Abstract
In vivo mapping of transcription-factor binding to the transcriptional output of the regulated gene is hindered by probabilistic promoter occupancy, the presence of multiple gene copies, and cell-to-cell variability. We demonstrate how to overcome these obstacles in the lysogeny maintenance promoter of bacteriophage lambda, PRM. We simultaneously measured the concentration of the lambda repressor CI and the number of mRNAs from PRM in individual E. coli cells, and used a theoretical model to identify the stochastic activity corresponding to different CI binding configurations. We found that switching between promoter configurations is faster than mRNA lifetime, and that individual gene copies within the same cell act independently. The simultaneous quantification of transcription factor and promoter activity, followed by stochastic theoretical analysis, provides a tool that can be applied to other genetic circuits.
Sequence-specific transcription factors drive the diversity of cell phenotypes in development and homeostasis (1). For each target gene, alternative transcription-factor binding configurations (by different transcription factors or by multiple copies of the same one) result in varied transcriptional outputs, in turn leading to alternative cell fates and behaviors (2, 3). Elucidating the relations between transcription-factor configurations (which can number in the hundreds (4–6)) and the resulting transcriptional activity remains a challenge. Application of traditional genetic and biochemical approaches usually requires a genetically modified system or assays of purified components in vitro (7). Ideally, however, one would like to map transcription-factor configuration to promoter activity inside the cell, with minimal perturbation to the endogenous system.
Multiple factors hinder such direct measurement. First, individual cells vary in both transcription-factor concentration and the resulting transcriptional activity (8, 9); averaging over many cells thus filters out details of the regulatory relation. Second, even within the single cell, more than one copy of the regulated gene is typically present, with each copy individually regulated (10). Finally, even at the level of a single gene copy, multiple binding configurations are possible at a given transcription-factor concentration (11, 12). The relative probabilities of these different configurations and the rate of switching between them will define the stochastic activity of the regulated promoter (13).
