Abstract
The ability to collect millions of molecular measurements from patients is a now a reality for clinical medicine. This reality has created the challenge of how to integrate these vast amounts of data into models that accurately predict complex pathophysiology and can translate this complexity into clinically actionable outputs. Integrative informatics and data-driven approaches provide a framework for analyzing large-scale datasets and combining them into multiscale models that can be used to determine the key drivers of disease and identify optimal therapies for treating tumors. In this perspective we discuss how an integrative modeling approach is being used to inform individual treatment decisions, highlighting a recent case report that illustrates the challenges and opportunities for personalized oncology.
Keywords: drug repositioning, genomics, multiple myeloma, multiscale modeling, networks, personalized medicine
The ability to deliver truly personalized, precise and predictive medicine depends on having tools to measure, model and modulate the complex and adaptive systems underlying human physiology and disease [1]. An immense number of interactions within and between cells of body tissues both define and regulate the complex biological states of individual physiology. Cells and tissue communicate information across multiple interacting layers (e.g., DNA, RNA, protein, metabolic) to regulate biological processes and maintain homeostasis in response to local and global stimuli. Technologies such as high-throughput genomic sequencing and real-time imaging techniques provide the ability to collect, quantify and digitize system-wide measurements of complex biological states with increased resolution and precision [2,3]. Multiple opportunities now exist in personalized medicine for developing innovative methods and systems that can integrate and model the growing digital universe of molecular and clinical data we collect from individuals [4–8]. Some of the key challenges for realizing these innovations include how to (i) more accurately understand how information flows, and is regulated, in human biological systems, (ii) better represent and describe the health status of an individual at any given time and (iii) provide analytical tools to identify the best course of action for a given intervention based on all available data. Addressing these challenges requires the convergence of medicine, science and technology, with researchers, providers and patients working together toward precision medicine.
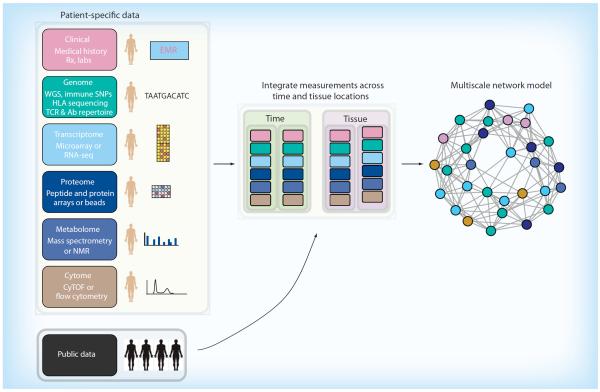
Our strategy for implementing personalized medicine is centered on a systems approach to human biology aimed at capturing as much genetic, molecular, cellular, physiological, clinical and historical data as possible for an individual and integrating this information into multiscale network models (Figure 1). Instead of assuming we know what elements are important for a given medical condition, we use the data to derive patterns of interactions that might drive a particular condition [9–11], and combine this knowledge with expertise from physicians to improve patient-specific care [12]. In this perspective, we discuss some of the challenges for personalized cancer medicine, describe our approach, highlight a case study with multiple myeloma and suggest a roadmap for incorporating multiscale network modeling into personalized oncology.
Figure 1. Integration of measurements from diverse data types into a personalized multiscale network model.
Advances in high-throughput modalities capture patient specific information from various data types including different tissue locations and over time. Sophisticated statistical models offer a framework for incorporating patient-specific data with public data to generate a comprehensive multiscale network model, which reflects the current health status of a patient and can be updated/improved with additional data.
Challenges & opportunities for personalized oncology
It is clear now that cancer is not a single disease but rather is a growing constellation of subtypes mapped by a large catalog of somatic mutations [13] and characterized by tremendous molecular heterogeneity [14]. This diversity is driven by genetically distinct cancerous cells that expand and co-evolve within a complex tumor microenvironment that adapts to cytokine signaling, the immune response, therapeutic interventions and numerous other intrinsic and extrinsic factors. The process of evolutionary selection under continuously changing environmental influences generates unique tumor cell lineages that present multiple challenges for how best to predict cancer progression and treat an individual. Whereas our current ability to determine the disease course for any given patient remains generally limited, molecular profiling combined with sophisticated modeling offer approaches for capturing biological complexity in its entirety and producing precise predictions required for personalized cancer therapies.
