Summary
This study describes the esophageal cancer methylation landscape and its impact on gene expression. Genes aberrantly methylated suggest a mechanism that could lead to genomic instability and chromothripsis. A CpG island methylator phenotype-like subtype with potentially worse clinical outcome was also identified.
Abstract
The incidence of esophageal adenocarcinoma (EAC) has risen significantly over recent decades. Although survival has improved, cure rates remain poor, with <20% of patients surviving 5 years. This is the first study to explore methylome, transcriptome and ENCODE data to characterize the role of methylation in EAC. We investigate the genome-wide methylation profile of 250 samples including 125 EAC, 19 Barrett’s esophagus (BE), 85 squamous esophagus and 21 normal stomach. Transcriptome data of 70 samples (48 EAC, 4 BE and 18 squamous esophagus) were used to identify changes in methylation associated with gene expression. BE and EAC showed similar methylation profiles, which differed from squamous tissue. Hypermethylated sites in EAC and BE were mainly located in CpG-rich promoters. A total of 18575 CpG sites associated with 5538 genes were differentially methylated, 63% of these genes showed significant correlation between methylation and mRNA expression levels. Pathways involved in tumorigenesis including cell adhesion, TGF and WNT signaling showed enrichment for genes aberrantly methylated. Genes involved in chromosomal segregation and spindle formation were aberrantly methylated. Given the recent evidence that chromothripsis may be a driver mechanism in EAC, the role of epigenetic perturbation of these pathways should be further investigated. The methylation profiles revealed two EAC subtypes, one associated with widespread CpG island hypermethylation overlapping H3K27me3 marks and binding sites of the Polycomb proteins. These subtypes were supported by an independent set of 89 esophageal cancer samples. The most hypermethylated tumors showed worse patient survival.
Introduction
The incidence of esophageal adenocarcinoma (EAC) has risen >600% in the last 30 years (1). EAC has a poor outcome with only 13–20% of patients surviving 5 years (2). Barrett’s esophagus (BE) is a precancerous precursor of EAC (3). Previous studies have made significant inroads into identifying the common somatic point mutations (1,4), copy number alterations (1), mutational signatures (5) and high incidence of genomic catastrophes (6) associated with EAC. Aberrant DNA methylation is a known hallmark of cancer, but genome-wide patterns of DNA methylation in EAC are yet to be fully characterized. It is well established that tumors arise through the accumulation of genetic and epigenetic aberrations and that these patterns of somatic changes differ from cancer to cancer. DNA methylation mostly occurs at cytosine residues in the CG dinucleotides context (CpG sites), genomic regions rich in these CpG sites are termed CpG islands (7). Aberrant methylation has been implicated in tumor initiation and progression in several cancer types, with hypermethylation of CpG islands and promoter regions associated with transcriptional silencing of tumor suppressor genes. Conversely, hypomethylation is associated with overexpression of oncogenes and genomic instability, although the mechanisms are yet to be completely understood (8).
In BE and EAC, DNA methylation studies have been limited to a small number of CpG sites (up to 27578) (9–12) and focused on identifying differentially methylated sites. Although these studies suggest that DNA methylation is an important and early event in the development and progression of EAC, further characterization of the genomic context in large cohorts remains less explored. Considering that epigenetic regulation adds an extra layer of complexity in cancer development, further studies to gain more insight about the landscape of DNA methylation in EAC are required to better understand this complex disease.
In this study, we explore methylome, transcriptome and ENCODE data to characterize the role of DNA methylation in EAC. We also assessed whether genome-wide methylation patterns confer new insights into tumorigenesis and the potential for patient stratification that might be relevant for future personalized treatment.
Material and methods
Cohort
Written informed consent was obtained from all patients enrolled in this study, with approval from the Princess Alexandra Hospital and QIMR Berghofer Medical Research Institute research ethics committees (PAH-HREC-2007/068, HREC/10/QPAH/152, HREC/11/QPAH/529 and QIMRP514).
A total of 250 samples were used: 125 EAC, 19 BE and 85 nontumor squamous esophagus (NSE), which include 64 esophageal squamous mucosa at least 5cm proximal to BE (n = 2) or EAC (n = 62), 11 control samples (biopsies of squamous esophagus from healthy individuals), 10 nondysplastic squamous esophageal mucosa from patients with gastroesophageal reflux disease and 21 gastric tissue (>5cm distal to EAC).
Sample preparation
Tissue samples were either obtained as fresh-frozen sections (snap frozen in liquid nitrogen) or from research endoscopic biopsies collected in RNAlater (LifeTechnologies). Biopsies were directly placed in RNAlater and stored at 4°C for 48h then tissue was removed from solution and stored at −70°C.
Fresh-frozen tumors and biopsies were assessed by an experienced gastrointestinal pathologist and classified based on the American Joint Committee on Cancer staging system. From each frozen sample, a 7 µm section was used to make a hematoxylin and eosin slide, followed by two 30 µm sections for DNA extraction; this was repeated to obtain a minimum of six unstained sections with serial hematoxylin and eosin. The hematoxylin and eosin slides were used to assess tumor percentage and guide macrodissection. Histopathology review of biopsy samples was carried out on an immediately adjacent biopsy at the same esophageal level to confirm the presence of BE or EAC. Samples with a minimum of 50% tumor tissue as estimated by the pathologist were included in the study. DNA and RNA were extracted using the phenol–chloroform–isoamyl alcohol method (13). In addition, for 22 tumor/matched squamous pairs, DNA and RNA were extracted using AllPrep DNA/RNA Mini Kits (Qiagen, Valencia, CA). DNA was quantified using a Qubit® dsDNA BR Assay Kit (Invitrogen, Carlsbad, CA).
Bisulfite conversion and 450K methylation arrays
Genomic DNA (500ng) was bisulfite converted using EZ DNA methylation Kits (Zymo Research) according to the manufacturer’s protocol for Illumina Methylation arrays. Bisulfite converted DNA was whole genome amplified and hybridized to Infinium Human Methylation 450K BeadChips (Illumina) according to the manufacturer’s protocol. Arrays were scanned using an iScan (Illumina). GenomeStudio v 2011.1 (Illumina) with methylation module (v 1.6.1) was used to process the raw image data.
