Abstract
Most species are structured and influenced by processes that either increased or reduced gene flow between populations. However, most population genetic inference methods assume panmixia and reconstruct a history characterized by population size changes. This is potentially problematic as population structure can generate spurious signals of population size change through time. Moreover, when the model assumed for demographic inference is misspecified, genomic data will likely increase the precision of misleading if not meaningless parameters. For instance, if data were generated under an n-island model (characterized by the number of islands and migrants exchanged) inference based on a model of population size change would produce precise estimates of a bottleneck that would be meaningless. In addition, archaeological or climatic events around the bottleneck's timing might provide a reasonable but potentially misleading scenario. In a context of model uncertainty (panmixia versus structure) genomic data may thus not necessarily lead to improved statistical inference. We consider two haploid genomes and develop a theory that explains why any demographic model with structure will necessarily be interpreted as a series of changes in population size by inference methods ignoring structure. We formalize a parameter, the inverse instantaneous coalescence rate, and show that it is equivalent to a population size only in panmictic models, and is mostly misleading for structured models. We argue that this issue affects all population genetics methods ignoring population structure which may thus infer population size changes that never took place. We apply our approach to human genomic data.
Introduction
Most species are structured and do not behave as panmictic populations (Wakeley, 1999; Harpending and Rogers, 2000; Goldstein and Chikhi, 2002; Charlesworth et al., 2003; Harding and McVean, 2004). They have been influenced by habitat fragmentation, expansion or reconnection events that either increased or reduced the amount of gene flow between local populations, as a result of climatic or anthropogenic events (Goossens et al., 2006; Quéméré et al., 2012). Although genomic data offer the possibility to reconstruct with increasing precision major events in that complex history (Gutenkunst et al., 2009; Li and Durbin, 2011; Sheehan et al., 2013; Schiffels and Durbin, 2013; Liu and Fu, 2015), it is computationally very difficult to account for population structure. As a consequence, many inferential methods tend to ignore population structure (Li and Durbin, 2011; Sheehan et al., 2013; Liu and Fu, 2015). This is potentially problematic because an increasing number of studies have shown that population structure generates spurious signals of changes in population size, even when populations were stationary (Wakeley, 1999, 2001; Nielsen and Beaumont, 2009; Chikhi et al., 2010; Peter et al., 2010; Heller et al., 2013; Paz-Vinas et al., 2013; Mazet et al., 2015). Here, we provide a simple theoretical framework that explains why any inferential method ignoring population structure will always infer population size changes as soon as populations are actually structured. In other words, this theory explains why any real demographic history, with or without structure, will necessarily and optimally be interpreted as a series of changes in population size by methods ignoring population structure.
We consider the case of two haploid genomes and we study T2, the coalescence time for a sample of size two (that is, the time to the common ancestor of two randomly sampled sequences (Herbots, 1994; Griffiths and Tavaré, 1994; Mazet et al., 2015)). We predict the history that any coalescent-based population genetics methods ignoring structure will try to reconstruct. We introduce a parameter that we call the inverse instantaneous coalescence rate (IICR). As coalescence rates are expected to be inversely related to effective population sizes, it may seem natural to see the IICR as an ‘instantaneous population size'. However, we stress that the IICR is equivalent to a population size only in panmictic models. For models incorporating population structure the IICR exhibits a temporal trajectory that can be strongly disconnected from the real demographic history (that is, identifying a decrease when the population size was actually constant or increasing).
We apply our approach to simulated data and use the pairwise sequentially Markovian coalescent (PSMC) method (Li and Durbin, 2011) as a reference method because it allows to reconstruct the history of a population or species from one single diploid genome. In addition, this method has been applied to a wide array of vertebrate species including reptiles (Green et al., 2014), birds (Zhan et al., 2013; Hung et al., 2014) and mammals such as primates (Prado-Martinez et al., 2013; Zhou et al., 2014), pigs (Groenen et al., 2012) and pandas (Zhao et al., 2013), and its outputs have been and typically are interpreted in terms of population size changes. However, our results are general and not specifically related to that particular method.
We then apply our approach to human data and show that an alternative model involving a minimum of three changes in migration rates can explain the PSMC results obtained by Li and Durbin (2011). The scenario that we infer represents an alternative to the population crashes and increases depicted in various population genetic studies, but is strikingly in phase with fossil data and provides a more realistic framework as several authors have suggested (Goldstein and Chikhi, 2002; Harding and McVean, 2004). Altogether, we call for a major reevaluation of what genomic data can actually tell us about the demographic history of our species. Beyond our species we argue that genomic data should be reinterpreted as a consequence of changes in levels of connection rather than simple changes in population size (see also Wakeley, 1999, 2001 and Harding and McVean, 2004 for interesting models incorporating structure).
Materials and methods
Coalescence time for a sample of size 2 in a model of population size change
We consider a model of arbitrary population size change, where N(t) represents the population size (N, in units of genes or haploid genomes) as a function of time (t) scaled by the number of genes (that is, in units of coalescence time, corresponding to 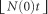 generations). We consider that t=0 is the present, and positive values represent the past. As N represents the population size in terms of haploid genomes, the number of individuals will be N/2 for diploid species. We can then apply the generalization of the coalescent in populations of variable size (Griffiths and Tavaré, 1994; Donnelly and Tavaré, 1995; Tavaré, 2004). If we denote by λ(t) the ratio
generations). We consider that t=0 is the present, and positive values represent the past. As N represents the population size in terms of haploid genomes, the number of individuals will be N/2 for diploid species. We can then apply the generalization of the coalescent in populations of variable size (Griffiths and Tavaré, 1994; Donnelly and Tavaré, 1995; Tavaré, 2004). If we denote by λ(t) the ratio  , we can then compute the probability density function (pdf)
, we can then compute the probability density function (pdf) 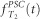 of the coalescence time T2 of two genes sampled in the present-day population. Indeed, the probability that two genes will coalesce at a time greater than t is
of the coalescence time T2 of two genes sampled in the present-day population. Indeed, the probability that two genes will coalesce at a time greater than t is
Given that
we can write the pdf as
Consequently, if we know the pdf of the coalescence time T2, the corresponding population size change function λ(t) can be computed as:
This equation may be seen as a simple rearrangement of previously known results (Griffiths and Tavaré, 1994; Tavaré, 2004), which we cited above, and to some extent it is. However, it practically means that if we only had access to a finite set of T2 values we could in theory infer the history λ(t) by simply computing this ratio. In the case of a model of population size change, this computation is by definition giving us the actual history of population size change. We show below how this ratio can be computed for any demographic scenario for which T2 distributions can be derived or simulated. And it is this computation for other models that significantly changes the outlook to genetic data and coalescence rates.