Discussions and applications of personalized medicine to date revolve largely around DNA genotype information, but more sophisticated approaches to personalized medicine require integrating DNA information with other molecular measures [2,15,16]. Knowing the genotype of an individual allows physicians to assess the consequences of drug metabolism [17], estimate a patient's risk factor for a number of conditions and better manage certain conditions such as specific cancers [18,19]. Genome-wide genotyping reveals the entirety of genetic information for an individual, yet this only provides a partial and largely static picture of individual biology. To capture and model a more complete representation of individual health and disease, DNA information must be complimented with additional functional genomic and dynamic molecular phenotypes.
Applying new technological developments to personalized oncology
A number of advanced technologies now provide the tools to collect massive amounts of functional data (e.g., comprehensive transcriptional profiling or high-resolution phenotyping) from collections or even single cells. DNA sequencing on next generation platforms provides a method to monitor the immune system as a diagnostic tool for leukemia [20], as well as to profile the T cell antigen receptor [21] or antibody repertoires [22]. Extraction of RNA from cells followed by high-throughput sequencing (RNA-seq) is a powerful technique to quantify transcriptomes, determine variation at the single nucleotide level, discover gene fusions and identify splice variants [23,24]. Single cell mass cytometry can simultaneously probe more than 40 different cellular markers (surface and intracellular proteins) to characterize phenotypic and functional changes in tumor and normal cell with unprecedented resolution and precision [25]. Each of these next-generation technologies captures a systems-wide snapshot of a single scale and type of data. Integrating these modalities into comprehensive multiscale models yields better outcomes for individual patients, whether through improved predictive power for cancer survival [12] or increased ability to identify informative mutations with clinically actionable treatment options [26].
Some of the most compelling developments in leveraging new technologies for personalized medicine have occurred in Hematology–Oncology [27]. In a landmark paper, whole-genome sequencing was performed on paired genomes from acute myeloid leukemia and normal cells, and the results were used to identify somatic mutations [28]. It is worth noting the speed at which the field is progressing as a follow-up study found a novel mutation in DNMT3A upon resequencing of the same sample using updated sequencing techniques and informatics tools [29]. The use of advanced technologies such as next-generation sequencing (NGS) to inform the selection of targeted therapies is already showing promise for patients [30]. More recently, NGS has shown potential for guiding therapeutic choices, either by tracking clonal architecture and tumor evolution [31,32], or by identifying specific translocations in defined molecular subgroups of multiple myeloma that can be targeted with treatments such as proteasome inhibitors and FGFR3 and MMSET inhibitors [33]. In a recent case, targeted sequencing of 25 cancer-related genes identified a codon deletion in KIT that has been associated with imatinib sensitivity, and subsequent treatment with imatinib resulted in stabilization of disease [34]. More broadly, a multipronged strategy that integrates whole genome and exome sequencing with transcriptional profiling identified genomic alterations in four individuals that suggested potential pathways in each for patient-specific targeted interventions using approved or investigational drugs [26].
Recognition that cancer arises from a single malignant cell has led oncologists to adopt novel techniques to better characterize and classify cellular phenotypes [35,36]. Deep phenotyping of cancer cells using mass cytometry (CyTOF) offers broader diagnostic and monitoring capabilities over conventional methods such as flow cytometry. Originally developed for single cell measurements akin to flow cytometry, the metal isotope labeling technology has recently been combined with laser ablation imaging to vastly improve the spatial resolution and cellular characterization by immunohistochemistry [37,38]. The ability to characterize the state of a tumor and determine the cellular heterogeneity accurately is significant because matching the tumor with treatment results in better remission and long-term survival [39–46].
Advances in molecular labeling technologies are providing better readouts of the proteomic state at single-cell resolution and NGS technologies are helping characterize the genomic state of single cells. The persistence of minimal residual disease following treatment of lymphoblastic leukemia is a strong indicator for relapse and sequencing much higher sensitivity for detecting cancerous clones, which allows enhanced monitoring and treatment refinement capabilities [20,47,48]. Analyzing genome-wide patterns of mRNA expression can determine drivers of cancer [49–51], predict potential therapeutic options [52,53] and monitor molecular responses to treatment [54]. Malignant tumors and leukemia cells can now be sequenced to identify specific genetic variants that would suggest choosing one tyrosine kinase inhibitor over another [55]. As the costs of advanced technologies and sequencing continue to drop, deep profiling of single cells (tumor and normal), quantification and analysis of cell-free circulating tumor DNA [56,57] and molecular matching of compound profiles to disease profiles [53,58] will become standard of care.