Methylation data were background subtracted in GenomeStudio and normalized by beta-mixture quantile normalization to adjust for the type II bias. Normalization was done using the wateRmelon Bioconductor R package (14). All samples were of high quality, with >85% of probes having a detection P value <0.05. Data were filtered to remove probes located on X and Y chromosomes (n = 11713), probes with >1 missing value (n = 33144), probes with single-nucleotide polymorphisms in the CpG site or in the last 10 bases of the probe, probes which mapped to repeats (n = 67152) (15,16) and probes with detection P value >0.05 in more than 25% of samples (n = 7). A total of 372817 probes were used for analysis. The methylation array data have been deposited in the Gene Expression Omnibus (accession number: GSE72874).
Gene expression profiling
RNA was obtained from 70 samples, 48 EAC, 4 BE and 18 squamous esophagus from EAC patients (Supplementary Table 1, available at Carcinogenesis Online). Total RNA (500ng) was amplified and labeled using an Illumina TotalPrep RNA Amplification Kit (LifeTechnologies). Amplified RNA (750ng) was hybridized to Illumina human HT12 (V4) arrays following the manufacturer’s protocol and scanned with an iScan (Illumina). The intensities were extracted using the GenomeStudio software (Illumina) and background corrected, log2-transformed and quantile normalized using Lumi package (17). The gene expression array data have been deposited into the Gene Expression Omnibus (accession number: GSE72874).
TCGA methylation data of stomach, esophageal and colon cancer
Publically available 450K methylation data of 395 stomach adenocarcinomas, 295 colon adenocarcinomas and 185 esophageal cancers [89 EAC and 96 esophageal squamous cell carcinomas (ESCC)] were obtained from the TCGA Data Portal (https://tcga-data.nci.nih.gov/) on 10 December 2015 (18,19). Level 1 DNA methylation data (HumanMethylation450) obtained from TCGA (tcga-data.nci.nih.gov/tcga/) was imported into R using the minfi (20) package, along with the EAC dataset generated in-house. The raw methylation values were background corrected using the minfi function preprocess Illumina. Methylation data were quality controlled and normalized with beta-mixture quantile as described above. Epstein–Barr virus information for stomach tumors was obtained from http://gdac.broadinstitute.org/runs/awg_stad__2013_09_30/samples_summary_report/.
Data analysis
Differentially methylated probes in EAC and BE
Differentially methylated probes in EAC were identified using linear mixed effect regression models, which included EAC diagnoses (yes/no) as a fixed effect predictor and patient as a random effect (beta value = EAC diagnoses + patient). In this analysis, 125 tumors were compared with 85 NSE. A second linear mixed effect regression model identified methylation changes associated with BE by using BE status (yes/no) as a fixed predictor and patient as a random effect (beta value = BE status + patient). This analysis compared 19 BE with 85 NSE samples. Methylation differences between 125 tumor and 19 BE were identified for each probe using linear regression (beta value = sample type BE or EAC).
Clustering of data
Methylation data of EAC, BE and NSE were grouped using unsupervised hierarchical clustering (Ward’s method, Euclidian distance) based on the top 5000 most variable probes across three sample types. Hilbert-curve transformation was used to identify regions of the genome that contained clusters of differential methylation, which allows visualization of methylation data in a 2D plot, while preserving genome locality.
Pathway analysis
To determine potential biological relevance of aberrant methylation in EAC and BE, pathway analyses were performed using the MetaCore package (Thomson Reuters). Genes with promoter-associated differential methylation [false discovery rate (FDR) < 0.01 and mean beta value difference ≥ 0.20] between EAC or BE and NSE were used for pathway analysis. The relationship between probes and genes was obtained using the vendor annotation (Illumina). Pathways with FDR <0.001 are reported.
Correlation of methylation and gene expression data
Methylation and gene expression probes were mapped using the vendor lookup table. Pearson correlations were calculated and correlation P values were corrected for FDR. The correlation was considered significant when q value <0.05.
Identification of tumor subtypes
To identify potential methylation EAC subtypes, clustering was performed using the 5000 most variable probes (in CpG islands) from the 125 EAC. Probes were not highly methylated in NSE (mean beta in NSE < 0.5) and do not contain missing values. To minimize the potential influence of tumor content of each sample or batch effects, beta values were dichotomized using a beta value of >0.3 as a threshold for positive DNA methylation. Subtypes were defined by unsupervised hierarchical clustering (Ward’s method, binary distance) of the dichotomized data (Supplementary Figure 1A, available at Carcinogenesis Online). For visualization, the dichotomized data were back-transformed to the original beta values (Supplementary Figure 1B, available at Carcinogenesis Online). To identify samples in the top and lowest quantiles levels of methylation, the samples within each subtype were ordered by the total number of hypermethylated CpGs. EAC subtypes were compared with clinical features, including alcohol consumption, smoking status and survival (Supplementary Table 2, available at Carcinogenesis Online).
Methylation of CpG island methylator phenotype marker genes
Methylation status of several genes has been proposed as markers for the CpG island methylator phenotype (CIMP), including MLH1, CDKN2A and MGMT in gastric cancer (21); CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1 in colorectal cancer (22) and KCNK13, SLIT1, RAB31, FOXL2, B3GAT2, FAM78A, MYOCD, KCNC1, FSTL1 and SLC6A4 also in colorectal cancer (23). We computed for each of these 16 marker genes the average methylation of probes that showed variance in beta values across all tumors >0.03 and that were located in CpG-rich promoters.
ENCODE data
Differentially methylated loci (FDR < 0.01 and mean beta value difference ≥ 0.20) were mapped to ENCODE ChIP-seq data (24,25) derived from four cell lines: H1-hESC, K562, NHLF and NHDF-Ad. Essentially, ChIP-seq peaks were downloaded from the UCSC Genome Browser using the R package rtracklayer (26). Peaks with a score of <600 were excluded. For each ChIP-seq dataset, the proportion of differentially methylated sites overlapping with a peak was determined.
EAC, stomach and colon adenocarcinomas methylation profiles
Methylation levels of our EAC and TCGA data were clustered using unsupervised clustering (Ward’s method, binary distance) using the top 5000 most variable probes in CpG islands across all cancers. Variance was computed across all tumor samples. To minimize potential influence of tumor cellularity and batch effects, beta values were dichotomized using a beta value of >0.3 as a threshold for positive DNA methylation. For visualization, the dichotomized data were back-transformed to the original beta values.