We simultaneously measured, in individual cells, the concentration of a transcription factor and the number of mRNAs produced from the regulated gene. We also measured how the gene copy number changes through the cell cycle. We then analyzed the full single-cell data using a theoretical model, which allowed us to identify the contributions of different transcription-factor binding configurations to the stochastic activity of the promoter.
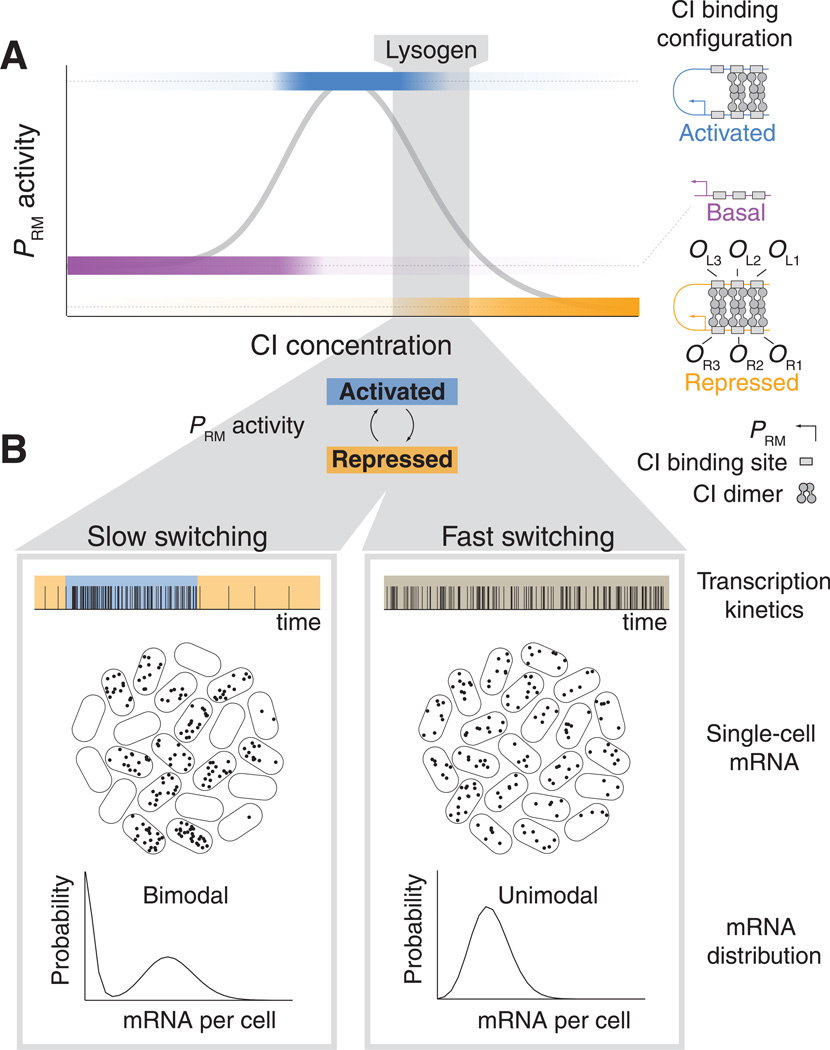
Specifically, we examined the lysogeny maintenance promoter of phage lambda, PRM. The regulation of this promoter by its own gene product, the lambda repressor (CI), is a paradigm for how alternative binding configurations drive transcriptional activity and the resulting cell fate—stable lysogeny or lytic induction resulting in cell death (7). The number of possible CI configurations is very large (>100 (4, 5)). Briefly, as CI concentration increases, CI dimers gradually occupy three proximal (OR1–3) and three distal (OL1–3) operator sites, leading first to activation, then repression, of PRM (Fig. 1A). Cooperative CI binding, and looping of DNA between the OR and OL sites, play important roles in shaping the PRM(CI) regulatory curve (14).
Fig. 1. Schematic: PRM regulation by CI.
(A) As the concentration of CI increases, the probabilities of different binding configurations of CI dimers at the OR and OL operators change (color shading), resulting in varying PRM activity (gray curve). Three configurations, expected to be the most probable, are depicted. In lysogenic cells, PRM has comparable probabilities of being in the activated and repressed promoter states (gray shading). (B) The rate of switching between activated and repressed states drives the stochastic activity of PRM in lysogenic cells. Two alternative hypotheses are illustrated: If switching is slow relative to the mRNA lifetime (left), two subpopulations of cells will exist, with low and high mRNA levels. If switching is fast (right), the mRNA distribution in the population will be unimodal.
In a lysogen (a bacterium carrying a prophage), CI concentration is believed to be such that PRM fluctuates between the activated and repressed states (15) (Fig. 1A), and this has been suggested to stabilize the lysogenic state against random fluctuations in CI levels (14). However, the nature of the lysogenic “mixed state” (activated/repressed) is unknown: Are the promoter fluctuations slow enough, such that two distinct cell populations coexist, exhibiting high and low PRM expression respectively? Alternatively, are promoter fluctuations fast, such that all cells exhibit an intermediate, well-defined, level of PRM expression (Fig. 1B)?
