41 869–875 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (2008). Respiratory syncytial virus activity – United States, July 2007–December 2008 MMWR Morb Mortal Wkly Rep 57 1355–1358 . [PubMed] [Google Scholar]
- CDC (2013). Respiratory syncytial virus activity – United States, July 2011–January 2013 MMWR Morb Mortal Wkly Rep 62 141–144 . [PMC free article] [PubMed] [Google Scholar]
- Cheng A.M., Byrom M.W., Shelton J., Ford L.P. (2005). Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis Nucleic Acids Res 33 1290–1297 10.1093/nar/gki200 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra R., Dubey R., Saini N. (2010). Cooperative and individualistic functions of the microRNAs in the miR-23a∼27a∼24-2 cluster and its implication in human diseases Mol Cancer 9 232 10.1186/1476-4598-9-232 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date D., Das R., Narla G., Simon D.I., Jain M.K., Mahabeleshwar G.H. (2014). Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization J Biol Chem 289 10318–10329 10.1074/jbc.M113.526749 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Faria I.C.J., de Faria E.J., Toro A.A.D.C., Ribeiro J.D., Bertuzzo C.S. (2008). Association of TGF-beta1, CD14, IL-4, IL-4R and ADAM33 gene polymorphisms with asthma severity in children and adolescents J Pediatr (Rio J) 84 203–210 . [DOI] [PubMed] [Google Scholar]
- Eis P.S., Tam W., Sun L., Chadburn A., Li Z., Gomez M.F., Lund E., Dahlberg J.E. (2005). Accumulation of miR-155 and BIC RNA in human B cell lymphomas Proc Natl Acad Sci U S A 102 3627–3632 10.1073/pnas.0500613102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J., Lynch O.T., Suessmuth Y., Qian P., Boyd C.R., Burrows J.F., Buick R., Stevenson N.J., Touzelet O., other authors (2007). Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase J Virol 81 3428–3436 10.1128/JVI.02303-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong C.Y., Pang L., Holland E., Knox A.J. (2000). TGF-beta1 stimulates IL-8 release, COX-2 expression, and PGE2 release in human airway smooth muscle cells Am J Physiol Lung Cell Mol Physiol 279 L201–L207 . [DOI] [PubMed] [Google Scholar]
- Foronjy R.F., Dabo A.J., Taggart C.C., Weldon S., Geraghty P. (2014). Respiratory syncytial virus infections enhance cigarette smoke induced COPD in mice PLoS One 9 e90567 10.1371/journal.pone.0090567 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. (2009). Most mammalian mRNAs are conserved targets of microRNAs Genome Res 19 92–105 10.1101/gr.082701.108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardo R., Chanez P., Gjomarkaj M., La Grutta S., Bonanno A., Montalbano A.M., Di Sano C., Albano G.D., Gras D., other authors (2013). The role of transforming growth factor-β1 in airway inflammation of childhood asthma Int J Immunopathol Pharmacol 26 725–738 . [DOI] [PubMed] [Google Scholar]
- Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. (2011). Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs Nat Struct Mol Biol 18 1139–1146 10.1038/nsmb.2115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J.D., Ornoff D.M., Igo H.A., Zeng J.Y., Imani F. (2009). Cell cycle arrest by transforming growth factor beta1 enhances replication of respiratory syncytial virus in lung epithelial cells J Virol 83 12424–12431 10.1128/JVI.00806-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami R., Majumdar T., Dhar J., Chattopadhyay S., Bandyopadhyay S.K., Verbovetskaya V., Sen G.C., Barik S. (2013). Viral degradasome hijacks mitochondria to suppress innate immunity Cell Res 23 1025–1042 10.1038/cr.2013.98 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. (2007). MicroRNA targeting specificity in mammals: determinants beyond seed pairing Mol Cell 27 91–105 10.1016/j.molcel.2007.06.017 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C.B., Weinberg G.A., Iwane M.K., Blumkin A.K., Edwards K.M., Staat M.A., Auinger P., Griffin M.R., Poehling K.A., other authors (2009). The burden of respiratory syncytial virus infection in young children N Engl J Med 360 588–598 10.1056/NEJMoa0804877 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallak L.K., Spillmann D., Collins P.L., Peeples M.E. (2000). Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection J Virol 74 10508–10513 10.1128/JVI.74.22.10508-10513.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa S., Kojima T., Obata K., Okabayashi T., Yokota S., Nomura K., Obonai T., Fuchimoto J., Himi T., other authors (2013). Marked induction of matrix metalloproteinase-10 by respiratory syncytial virus infection in human nasal epithelial cells J Med Virol 85 2141–2150 . [DOI] [PubMed] [Google Scholar]