Results
The clinical features for the 250 samples, which includes 19 BE (11 with synchronous EAC and 8 from non-EAC patients), 125 EAC, 85 NSE (11 squamous esophagus from healthy individuals, 10 squamous esophagus from patients with gastroesophageal reflux disease and 64 adjacent squamous esophagus from EAC or BE patients) and 21 stomach samples, are summarized in Supplementary Table 2, available at Carcinogenesis Online.
The methylation landscape of EAC and BE
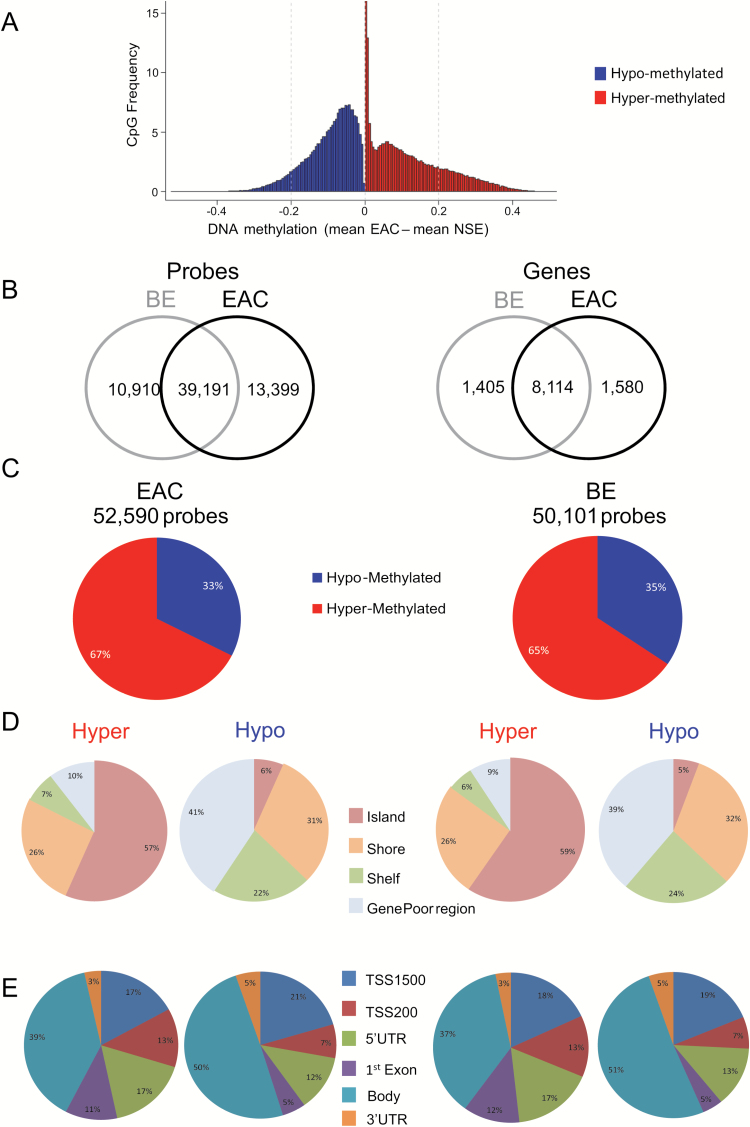
A total of 52590 CpG sites were differentially methylated (FDR < 0.01 and average beta value difference ≥ 0.20) in EAC compared with NSE (Supplementary Table 3, available at Carcinogenesis Online; Figure 1A), and 50101 sites were differentially methylated in BE compared with NSE (Supplementary Table 4, available at Carcinogenesis Online). Interestingly, 74.5% of the probes aberrantly methylated in EAC were also aberrantly methylated in BE (Figure 1B). However, only 2024 sites were differentially methylated between EAC and BE (Supplementary Table 5, available at Carcinogenesis Online). The majority of differentially methylated sites were hypermethylated in EAC and BE, 67 and 65%, respectively (Figure 1C) were preferentially located in CpG islands and promoter regions (TSS1500, TSS200, 5′ untranslated regions and first exon) (Figure 1D and E). The majority of hypomethylated sites were situated outside from CpG islands (shelf and gene poor-regions) and in the body of genes (Figure 1D and E). Hypermethylated sites in CpG islands were located closer to the transcription start site (TSS) than hypomethylated sites (P = 10−5, Wilcoxon test) (Supplementary Figure 2A, available at Carcinogenesis Online). No difference was observed in the absolute distance to the TSS for sites hypomethylated or hypermethylated located outside of CpG islands; however, hypomethylated sites tended to be located more downstream of the TSS than the background distribution (Supplementary Figure 2B, available at Carcinogenesis Online). Hilbert-curve transformation identified clusters of hypermethylated loci within the genome (Supplementary Figure 3, available at Carcinogenesis Online). The largest cluster of hypermethylated CpG sites was an 110 kbp region on chr2 which harbors a cluster of HOX genes (Supplementary Figure 4, available at Carcinogenesis Online). HOX proteins regulate numerous processes including apoptosis, differentiation, motility and angiogenesis playing an important role in tumorigenesis. The HOX genes cluster on chromosome 2 is extensively hypermethylated in both EAC and BE (Supplementary Figure 4, available at Carcinogenesis Online).
Figure 1.
Global overview of DNA methylation patterns in BE and EAC. (A) Range of beta value differences of differentially methylated probes (FDR < 0.01) in EAC compared with NSE. (B) Number of differentially methylated probes and genes in EAC and BE (FDR < 0.01 and beta value difference ≥ 0.20). (C) Number of probes differentially methylated in EAC and BE and percentage hypomethylated and hypermethylated probes. (D) Pie charts show the percentage of hypomethylated and hypermethylated probes across different genomic regions (CpG islands, Shore probes located less 2kb from CpG island, Shelf = probes located >2 k from CpG islands and Gene Poor Region = probes not in island or annotated genes). (E) Pie charts show the percentage of hypomethylated and hypermethylated probes at gene level [TSS1500 and TSS200—probes located within 1500 and 200bp from TSS, respectively; 5′ untranslated regions (UTR); first exon; Body and 3′UTR].