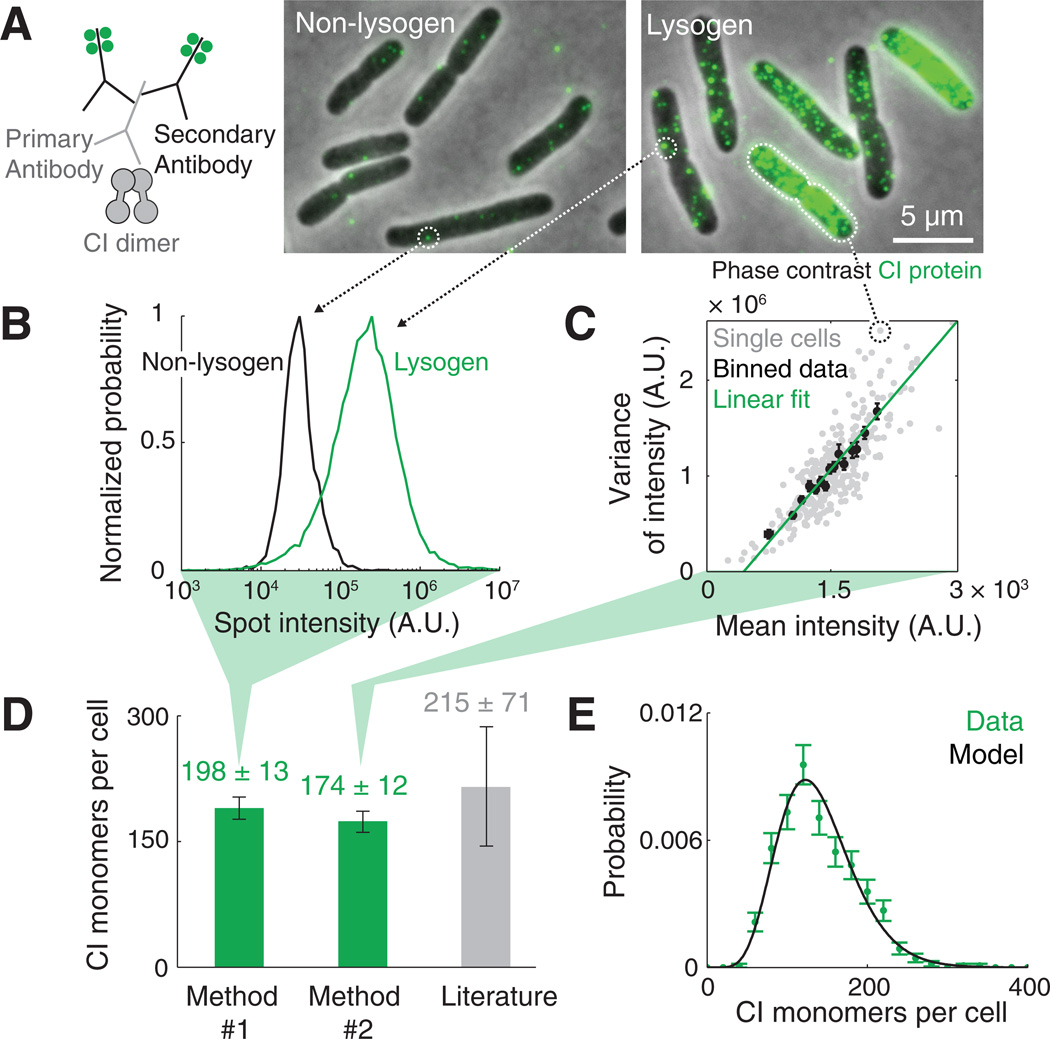
To measure CI concentration in individual cells, we used antibody labeling (immunofluorescence). Lysogenic cells (see table S1) exhibited a strong CI signal whereas non-lysogenic (uninfected) cells showed only a weak background signal (Fig. 2A, fig. S1). To verify that the antibody signal reliably represents CI levels, we expressed a CI-yellow fluorescent protein (YFP) fusion protein (16) in non-lysogenic cells and compared the YFP fluorescence to the CI antibody signal in each cell. The two signals were linear with each other (fig. S2A), and single-molecule imaging revealed that most YFP molecules were colocalized with a CI antibody, as expected (fig S2B).
Fig. 2. Measuring the number of CI molecules in individual cells.
(A) CI proteins were labeled using antibodies to CI and fluorescently-labeled secondary antibodies (left). Under the microscope, lysogenic E. coli cells exhibited a strong CI signal (right) whereas non-lysogens showed a weak background signal (center). (B) Method #1 for measuring the number of CI proteins per cell. The typical fluorescence of a single CI dimer was obtained from the spot intensity distribution in lysogenic cells (green, N = 23631 spots), distinguishable from that of the negative sample (black, N = 1764 spots). (C) Method #2 for measuring the number of CI proteins per cell. The variance versus the mean of pixel intensity in individual cells (gray, N = 324) was fitted to a linear function (green). The slope of this line was used to estimate the fluorescent intensity of a single CI dimer. (D) The estimated number of CI molecules in a lysogen, obtained using the two single-cell methods (green, mean ± SEM from 6 experiments, 327 to 704 cells each). Also shown is the value reported in the literature (gray, mean ± SD from three studies (19–21)). (E) The distribution of CI copy number in lysogenic cells (green; N = 560 cells). The data is described well by a gamma distribution (black).