- Hoffman S.J., Laham F.R., Polack F.P. (2004). Mechanisms of illness during respiratory syncytial virus infection: the lungs, the virus and the immune response Microbes Infect 6 767–772 10.1016/j.micinf.2004.03.010 . [DOI] [PubMed] [Google Scholar]
- Hoshino M., Nakamura Y., Sim J.J. (1998). Expression of growth factors and remodelling of the airway wall in bronchial asthma Thorax 53 21–27 10.1136/thx.53.1.21 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotard A.L., Lee S., Currier M.G., Crowe J.E., Jr, Sakamoto K., Newcomb D.C., Peebles R.S., Jr, Plemper R.K., Moore M.L. (2015). Identification of residues in the human respiratory syncytial virus fusion protein that modulate fusion activity and pathogenesis J Virol 89 512–522 10.1128/JVI.02472-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell J.E., McAnulty R.J. (2006). TGF-beta: its role in asthma and therapeutic potential Curr Drug Targets 7 547–565 10.2174/138945006776818692 . [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009). Systematic and integrative analysis of large gene lists using david bioinformatics resources Nat Protoc 4 44–57 10.1038/nprot.2008.211 . [DOI] [PubMed] [Google Scholar]
- Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., other authors (2009). string 8 – a global view on proteins and their functional interactions in 630 organisms Nucleic Acids Res 37 (Database), D412–D416 10.1093/nar/gkn760 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Zhou H., Cheng X., Tang R., Munoz M., Nguyen N. (2000). Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo Virology 273 210–218 10.1006/viro.2000.0393 . [DOI] [PubMed] [Google Scholar]
- Kidane Y.H., Lawrence C., Murali T.M. (2013). The landscape of host transcriptional response programs commonly perturbed by bacterial pathogens: towards host-oriented broad-spectrum drug targets PLoS One 8 e58553 10.1371/journal.pone.0058553 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X., Zhang W., Lockey R.F., Auais A., Piedimonte G., Mohapatra S.S. (2007). Respiratory syncytial virus infection in Fischer 344 rats is attenuated by short interfering RNA against the RSV-NS1 gene Genet Vaccines Ther 5 4 10.1186/1479-0556-5-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelkin A., Belyakov I.M., Yang L., Berzofsky J.A., Collins P.L., Bukreyev A. (2006). The NS2 protein of human respiratory syncytial virus suppresses the cytotoxic T-cell response as a consequence of suppressing the type I interferon response J Virol 80 5958–5967 10.1128/JVI.00181-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzaniak M.A., Zumstein M.T., Gerez J.A., Picotti P., Helenius A. (2013). Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein PLoS Pathog 9 e1003309 10.1371/journal.ppat.1003309 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt-Jones E.A., Popova L., Kwinn L., Haynes L.M., Jones L.P., Tripp R.A., Walsh E.E., Freeman M.W., Golenbock D.T., other authors (2000). Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus Nat Immunol 1 398–401 10.1038/80833 . [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets Cell 120 15–20 10.1016/j.cell.2004.12.035 . [DOI] [PubMed] [Google Scholar]
- Li W., Lewis-Antes A., Huang J., Balan M., Kotenko S.V. (2008). Regulation of apoptosis by type III interferons Cell Prolif 41 960–979 10.1111/j.1365-2184.2008.00558.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Kawamura K., Ma G., Iwata F., Numasaki M., Suzuki N., Shimada H., Tagawa M. (2010). Interferon-lambda induces G1 phase arrest or apoptosis in oesophageal carcinoma cells and produces anti-tumour effects in combination with anti-cancer agents Eur J Cancer 46 180–190 10.1016/j.ejca.2009.10.002 . [DOI] [PubMed] [Google Scholar]
- Li W., Huang X., Liu Z., Wang Y., Zhang H., Tong H., Wu H., Lin S. (2012). Type III interferon induces apoptosis in human lung cancer cells Oncol Rep 28 1117–1125 . [DOI] [PubMed] [Google Scholar]