Instantaneous coalescence rate for a sample of size 2
If we consider now the coalescence time of two genes sampled in a population under an arbitrary model, whichever model this may be (structured or not, with population size change or not, and so on), and if we assume that we know its pdf,  , it is straightforward to compute the ratio λ(t) of Equation (4)
, it is straightforward to compute the ratio λ(t) of Equation (4)
Let us now denote  . We then have by definition
. We then have by definition 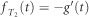 , hence
, hence
from where we get, as g(0)=1,
It therefore follows that the pdf
 can always be written as
can always be written as
even if the so-computed function λ(t) has nothing to do with any population size change.
In other words, for any given model, there always exists a function λ(t) that explains the coalescence time distribution of this model for a sample of size two,  . The pdf of T2 can thus always be written as a function of λ(t) as in Equation (8), exactly as if the model under which the data were produced was only defined by population size changes. This function λ(t) is a fictitious or spurious population size change function whose coalescence time T2 would mimic perfectly the demographic model.
. The pdf of T2 can thus always be written as a function of λ(t) as in Equation (8), exactly as if the model under which the data were produced was only defined by population size changes. This function λ(t) is a fictitious or spurious population size change function whose coalescence time T2 would mimic perfectly the demographic model.
Now, if we define μ(t) as
it should be natural to see μ(t) as an instantaneous coalescence rate, as it represents the probability that two lineages that have not yet coalesced at time t (as expressed by the denominator) will do so in an infinitesimal amount of time starting at t (as expressed in the numerator). Another way to realize it is to use theoretical results and terminology from reliability theory. If we note that T2 can be seen as a lifetime, then we can also note that the quantity  , known as the hazard function or failure rate in the reliability engineering community, represents the instantaneous rate of failure of a system at time t (see, for instance, Ruegg, 1989 or Klein and Moeschberger, 2003). The term instantaneous is central and we show in the next section that it is crucial for the interpretation of structured models.
, known as the hazard function or failure rate in the reliability engineering community, represents the instantaneous rate of failure of a system at time t (see, for instance, Ruegg, 1989 or Klein and Moeschberger, 2003). The term instantaneous is central and we show in the next section that it is crucial for the interpretation of structured models.
Linking population structure and population size change
We now consider a model of population structure such as the classical symmetric n-island model (Wright, 1931), where we have a set of n islands (or demes) of constant size N, interconnected by gene flow with a migration rate m, where 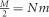 is the number of immigrants (genes) in each island every generation. The total number of genes or haploid genomes in the whole metapopulation is nN and it is therefore constant. Again, N is the number of haploid genomes, and N/2 the number of diploid individuals.
is the number of immigrants (genes) in each island every generation. The total number of genes or haploid genomes in the whole metapopulation is nN and it is therefore constant. Again, N is the number of haploid genomes, and N/2 the number of diploid individuals.
Under this model we can write the pdf for T2 (see Herbots, 1994; Wilkinson-Herbots, 1998; and Mazet et al., 2015 for details and notation and Bahlo and Griffiths, 2001 for related results and Charlesworth et al., 2003 for an insightful review) by considering the cases when the two genes are sampled from the same (s) or from different (d) demes:
where
and where −α and −β are the roots of the polynomial
whose discriminant is Δ=(1+nγ)2−4γ, and therefore
and
with 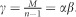
Now let us consider a hypothetical demographic history characterized by population size changes but without any population structure. For that history to explain the data generated by a model of population structure, this hypothetical demographic history will correspond to the function λ(t) as defined by Equation (5). Thus, in the case of two haploid genomes sampled in the same deme (a most reasonable assumption for a diploid individual) we get:
 |
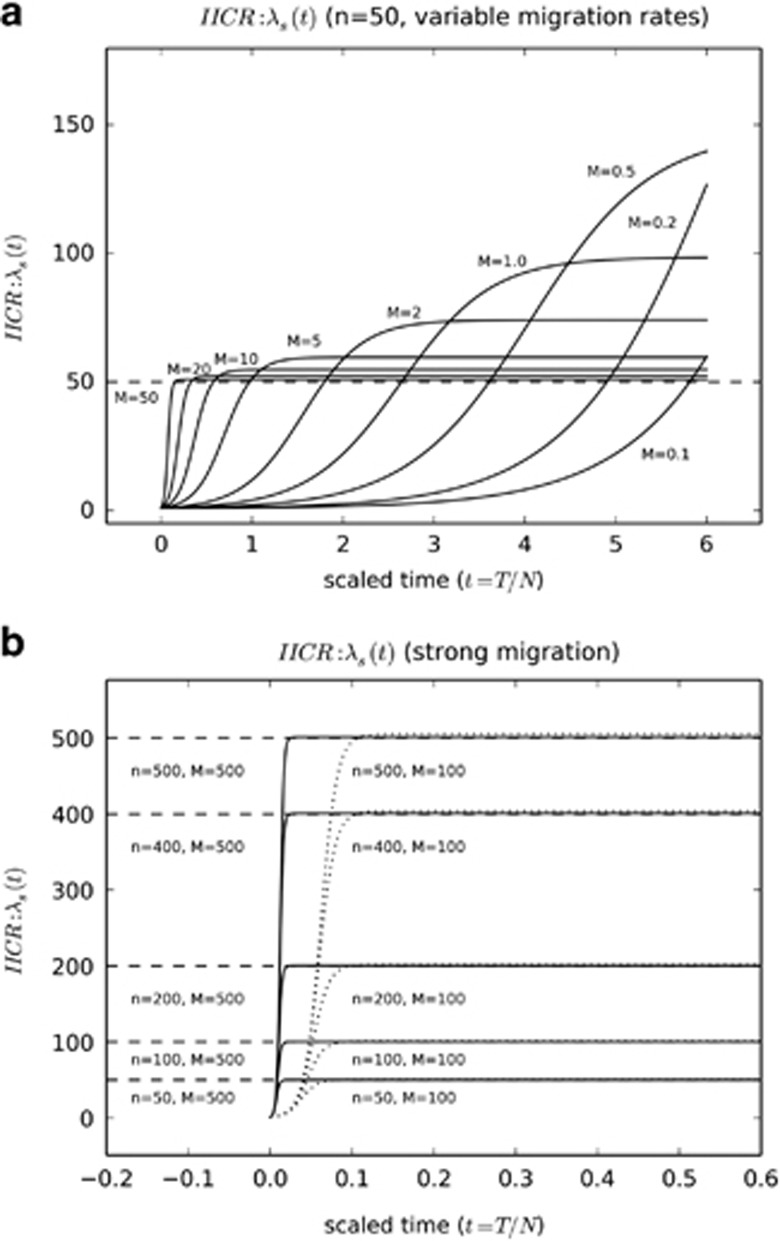
It is then trivial to compute the function λs(t) for any set of parameters n and M. For instance, Figure 1a shows the corresponding curves for n=50 and M values between 0.1 and 50. As expected (Chikhi et al., 2010; Mazet et al., 2015), we observe a (fictitious) population decrease from a large hypothetical ancestral population of size  to a smaller hypothetical current population of size
to a smaller hypothetical current population of size  . Note that λs(t) is a population size ratio that does not provide absolute values of the effective population size. In our case, it is however trivial to show that for t sufficiently close to 0, we find that λs(t)=1 and hence it follows that
. Note that λs(t) is a population size ratio that does not provide absolute values of the effective population size. In our case, it is however trivial to show that for t sufficiently close to 0, we find that λs(t)=1 and hence it follows that  , the size of a deme. Indeed, at the time of sampling, the coalescence history for two genes sampled from the same deme is mostly dependent on the size of the local deme. Interestingly, this is true for any value of M. Figure 1 indicates that as M becomes larger,
, the size of a deme. Indeed, at the time of sampling, the coalescence history for two genes sampled from the same deme is mostly dependent on the size of the local deme. Interestingly, this is true for any value of M. Figure 1 indicates that as M becomes larger,  becomes closer to nN, represented by the horizontal dashed line. This is expected: when the migration rate increases the whole set of populations behaves less and less like a structured model and increasingly like a single random mating population of size nN. Several authors have shown that under the strong migration condition, it is possible to define a coalescent effective population size toward which the structured population tends (Sjödin et al., 2005; Wakeley and Sargsyan, 2009). Figure 1b shows indeed that when M is very high (M=100 and M=500), the n-island model behaves as a population characterized by a constant size until the very recent past. For instance, when M=500, λs(t) only drops at time t=0.02 that for N=100 would correspond to 2 generations ago. In other words, the strong migration assumption implicitly assumes that the bottleneck seen in our results is so recent that it can be neglected. Using the terminology introduced by Wakeley (1999), it assumes that the scattering phase is very short. Altogether, our results provide a more general framework that allows us to easily incorporate the strong migration assumptions.