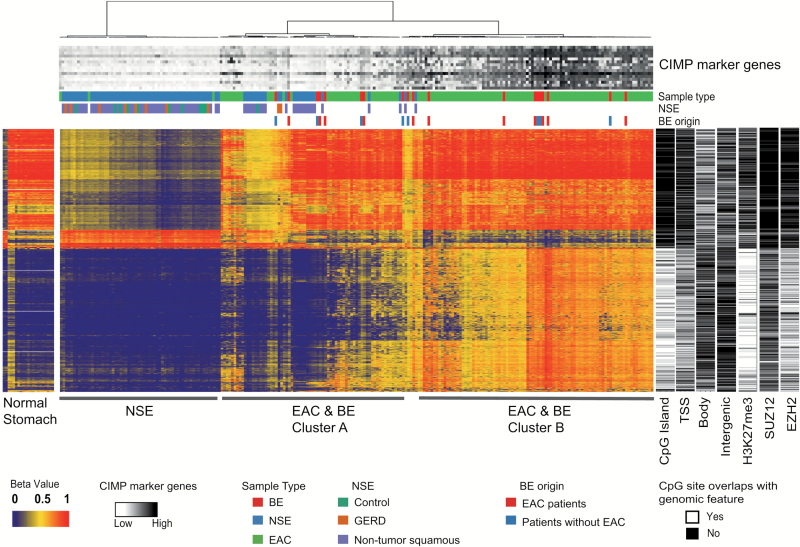
Methylation patterns of the top 5000 most variable probes across EAC, BE and NSE were able to separate EAC and BE from NSE, but not EAC from BE (Figure 2). Samples clustered into three groups, one containing mostly NSE and two groups (Clusters A and B) comprised mostly of EAC and BE. In our cohort, 58% (n = 11/19) of the BE samples were from patients with synchronous EAC; however, we saw no separation of BE samples if they were collected from patients with EAC versus without EAC. The methylation profile of samples in Cluster A shows similarities to the methylation profile of the columnar gastric mucosa (stomach tissue) (Figure 2), suggesting metaplastic transformation of the esophageal lining from normal squamous epithelium into columnar epithelium, which is characteristic of BE and EAC. Cluster B contains EAC and BE samples that are characterized by hypermethylation of sites located in CpG islands (Figure 2). Tumors with CpG islands hypermethylation were first described in colon cancer and are referred to as ‘CpG island methylator phenotype’ (CIMP). Therefore, Cluster B contains EAC samples that show a CIMP-like methylation pattern.
Figure 2.
Unsupervised clustering of 125 EAC, 19 BE and 85 NSE (NSE includes 64 adjacent morphologically normal mucosa from patients with BE or EAC, 10 gastroesophageal reflux disease and 11 control samples from healthy individuals). Cluster shows clear separation between NSE and EAC and BE, and suggests two EAC and BE subtypes (Clusters A and B). Rows in the heatmap represent CpG sites and columns represent samples. Heatmaps on the right side (black and white) show if location of CpG sites overlaps genomic features. White depicts overlap of CpG sites (probes) with ChIP-seq peaks (score > 600) for H3K27me3 histone marks and binding sites of SUZ12 and EZH2 in cell line H1-hESC and black indicates no overlap. Peaks of ENCODE ChIP-seq data were downloaded from the UCSC Genome Browser. Methylation profiles of 21 normal stomach samples are shown in a separate heatmap with CpG sites presented in the same order as in the unsupervised clustering. Methylation level of 16 CIMP marker genes is presented in a gray scale at the top of the hierarchical clustering. Methylation level of CIMP marker genes are represented as average beta value of probes that showed variance in beta values across all tumors >0.03 and that were located in CpG-rich promoters of genes.
Association between methylation and gene expression
Differentially methylated CpG sites in EAC and BE compared with NSE were associated with 9694 and 9519 genes, respectively (Figure 1B; Supplementary Tables 3 and 4, available at Carcinogenesis Online). From those, 83% were differentially methylated in both comparisons. Previously, 274 genes were reported as aberrantly methylated in EAC and/or BE methylation studies (9–11,27–37), of which 245 (89.4%) were also aberrantly methylated in our cohort (Supplementary Table 6, available at Carcinogenesis Online). We combined methylation and gene expression data for 70 samples (4 BE, 18 squamous esophagus from EAC patients and 48 EAC). In total, 52590 CpG sites were differentially methylated in EAC compared with NSE; 35045 sites were associated with genes. Of these, 18575 CpG sites were associated with 5538 genes expressed above the arrays’ detection limit. In total, 10598 of the CpG sites (57% of testable sites) and 3502 genes (63%) showed significant correlation between methylation and gene expression (FDR < 0.05, Pearson correlation) (Supplementary Tables 7 and 8, available at Carcinogenesis Online). These CpG sites were mainly located in promoter regions (53.5%), while 52% of CpG sites with a positive correlation with gene expression were located in the body of genes. Hypomethylated CpG sites located at promoter regions with positive correlation with gene expression were generally located further downstream of the TSS compared with negatively correlated CpG sites (Supplementary Figure 5, available at Carcinogenesis Online).
Pathways aberrantly methylated in EAC and BE
Pathway analyses were performed using genes with promoter-associated aberrant methylation, which comprised a total of 6521 and 6299 genes in EAC and BE, respectively (Supplementary Tables 9 and 10, available at Carcinogenesis Online). Enriched pathways included known cancer signaling pathways, some previously described as aberrantly methylated in EAC: cell adhesion, regulation of epithelial-to-mesenchymal transition and TGF and WNT signaling.
The highest ranked pathway affected by aberrant methylation, which has not previously been described in EAC, was neurophysiological process dynein–dynactin motor complex in axonal transport in neurons (Supplementary Table 9, available at Carcinogenesis Online), members of this pathway are involved in the neurotrophin–Trk signaling. Other members of the highest ranked pathway in EAC are involved in the regulation of chromosomal segregation and spindle assembly. Some of these genes were also aberrantly methylated in BE (Supplementary Table 8, available at Carcinogenesis Online). We identified 34 genes aberrantly methylated reported previously to be involved in spindle assembly and chromosome segregation (Supplementary Table 11, available at Carcinogenesis Online). Twenty-three out 34 genes showed detectable expression (detected in at least 20% of the samples) and 12/23 genes showed significant correlation between methylation levels and gene expression (FDR < 0.05) (Supplementary Table 12 and Supplementary Figure 6, available at Carcinogenesis Online). Here, members of the WNT signaling pathway were also aberrantly methylated (Supplementary Table 13, available at Carcinogenesis Online), and some showed significant correlation with gene expression (Supplementary Tables 13, 14 and Supplementary Figure 7, available at Carcinogenesis Online).