To convert the antibody signal to CI concentration in each cell, we needed to know the fluorescence value corresponding to a single antibody-labeled CI molecule (a CI dimer, which is the dominant species in the cell (17)). To obtain this calibration constant, we used two methods (18) (Fig. 2, B and C): In the first method, we used automated image analysis to identify individual fluorescent particles (spots, fig. S3). These spots displayed a well-defined intensity value, distinct from the corresponding signal found in negative samples (Fig. 2B). We identified the positive-sample spot intensity as corresponding to individual CI dimers (fig. S4A) (each one decorated by ~20 florescent dyes, due to the stoichiometry of antibody labeling, figs. S5), and used it to convert cell fluorescence to CI concentration. In the second method, we used the fact that the Poisson statistics of random protein positions within the cell lead to a linear relation between the fluorescence mean and the pixel-to-pixel variance within each cell (Fig. 2C, fig. S6). Measuring the slope of this line allowed us to identify the fluorescence corresponding to a single labeled protein (fig. S7). Using either method to estimate CI concentration in lysogens gave similar results (Fig. 2D and fig. S4B). These measured values also agreed with those reported in the literature (19–21)(Fig. 2D and table S2).
The two imaging-based methods allowed us to measure CI numbers in individual lysogenic cells (Fig. 2E). Fitting the CI distribution to a stochastic model of protein production (22) indicated that, on average, the ~200 CI monomers in the cell are produced in ~10 random bursts, of ~20 proteins each, during the 30-min cell cycle (table S3). The estimated burst frequency is consistent with a (more accurate) value that we obtained from cI mRNA statistics (Fig. 3). It is also consistent with the measured stability of the lysogenic state (which depends exponentially on the CI burst frequency (23)).
Fig. 3. Measuring the transcriptional activity of a single PRM copy.
(A) (Left) cI mRNA in lysogenic cells was labeled using single molecule fluorescence in situ hybridization (smFISH). (Right) The measured distribution of cI mRNA number per cell from the whole population (gray, N = 2893 cells) consists of contributions from cells containing 2 and 4 gene copies (black; see panels B–D). α is the fraction of cells with two copies of the PRM-cI gene. (B) Estimating the number of PRM-cI gene copies in lysogenic cells. TetR-YFP binds to an array of tetO sites inserted next to the gene locus, resulting in visible foci (left). (Right) Newborn cells (length percentile 5 to 20, “short”, red, N = 493) contained two copies of the PRM-cI locus, whereas cells about to divide (length percentile 80 to 95, “long”, blue, N = 493) contained four copies. Error bars indicate SEM. (C) The measured distributions of cI mRNA numbers for short (left) and long (right) cells. Both were well fitted by a model assuming independent stochastic activity of each gene copy. (D) The theoretical fit from panel C was used to reconstruct the cI mRNA distribution from a single gene copy. This distribution was then used to predict the distribution for the whole-population (panel A).
To measure the activity of the PRM promoter in individual lysogenic cells, we used single-molecule fluorescence in situ hybridization (smFISH) (24, 25) to label and count cI mRNAs, produced from PRM (Fig. 3A). Fluorescent spots were identified using an automated algorithm (25) (fig. S3), and the fluorescence intensity corresponding to a single mRNA was identified (fig. S8). We used this intensity to convert the total spot intensity in each cell to the number of cI mRNAs (25). The copy-number distribution of cI mRNA in a lysogen (Fig. 3A) represents the combined contribution from multiple copies of the PRM-cI gene in each cell (26). To identify the contribution of a single gene copy, we first examined how the cI gene copy number varies during the cell cycle. We engineered an array of 140 Tet operators (tetO) (27) into the gal locus of E. coli (~16 kb away from the lambda integration site). The gene locus was detected through the binding of a Tet repressor (TetR)-YFP fusion (27) (Fig. 3B). We used automated image analysis to count the number of YFP foci in each cell. Gating the cell population by length, we found that newborn cells had on average 2.1 ± 0.1 (mean ± SEM) foci per cell. Cells about to divide had 4.0 ± 0.1 foci per cell (Fig. 3B). These values are in good agreement with the expected copy number of the cI locus under our experimental conditions (26). We used these measured copy numbers to delineate the transcriptional activity of individual gene copies. If the stochastic activity of each copy is independent of the other copies in the same cell, then the cI mRNA distribution for cells having two gene copies will be given by the auto-convolution of the distribution for a single gene copy (a distribution that we cannot measure directly). Similarly, the mRNA distribution for 4-copy cells will be equal to the 1-copy distribution taken to the 4th convolution power. The experimental histograms agreed well with these predictions (Fig. 3C and fig. S9). Furthermore, knowing the fraction of cells in the population that have 2 and 4 copies allowed us to then predict the cI mRNA distribution for the whole population. The predicted distribution agreed well with the experimentally measured one (Fig. 3A).