becomes closer to nN, represented by the horizontal dashed line. This is expected: when the migration rate increases the whole set of populations behaves less and less like a structured model and increasingly like a single random mating population of size nN. Several authors have shown that under the strong migration condition, it is possible to define a coalescent effective population size toward which the structured population tends (Sjödin et al., 2005; Wakeley and Sargsyan, 2009). Figure 1b shows indeed that when M is very high (M=100 and M=500), the n-island model behaves as a population characterized by a constant size until the very recent past. For instance, when M=500, λs(t) only drops at time t=0.02 that for N=100 would correspond to 2 generations ago. In other words, the strong migration assumption implicitly assumes that the bottleneck seen in our results is so recent that it can be neglected. Using the terminology introduced by Wakeley (1999), it assumes that the scattering phase is very short. Altogether, our results provide a more general framework that allows us to easily incorporate the strong migration assumptions.
Figure 1.
Inferred population size changes for n-island models with constant size. This figure shows λs(t) for different values of M, the number of migrants, and n, the number of islands. In (a) we assumed an island model with n=50, and varied M, the number of migrants between 0.1 and 50. In (b) we varied n between 50 and 500 and used two large values for M, namely 100 and 500. For both panels, the y axis is scaled by N and the horizontal dashed lines correspond to nN, the total population size. In all cases, λs(t) identifies a population decrease.
Coming back to Figure 1a, we also note that as M decreases, the fictitious bottleneck becomes older and the ancestral population becomes larger, for a constant value of n, the number of islands. We can derive the asymptotic coalescent effective size of this n-island model by computing the limit of λ(t) when t goes to infinity, and find that, as 0<β<α,
where we recall that −β was the largest of the roots found above (Equation 15). By developing Equation (15), we find
Here we can see that for large values of M, λs(t) is close to
This is the nucleotide diversity effective size computed in Nei and Takahata (1993) for the n-island model.
If we now perform the same analyses and computations for the case where the haploid genomes are sampled from different demes, it leads to the following result:
Here the population dynamics is inverted, and we observe a fictitious population expansion. Figure 2 shows some plots of λd(t) for different values of M. This is in agreement with several previous studies that noted that when sampling is carried out across demes, the bottleneck signal either disappears or can be replaced by a population expansion signal (Peter et al., 2010; Chikhi et al., 2010; Heller et al., 2013). We note that 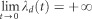 . The two lineages being in different demes at time t=0, it is by definition impossible for them to coalesce in the very recent past, as a migration event has first to occur. Let us note also that
. The two lineages being in different demes at time t=0, it is by definition impossible for them to coalesce in the very recent past, as a migration event has first to occur. Let us note also that  as for λs.
as for λs.
Figure 2.
Inferred population size changes for n-island models and samples from different demes. This figure shows λd(t) for different values of M, the number of migrants. The number of islands was assumed to be n=50. Samples come from different islands. In all cases, λd(t) identifies a population increase.
Our results, as expressed by equations (16) and (20), stress the difficulty in defining an effective size for a structured population, because a structured population has properties that a stationary nonstructured population does not have. It behaves like a nonstructured population that changes in size. The IICR is therefore what connects the two (structured and panmictic) models. As a consequence, there is no overwhelming reason to summarize its properties by one single number when it actually is defined either by a number of islands and a migration rate or by a full trajectory of effective sizes. We point toward the studies of Sjödin et al. (2005) and Wakeley and Sargsyan (2009) for models and conditions under which an effective size can be defined. What we wish to stress is that the theory presented here provides a general framework for explaining and predicting population size changes that population genetics methods will infer. Below, we illustrate how this can be applied to simple and complex structured models and we also predict the population size changes that methods ignoring structure will infer. Given that λ(t) does not necessarily correspond to actual changes in Ne (effective size), we introduce the inverse instantaneous coalescence rate or IICR that we will use for the rest of the manuscript instead of λ(t). The reason for this is that the IICR is only equivalent to an instantaneous coalescent Ne in the case of models without structure. For other models, it is, in the absence of a better term, the inverse of an instantaneous coalescence rate. The IICR is of course by definition a function of time and implicitly leads us to consider a trajectory rather than a single value even for constant size models such as the n-island model.
Application to simulated and real data
In order to illustrate how an observed distribution of T2 values can be used to infer the IICR we carried out simulations under structured and unstructured scenarios. Data were simulated using the ms software (Hudson, 2002). For each scenario, we simulated independent values of T2 and used them to estimate the IICR at various time points ti, as follows:
where  is the estimated or empirical cumulative distribution function of T2 and
is the estimated or empirical cumulative distribution function of T2 and 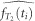 is an estimated approximation of its density around ti. The two scenarios of population size change without structure were simulated with the following ms commands: ms 2 100 -T -L -G -16.094 -eG 0.1 0.0 for the exponential population size change (Figure 3a) and ms 2 100 -T -L -eN 0.01 0.1 -eN 0.06 1 -eN 0.2 0.5 -eN 1 1 -eN 2 2 (Figure 3b) for the stepwise population size change.
is an estimated approximation of its density around ti. The two scenarios of population size change without structure were simulated with the following ms commands: ms 2 100 -T -L -G -16.094 -eG 0.1 0.0 for the exponential population size change (Figure 3a) and ms 2 100 -T -L -eN 0.01 0.1 -eN 0.06 1 -eN 0.2 0.5 -eN 1 1 -eN 2 2 (Figure 3b) for the stepwise population size change.