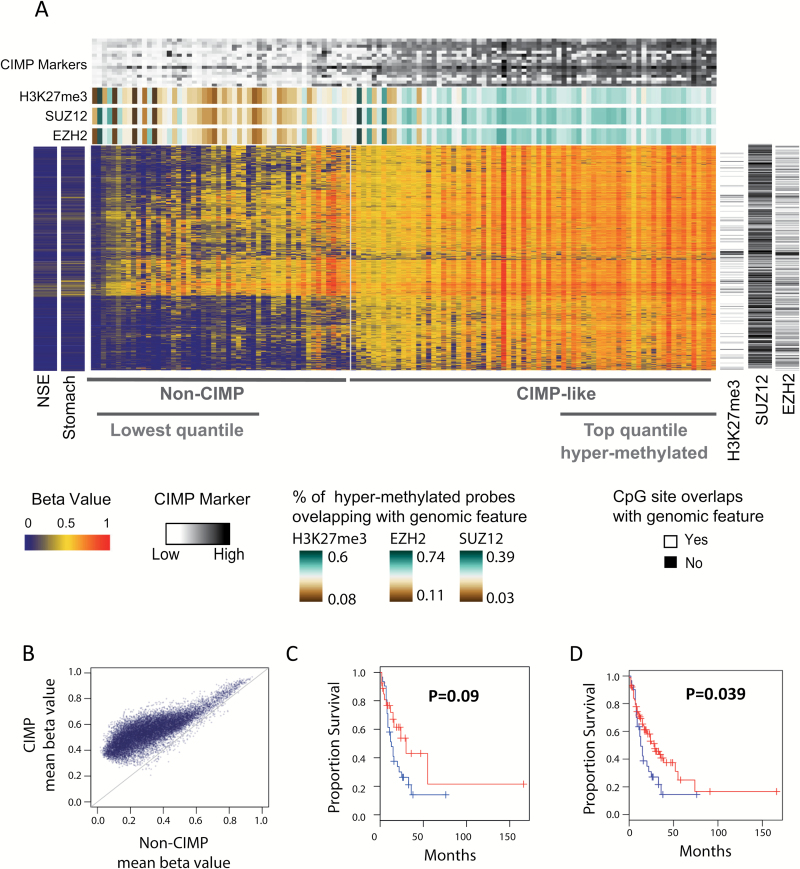
Tumor subtypes within EAC
Clustering of NSE, BE and EAC suggested two groups of BE and EAC (Clusters A and B; Figure 2). To better understand the EAC groups, we performed unsupervised clustering of EAC samples (n = 125) using the 5000 most variable probes located in CpG islands (Figure 3A). Unsupervised clustering resulted in two clearly distinct cluster (Supplementary Figure 1, available at Carcinogenesis Online; Figure 3A), which confirmed the two EAC molecular subtypes observed in Figure 2. The CIMP-like group is characterized by extensive hypermethylation of CpG islands with hypermethylation of CIMP marker genes identified in other studies (Figure 3A). Methylation was higher in the CIMP-like EACs (Figure 3B) and showed a strong enrichment for hypermethylation in CpG islands (P < 10−15) when compared with non-CIMP tumors (Supplementary Figure 8, available at Carcinogenesis Online). Extensively hypermethylated EACs tend to have poorer patient survival (Figure 3C and D). The most hypermethylated tumors (n = 29, subgroup of the CIMP-like tumors) showed a trend of worse patient survival (P = 0.09) compared with the least hypermethylated (n = 29, subgroup of the non-CIMP tumors) (Figure 3C). The most hypermethylated tumors did show a significantly worse patient survival when compared with all other tumors (n = 94) (P = 0.039) (Figure 3D), an observation that requires further validation in other cohorts.
Figure 3.
Unsupervised clustering of 125 EAC using methylation levels of the 5000 most variable probes located in CpG islands. (A) Heatmap suggests two subtypes: CIMP-like with high frequency of hypermethylated sites overlapping histone marks H3K27me3 and binding sites of EZH2 and SUZ12. CIMP-like also showed high levels of methylation associated to CIMP marker genes identified in other cancers. Rows in the heatmap represent CpG sites and columns represent samples. Methylation of 16 CIMP marker genes is presented in a grey scale at the top of the hierarchical clustering. Methylation of CIMP marker genes is represented as average beta value of probes that showed variance in beta values across all tumors >0.03 and that were located in CpG-rich promoters of genes. Heatmaps in the right side (black and white) show location of CpG sites in the genome. Overlap of CpG sites with ChIP-seq peaks (score > 600) for H3K27me3 histone marks and binding sites of SUZ12 and EZH2 is depicted in white and black indicates no overlap. The heat map in green to brown scale represents the percentage of hypermethylated probes that overlap with H3K27me3 histone marks and binding sites of SUZ12 and EZH2. Average methylation of NSE and stomach samples are shown in a separate heatmap (left) with CpG sites presented in the same order as in the unsupervised clustering. (B) Scatterplot comparing methylation in CIMP-like and non-CIMP EACs. Each dot represents the average beta value of probes hypermethylated in EAC compared with NSE (FDR < 0.01 and average beta value difference ≥ 0.20) in the CIMP-like versus the non-CIMP subtype. Methylation is generally higher in CIMP-like than in the non-CIMP tumors. (C) Kaplan–Meier plot of overall survival comparing the top quantile most hypermethylated tumors (n = 29 in blue) to the lowest quantile tumors with least hypermethylation (n = 29 in red). (D) Kaplan–Meier plot of overall survival comparing the top quantile most hypermethylated tumors (n = 29 in blue) to all other tumors (n = 94 in red).