Analyzing the single-gene mRNA distribution (Fig. 3D) revealed that a single copy of PRM produces a burst of cI mRNA every ~6 min on average (table S4). When accounting for the presence of 2 to 4 gene copies per cell (Fig. 3B), this value is consistent with the burst frequency estimated from the CI protein histogram (Fig. 2E). Comparing the protein and mRNA data also allowed us to directly calculate the number of CI proteins produced from each cI mRNA, ~6 on average (table S3). This value is in good agreement with a previous theoretical calculation (23).
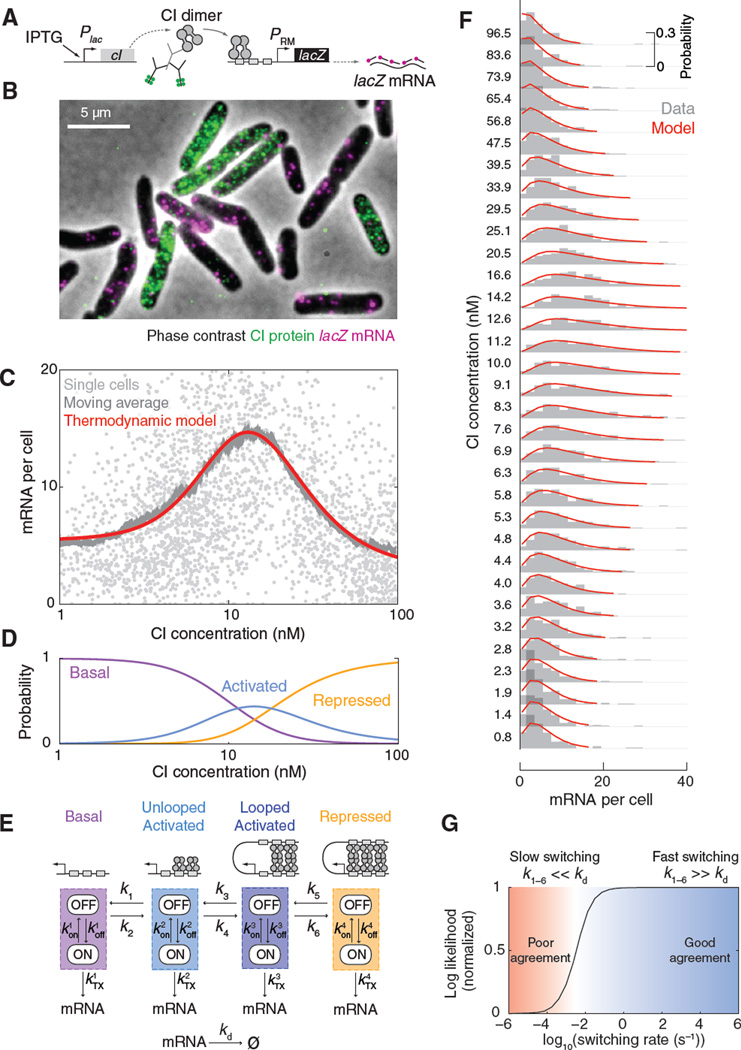
To measure the regulatory relation between CI concentration and PRM activity, we used a reporter system in which the autoregulatory feedback from CI to PRM existing in the lysogen is broken: CI is expressed from an inducible promoter, while PRM transcribes the lacZ gene rather than cI (14) (Fig. 4A). To simultaneously measure CI concentration and PRM activity in the same cell, we combined immunofluorescence (using antibody to CI) with smFISH (using lacZ probes)(18)(Fig. 4B and fig. S10), and measured the corresponding protein and mRNA numbers as described above. Performing this measurement over a range of CI levels, then plotting lacZ mRNA numbers versus CI concentration from many individual cells, produced highly scattered data (Fig. 4C), as expected from the stochasticity of the regulation and transcription processes (9). Averaging within finite windows of CI concentration revealed the mean regulatory relation between CI and PRM, known as the gene regulation function (16)(Fig. 4C, fig. S11). The shape of the regulation function agreed with that from previous reports, with PRM activity first increasing, then decreasing, with CI concentration (4, 14, 28). However, our measurement provides the absolute numbers for both the input (CI concentration) and output (mRNA numbers), rather than relative expression levels (4, 5, 14, 28). The absolute values are crucial for the subsequent steps in our analysis of PRM regulation.