Figure 3.
Inferred population size changes for populations without structure. For both panels the x axis represents time in generations, whereas the y axis represents population size in units of 104 diploids (an IICR of 0.5 corresponds to 500 × 10=5000 diploid genomes). (a) A panmictic population that experienced an exponential decrease from a previously constant size ancestral population. The solid blue line (theoretical IICR) was obtained using Equation (4). The dashed line represents the simulated demographic history and corresponds to the total number of haploid genomes (the actual size). The stepwise red solid curve (estimated IICR) was obtained using the simulated T2 values and Equation (21). (b) A history of stepwise population size changes is shown. The color codes are identical to (a). A full color version of this figure is available at the Heredity journal online.
In addition, for the scenarios involving population structure (Figures 4 and 5) we simulated both T2 values and DNA sequences assuming an n-island model with n=10 demes of size of N=1000 haploid genomes each (that is, 500 diploids), and a mutation rate of μ=10−8. We then computed the empirical IICR from the T2 values, and did a PSMC analysis using the corresponding DNA sequences. The ms commands used to produce the data for a model with three changes in migration rates were ms 2 100 -t 600 -r 120 30000000 -I 10 2 0 0 0 0 0 0 0 0 0 1 -eM 3 5 -eM 6 0.8 -eM 15 5 -p 8 and ms 2 100 -t 600 -r 120 30000000 -I 10 2 0 0 0 0 0 0 0 0 0 1 -eN 1 0.5 -p 8 for a model in which deme sizes doubled (and hence the metapopulation too). We also simulated scenarios with a 10- and a 50-fold deme size increase. We either kept M, the number of migrants, or m, the migration rate, constant after the changes in N (Supplementary Figures S1 and S2). In addition, we simulated a scenario where the deme size varied according to a complex step function, and inferred the IICR under various migration rates (see Supplementary Figure S3).
Figure 4.
Inferred population size changes under population structure and two sampling schemes. This figure shows the predicted population size changes that will be inferred for an n-island model under the assumption that populations are not structured. For both panels the x axis represents time in generations, whereas the y axis represents real or inferred population size in units of 104 diploid genomes. We simulated an n-island model with n=10 and M=1 and computed the theoretical IICR using Equation (4), and the estimated IICR using the simulated T2 values and Equation (21). The color codes are identical to Figure 3. The green solid lines represent the history inferred by the PSMC. (a) The results when the two haploid genomes are sampled in the same deme are shown. In (b) they come from different demes. The constant size of the metapopulation at y=0.5 corresponds to 5000 diploid genomes or 10 islands of size 500 diploids. A full color version of this figure is available at the Heredity journal online.
Figure 5.
Inferred population size changes under population structure with changes in migration rates or deme size. The x axis represents time in generations, whereas the y axis represents real or inferred population size in units of 104 diploid genomes. Color codes are identical to Figure 4. Data were simulated under an n-island model with n=10. In (a) the population size was constant in size with each deme having a size N=1000 haploid genomes (500 diploids) but three changes in migration rate occured at T3=30 000, T2=12 000 and T1=6000 generations in the past. Before T3 the migration rate was M3=5. At T3 it changed to M2=0.8 and remained constant until T2, and then changed to M1=5 at T1. After that it remained at M=1 until the present. In (b) all the demes doubled in size from 500 to 1000 haploids (or 250 to 500 diploids) at T=2000 generations and migration was constant with M=1. A full color version of this figure is available at the Heredity journal online.
For the comparisons with the analyses of the human data we assumed the mutation rate used by Li and Durbin (2011), namely μ=2.5 × 10−8. These authors note that the PSMC is not expected to give reliable estimates of recent population sizes (that is, <10 kyr in humans), and we therefore carried out simulations with and without a recent demographic expansion following the Neolithic transition. The simulations incorporating a recent increase in deme size in humans produce PSMC and IICR profiles similar to the PSMC estimations on human data, whereas the lack of a recent increase produces a curve that is flat in the recent past (see Supplementary Figure S4). For simplicity, the genomic data for the scenario with three migration rate changes were simulated assuming n=10 demes. The ms command used was ms 2 100 -t 1590 -r 318 30000000 -I 10 2 0 0 0 0 0 0 0 0 0 0.55 -eM 4.5 4 -eM 18.0 0.55 -eM 47.5 0.85. This command simulates an n-island model of n=10 islands, of size N=1060 haploid genomes or 530 diploids. A generation time of 25 years and a mutation rate μ=2.5 × 10−8 were assumed as in Li and Durbin (2011). Following these authors we simulated 100 independent 30 Mb long ‘chromosomes' that were then used together to represent the full 3 Gb long human genome. Under that scenario, the scaled mutation is θ=4 × 530 × 2.5 × 10−8 × 30 × 106 =1590. Given that each island has 530 diploid individuals, the metapopulation is composed of 5300 diploid individuals. In ms commands, the migration rate and time are scaled in units of the diploid deme size. The number of migrants exchanged was M=0.55 in the recent past and M=0.85 in the most ancient past, and changed at various times indicated by the eM flag in the ms command. Going from the past to the present, the ms commands thus simulate the following demographic events: M decreased from 0.85 to 0.55 at ∼47.5 × 4 × 530 × 25=2 517 500 years ago, then M increased from 0.55 to 4.00 at ∼18 × 4 × 530 × 25=954 000 years ago and finally M decreased 4.5 × 4 × 530 × 25=2 38 500 years ago from 4.00 to 0.55. After that M remained constant. Moreover, in addition to scenarios where the deme size never changed we also simulated scenarios with a rapid increase in deme size 0.25 × 4 × 530 × 25=13 250 years ago by a factor 40, to represent the Neolithic transition. The figure without this change is in the Supplementary Material, Figure S4.
