Abstract
We present the annotation of the draft genome sequence of Oscheius sp. TEL-2014 (Genbank accession number KM492926). This entomopathogenic nematode was isolated from grassland in Suikerbosrand Nature Reserve near Johannesburg in South Africa. Oscheius sp. Strain TEL has a genome size of 110,599,558 bp and a GC content of 42.24%. The genome sequence can be accessed at DDBJ/EMBL/GenBank under the accession number LNBV00000000.
Keywords: Oscheius, Entomopathogenic nematodes, Whole-genome sequencing, Genome assembly, Genome annotation
| Specifications | |
|---|---|
| Organism | Oscheius sp. |
| Strain | TEL-2014 |
| Sequencer or array type | Sequencer; Illumina HiSeq |
| Data format | Processed |
| Experimental factors | Nematode strains |
| Experimental features | Draft genome sequence of Oscheius sp. TEL-2014, assembly and annotation |
| Consent | N/A |
| Sample source location | Grassland in Suikerbosrand Nature Reserve near Johannesburg in South Africa |
1. Direct link to deposited data
2. Experimental design, materials and methods
Nematodes are very diverse, with entomopathogenic nematodes (EPNs) receiving a lot of interest in nematology and entomology studies because of their ability to infect and kill insects [1] . EPNs reside naturally in the soil and are obligate parasites of a wide variety of insect species. They have evolved symbiotic relationships with insect pathogenic bacteria [2] . Three genera of EPNs which are able to fulfill the role of vectors for entomopathogenic bacteria have been identified and reported on so far. They include species belonging to the genera Heterorhabditis, Steinernema, and Oscheius. Their ability to infect insects is dependent on their symbiotic association with pathogenic bacteria belonging to the genera Photorhabdus, Xenorhabdus, and Serratia, respectively [3], [4], [5], [6]. The Oscheius genus was acknowledged as an independent genus [7] and later [8] supported the recognition of Oscheius as an autonomous genus. Examples of described Oscheius species include Oscheius maqbooli [8] , Oscheius shamimi [9] , Oscheius carlianonsis [10] and Oscheius amsactae [7] . Not all nematodes belonging to the Oscheius genus are entomopathogenic [6] such as O. insectivorus. This is mainly because of their inability to infect insects and cause mortality. This may be due to the absence of a symbiotic pathogenic bacteria association, which is the main characteristic of insect-killing nematodes.
Several insect pathogenic strains or species of Serratia bacteria have also developed successful endosymbiotic relationships with some nematodes belonging to the Caenorhabditis and Oscheius species [3], [11]. For example, Serratia species SCBI associated with Caenorhabditis briggasae isolated from South Africa in the KwaZulu-Natal province was found to have entomopathogenic potential as it caused mortality of the insect larvae Galleria mellonella [12] .
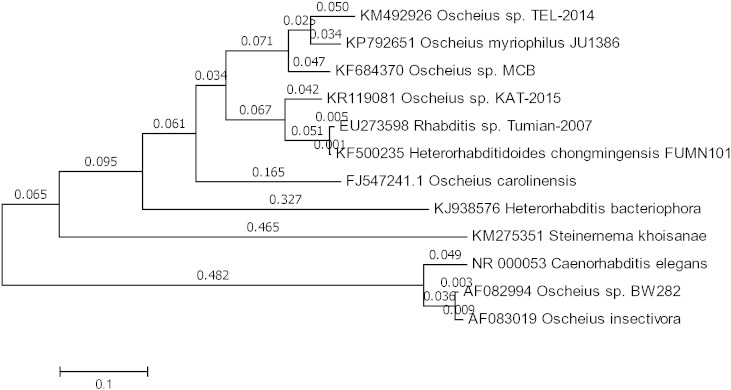
In this paper we discuss the whole genome draft of the Oscheius sp. TEL-2014, which was found to be symbiotically associated with Serratia sp. TEL [13], [14]. The infective juveniles of this entomopathogenic nematode were collected from freshly prepared White traps and surface sterilised with 0.1% sodium hypochlorite for 3 h in sterile 1.5 ml Eppendorf tubes. The nematodes were rinsed 3 times with sterile distilled water under sterile conditions in a laminar flow hood. Whole genomic DNA was extracted from the sterile nematodes using a protocol adopted from Puregene® DNA Purification Kit, Gentra systems 2003. 0.5% agarose gel was prepared in order to confirm the quality and integrity of the extracted DNA. A polymerase chain reaction was employed to amplify the 18S rDNA region using TW81 Forward Primer 5′-GCGGATCCGTTTCCGTAGGTGAACCTGC -3′, Tm (°C) = 71.94 and AB28 Reverse Primer 5′-GCGGATCCATATGCTTAAGTTCAGCGGGT -3′, Tm (°C) = 68.87. The same primers were used for the sequencing of this gene. The sequence obtained was subjected to NCBI BLAST under the default settings for highly similar alignments. The analysis revealed that among all the matches for the 18S rDNA gene sequences, the unknown sequence differed sufficiently from other submissions and thus the