Fig. 4. PRM regulation at the single-cell level.
(A) The reporter system used for measuring PRM activity. (B) Cells labeled for CI protein (immunofluorescence, green) and PRM-lacZ mRNA (smFISH, magenta). (C) The PRM(CI) gene regulation function. The single-cell data (light gray, N = 2941 cells) was filtered using a moving average (dark gray, 200 cells/bin). The averaged curve was well fitted by the thermodynamic model (red). (D) The calculated probabilities of different promoter activity states as a function of CI concentration: basal (purple), activated (looped and unlooped, blue) and repressed (orange). (E) A stochastic model for PRM kinetics. Each promoter activity state was modeled using stochastic ON/OFF transcription kinetics. The promoter stochastically transitions between activity states in a CI-dependent manner. (F) The stochastic model successfully described the PRM(CI) single-cell data. The experimental data (panel C) was binned into 100-cell histograms (gray) and fitted to the model (red) using maximum likelihood estimation. (G) Consistency of measured mRNA statistics with rapid switching between promoter states. Solving the stochastic model for different switching rates, and fitting the model results to the measured mRNA statistics, resulted in a good fit for fast switching (right, blue); slow switching yielded a poor fit (left, red).
As the first step in this analysis, we wrote down a theoretical model, in which the probabilities of different CI binding configurations are given by their thermodynamic weights (15) (fig. S12A). This thermodynamic model successfully reproduced the regulation function (Fig. 4C and fig. S13). In performing this procedure, most free energy values used in the model were identical to those reported (15)(table S5). The model also provided the probabilities of observing the different promoter activity states—basal, activated [with the DNA between OR and OL either looped or unlooped (15)], and repressed—as a function of CI concentration (Fig. 4D). The overlap between the different states underlines the challenge in identifying the transcriptional signature of a single promoter state: For example, the probability of PRM being in the activated state never surpasses ~50%.
To reveal the activity of individual promoter states, we introduced a stochastic version of the theoretical model (Fig. 4E and fig. S12). In the model, the CI binding configurations are grouped based on the expected promoter activity: basal, activated unlooped, activated looped and repressed (15). Each promoter activity state is described by stochastic bursty kinetics of mRNA production (29). PRM stochastically switches between its four activity states. The switching rates are initially unknown, but the thermodynamic model above provides us with the equilibrium constant (ratio between switching left and right) for each pair of states, at a given CI concentration. For each set of parameters, the stochastic model can be solved to yield the expected mRNA copy-number distribution for the population of multi-copy cells.
We used the stochastic model to analyze the full PRM(CI) single-cell data set (Fig. 4C above). Applying maximum-likelihood estimation, we found good agreement between the experimental and theoretical mRNA distributions over the full range of CI concentrations (Fig. 4F, fig. S14 and movie S1). The fitting procedure allowed us to extract the mRNA statistics corresponding to the different activity states of PRM (fig. S15). The calculated distributions were in good agreement with those obtained using genetic controls: Cells expressing no CI (basal), overexpressing CI in wild-type PRM (repressed) and in a mutant lacking the OL operator (activated unlooped) (14)(fig. S15B and table S6). The stochastic kinetics of each promoter state exhibited a similar relation between expression level and burst size to that measured in other E. coli promoters (29)(fig. S15C).