- Liesman R.M., Buchholz U.J., Luongo C.L., Yang L., Proia A.D., DeVincenzo J.P., Collins P.L., Pickles R.J. (2014). RSV-encoded NS2 promotes epithelial cell shedding and distal airway obstruction J Clin Invest 124 2219–2233 10.1172/JCI72948 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Z., Tran K.C., Teng M.N. (2009). Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I J Virol 83 3734–3742 10.1128/JVI.02434-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Friggeri A., Yang Y., Park Y.J., Tsuruta Y., Abraham E. (2009). miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses Proc Natl Acad Sci U S A 106 15819–15824 10.1073/pnas.0901216106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M.S., Brazas R.M., Holtzman M.J. (2005). Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness J Virol 79 9315–9319 10.1128/JVI.79.14.9315-9319.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveday E.K., Diederich S., Pasick J., Jean F. (2015). Human microRNA-24 modulates highly pathogenic avian-origin H5N1 influenza A virus infection in A549 cells by targeting secretory pathway furin J Gen Virol 96 30–39 10.1099/vir.0.068585-0 . [DOI] [PubMed] [Google Scholar]
- Luna C., Li G., Qiu J., Epstein D.L., Gonzalez P. (2011). MicroRNA-24 regulates the processing of latent TGFβ1 during cyclic mechanical stress in human trabecular meshwork cells through direct targeting of FURIN J Cell Physiol 226 1407–1414 10.1002/jcp.22476 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luongo C., Winter C.C., Collins P.L., Buchholz U.J. (2013). Respiratory syncytial virus modified by deletions of the NS2 gene and amino acid S1313 of the L polymerase protein is a temperature-sensitive, live-attenuated vaccine candidate that is phenotypically stable at physiological temperature J Virol 87 1985–1996 10.1128/JVI.02769-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I., Lombardía L., García-Barreno B., Domínguez O., Melero J.A. (2007). Distinct gene subsets are induced at different time points after human respiratory syncytial virus infection of A549 cells J Gen Virol 88 570–581 10.1099/vir.0.82187-0 . [DOI] [PubMed] [Google Scholar]
- Mastrangelo P., Hegele R.G. (2013). RSV fusion: time for a new model Viruses 5 873–885 10.3390/v5030873 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata M., Sarrion I., Armengot M., Carda C., Martinez I., Melero J.A., Cortijo J. (2012). Respiratory syncytial virus inhibits ciliagenesis in differentiated normal human bronchial epithelial cells: effectiveness of N-acetylcysteine PLoS One 7 e48037 10.1371/journal.pone.0048037 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann K.L., Imani F. (2007). Transforming growth factor beta enhances respiratory syncytial virus replication and tumor necrosis factor alpha induction in human epithelial cells J Virol 81 2880–2886 10.1128/JVI.02583-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., Lee S., Hotard A.L., Moore M.L. (2014). Refining the balance of attenuation and immunogenicity of respiratory syncytial virus by targeted codon deoptimization of virulence genes MBio 5 e01704–e01714 10.1128/mBio.01704-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mgbemena V., Segovia J., Chang T., Bose S. (2011). Krüppel-like factor 6 regulates transforming growth factor-β gene expression during human respiratory syncytial virus infection Virol J 8 409 10.1186/1743-422X-8-409 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mgbemena V., Segovia J.A., Chang T.H., Tsai S.Y., Cole G.T., Hung C.Y., Bose S. (2012). Transactivation of inducible nitric oxide synthase gene by Kruppel-like factor 6 regulates apoptosis during influenza A virus infection J Immunol 189 606–615 10.4049/jimmunol.1102742 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mgbemena V., Segovia J., Chang T.-H., Bose S. (2013). KLF6 and iNOS regulates apoptosis during respiratory syncytial virus infection Cell Immunol 283 1–7 10.1016/j.cellimm.2013.06.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore E.C., Barber J., Tripp R.A. (2008). Respiratory syncytial virus (RSV) attachment and nonstructural proteins modify the type I interferon response associated with suppressor of cytokine signaling (SOCS) proteins and IFN-stimulated gene-15 (ISG15) Virol J 5 116 10.1186/1743-422X-5-116 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.L., Stokes K.L., Hartert T.V. (2013). The impact of viral genotype on pathogenesis and disease severity: respiratory syncytial virus and human rhinoviruses Curr Opin Immunol 25 761–768 10.1016/j.coi.2013.09.016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Morio T., Ito S., Morimoto A., Ota S., Mizuta K., Iwata T., Hara T., Saji T. (2014). Risks and prevention of severe RS virus infection among children with immunodeficiency and Down's syndrome J Infect Chemother 20 455–459 10.1016/j.jiac.2014.05.001 . [DOI] [PubMed] [Google Scholar]