Differentially methylated sites in EAC were mapped to histone marks and transcription factor binding sites identified in embryonic stem cells (H1-hESC—ENCODE) (25). The CIMP-like subtype showed a bias for hypermethylation of loci overlapping H3K27me3 histone marks (P < 10−15) and binding sites of EZH2 (P < 10−15) and SUZ12 (P < 10−15) in embryonic stem cells (H1-hESC) (Figure 3 and Supplementary Figure 8, available at Carcinogenesis Online). This enrichment was weaker in non-CIMP tumors (Supplementary Figure 8, available at Carcinogenesis Online). Similar results were obtained comparing Chip-Seq data for three other cell lines K562, NHLF and NHDF-Ad (data not shown).
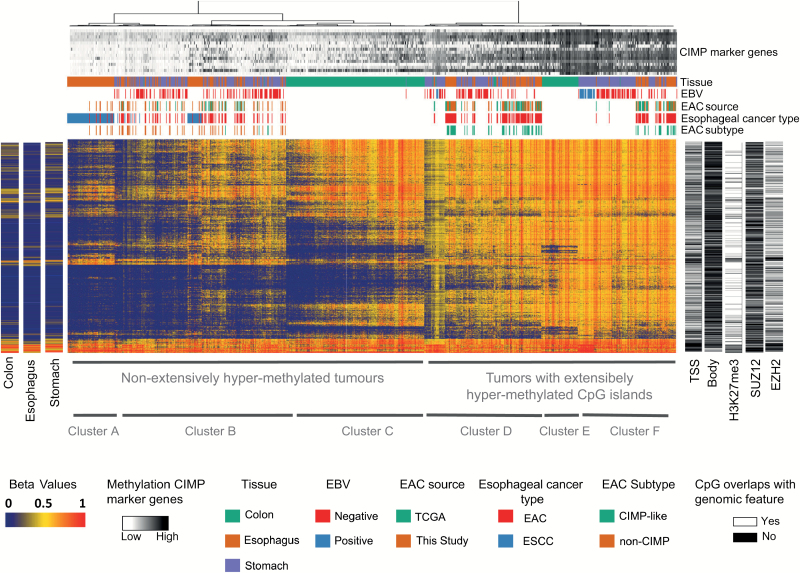
EAC, colon and stomach adenocarcinomas show similarities in methylation profiles
A meta-analysis of our EAC data (n = 125) and TCGA data from colon (n = 295), stomach (n = 395) adenocarcinomas and esophageal cancers (n = 185, 89 EAC and 96 ESCC) indicated that these gastrointestinal cancers may share a similar hypermethylation signature independent of their site of origin along the gastrointestinal tract (Figure 4). EAC, stomach and colon cancer formed two broad molecular subgroups, the first group contained CIMP-like tumors with extensive hypermethylation of CpG islands and hypermethylation of CIMP marker genes (Figure 4). Almost all CIMP-like EACs (68 out of 72) in our initial analysis (Figure 3) were members of this group. This supports the presence of a CIMP phenotype in EAC. The presence of a CIMP-like subtype in EAC was replicated using 89 EACs from TCGA (Figure 4). From 89 TCGA EAC samples, 74% showed high methylation of CIMP markers and cluster with CIMP-like tumors identified in Figure 3. In contrast, all 96 ESCC tumors from TCGA clustered with non-CIMP EAC tumors most in a separate cluster (Cluster A), suggesting no evidence of CIMP phenotype in ESCC. In accordance with the results obtained for our samples (Figure 3), the CIMP-like subtype showed a strong enrichment for hypermethylation of CpG sites overlapping H3K27me3 histone marks and binding sites of the Polycomb complex 2 proteins. Clustering revealed several subgroups of tumors. Interestingly, EAC and stomach cancer showed a more similar methylation pattern (subclusters A, B, D and F in Figure 4) than colon tumors, which formed two separate subclusters (Clusters C and E). This may reflect the tissue of origin of this cancer.
Figure 4.
Unsupervised clustering of 125 EAC (present study) and TCGA 450K methylation data from 395 stomach, 295 colon adenocarcinomas and 185 esophageal cancers (89 EAC and 96 ESCC). Mean methylation of nontumor squamous esophagus (n = 64) and TCGA data of nontumor stomach (n = 2) and nontumor colon (n = 28) is given as separate heatmap on the left with probes in the same order as in the unsupervised clustering. Stomach adenocarcinomas and EAC showed strikingly similar methylation profiles and formed mixed clusters (B, D and F). ESCC tumors formed mainly a separate (Cluster A), and no ESCC clustered with extensively hypermethylated tumors (CIMP-like tumors). Colon adenocarcimas displayed a more distinct methylation profile and formed separate clusters (C and E). Clusters D, E and F showed a group of tumors with extensive CpG island hypermethylation suggesting a potential common mechanism independent of tissue of origin. Methylation data were grouped using unsupervised clustering (Ward’s method, binary distance) based on the top 5000 most variable probes located in CpG islands. Variance was computed across all tumor samples. To minimize potential influence of tumor cellularity or batch effects, beta values were dichotomized using a beta value of >0.3 as a threshold for positive DNA methylation. After clustering the dichotomized values were back-transformed to the original beta values. Rows in the heatmap represent CpG sites and columns represent samples. Heatmaps in the right side (black and white) show location of CpG sites in the genome. Overlap of CpG sites with H3K27me3 histone marks and binding sites of SUZ12 and EZH2 is depicted in white and black indicates no overlap. Methylation of 16 CIMP marker genes is presented in a gray scale at the top of the hierarchical clustering, which are represented as average beta value of probes that showed variance in beta values across all tumors >0.03 and that were located in CpG-rich promoters of genes.
Discussion
EAC is treated as a single disease; however, increasing evidence suggests that EAC is complex with different molecular subtypes. We and others have shown that EAC tumors show a wide range of mutation rates (1,5,6), differences in the burden of copy number and structural variations (1,6) and approximately one third of EAC tumors present evidence of chromothripsis (6). But the role of DNA methylation in EAC tumorigenesis is poorly understood and needs further exploration. Here, we performed the first genome-wide methylation study of EAC and its impact on gene expression to better understand this complex disease.
EAC and BE presented hypermethylation of sites located CpG islands and promoter regions when compared with squamous esophagus. Although the majority of hypomethylated CpG sites were located outside of CpG islands and in the body of genes, patterns reported previously in other cancers (21,38). Unsupervised clustering showed good separation between squamous esophagus and BE and EAC, but not between EAC and BE, confirming previous reports that aberrant methylation is an early event in metaplasia–dysplasia–neoplasia progression (9–12).