Despite the fact that the measured mRNA distribution at each CI concentration represents a mixture of multiple promoter states, each of the histograms is unimodal, and can be described by a simple kinetic model with a single burst size and frequency (Fig. 4F and fig. S16). The parameter that determines the shape of the “mixed state” mRNA distribution is the rate of switching between promoter states (Fig. 1B above). Previous in vitro studies of OR-OL looping suggested that the switching between looped and unlooped promoter configurations is fast (~seconds) (30), but similar studies of looping in the cell left the question open (31). Our stochastic model predicts that if promoter switching is very slow relative to mRNA lifetime (here ~2 min (29)), the observed mRNA distribution will be the weighed sum of the underlying single-promoter-state distributions. Our experimental data strongly disagreed with this prediction (Fig. 4G). On the other hand, if switching is fast, the observed distribution will be given by a (weighed) convolution of the underlying single-promoter-state distributions, and, if the underlying states can each be described by simple bursty kinetics, the new mixed state can be as well. This is indeed what we observed (Fig. 4, F and G; fig. S16). Thus, PRM switches rapidly between different promoter states, resulting in a stochastic signature that (at a given CI concentration) is indistinguishable from that of a single promoter state, but with renormalized kinetic parameters. Our finding of rapid switching explains why, in the lysogen, we did not detect distinct “active” and “repressed” populations in either the protein (Fig. 2E) or mRNA (Fig. 3A) histograms, but instead both data indicated a single, well-defined promoter activity.
Precise single-cell measurements, accompanied by theoretical analysis, can reveal new features even in well-studied model systems. When combined with genetic and synthetic-biology approaches (13), this strategy may allow prediction of the stochastic characteristics of promoter activity, a prediction which remains a challenge to our understanding of gene regulation (9, 32).
Supplementary Material
Acknowledgments
We are grateful to the following people for generous advice and for providing reagents: I. Dodd, M. Elowitz, L. Finzi, H. Garcia, T. Gregor, T. Kuhlman, L. McLane, R. Phillips, A. Raj, E. Rothenberg, A. Sanchez, K. Shearwin, R. Singer, S. Skinner, L-H. So, A. Sokac, L. Zeng and C. Zong. Work in the Golding lab is supported by grants from NIH (R01 GM082837), NSF (PHY 1147498, PHY 1430124 and PHY 1427654), The Welch Foundation (Q-1759) and The John S. Dunn Foundation (Collaborative Research Award). H.X. is supported by the Burroughs Wellcome Fund Career Award at the Scientific Interface. We gratefully acknowledge the computing resources provided by the CIBR Center of Baylor College of Medicine.
References and Notes
- 1.Pulverer B. Getting specific. Nat. Rev. Mol. Cell. Biol. 2005;6:S12–S13. [Google Scholar]
- 2.Ptashne M, Gann A. Genes & signals. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- 3.Davidson EH, Levine MS. Properties of developmental gene regulatory networks. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20063–20066. doi: 10.1073/pnas.0806007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson LM, Yang H. DNA looping can enhance lysogenic CI transcription in phage lambda. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5827–5832. doi: 10.1073/pnas.0705570105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui L, Murchland I, Shearwin KE, Dodd IB. Enhancer-like long-range transcriptional activation by lambda CI-mediated DNA looping. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2922–2927. doi: 10.1073/pnas.1221322110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson EH. The regulatory genome : gene regulatory networks in development and evolution. Burlington, MA ; San Diego: Academic; 2006. p. xi.p. 289. [Google Scholar]
- 7.Ptashne M. A genetic switch. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, Nerw York: 2004. [Google Scholar]