species was registered as a novel entomopathogenic nematode based on the originality of the 18S rDNA sequence. The nematode was then assigned the name Oscheius sp. TEL-2014. A phylogenetic tree was constructed using MEGA 6 to show the evolutionary relationship of Oscheius sp. TEL-2014 with selected species from genera Oscheius, Steinernema and Heterorhabditis shown in Fig. 1 . Genomic DNA paired-end libraries were generated with the Nextera DNA sample preparation kit (Illumina) and indexed using the Nextera index kit (Illumina). Paired-end (2*125 bp) sequencing was performed on a Illumina Hiseq 2500 using the Illumina SBS v4 chemistry at the Agricultural Research Council Biotechnology Platform. Quality control was done using FastQC version 0.11.3 and adapter trimming was performed using Trimmomatic version 0.32. The genome was assembled using Velvet version 1.2.10, generating 53,190 contigs. The largest contig was 146,289 bp long. QUAST version 3.1 was used for the assessment of the assembly and BUSCO version 1.1 was used to assess the completeness of the assembly. The N50 value was 3 019 and the total genome size was 110,599,558 bp, which is on the same size range as Caenorhabditis elegans. BUSCO revealed that the Osheius. sp. TEL-2014 genome draft in this study has 44% completeness when using kmer 29. The genome might contain some amount of novel genes and genomic regions which BUSCO would not have been able to align to the reference sequence of C. elegans. Repetitive DNA sequences were masked and identified using Repeat Masker version 3.3.0. The number of bases masked was 425 3249 bp. Retroelements, long terminal repeats (LTR elements), DNA transposons, Small RNA, Satellites and Simple repeats were some of the features identified. 49 947 genes were predicted using blastx and AUGUSTUS version 2.5.5 Protein sequences of the predicted genes were subjected to SwissProt and NCBI BLASTP to identify the proteins.
Fig. 1.
The evolutionary history of several species of Oscheius was centred on the analysis of 18S rDNA ITS region inferred by using the Maximum Likelihood method based on the Tamura-Nei model. The bootstrap consensus tree inferred from 1000 replications and tree is drawn to scale, with branch lengths measured in the number of substitutions per site (next to the branches). Evolutionary analyses were conducted in MEGA6. Oscheius sp. TEL-2014 is closely related to O. myriophilus.
These gene prediction and protein identification tools have revealed the presences of protein domains, hypothetical protein and other proteins also found in nematodes. For example, the WD40 repeat domain ( Fig. 2 ) was predicted and found on position 5208 (start) to 5385 (end). This protein was identified from Haemonchus contortus also known as Barber pole worm which also belongs to the phylum Nematoda, Chromadorea, Rhabditida.
Fig. 2.
WD40 repeat domain found in Oscheius sp. TEL-2014.
(Images and information were obtained from NCBI Conserved Protein Domain Database).
Another protein predicted in Oscheius sp. TEL-2014 is a hypothetical protein CAEBREN_28360, also found in Caenorhabditis brenneri genome. This protein may be further hypothesised to be involved in nematodes chemotaxis and behaviour.
Topoisomerase II large subunit originally found in Escherichia phage PBECO 4 was predicted to be present I the Oscheius nematodes genome. Histidine kinase-like ATPases is one of the domains present in this protein.
A Histidine kinase-like ATPases was predicted to be present in Oscheius nematodes. The TOPRIM superfamily also comprises of numerous ATP-binding proteins such as histidine kinase, DNA gyrase B, topoisomerases, heat shock protein HSP90, phytochrome-like ATPases and DNA mismatch repair proteins ( Fig. 3 ). The heat shock protein HSP90 may be hypothesised to be involved in desiccation tolerance of these entomopathogenic nematodes.
Fig. 3.
TOPRIM superfamily found in Oscheius sp. TEL-2014.
The genome data described in the present study offers a valuable platform for future studies of Oscheius nematodes and possesses momentous importance in the agricultural industries and scientific research. More features of the genome will be identified and analysed using more annotation tools.