Pathway analysis identified enrichment of known cancer signaling pathways previously described as aberrantly methylated in EAC: cell adhesion (9,11), regulation of epithelial-to-mesenchymal transition and TGF and WNT signaling (11). Here the top pathway affected by aberrant methylation in EAC was ‘neurophysiological process dynein–dynactin motor complex in axonal transport in neurons’. Members of this pathway are involved in the neurotrophin–Trk signaling, which was previously reported to be aberrantly methylated in EAC (11). Mutations and rearrangements of TRK genes are only sporadically seen in human cancers, but recent studies indicate that expression of TrkB contributes to tumor pathology (39). Other members of the highest ranked pathway which were aberrantly methylated in EAC are involved in chromosomal segregation and spindle assembly, these include the BUB3, AURKA, DYNC1I1 and DCTN2 and CHFR genes. CHFR is involved in the mitotic G2-M checkpoint and has been reported hypermethylated in lung cancer (40), EAC (41) and gastric cancer (42). Here, members of the WNT signaling pathway were also aberrantly methylated, inhibition of WNT signaling has recently been reported to increase microtubule assembly rates, abnormal mitotic spindle formation and induction of aneuploidy and generation of lagging chromosomes (43). The complexity and current knowledge of the mitotic apparatus has been reviewed by Vitale et al. (44), who highlighted that the molecular mechanisms involved in mitotic aberration, cell death and genomic stability are still largely elusive. They suggested that cancer cells develop strategies to breach aneuploidy, thus avoiding mitotic catastrophes and death. Mutations of several mitotic checkpoint proteins have been found in different cancers but are not common. Epigenetic events are thought to be one of the mechanisms to avoid cell death after mitotic catastrophes (44) and interestingly alteration in the levels of expression of the mitotic checkpoint genes is more common (45). Therefore, we suggest that epigenetic dysregulation may account, in part, for gene expression changes observed in those pathways. It is known that EAC tumors contain a high frequency of chromosomal rearrangements, aneuploidy (46,47) and chromosome catastrophes that result in amplification of oncogenes (6). Therefore, aberrant methylation of genes involved in chromosomal segregation and spindle assembly deserves further investigation due their role in the mitotic apparatus that can lead to chromosomal instability and chromothripsis (48,49).
Identification of EAC subtypes may give important insights into tumorigenesis and represents an important step toward patient-specific personalized treatment. Here, we identified two groups of tumors, one of which showed high levels of methylation in CpG islands similar to the CIMP phenotype identified in other cancers. EAC CIMP-like showed enrichment of hypermethylation of sites overlapping the H3K27me3 histone mark and binding sites of the Polycomb complex 2 (PRC2) proteins. This pattern was previously described in gastric cancer (21) and could suggest that DNA methylation may replace Polycomb-based repression near key genes possibly reducing their regulatory plasticity (50). In other cancers, the CIMP phenotype has been associated to somatic mutation such BRAF in colorectal cancer (23) and IDH1 in gliomas (51). In gastric cancer, two CIMP subtypes have been identified, one of which was associated with Epstein–Barr virus-positive tumors and the other CIMP group associated with microsatellite instability (18). Future multi-omic studies of EAC are required to show an association of the CIMP-like phenotype and other biological features. However, our data suggested that tumors with extensive CpG island hypermethylation are associated with poorer clinical outcome.
In summary, this study described the methylation landscape of EAC and its impact on gene expression, these data suggest an orchestrated epigenetic deregulation with potentially a much stronger role in EAC carcinogenesis than anticipated. Methylation patterns revealed two subtypes, one similar to the CIMP phenotype which was potentially associated with worse clinical outcome. These findings provide new insights into the contribution of epigenetics to EAC carcinogenesis. Further molecular characterization of the subtypes identified here with integration of additional information such copy number changes, structural variants and somatic mutations may lead to better stratification of EAC subtypes with clinical implications.
Supplementary Material
Supplementary Figures 1–8 and Supplementary Tables 1–14 can be found at http://carcin.oxfordjournals.org/
Funding
Australian National Health and Medical Research Council (NHMRC; grants APP1021403, 631701, 199600 and 552429); Smart Futures National and International Research Alliances Program (2008004333); National Cancer Institute (5 RO1 CA 001833-02) (its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute); Keith Boden Fellowship to K.N.; NHMRC Principal Research Fellow (1025427) to S.M.G.; Principal Research Fellowship (APP1058522) and Senior Principal Research Fellowship from NHMRC to D.C.W. and N.K.H.; German Academic Exchange Service (DAAD) to S.B.; Translational Research Institute is supported by a grant from the Australian Government.