- 8.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez A, Golding I. Genetic determinants and cellular constraints in noisy gene expression. Science. 2013;342:1188–1193. doi: 10.1126/science.1242975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levesque MJ, Ginart P, Wei Y, Raj A. Visualizing SNVs to quantify allele-specific expression in single cells. Nat. Methods. 2013;10:865–867. doi: 10.1038/nmeth.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shea MA, Ackers GK. The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J Mol. Biol. 1985;181:211–230. doi: 10.1016/0022-2836(85)90086-5. [DOI] [PubMed] [Google Scholar]
- 12.Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 13.Jones DL, Brewster RC, Phillips R. Promoter architecture dictates cell-to-cell variability in gene expression. Science. 2014;346:1533–1536. doi: 10.1126/science.1255301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB. Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes. Dev. 2001;15:3013–3022. doi: 10.1101/gad.937301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodd IB, Shearwin KE, Perkins AJ, Burr T, Hochschild A, Egan JB. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes. Dev. 2004;18:344–354. doi: 10.1101/gad.1167904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 17.Koblan KS, Ackers GK. Energetics of subunit dimerization in bacteriophage lambda cI repressor: linkage to protons, temperature, and KCl. Biochemistry. 1991;30:7817–7821. doi: 10.1021/bi00245a022. [DOI] [PubMed] [Google Scholar]
- 18.Xu H, Sepúlveda LA, Figard L, Sokac AM, Golding I. Combining protein and mRNA quantification to decipher transcriptional regulation. Nat. Methods. 2015;12:739–742. doi: 10.1038/nmeth.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensel Z, Feng H, Han B, Hatem C, Wang J, Xiao J. Stochastic expression dynamics of a transcription factor revealed by single-molecule noise analysis. Nat. Struct. Mol. Biol. 2012;19:797–802. doi: 10.1038/nsmb.2336. [DOI] [PubMed] [Google Scholar]
- 20.Levine A, Bailone A, Devoret R. Cellular levels of the prophage lambda and 434 repressors. J Mol. Biol. 1979;131:655–661. doi: 10.1016/0022-2836(79)90014-7. [DOI] [PubMed] [Google Scholar]
- 21.Reichardt L, Kaiser AD. Control of lambda repressor synthesis. Proc. Natl. Acad. Sci. U. S. A. 1971;68:2185–2189. doi: 10.1073/pnas.68.9.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman N, Cai L, Xie XS. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys. Rev. Lett. 2006;97:168302. doi: 10.1103/PhysRevLett.97.168302. [DOI] [PubMed] [Google Scholar]
- 23.Zong C, So LH, Sepúlveda LA, Skinner SO, Golding I. Lysogen stability is determined by the frequency of activity bursts from the fate-determining gene. Mol. Syst. Biol. 2010;6:440. doi: 10.1038/msb.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skinner SO, Sepúlveda LA, Xu H, Golding I. Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nat. Protoc. 2013;8:1100–1113. doi: 10.1038/nprot.2013.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nordstrom K, Dasgupta S. Copy-number control of the Escherichia coli chromosome: a plasmidologist's view. EMBO Rep. 2006;7:484–489. doi: 10.1038/sj.embor.7400681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joshi MC, Bourniquel A, Fisher J, Ho BT, Magnan D, Kleckner N, Bates D. Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2765–2770. doi: 10.1073/pnas.1019593108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis D, Le P, Zurla C, Finzi L, Adhya S. Multilevel autoregulation of lambda repressor protein CI by DNA looping in vitro. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14807–14812. doi: 10.1073/pnas.1111221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.So LH, Ghosh A, Zong C, Sepúlveda LA, Segev R, Golding I. General properties of transcriptional time series in Escherichia coli. Nat. Genet. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manzo C, Zurla C, Dunlap DD, Finzi L. The effect of nonspecific binding of lambda repressor on DNA looping dynamics. Biophys J. 2012;103:1753–1761. doi: 10.1016/j.bpj.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hensel Z, Weng X, Lagda AC, Xiao J. Transcription-factor-mediated DNA looping probed by high-resolution, single-molecule imaging in live E coli cells. PLoS Biol. 2013;11:e1001591. doi: 10.1371/journal.pbio.1001591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coulon A, Chow CC, Singer RH, Larson DR. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat. Rev. Genet. 2013;14:572–584. doi: 10.1038/nrg3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.