Impact of targeted therapeutics on personal cancer medicine
Targeted cancer therapeutics — chemical compounds or monoclonal antibodies — manipulate a specific aspect of cellular processes that is integral to tumor survival. These therapies have radically altered medical practice and offer concrete examples of personalizing treatment through the use of molecular matching between drug and tumor. To date, there are approximately 50 compounds with indications for specific types of cancers with dozens of additional drugs being evaluated in targeted therapy clinical trials [59]. These treatments are effective in part because they address the consequence of a single mutation being the key driver in cancer. Unfortunately, in many cases, these initial successes ultimately fail because mutations assuredly arise in cancerous cells. These mutations permit cell cancer survival and growth by (i) altering protein structure to subvert drug binding while retaining protein function [60], (ii) using alternative cellular pathways [61] or (iii) modifying gene expression patterns to compensate for drug-induced function loss (e.g., copy number variant changes amplify gene expression) [62]. The next generation of targeted therapeutics for personalized medicine must be designed to account for tumor complexity and heterogeneity, and thus be able to modulate multiple driver mutations and pathways simultaneously.
Combination therapy for cancer treatment continues to hold great promise for addressing tumor complexity and drug resistance, and will play a larger role in the therapeutic strategy for personalized oncology. Evidence now strongly indicates that treatment with combination therapy increases survival in a variety of cancers [63]. The wealth of treatment options — small molecules, monoclonal antibodies, immunotherapy, bispecific antibodies or synthetic biology — suggests numerous possible combinations to draw from so patients might realize substantial therapeutic benefits. Initial efforts at combining the anti-CD20 antibody (Rituximab) and the anti-CD47 antibody led to the complete elimination of lymphoma in human non-Hodgkin lymphoma-engrafted mice [64]. Preclinical trials of tumor models point toward enhanced tumor elimination using combinatorial immunotherapy with anti-CTLA4 and a targeted secondary antibody to block co-stimulation [65]. The merging of improved target guidance based on more refined tumor characterization with the large potential in combinatorial therapeutic diversity will continue to drive better outcomes for patients.
Computational & collaborative approach to personalized medicine
Our approach to personalized medicine is to construct comprehensive, multiscale network models that accurately represent the regulatory interactions between genes and transcripts with tumor cells collected from an individual [66–71]. These networks are mathematical models that depict relationships between and among data types (e.g., associations between gene variants, transcripts, proteins, cells, etc.) that comprise the network. Conditional correlations are used to assign like-lihoods to each interaction (association strength) and determine if a causal relationship is supported by the data [69]. Additional data sources (e.g., transcription factors, metabolites, proteomics, etc.) can be added as prior information in the parameters for constructing a multiscale network graph [4]. While these integrative models are constructed in a data-driven manner without a specific a priori hypothesis, they produce specific predictions about functional relationships between parameters of the data that often suggest new roles for these components [9] and, more importantly, propose particular hypotheses that can be validated experimentally. Rather than imposing strong preconceived constraints on the models, these approaches provide an unbiased method for identifying statistically significant and robust patterns in the data with some relationship to disease or biology. The models are then validated based on how well they recover known biology and provide new insights.
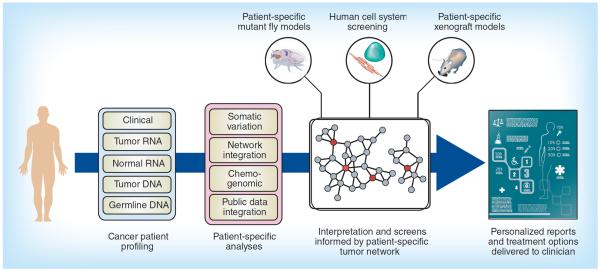
The foundation for a personalized cancer treatment program is the ability to identify patient-specific profiles from a vast sea of genetic and molecular data. This process is now possible because of the confluence of inexpensive, high-throughput diagnostic tools and computational processing power that can look for important molecular signatures by rapidly sifting through billions of data points. Our approach is to provide personalized cancer therapy recommendations by applying state-of-the art informatics methods that construct individual disease profiles and match them to the available therapeutic options (Figure 2). We also have an option of incorporating experimental feedback and validation using advanced preclinical models that most closely mimic an individual's tumor. This type of experimental modeling and validation currently factors into patient care for specific cases with slightly less urgency or when alternative options have been exhausted.