Supplementary Material
Acknowledgements
The results shown here are in part based on data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BE
Barrett’s esophagus
- CIMP
CpG island methylator phenotype
- EAC
esophageal adenocarcinoma
- ESCC
esophageal squamous cell carcinomas
- FDR
false discovery rate
- NSE
nontumor squamous esophagus
- TSS
transcription start site
References
- 1. Dulak A.M., et al. (2013) Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet., 45, 478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jemal A., et al. (2010) Cancer statistics, 2010. CA Cancer J. Clin., 60, 277–300. [DOI] [PubMed] [Google Scholar]
- 3. Cook M.B., et al. (2007) Risk of mortality and cancer incidence in Barrett’s esophagus. Cancer Epidemiol. Biomark. Prev., 16, 2090–2096. [DOI] [PubMed] [Google Scholar]
- 4. Weaver J.M., et al. (2014) Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat. Genet., 46, 837–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alexandrov L.B., et al. (2013) Signatures of mutational processes in human cancer. Nature, 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nones K., et al. (2014) Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat. Commun., 5, 5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laird P.W. (2003) The power and the promise of DNA methylation markers. Nat. Rev. Cancer, 3, 253–266. [DOI] [PubMed] [Google Scholar]
- 8. Jones P.A., et al. (2002) The fundamental role of epigenetic events in cancer. Nat. Rev. Genet., 3, 415–428. [DOI] [PubMed] [Google Scholar]
- 9. Xu E., et al. (2013) Genome-wide methylation analysis shows similar patterns in Barrett’s esophagus and esophageal adenocarcinoma. Carcinogenesis, 34, 2750–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alvarez H., et al. (2011) Widespread hypomethylation occurs early and synergizes with gene amplification during esophageal carcinogenesis. PLoS Genet., 7, e1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaz A.M., et al. (2011) DNA methylation profiling in Barrett’s esophagus and esophageal adenocarcinoma reveals unique methylation signatures and molecular subclasses. Epigenetics, 6, 1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith E., et al. (2008) Similarity of aberrant DNA methylation in Barrett’s esophagus and esophageal adenocarcinoma. Mol. Cancer, 7, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ott P.A., et al. (2014) Vaccines and melanoma. Hematol. Oncol. Clin. North Am., 28, 559–569. [DOI] [PubMed] [Google Scholar]
- 14. Pidsley R., et al. (2013) A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics, 14, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Naeem H., et al. (2014) Reducing the risk of false discovery enabling identification of biologically significant genome-wide methylation status using the HumanMethylation450 array. BMC Genomics, 15, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y.A., et al. (2013) Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics, 8, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Du P., et al. (2008) lumi: a pipeline for processing Illumina microarray. Bioinformatics, 24, 1547–1548. [DOI] [PubMed] [Google Scholar]
- 18. Cancer Genome Atlas Research Network (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature, 513, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cancer Genome Atlas Network (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature, 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aryee M.J., et al. (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics, 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zouridis H., et al. (2012) Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci. Transl. Med., 4, 156ra140. [DOI] [PubMed] [Google Scholar]
- 22. Weisenberger D.J., et al. (2006) CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet., 38, 787–793. [DOI] [PubMed] [Google Scholar]
- 23. Hinoue T., et al. (2012) Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res., 22, 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Consortium E.P. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosenbloom K.R., et al. (2013) ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res., 41, D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lawrence M., et al. (2009) rtracklayer: an R package for interfacing with genome browsers. Bioinformatics, 25, 1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eads C.A., et al. (2000) Fields of aberrant CpG island hypermethylation in Barrett’s esophagus and associated adenocarcinoma. Cancer Res., 60, 5021–5026. [PubMed] [Google Scholar]
- 28. Clément G., et al. (2006) Methylation of APC, TIMP3, and TERT: a new predictive marker to distinguish Barrett’s oesophagus patients at risk for malignant transformation. J. Pathol., 208, 100–107. [DOI] [PubMed] [Google Scholar]
- 29. Tischoff I., et al. (2007) Methylation of SOCS-3 and SOCS-1 in the carcinogenesis of Barrett’s adenocarcinoma. Gut, 56, 1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng D.F., et al. (2009) DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett’s adenocarcinoma. Gut, 58, 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin Z., et al. (2008) Hypermethylation of the AKAP12 promoter is a biomarker of Barrett’s-associated esophageal neoplastic progression. Cancer Epidemiol. Biomark. Prev., 17, 111–117. [DOI] [PubMed] [Google Scholar]
- 32. Jin Z., et al. (2008) Hypermethylation of the somatostatin promoter is a common, early event in human esophageal carcinogenesis. Cancer, 112, 43–49. [DOI] [PubMed] [Google Scholar]
- 33. Jin Z., et al. (2007) Hypermethylation of the nel-like 1 gene is a common and early event and is associated with poor prognosis in early-stage esophageal adenocarcinoma. Oncogene, 26, 6332–6340. [DOI] [PubMed] [Google Scholar]
- 34. Jin Z., et al. (2007) Hypermethylation of tachykinin-1 is a potential biomarker in human esophageal cancer. Clin. Cancer Res., 13, 6293–6300. [DOI] [PubMed] [Google Scholar]
- 35. Brock M.V., et al. (2003) Prognostic importance of promoter hypermethylation of multiple genes in esophageal adenocarcinoma. Clin. Cancer Res., 9, 2912–2919. [PubMed] [Google Scholar]
- 36. Moinova H., et al. (2012) Aberrant vimentin methylation is characteristic of upper gastrointestinal pathologies. Cancer Epidemiol. Biomark. Prev., 21, 594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zou H., et al. (2005) Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barrett’s esophagus. Int. J. Cancer, 116, 584–591. [DOI] [PubMed] [Google Scholar]
- 38. Nones K., et al. (2014) Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int. J. Cancer, 135, 1110–1118. [DOI] [PubMed] [Google Scholar]
- 39. Thiele C.J., et al. (2009) On Trk—the TrkB signal transduction pathway is an increasingly important target in cancer biology. Clin. Cancer Res., 15, 5962–5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guo M., et al. (2015) CHFR methylation strongly correlates with methylation of DNA damage repair and apoptotic pathway genes in non-small cell lung cancer. Discov. Med., 19, 151–158. [PubMed] [Google Scholar]
- 41. Soutto M., et al. (2010) Epigenetic and genetic silencing of CHFR in esophageal adenocarcinomas. Cancer, 116, 4033–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang M., et al. (2014) Association between CHFR methylation and chemosensitivity of paclitaxel in advanced gastric cancer. Med. Oncol., 31, 907. [DOI] [PubMed] [Google Scholar]
- 43. Stolz A., et al. (2015) Wnt-mediated protein stabilization ensures proper mitotic microtubule assembly and chromosome segregation. EMBO Rep., 16, 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vitale I., et al. (2011) Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol., 12, 385–392. [DOI] [PubMed] [Google Scholar]
- 45. Weaver B.A., et al. (2006) Does aneuploidy cause cancer? Curr. Opin. Cell Biol., 18, 658–667. [DOI] [PubMed] [Google Scholar]
- 46. Maley C.C., et al. (2006) Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet., 38, 468–473. [DOI] [PubMed] [Google Scholar]
- 47. Davison J.M., et al. (2014) The degree of segmental aneuploidy measured by total copy number abnormalities predicts survival and recurrence in superficial gastroesophageal adenocarcinoma. PLoS One, 9, e79079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bharadwaj R., et al. (2004) The spindle checkpoint, aneuploidy, and cancer. Oncogene, 23, 2016–2027. [DOI] [PubMed] [Google Scholar]
- 49. Duijf P.H., et al. (2013) The cancer biology of whole-chromosome instability. Oncogene, 32, 4727–4736. [DOI] [PubMed] [Google Scholar]
- 50. Gal-Yam E.N., et al. (2008) Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc. Natl Acad. Sci. USA, 105, 12979–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Noushmehr H., et al. (2010) Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell, 17, 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.