Results
Predicting the inferred demographic history of nonstructured and structured populations: illustrations by simulations
Figure 3 shows the results for nonstructured populations that were subjected to various histories of population size change. The left-hand panel shows a population that experienced an exponential decrease from a previously constant size ancestral population. As expected, the blue solid line obtained using the full theoretical T2 distribution is identical to the simulated history of population size changes (that is, the real population size changes). The stepwise red solid line represents the empirical IICR. The number of ti values or steps can be changed depending on the precision that one wishes to reach and the total number of T2 values. We chose values similar to those typically used in recent genomic studies for comparison (Zhao et al., 2013; Zhan et al., 2013; Zhou et al., 2014) but a much greater precision can be achieved under our framework. The right-hand panel shows similar results but for a population that went through various stepwise population size changes. This shows the remarkable match between the theoretical and empirical IICR curves and the simulated history. When a population is not structured the IICR will exactly match the real history in terms of population size changes.
Figure 4 is similar to Figure 3 but with structured populations: we sampled two haploid genomes under the n-island model, with n=10 and M=1. Figure 4a shows the results when the genomes were sampled in the same deme (a single diploid individual), whereas Figure 4b shows the results when the two haploid genomes were sampled in different demes. These figures show again that the empirical and theoretical IICR distributions match each other. Moreover, they predict the population size change history inferred by the PSMC. This suggests that the PSMC does not infer a population size change but the IICR and estimates it rather well. Finally, the IICR and the PSMC identify a (spurious) population decrease or increase depending on the sampling scheme, even though the total number of haploid genomes was constant (horizontal dashed line representing the real population size). These results are in agreement with several studies showing that different sampling strategies applied to the same set of populations may lead to infer quite distinct demographic histories (Chikhi et al., 2010; Heller et al., 2013), even though they used different methods. Whereas the effect described by Heller et al. (2013) was observed using the Bayesian Skyline Plot method (Drummond et al., 2005), Chikhi et al. (2010) used the msvar approach of Beaumont (1999).
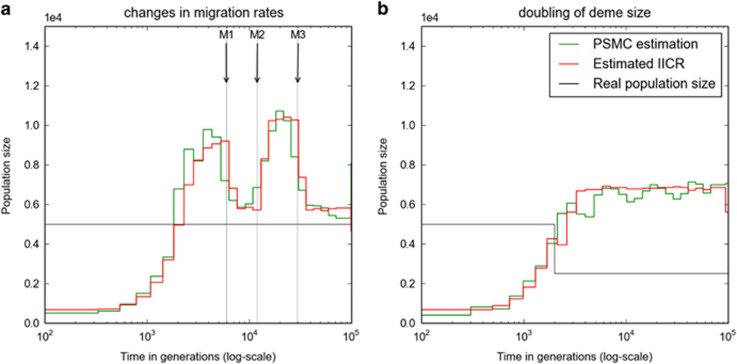
Although Figures 3 and 4 illustrate and validate the theory developed in previous sections using two models (the n-island and population size change) for which the T2 distribution is known, our approach to estimate the IICR is still valid when we have values of T2 but the distribution is not known. This can happen for models that can be simulated but for which no analytical results exist (Figure 5). In Figure 5a, we considered an n-island model with n=10 demes where the total population size remained constant (each deme had a size of N=1000 haploid genomes or N/2=500 diploids) but migration rates changed at three different moments in the last 30 000 generations, as indicated by the vertical arrows. This scenario mimics a set of populations whose connectivity is changing because of fragmentation or reconnection of habitat either due to climatic or anthropogenic effects (Goossens et al., 2006; Quéméré et al., 2012). The demographic history reconstructed by the PSMC matches again the history predicted by the empirical IICR, but it is strikingly different from the actual size of the metapopulation (horizontal line). Whereas the total population size was constant throughout, the reconstructed history suggests that the population expanded and contracted on at least two occasions. A more serious issue arises from the fact that the population size changes inferred by the PSMC do not appear to match the times at which the migration rates changed, at least at the level of precision provided by the PSMC. For instance, the last change in migration rate, M1, occurred 6000 generations in the past. Instead, the PSMC infers a population expansion and contraction after that event. Figure 5b corresponds to a scenario in which the size of all demes doubled 2000 generations before the present. Here the striking result comes from the fact that whereas the population size doubled (black broken line), the IICR and PSMC would suggest a continuous population decrease over a very long period, whose timing has again little to do with the actual history of the population. The population size change is thus missed by the PSMC. See Supplementary Figures S1 and S2 for cases where the population increased by a factor 10 and 50 and where either M or m was constant. Altogether, this figure and the associated Supplementary Figures suggest that changes in migration patterns or changes in deme size may be misinterpreted by population genetics methods that ignore population structure, and that there is a need for methods able to distinguish population structure from population size change (see Peter et al., 2010; Chikhi et al., 2010; Heller et al., 2013; Mazet et al., 2015).
A tentative reinterpretation of human past demography: on the importance of being structured
In their study, Li and Durbin (2011) applied the PSMC to genomic data obtained from humans and inferred a history of population size changes. As demonstrated above, what the PSMC estimates is the IICR that does not necessarily correspond to real population size changes, but may also arise from a model with changes in migration rates. To illustrate this we applied our approach to identify an island model with constant population size reproducing closely the IICR obtained by Li and Durbin (2011). For simplicity we arbitrarily assumed that the number of islands was n=10, and that there were three changes in migration rates as this is the minimum number of changes required to obtain an IICR curve with two humps, assuming a constant deme size. We propose a history in which migration rates (Mi, i=1, 2, 3, 4) changed at three moments (Ti, i=1, 2, 3), and where M1 corresponds to the number of migrants exchanged between demes each generation during the period between the present and T1. More specifically, we found a change in migration rates (from M4=0.85 to M3=0.55) at ∼T3=2.52 million years (Myr) ago, then a major increase (from M3=0.55 to M2=4) at ∼T2=0.9–1.0 Myr and finally a major decrease (from M2=4 to M1=0.55) at ∼T1=0.23–0.25 Myr ago. In other words, our results would suggest changes in connectivity at the start of the Lower Pleistocene (dated at 2.58 Myr) that corresponds to the emergence of the genus Homo. The most striking change corresponds to major increase in connectivity just before the transition between the Lower and Middle Pleistocene (dated at 0.78 Myr). We find that the Middle Pleistocene is characterized by high and sustained gene flow. Finally, connectivity abruptly decreases at 210–230 kyr ago just before the earliest remains of anatomically modern humans Homo sapiens at ca. 200 kyr.
Discussion
The IICR and the PSMC
In this study we have shown that it is always possible to find a demographic history involving only population size changes that perfectly explains any distribution of coalescence times T2, even when this distribution was actually generated by a model in which there was no population size change. To illustrate this we first focused on a simple n-island model for which the pdf of T2 can be derived, and obtained an analytic formula of the fictitious population size change history, named IICR, as a function of the number of islands and the migration rate of the model. We also showed that the IICR can be computed for any (neutral) model from any observed distribution of T2 values. We showed that the empirical and theoretical IICRs were identical when the latter could be obtained. We then obtained the empirical IICR under models involving changes in migration rates or in deme size. This suggests that, at least for a sample of size 2, even an infinite amount of genetic data from independent loci alone may not allow to distinguish structure and population size change models. In addition, the history of population size changes in Figure 5 would suggest that four demographic changes occured, two expansions and two contractions, whereas only three changes of the migration rate were actually simulated.