Figure 2. Schematic of the processing steps in a personalized cancer therapy pipeline.
The integration of clinical information with genome-wide expression data (from tumor and normal cells) and complete genotyping provides the raw data for characterization of an individual's tumor. A standard set of analyses examines these data sources for actionable anomalies and creates a patient-specific profile, which is compared with a collection of reference databases (e.g., compound libraries, chemogenomic information and cancer biology) to construct a patient-specific tumor network. Key driver analyses and identification of enriched subnetworks inform in vitro screening processes and produce recommendations for personalized treatment options.
This entire process of collecting, analyzing and testing individual samples to come up with the best model of their disease to drive personal care is a large multidisciplinary effort to patient-centric care. Our tumor board consists of domain-specific experts from clinical genetics, clinical oncology, genomics, bioinformatics and drug repurposing. This group collaborates to generate a personalized report indicating what we believe will be the best treatment for our patient. The integrative approach to evidence-based medicine uses many platforms ranging from molecular to clinical to computational. It is our philosophy that one piece of evidence in isolation is not sufficient to make a clinical decision. Based on this principle, our selection of a therapy is multifactorial and stems from our patient's clinical state, circumstances, values and opinions, as well as our pooled knowledge in our given areas of research and expertise.
Matching drug & disease profiles in personalized oncology
A major challenge for personalized oncology is selecting the right drug for the right patient at the right time. Solutions to this challenge aim to identify the best match between two complex and diverse data sets: an individual's disease signature, and a drug response signature. To be effective, this matching process must consider the universe of therapeutically relevant compounds, which could be large, and be performed in a timeframe that provides clinical utility. One strategy for meeting this challenge is to build algorithms and systems that standardize these complex datasets into compatible ranges so they are amenable to statistical tests that can identify the best match in an unbiased and systematic fashion. This general approach provides a procedure to rapidly scan through a library of compounds and select a drug to `treat' a patient's specific disease signature.
In order to streamline the drug-to-tumor matching process, drug response profiles are collected and curated from a wealth of publicly available data (e.g., the compendium of genome-wide transcriptional changes in response to drug perturbations [72], curated drug-gene feature databases [73] and drug sensitivity databases compiled from high-throughput screening of cancer cell lines [74,75]). These response profiles are matched in silico to tumor-specific molecular profiles of a patient to produce a ranked list of drugs, which are integrated into a single list of recommendations. We combine this list of recommendations with available public data on diseases (e.g., GWAS catalog [76]), thousands of other tissue- and disease-specific transcriptional profiles (e.g., ArrayExpress [77] and Gene Expression Omnibus [78]), and genomic information (e.g., ENCODE [79]) to provide broader context for the specific patient. Physicians consider this background of information with the prioritized list of drugs in the decision making process for delivering care to a patient.
Case study in multiple myeloma
As an example of how integrative modeling can be applied to personalized oncology, we present a brief case study of multiple myeloma within this perspective. Multiple Myeloma (MM) is a neoplastic plasma-cell disorder, characterized by clonal proliferation of malignant plasma cells in the bone marrow, and is the second most common hematological malignancy [80]. Currently, myeloma is an incurable disease, with a median survival after diagnosis of approximately 6 years [81].
The treatment of multiple myeloma has been a challenge for decades. Typically, treatment of MM consists of a single drug or combination of drugs such as targeted therapy or chemotherapy, with or without steroids, and in certain cases, autologous stem cell transplant [82]. Four major classes of drugs are currently used in the treatment of MM: traditional chemotherapy; corticosteroids; immunomodulating agents including thalidomide, lenalidomide and pomalidomide and proteasome inhibitors including bortezomib and carfilzomib [83]. Often, a combination of these drugs are used based on factors such as the patient's age, the stage of the MM, patient's kidney function and the possibility of a future stem cell transplant [83]. Although, advancements in therapies over the past several years, have led to an increase in median overall survival after diagnosis [81], relapse is inevitable and the unpredictable. The natural course and biological variability among MM patients makes an individualized approach to therapy a very attractive option.