- Munday D.C., Emmott E., Surtees R., Lardeau C.H., Wu W., Duprex W.P., Dove B.K., Barr J.N., Hiscox J.A. (2010). Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus Mol Cell Proteomics 9 2438–2459 10.1074/mcp.M110.001859 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir S., Hillyer P., Le Nouën C., Buchholz U.J., Rabin R.L., Collins P.L., Bukreyev A. (2011). Respiratory syncytial virus interferon antagonist NS1 protein suppresses and skews the human T lymphocyte response PLoS Pathog 7 e1001336 10.1371/journal.ppat.1001336 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid M.A., Pauley K.M., Satoh M., Chan E.K. (2009). miR-146a is critical for endotoxin-induced tolerance: implication in innate immunity J Biol Chem 284 34590–34599 10.1074/jbc.M109.056317 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid M.A., Satoh M., Chan E.K. (2011a). Mechanistic role of microRNA-146a in endotoxin-induced differential cross-regulation of TLR signaling J Immunol 186 1723–1734 10.4049/jimmunol.1002311 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid M.A., Satoh M., Chan E.K. (2011b). MicroRNA in TLR signaling and endotoxin tolerance Cell Mol Immunol 8 388–403 10.1038/cmi.2011.26 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair H., Nokes D.J., Gessner B.D., Dherani M., Madhi S.A., Singleton R.J., O'Brien K.L., Roca A., Wright P.F., other authors (2010). Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis Lancet 375 1545–1555 10.1016/S0140-6736(10)60206-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura-López Y., Villegas-Sepúlveda N., Sarmiento-Silva R.E., Gómez B. (2011). Intrinsic apoptotic pathway is subverted in mouse macrophages persistently infected by RSV Virus Res 158 98–107 10.1016/j.virusres.2011.03.016 . [DOI] [PubMed] [Google Scholar]
- O'Connell R.M., Taganov K.D., Boldin M.P., Cheng G., Baltimore D. (2007). MicroRNA-155 is induced during the macrophage inflammatory response Proc Natl Acad Sci U S A 104 1604–1609 10.1073/pnas.0610731104 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Openshaw P.J., Chiu C. (2013). Protective and dysregulated T cell immunity in RSV infection Curr Opin Virol 3 468–474 10.1016/j.coviro.2013.05.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshansky C.M., Krunkosky T.M., Barber J., Jones L.P., Tripp R.A. (2009a). Respiratory syncytial virus proteins modulate suppressors of cytokine signaling 1 and 3 and the type I interferon response to infection by a toll-like receptor pathway Viral Immunol 22 147–161 10.1089/vim.2008.0098 . [DOI] [PubMed] [Google Scholar]
- Oshansky C.M., Zhang W., Moore E., Tripp R.A. (2009b). The host response and molecular pathogenesis associated with respiratory syncytial virus infection Future Microbiol 4 279–297 10.2217/fmb.09.1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papic N., Maxwell C.I., Delker D.A., Liu S., Heale B.S., Hagedorn C.H. (2012). RNA-sequencing analysis of 5′ capped RNAs identifies many new differentially expressed genes in acute hepatitis C virus infection Viruses 4 581–612 10.3390/v4040581 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastey M.K., Crowe J.E., Jr, Graham B.S. (1999). RhoA interacts with the fusion glycoprotein of respiratory syncytial virus and facilitates virus-induced syncytium formation J Virol 73 7262–7270 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley K.M., Satoh M., Pauley B.A., Dominguez-Gutierrez P.R., Wallet S.M., Holliday L.S., Cha S., Reeves W.H., Chan E.K. (2010). Formation of GW/P bodies as marker for microRNA-mediated regulation of innate immune signaling in THP-1 cells Immunol Cell Biol 88 205–212 10.1038/icb.2009.84 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaia G., Gallelli L., D'Agostino B., Vatrella A., Cuda G., Fratto D., Renda T., Galderisi U., Piegari E., other authors (2007). Effects of TGF-beta and glucocorticoids on map kinase phosphorylation, IL-6/IL-11 secretion and cell proliferation in primary cultures of human lung fibroblasts J Cell Physiol 210 489–497 10.1002/jcp.20884 . [DOI] [PubMed] [Google Scholar]