The theory presented here is simple and general. It allows us to predict the IICR and state that any method ignoring population structure will try to estimate the IICR. In the case of complex demographic histories with population structure, interpreting the IICR as a population size or a ratio of population sizes can be misleading. To clarify the difference between the IICR and an effective population size we can consider the following rationale. If a structured population could be summarized by a single Ne then a change in gene flow should be matched by a simultaneous change in Ne. In that case, changes in Ne would be misleading (as the size would not change) but their timing might still be meaningful. For instance a ‘hump' inferred using diCal or the PSMC could be easily translated into a change in gene flow patterns. In such a case, we could reinterpret the changes in Ne by saying, for each hump, that gene flow decreased and then increased again. What the IICR shows is that it is not that simple. The fact that a structured model can only be summarized by a trajectory of spurious population sizes means that the timing of changes in migration rates will interact in a complex manner, hence generating IICR profiles that may be only loosely related with population-related events. This can be seen in Figures 5 and 6 (and the Supplementary Figures S1–S4).
Figure 6.
Human history with changes in migration rates. This figure shows, in red, the history of population size changes inferred by Li and Durbin (2011) from the complete diploid genome sequences of a Chinese male (YH) (Wang et al., 2008). The 10 green curves correspond to the IICR of 10 independent replicates of the same demographic history involving three changes in migration rates. The x axis represents time in years in a log scale, whereas the y axis represents real or inferred population size in units of diploid genomes. The times at which these changes occur are represented by the vertical arrows at 2.52 Myr ago, 0.95 Myr ago and 0.24 Myr ago. The blue shaded areas correspond to (1) the beginning of the Pleistocene (Pleist.) at 2.57–2.60 Myr ago, (2) the beginning of the Middle Pleistocene (Mid. Pleist.) at 0.77–0.79 Myr ago and (3) the oldest known fossils of anatomically modern humans (AMH) at 195–198 kyr ago. Following Li and Durbin (2011), we assumed that the mutation rate was μ=2.5 × 10−8 and that generation time was 25 years. We also kept their ratio between mutation and recombination rates. Each deme had a size of 530 diploids and the total number of haploid genomes was thus constant and equal to 10 600. A full color version of this figure is available at the Heredity journal online.
These results do not invalidate the use of panmictic models for the reconstruction of population history as long as population structure can indeed be neglected (Figure 3 and Supplementary Figure S3), but it certainly stresses the need for caution in the interpretation of this history. When Li and Durbin published their landmark study in 2011 (Li and Durbin, 2011), they showed for the first time that it was possible to reconstruct the demographic history of a population by using the genome of a single diploid individual. It was a remarkable feat based on the SMC model introduced by McVean and Cardin (2005). Its application to various species (Groenen et al., 2012; Prado-Martinez et al., 2013; Zhao et al., 2013; Zhan et al., 2013; Green et al., 2014; Hung et al., 2014; Zhou et al., 2014) has been revolutionary and led to the development of new methods (Sheehan et al., 2013; Schiffels and Durbin, 2013; Liu and Fu, 2015). However, the increasing number of studies pointing at the effect of population structure (Leblois et al., 2006; Nielsen and Beaumont, 2009; Chikhi et al., 2010; Heller et al., 2013; Paz-Vinas et al., 2013) or changes in population structure (Wakeley, 1999, 2001; Wakeley and Aliacar, 2001; Städler et al., 2009; Broquet et al., 2010; Heller et al., 2013; Paz-Vinas et al., 2013) in generating spurious changes in inferred population size suggested that new models should be analyzed that can incorporate population structure (Goldstein and Chikhi, 2002; Harding and McVean, 2004. For instance, Mazet et al. (2015) have recently shown that genomic data from a single diploid individual can be used to distinguish an n-island model from a model with a single population size change. Their likelihood-based approach uses the distribution of coalescence times for a sample of size two (T2). This study represents an interesting alternative as it should be possible to determine whether a model of population structure is more likely than a model of population size change to explain a particular data set. The approach of Mazet et al. (2015) is however limited to a very simple model of population size change. Demographic models inferred by several recent methods (Li and Durbin, 2011; Schiffels and Durbin, 2013; Sheehan et al., 2013; Liu and Fu, 2015) are not limited to one population size change. They are thus more realistic and, as we have shown here, this comes at a certain price. As they allow for several tens of population size changes, they mimic more precisely the genomic patterns arising from structured models. Therefore, they reconstruct a demographic history that can optimally explain any particular pattern of genomic variation only in terms of population size changes. As we have shown here, and until we can separate models (see below), this casts doubts on any history reconstructed from genomic data by the above-mentioned approaches. Indeed, if any pattern of (neutral) genomic variation can be interpreted efficiently in terms of population size changes, then how can we identify the cases where the observed genomic data were not generated by population size changes?
Li and Durbin (2011) acknowledged that one should be cautious when interpreting the changes inferred by their method. For instance, they showed (see their Supplementary Materials, Figure S5) that when one population of constant size N splits in two half-sized populations that later merge again, their method will identify a change of N even though N actually never changed. Still, their method is implicitly or explicitly used and interpreted in terms of population size changes, including by themselves. There are therefore several issues that need to be addressed. One issue is to determine whether it is possible to separate models of population size change from models of population structure (Mazet et al., 2015, and see perspectives below). When population structure can be ignored, our results actually contribute to the validation of the PSMC (Figure 3 and Supplementary Figure S3). We found that the PSMC performed impressively well and generally reconstructed the IICR with great precision. It is therefore at this stage one of the best methods (Sheehan et al., 2013; Schiffels and Durbin, 2013; Liu and Fu, 2015) published so far and remains a landmark in population genetics inference.
The IICR: toward a critical interpretation of effective population sizes
The concept of effective size is central to population genetics. It allows population geneticists to replace complex real-world populations by equivalent and simpler Wright–Fisher populations that would have the same ‘rate of genetic drift' (Wakeley and Sargsyan, 2009). The concept is however far from trivial and it is not always clear what authors mean when they mention the Ne of a particular species or population, as rightly noted by Sjödin et al. (2005) among others. Several Nes have been defined depending on the property of interest (inbreeding, variance in allele frequency over time and so on) and its relationship to genetic drift (Wakeley and Sargsyan, 2009). This is a complex issue that we do not aim at reviewing or discussing in detail here.
The IICR is related to the coalescent Ne (Sjödin et al., 2005; Wakeley and Sargsyan, 2009) but it is explicitly variable with time. Given that most species are likely to be spatially structured, interpreting the IICR as a simple (coalescent) effective size may generate serious misinterpretations.