Myeloma is characterized biologically by increased genomic instability and clonal evolution within the tumor over time. Most patients ultimately become refractory to further therapy and have a median survival of only 6–9 months in this situation [84]. Treatment selection for relapsed/refractory myeloma following treatment with all US FDA approved options and stem cell transplantation is challenging due to lack of active agents and patient comorbidities. Currently, patients with relapsed/refractory MM are treated empirically based on clinical trials or rechallenged with prior active chemotherapeutic agents or autologous stem cell transplant.
A significant amount of translational research exists for changing the therapeutic approach to multiple myeloma in order to improve patient outcomes [85,86]. For instance, the CoMMpass study [87] is a longitudinal, prospective observational study that aims to identify the molecular profiles and clinical characteristics in MM in order to stratify myeloma patients at initial diagnosis and at relapse. CD138+ tumor cells are analyzed by whole exome sequencing and RNA-seq, and then correlated with clinical characteristics. The overall rationale of the study is to develop a deeper understanding of the molecular basis of MM in order to devise a strategy for administering target therapeutics and personalized cancer care to patients with MM. Over the course of each patient's treatment, there is ongoing molecular profiling to evaluate the relationship between treatment regimens and patient outcomes [85]. The study has confirmed frequent mutations in KRAS, NRAS, DIS3 and P53 and less frequently in BRAF, TRAF3, CYLD, RB1 and PRDM1, which may have biological and therapeutic potential in MM [85]. The hope is that the study will lead to successful drug development based on these novel biological insights and improved patient care in multiple MM by using molecular profiles as a key to understanding mechanisms of disease, drug response and patient relapse [85]. However, while this study will provide an abundance of valuable information, it does not currently address the clinical challenge of treating relapsed myeloma patients. We hypothesize that a genomic, data-driven drug-repurposing program could be an effective way of treating patients who may be resistant to the standard, traditional therapies. The following clinical vignette is one example of how drugs that are not typically used to treat multiple myeloma may be effectively repositioned in relapsed/refractory myeloma.
A 54 year old male was diagnosed with IgA lambda plus lambda, Durie–Salmon stage IIA MM. He initially presented with back pain in April 2009 and upon evaluation was found to have a T12 compression fracture. The evaluation also revealed an elevated total protein of 10.6 g/dl with a globulin of 6.9 g/dl, an IgA level of 4662 mg/dl, free lambda of 1190 mg/l and a total proteinuria of 13.87 mg/day. A bone marrow biopsy on April 17th, 2009 showed a 90% clonal plasma cells.
Table 1 gives an overview of the patient's various therapies, remissions and relapses after his initial diagnosis in April 2009. At this point, as the patient continued to relapse, even after a bone marrow transplant, and on account of his recurrent malignant pleural effusions, we thought there might be a dominant resistant clone and started considering a genomic approach. To this end, we used a molecular matching approach described previously [58,88] to generate a list of top drug candidates that matched the RNA-seq profile of the CD138+ cells from patient's tumor. This approach identified two cytidine analogues, azacitidine and decitabine, as top candidates, both of which are indicated for myelodysplastic syndromes (Table 2). When we followed up on the computational prediction, we found preclinical and clinical evidence supporting the use of azacitidine as a possible treatment for multiple myeloma. One study suggested that azacitidine had less frequent episodes of grade 3 or 4 cytopenia and fewer infectious episodes when compared with decitabine [89]. The 5T33MM murine model of multiple myeloma showed improved survival when treated with azacitidine [90]. More recently, a Phase I study found that azacitidine in combination with lenalidomide and dexamethasone was well tolerated in relapsed/refractory MM patients who had previously received multiple lines of therapy treatment for their disease [91]. Responses to this combination are in the same range reported for a combination of pomalidomide + dexamethasone or carfizomib in comparable patient populations [91]. Hence, a combination of genomic data, evidence-based medicine and clinical judgment, led us to choose azacitidine as the top drug candidate for our patient's recurrent multiple myeloma.
Table 1.