- Piedimonte G. (2002). Pathophysiological mechanisms for the respiratory syncytial virus-reactive airway disease link Respir Res 3 (Suppl 1), S21–S25 10.1186/rr185 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedimonte G. (2003). Contribution of neuroimmune mechanisms to airway inflammation and remodeling during and after respiratory syncytial virus infection Pediatr Infect Dis J 22 (Suppl), S66–S75 10.1097/01.inf.0000053888.67311.1d . [DOI] [PubMed] [Google Scholar]
- Psarras S., Papadopoulos N.G., Johnston S.L. (2004). Pathogenesis of respiratory syncytial virus bronchiolitis-related wheezing Paediatr Respir Rev 5 (Suppl A), S179–S184 10.1016/S1526-0542(04)90034-6 . [DOI] [PubMed] [Google Scholar]
- Qin L., Peng D., Hu C., Xiang Y., Zhou Y., Tan Y., Qin X. (2014). Differentiation of Th subsets inhibited by nonstructural proteins of respiratory syncytial virus is mediated by ubiquitination PLoS One 9 e101469 10.1371/journal.pone.0101469 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Liu T., Pang L., Li K., Garofalo R.P., Casola A., Bao X. (2011). A novel mechanism for the inhibition of interferon regulatory factor-3-dependent gene expression by human respiratory syncytial virus NS1 protein J Gen Virol 92 2153–2159 10.1099/vir.0.032987-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlender J., Bossert B., Buchholz U., Conzelmann K.K. (2000). Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response J Virol 74 8234–8242 10.1128/JVI.74.18.8234-8242.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte L.N., Westermann A.J., Vogel J. (2013). Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing Nucleic Acids Res 41 542–553 10.1093/nar/gks1030 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Raby B.A., Hunninghake G.M., Soto-Quirós M., Avila L., Murphy A.J., Lasky-Su J., Klanderman B.J., Sylvia J.S., other authors (2009). Variants in TGFB1, dust mite exposure, and disease severity in children with asthma Am J Respir Crit Care Med 179 356–362 10.1164/rccm.200808-1268OC . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann K.M., Tran K.C., Chi B., Rabin R.L., Collins P.L. (2004). Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected] J Virol 78 4363–4369 10.1128/JVI.78.8.4363-4369.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann K.M., Tran K.C., Collins P.L. (2005). Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines J Virol 79 5353–5362 10.1128/JVI.79.9.5353-5362.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockman L.J., Curns A.T., Anderson L.J., Fischer-Langley G. (2012). Respiratory syncytial virus-associated hospitalizations among infants and young children in the United States, 1997–2006 Pediatr Infect Dis J 31 5–9 10.1097/INF.0b013e31822e68e6 . [DOI] [PubMed] [Google Scholar]
- Straub C.P., Lau W.H., Preston F.M., Headlam M.J., Gorman J.J., Collins P.L., Spann K.M. (2011). Mutation of the elongin C binding domain of human respiratory syncytial virus non-structural protein 1 (NS1) results in degradation of NS1 and attenuation of the virus Virol J 8 252 10.1186/1743-422X-8-252 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugrue R.J., Brown C., Brown G., Aitken J., McL Rixon H.W. (2001). Furin cleavage of the respiratory syncytial virus fusion protein is not a requirement for its transport to the surface of virus-infected cells J Gen Virol 82 1375–1386 . [DOI] [PubMed] [Google Scholar]
- Sun Q., Zhang Y., Yang G., Chen X., Zhang Y., Cao G., Wang J., Sun Y., Zhang P., other authors (2008). Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation Nucleic Acids Res 36 2690–2699 10.1093/nar/gkn032 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov K.D., Boldin M.P., Chang K.-J., Baltimore D. (2006). NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses Proc Natl Acad Sci U S A 103 12481–12486 10.1073/pnas.0605298103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.R., Yang T., Liu S.P., Xiang Y., Qu F., Liu H.J., Qin X.Q. (2008). Pulmonary peptidergic innervation remodeling and development of airway hyperresponsiveness induced by RSV persistent infection Peptides 29 47–56 10.1016/j.peptides.2007.10.020 . [DOI] [PubMed] [Google Scholar]