3. Nucleotide sequence accession numbers
This whole-genome shotgun project has been deposited at DDBJ/
EMBL/GenBank under the accession LNBV00000000.
Conflict of interest
The authors declare that there is no conflict of interest on any work published in this paper.
Acknowledgments
I, Tiisetso Elizabeth Lephoto received an Innovation Doctoral Scholarship from the NRF National Research Foundation (NRF) with the grant number [SFH1208147793] and a Wits Postgraduate Merit Award (PMA) from the University of the Witwatersrand. Thanks to Gauteng Department of Agriculture and Rural Development (GDARD) (GDARO 12TS) for funding the research project. Thanks to the Agricultural Research Council (ARC) Biotechnology platform for Illumina technology sequencing services.
References
- 1.Hatting J., Patricia Stock S., Hazir S. Diversity and distribution of entomopathogenic nematodes (Steinernematidae, Heterorhabditidae) in South Africa. J. Invertebr. Pathol. 2009;102:120–128. doi: 10.1016/j.jip.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Malan A.P., Nguyen K.B., Addison M.F. Entomopathogenic nematodes (Steinernematidae and Heterorhabditidae) from the southwestern parts of South Africa. Afr. Entomol. 2006;12:65–69. [Google Scholar]
- 3.Abebe E., Jumba M., Bonner K., Gray V., Morris K., Thomas W.K. An entomopathogenic Caenorhabditis briggsae. J. Exp. Biol. 2010;213(18):3223–3229. doi: 10.1242/jeb.043109. [DOI] [PubMed] [Google Scholar]
- 4.Adams B.J., Fodor A., Koppenhöfer H.S., Stackebrandt E., Stock S.P., Klein M.G. Biodiversity and systematics of nematode–bacterium entomopathogens. Biol. Control. 2006;37:32–49. [Google Scholar]
- 5.Torres-Barragan A., Suazo A., Buhler W.G., Cardoza Y.J. Studies on the entomopathogenicity and bacterial associates of the nematode Oscheius carolinensis. Biol. Control. 2011;59:123–129. [Google Scholar]
- 6.Pervez R., Eapen S.J., Devasahayam S., Jacob T.K. A new species of entomopathogenic nematode Oscheius gingeri sp. n. (Nematoda: Rhabditidae) from ginger rhizosphere. Arch. Phytopathol. Plant Protect. 2012;46(5):526–535. [Google Scholar]
- 7.Tabassum K.A., Shahina F. Oscheius maqbooli n. sp. and observations on three known rhabditid species (Nemata: Rhabditida) from sugarcane fields of Balochistan, Pakistan. Pak. J. Nematol. 2002;20:1–21. [Google Scholar]
- 8.Ali S.S., Pervez R., Andrabi R., Sharma R., Verma V. Oscheius amsactae n. sp. (Nematoda:Rhabditida), a necromenic associate of red hairy caterpillar, Amsacta moori (Lepidoptera:Arctiidae) from Kanpur, India. Arch. Phytopathol. Plant Protect. 2011;44(9):871–881. [Google Scholar]
- 9.Tahseen Q., Nisa S.U. Embryology and gonad development in O. shamimi sp. n. (Nematoda:Rhabditida) Nematology. 2006;8:211–221. [Google Scholar]
- 10.Weimin Y.E., Barragan A.T., Cardoza Y.Z. Oscheius carolinensis n. sp. (Nematoda: Rhabditidae), a potential entomopathogenic nematode from vermicompost. Nematology. 2010;12(1):121–135. [Google Scholar]
- 11.Abebe E., Akele F.A., Morrison J., Cooper V., Thomas W.K. An insect pathogenic symbiosis between a Caenorhabditis and Serratia. Virulence. 2011;2(2):158–161. doi: 10.4161/viru.2.2.15337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abebe-Akele F., Tisa L.S., Cooper V.S., Hatcher P.j, Abebe E., Thomas W.K. Genome sequence and comparative analysis of a putative entomopathogenic Serratia isolated from Caenorhabditis briggsae. BMC Genomics. 2015;16:531. doi: 10.1186/s12864-015-1697-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lephoto T.E., Featherston J., Gray V.M. Draft whole-genome sequence of Serratia sp. strain TEL, associated with Oscheius sp. TEL-2014 (nematoda: rhabditidae) isolated from a grassland in South Africa. Genome Announc. 2015;3(4) doi: 10.1128/genomeA.00747-15. (e00747–15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lephoto T.E., Gray V.M. Genome sequencing and annotation of Serratia sp. strain TEL. Genomics Data. 2015;6:54–56. doi: 10.1016/j.gdata.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]