The IICR is a trajectory of instantaneous ‘population sizes' that fully explains complex models without loss of information. The circumstances under which this trajectory can indeed be appropriately summarized by one effective population size are still to be determined and will depend on the questions asked and the amount of markers used. For instance, for ‘strong migration scenarios' (M=500 and M=100) the inferred population size changes are recent and abrupt, and the period during which the population was stationary will be significant in generating patterns of genetic diversity (Wakeley, 1999, 2001; Wakeley and Aliacar, 2001; Charlesworth et al., 2003; Wakeley and Sargsyan, 2009). However, even for such cases of low genetic differentiation (FST≈1/2001=0.0005 and FST≈1/401=0.0025, respectively), the spurious population size drop could perhaps be detected with genomic information. For M=100 the population size decrease starts between t=0.05 and t=0.10, which for N=100 to N=1000 could correspond to values between 5 and 100 generations ago, respectively. In other words, an n-island model may actually behave differently from a Wright–Fisher model even under some ‘strong migration' conditions. The approximation will therefore be valid for some questions and data sets, and invalid for others (Charlesworth et al., 2003; Wakeley and Sargsyan, 2009). Note also that for very low migration rates (M=0.1, M=0.2, corresponding to very high FST≈0.71 and FST≈0.56, respectively) the recent history is also characterized by a stationary IICR. Most genes will then coalesce within demes and only a small proportion will provide information on the ancient IICR values and therefore on population structure (see Mazet et al., 2015).
The IICR and the complex history of species: toward a critical reevaluation of population genetics inference
The PSMC has now been applied to many species, generating curves that are very similar to those represented in Figure 5. In Figure 5a, the population size changes detected by the PSMC were not correlated in a simple manner to the changes in gene flow or deme size. This is likely the result of two factors. First, a structured population cannot always be summarized by a single number. Second, the PSMC requires a discretized distribution of time that may lead to missing abrupt changes such as those simulated here. For real data sets where changes in migration rates or in population size may be smoother, this may not be so problematic. For the human data, assuming a simple model of population structure, we inferred periods of change in gene flow that correspond to major transitions in the recent human evolutionary history, including the emergence of anatomically modern humans. Given that humans are likely to have been subjected to a complex history of spatial expansions and contractions and changes in the levels of gene flow (Wakeley, 1999, 2001; Harpending and Rogers, 2000; Goldstein and Chikhi, 2002; Harding and McVean, 2004), our results are necessarily simplistic but suggest that a reinterpretation of panmictic models may be needed and possible. Our results are at odds with a history of population crashes and increases depicted in various population genetic studies, but it is in phase with fossil data and provides a more realistic interpretation framework. We thus wish to call for a critical reappraisal of what can be inferred from genetic or genomic data. The histories inferred by methods ignoring structure represent a first approximation but they are unlikely to provide us with the information we need to better understand the recent evolutionary history of humans or other species. It is difficult to imagine that humans have been one single panmictic population whose size has changed over the last few million years (that is, since the appearance of the Homo genus). This does not minimize the achievement of the Li and Durbin (2011) study, but it does question how inference from genetic data are sometimes presented and interpreted.
Perspectives
We focused throughout this study on T2, the time to the most recent common ancestor for a sample of size two. For larger samples we can define Tk as the time during which there are k lineages. It would be important to determine whether, for structured models, the IICR estimated from the distribution of Tk varies significantly with k. If that were the case, that would suggest that it is possible to separate structure from population size change with the distributions of Tk for various k values. The reason for this is that population size change models should generate identical IICR for all Tk distributions, as they should all correspond to the same (real) history of population size change. To our knowledge the distribution of Tk for k>2 has not yet been explicitly derived for the n-island or other structured models (but see interesting studies such as Herbots, 1994; Wakeley and Aliacar, 2001; Wakeley, 2001; Nielsen and Wakeley, 2001).
One simple solution to this question is to simulate genetic data under a structured model of interest and then compare the simulated Tk distributions under that model and the Tk distributions of the corresponding model of population size change identified using the T2 distribution. Preliminary simulations suggest that the Tk distributions produce different IICRs, at least for some models of population structure. For instance, we predict that the analysis of human genomic data with the PSMC and MSMC (multiple sequentially Markovian coalescent) should produce different curves under a model of population structure but identical ones for a model of population size change. This prediction can be tested by comparing the PSMC and MSMC curves of Li and Durbin (2011) and Schiffels and Durbin (2013), respectively. Visual inspection of the corresponding figures suggests indeed that they are different, and therefore that our model of population structure is a valid alternative. However, we stress that an independent study is required. Indeed, the history reconstructed by these methods with real data is not very precise and the two curves are not easily comparable because they are expected to provide poor estimates at different moments. Any difference between the two analyses should thus be evaluated and validated with simulations.
Finally, one underlying assumption of our study is that the coalescent represents a reasonable model for the genealogy of the genes sampled. Given that the coalescent is an approximation of the true gene genealogy, and that there are species for which the coalescent may not be the most appropriate model (Wakeley and Sargsyan, 2009), we should insist that our results can, at this stage, only be considered for coalescent-like genealogies. The development of similar approaches for other genealogical models would definitely be a very interesting avenue of research.