Timeline of the patient events, treatments, and outcome status. MM status categories based on definitions from the International Myeloma Working Group Uniform Response Criteria.
| Date of relapse | Event | Therapy | MM status | Time |
|---|---|---|---|---|
| April 2009 – November 2009 | Initial diagnosis | VCD stopped → ASCT → Len maintenance | PR | 1 year |
| December 2010 | 1st relapse | VCD + Doxil VCD + Thal | PR CR | 4 months 16 months |
| VCD + Thal | CR | 16 months | ||
| August 2012 | 2nd relapse | DCEP × 1 | SD | 5 months |
| Cfz × 5 | PR | |||
| March 2013 | 3rd relapse | Pom + D | PD | <1 months |
| April 2013 | 4th relapse | SC Boost Mel/BCNU | PR | 1 months |
| May 2013 | 5th relapse | VDCEP + Adriamycin | PR | 1 months |
ASCT: Autologous stem cell transplant; BCNU: Carmustine; C: Cyclophosphamide; Cfz: Carfilzomib; CR: Complete remission; D: Dexamethasone; Doxil: Coated doxorubicin; E: Etoposide; Len: Lenalidomide; Mel: Melphalan; P: Cisplatin; PD: Progressive disease; Pom: Pomalidomide; PR: Partial remission; SC: Stem cells; SD: stable disease; Thal: Thalidomide; V: Bortezomib.
Data taken from [94].
Table 2.
Top compound recommendations based on computational matching and filtering.
| Drug | Score | Number of targets | Indication |
|---|---|---|---|
| Irinotecan | 1.00 | 8 | Metastatic colorectal cancer, small cell lung cancer |
| Azacitidine | 0.97 | 7 | Myelodysplastic syndromes |
| Daunorubicin | 0.73 | 10 | Acute nonlymphocytic leukemia and acute lymphocytic leukemia |
| Etoposide | 0.55 | 4 | Small cell lung cancer, lymphoma, nonlymphocytic leukemia and glioblastoma multiforme |
| Decitabine | 0.43 | 5 | Myelodysplastic syndromes |
| letrozole | 0.15 | 4 | Breast cancer |
| Sulindac | 0.11 | 4 | Bronchopulmonary dysplasia, colorectal neoplasms and melanoma |
| Carmustine | 0.10 | 4 | Glioma, non-Hodgkin lymphoma, glioblastoma |
We discussed our approach and findings with the patient in August 2013, and it was decided to start therapy with azacitidine (50 mg/m2) twice a week, and dexamethasone 40 mg weekly. Pomalidomide was added starting on day 15 at 2 mg × 21 doses. The patient was treated at Mount Sinai. He was given Azacytidine on a compassionate use after informed and written consent and as per Helsinki and Institutional guidelines. On September 9th, 2013, after the addition of pomalidomide, we saw a drop in his free lambda light chain from 5627 mg/l on September 13, 2013 to 1597 mg/l on September 26, 2013. This regimen, azacitidine twice a week with 2 mg of pomalidomide daily was continued until his last dose of azacitidine, which was received on October 7, 2013. That same day, the patient was admitted to the hospital for shortness of breath. The patient's overall condition worsened significantly and he eventually went into cardiac arrest and remained hypotensive and bradycardic following the episode. On October 9, 2013 the patient was released to hospice care and passed away later that day.
The patient's paraprotein levels dropped greater than 50% when treated with a combination of azacitidine, dexamethasone and pomalidomide, whereas he showed virtually no response when treated with a combination of pomalidomide and dexamethasone at an earlier relapse time point. Although the duration of response was short lived, this clinical case serves as an example of how genomic studies may guide novel drug selection for relapsed/refractory MM patients.
Lessons learned thus far
Although in its early stages of implementation, an integrative approach to the treatment of MM has already shed light on some general concepts that should be considered as we move forward in this rapidly developing field. The recent finding that MM tumors are highly heterogeneous has major implications for a targeted therapy to this disease, as well as many other types of cancer [85]. In order to determine the clinical benefit of specific, targeted drugs, it will be important to obtain serial bone marrow and blood samples from a patient throughout the course of their disease. These samples will allow us to track the number of clones present and the clonal evolution of the plasma cells over time, which helps guide the selection of appropriate treatment options in individual patients. As such, it is becoming increasingly apparent that physicians should start thinking about genomics early, perhaps even at initial diagnosis. This baseline will serve as a benchmark for tracking the evolution of the disease and allow for adjustments to the computational algorithms to be even more specific throughout disease progression.