- Tayyari F., Marchant D., Moraes T.J., Duan W., Mastrangelo P., Hegele R.G. (2011). Identification of nucleolin as a cellular receptor for human respiratory syncytial virus Nat Med 17 1132–1135 10.1038/nm.2444 . [DOI] [PubMed] [Google Scholar]
- Teng M.N., Collins P.L. (1999). Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein J Virol 73 466–473 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng M.N., Whitehead S.S., Bermingham A., St Claire M., Elkins W.R., Murphy B.R., Collins P.L. (2000). Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees J Virol 74 9317–9321 10.1128/JVI.74.19.9317-9321.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg N.J., Shepherd B., Crowe J.E.,, Jr. (2010). Transforming growth factor beta is a major regulator of human neonatal immune responses following respiratory syncytial virus infection J Virol 84 12895–12902 10.1128/JVI.01273-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg N.J., Hayward S.L., Crowe J.E.,, Jr. (2012). Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF-κB MBio 3 e00220-12 10.1128/mBio.00220-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp R.A., Jones L.P., Haynes L.M., Zheng H., Murphy P.M., Anderson L.J. (2001). CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein Nat Immunol 2 732–738 10.1038/90675 . [DOI] [PubMed] [Google Scholar]
- Vicencio A.G. (2010). Susceptibility to bronchiolitis in infants Curr Opin Pediatr 22 302–306 10.1097/MOP.0b013e32833797f9 . [DOI] [PubMed] [Google Scholar]
- Webster Marketon J.I., Corry J., Teng M.N. (2014). The respiratory syncytial virus (RSV) nonstructural proteins mediate RSV suppression of glucocorticoid receptor transactivation Virology 449 62–69 10.1016/j.virol.2013.11.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead S.S., Bukreyev A., Teng M.N., Firestone C.Y., St Claire M., Elkins W.R., Collins P.L., Murphy B.R. (1999). Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees J Virol 73 3438–3442 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P.F., Karron R.A., Madhi S.A., Treanor J.J., King J.C., O'Shea A., Ikizler M.R., Zhu Y., Collins P.L., other authors (2006). The interferon antagonist NS2 protein of respiratory syncytial virus is an important virulence determinant for humans J Infect Dis 193 573–581 10.1086/499600 . [DOI] [PubMed] [Google Scholar]
- Wu W., Munday D.C., Howell G., Platt G., Barr J.N., Hiscox J.A. (2011). Characterization of the interaction between human respiratory syncytial virus and the cell cycle in continuous cell culture and primary human airway epithelial cells J Virol 85 10300–10309 10.1128/JVI.05164-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Tran K.C., Teng M.N., Heesom K.J., Matthews D.A., Barr J.N., Hiscox J.A. (2012). The interactome of the human respiratory syncytial virus NS1 protein highlights multiple effects on host cell biology J Virol 86 7777–7789 10.1128/JVI.00460-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Tobin L.A., Camps J., Wangsa D., Yang J., Rao M., Witasp E., Awad K.S., Yoo N., other authors (2013). MicroRNA-24 regulates XIAP to reduce the apoptosis threshold in cancer cells Oncogene 32 2442–2451 10.1038/onc.2012.258 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zheng J., Zheng K., Hou Y., Zhao F., Zhao D. (2014). Respiratory syncytial virus NS1 protein degrades STAT2 by inducing SOCS1 expression Intervirology 57 65–73 . [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lei C.Q., Hu Y.H., Xia T., Li M., Zhong B., Shu H.B. (2014). Krüppel-like factor 6 is a co-activator of NF-κB that mediates p65-dependent transcription of selected downstream genes J Biol Chem 289 12876–12885 10.1074/jbc.M113.535831 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Thompson W.W., Viboud C.G., Ringholz C.M., Cheng P.Y., Steiner C., Abedi G.R., Anderson L.J., Brammer L., Shay D.K. (2012). Hospitalizations associated with influenza and respiratory syncytial virus in the United States, 1993–2008 Clin Infect Dis 54 1427–1436 10.1093/cid/cis211 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data