Acknowledgments
We are grateful to Amaury Lambert and Guillaume Achaz for constructive comments on earlier versions of this work, and to three referees for positive and useful criticisms that helped us improve the manuscript. We are also grateful to Mike Bruford and Mafalda Costa for handling our manuscript with great speed and efficiency. We are grateful to the genotoul bioinformatics platform Toulouse Midi-Pyrenees for providing computing and storage resources and to the LIA BEEG-B (Laboratoire International Associé–Bioinformatics, Ecology, Evolution, Genomics and Behaviour) (CNRS) for facilitating travel and collaboration between Toulouse and Lisbon. This work was partly performed using HPC resources from CALMIP (Grant 2012–projects 43 and 44) from Toulouse, France. This study was partly funded by the Fundação para a Ciência e Tecnologia (ref. PTDC/BIA- BIC/4476/2012), the Projets Exploratoires Pluridisciplinaires (PEPS 2012 Bio-Maths-Info) project, the LABEX entitled TULIP (ANR-10-LABX-41) as well as the Pôle de Recherche et d'Enseignement Supérieur (PRES) and the Région Midi-Pyrénées, France.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Bahlo M, Griffiths RC. (2001). Coalescence time for two genes from a subdivided population. J Math Biol 43: 397–410. [DOI] [PubMed] [Google Scholar]
- Beaumont MA. (1999). Detecting population expansion and decline using microsatellites. Genetics 153: 2013–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broquet T, Angelone S, Jaquiery J, Joly P, Lena JP, Lengagne T et al. (2010). Genetic bottlenecks driven by population disconnection. Conserv Biol 24: 1596–1605. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D, Barton NH. (2003). The effects of genetic and geographic structure on neutral variation. Annu Rev Ecol Evol Syst 34: 99–125. [Google Scholar]
- Chikhi L, Sousa VC, Luisi P, Goossens B, Beaumont MA. (2010). The confounding effects of population structure, genetic diversity and the sampling scheme on the detection and quantification of population size changes. Genetics 186: 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P, Tavaré S. (1995). Coalescents and genealogical structure under neutrality. Annu Rev Genet 29: 401–421. [DOI] [PubMed] [Google Scholar]
- Drummond A J, Rambaut A, Shapiro B, Pybus OG. (2005). Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22: 1185–1192. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Chikhi L. (2002). Human migrations and population structure: what we know and why it matters. Annu Rev Genomics Hum Genet 3: 129–152. [DOI] [PubMed] [Google Scholar]
- Goossens B, Chikhi L, Ancrenaz M, Lackman-Ancrenaz I, Andau P, Bruford MW et al. (2006). Genetic signature of anthropogenic population collapse in orangutans. PLoS Biol 4: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, Hickey G et al. (2014). Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346: 1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R, Tavaré S. (1994). Simulating probability distributions in the coalescent. Theor Popul Biol 46: 131–159. [Google Scholar]
- Groenen M, Archibald A, Uenishi H, Tuggle C, Takeuchi Y, Rothschild M et al. (2012). Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491: 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. (2009). Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet 5: el000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding RM, McVean G. (2004). A structured ancestral population for the evolution of modern humans. Curr Opin Genet Dev 14: 667–674. [DOI] [PubMed] [Google Scholar]
- Harpending H, Rogers A. (2000). Genetic perspectives on human origins and differentiation. Annu Rev Genomics Hum Genet 1: 361–385. [DOI] [PubMed] [Google Scholar]
- Heller H., Chikhi L, Siegismund HR. (2013). The confounding effect of population structure on Bayesian skyline plot inferences of demographic history. PLoS One 8: e62992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbots HMJD. (1994). Stochastic models in population genetics: genealogy and genetic differentiation in structured populations. PhD thesis; University of London (Queen Mary and Westfield College).
- Hudson RR. (2002). Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Hung CM, Shaner PJL, Zink RM, Liu WC, Chu TC, Huang WS et al. (2014). Drastic population fluctuations explain the rapid extinction of the passenger pigeon. Proc Natl Acad Sci USA 111: 10636–10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JP, Moeschberger ML. (2003) Survival Analysis: Techniques for Censored and Truncated Data. Springer Science & Business Media. [Google Scholar]
- Leblois R, Estoup A, Streiff R. (2006). Genetics of recent habitat contraction and reduction in population size: does isolation by distance matter? Mol Ecol 15: 3601–3615. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. (2011). Inference of human population history from individual whole-genome sequences. Nature 475: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fu YX. (2015). Exploring population size changes using SNP frequency spectra. Nat Genet 47: 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazet O, Rodriguez W, Chikhi L. (2015). Demographic inference using genetic data from a single individual: Separating population size variation from population structure. Theor Popul Biol 104: 46–58. [DOI] [PubMed] [Google Scholar]
- McVean GAT, Cardin NJ. (2005). Approximating the coalescent with recombination. Philos Trans R Soc Lond B Biol Sci 360: 1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Takahata N. (1993). Effective population size, genetic diversity, and coalescence time in subdivided populations. J Mol Evol 37: 240–244. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Beaumont MA. (2009). Statistical inferences in phylogeography. Mol Ecol 18: 1034–1047. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J. (2001). Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Vinas I, Quéméré E, Chikhi L, Loot G, Blanchet S. (2013). The demographic history of populations experiencing asymmetric gene flow: combining simulated and empirical data. Mol Ecol 22: 3279–3291. [DOI] [PubMed] [Google Scholar]
- Peter BM, Wegmann D, Excoffier L. (2010). Distinguishing between population bottleneck and population subdivision by a Bayesian model choice procedure. Mol Ecol 19: 4648–4660. [DOI] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B et al. (2013). Great ape genetic diversity and population history. Nature 499: 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quéméré E, Amelot X, Pierson J, Crouau-Roy B, Chikhi L. (2012). Genetic data suggest a natural prehuman origin of open habitats in northern Madagascar and question the deforestation narrative in this region. Proc Natl Acad Sci USA 109: 13028–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegg A. (1989) Processus Stochastiques: Avec Applications aux Phénoménés d'attente et de fiabilité vol. 6, PPUR Presses Poly Techniques. [Google Scholar]
- Schiffels S, Durbin R. (2013). Inferring human population size and separation history from multiple genome sequences. Nat Genet 8: 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan S, Harris K, Song YS. (2013). Estimating variable effective population sizes from multiple genomes: a sequentially Markov conditional sampling distribution approach. Genetics 194: 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödin P, Kaj I, Krone S, Lascoux M, Nordborg M. (2005). On the meaning and existence of an effective population size. Genetics 169: 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Städler T, Haubold B, Merino C, Stephan W, Pfaffelhuber P. (2009). The impact of sampling schemes on the site frequency spectrum in nonequilibrium subdivided populations. Genetics 182: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S. (2004) Part I: Ancestral inference in population genetics In Picard J ed Lectures on Probability Theory and Statistics. Springer. pp 1–188. [Google Scholar]
- Wakeley J. (1999). Nonequilibrium migration in human history. Genetics 153: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J. (2001). The coalescent in an island model of population sub-division with variation among demes. Theor Popul Biol 59: 133–144. [DOI] [PubMed] [Google Scholar]
- Wakeley J, Aliacar N. (2001). Gene genealogies in a metapopulation. Genetics 159: 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J, Sargsyan O. (2009). Extensions of the coalescent effective population size. Genetics 181: 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li H, Li Y, Tian G, Goodman L et al. (2008). The diploid genome sequence of an Asian individual. Nature 456: 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson-Herbots HM. (1998). Genealogy and subpopulation differentiation under various models of population structure. J Math Biol 37: 535–585. [Google Scholar]
- Wright S. (1931). Evolution in Mendelian populations. Genetics 16: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Pan S, Wang J, Dixon A, He J, Muller MG et al. (2013). Peregrine and saker falcon genome sequences provide insights into evolution of a predatory lifestyle. Nat Genet 45: 563–566. [DOI] [PubMed] [Google Scholar]
- Zhao S, Zheng P, Dong S, Zhan X, Wu Q, Guo X et al. (2013). Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat Genet 45: 67–71. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang B, Pan Q, Zhang J, Kumar S, Sun X et al. (2014). Whole genome sequencing of the snub-nosed monkey provides insights into folivory and evolutionary history. Nat Genet 46: 1303–1310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.