A concern that gets raised when integrating molecular profiles from public data generated on cancer cell lines (e.g., Connectivity Map or LINCS data) with profiles of primary tumor samples is whether these profiles can be combined. While the drug response profiles from public data are generated from non-CD138 positive cell lines, we have a number of examples where the molecular matching provides predictive power across tissue, cell type or even species (e.g., applies to mouse cells) [53,58,92]. Thus, we had reason to believe the suggested drug predictions would be reasonable therapeutic options. The decision was made in consultation with the primary oncologist, tumor review board and patient. Furthermore, the patient did receive therapeutic benefit from the treatment.
In order to treat patients efficiently and effectively using a targeted therapy, a few items must be considered. First, in order to expedite the process of selecting a drug and treating a patient, we will consider choosing drugs that have already been FDA approved. At these early stages personalized oncology, drugs that are already being used to treat cancer provide a quicker route to the clinic because the approval pathway is already established in the medical community. However, one of the major challenges still ahead is overcoming regulatory barriers to treating patients with these off-label drugs. Second, due to the multifactorial nature of patient responses, we will have multiple levels of validation in place to optimize our drug choice. These measures will include incorporating clinical data and trial data into the choice of therapy as well as experimental validation steps, such as in vitro and xenograft systems to test hypotheses generated by the computational pipeline (Figure 2).
Given that treatments are being tailored toward individual patients, we must address the question of how much of what we learn from a single patient (n of 1) will apply to others. This question is critical for any program in personalized cancer therapy and one way to deal with this challenge is through processes that engage in feedback. Cancer patients are closely monitored and patient measurements collected over time, which provides a process for a “successive learning pipeline” for each individual. In other words, we improve the patient-specific network model by updating the information as it becomes available (e.g., with each new visit). Another feedback mechanism comes from the collected wisdom of the crowds. Multiple centers, including our own, have accumulated biobank repositories with genomic data, which can be used as an appropriate reference to place the individual patient into context. This feedback loop would allow us to use the information obtained from each patient to improve outcomes for future patients with a similar disease profiles.
How this approach can be applied at other institutions
In order for the advances in precision cancer medicine, specifically drug repurposing, to translate into clinical practice, collaboration between institutions will be critical. Given the high response rates with initial therapy and availability of FDA approved drugs in the newly diagnosed patients, we propose designing a pilot study in the model of the CoMMpass trial for patients with relapsed multiple myeloma. In this trial, participants will obtain bone marrow and blood samples from 100–1000 patients with relapsed disease following standard therapy with FDA approved drugs. The marrow will be used to isolate tumor cells for the purpose of obtaining genomic information using next-generation sequencing. These patient specific genomic profiles will then allow us to use our drug-repurposing platform to generate a list of personalized therapeutic options. We will then return this data in real time to participating physicians and determine whether a patient's potential treatment was altered as a result of the suggested therapy generated by the computational pipeline. By implementing such a study, we would be able to identify which subset of patients with relapsed MM would benefit most from having a drug selected based on the computational repurposing program. Additionally, we could determine the fraction of relapsed MM patients where next-generation sequencing data, and drug repurposing would make a difference to their treatment. Although we realize the group of participating physicians would be somewhat self-selecting, we believe that a multi-center trial would certainly be beneficial in providing a more heterogeneous patient and physician population. This example of a pilot study could help us gain valuable insight into the biology of relapsed disease and the practicality and utility of a drug repurposing approach in relapsed MM.
Conclusion
Cancer represents a special challenge for medicine because each tumor is shaped by the individual in which the rogue cells arise and grow. The pursuit to understand the underlying biology driving cancer has created a culture that adopts the latest technologies to support patient care and strives to deliver personalized medicine. Technological advances over the past few decades have created tools to collect unprecedented amounts of molecular, physiological, environmental, and clinical data. Parallel advances in bioinformatics and computational sciences have produced the infrastructure and algorithms to integrate these diverse data sources into realistic and powerful models that are proving useful to diagnose and treat patients. Although personalized medicine might see initial successes in oncology, the approach serves as a prototype for other medical disciplines and promises to provide improved care for all patients.
Future perspective
The next series of advances in personalized oncology will be driven by improved models that link together more data and increased therapeutic options that cover a broader array of tumors. The mathematical models currently used in personalized oncology incorporate a small slice of the potential data relevant for understanding human health and disease. Expanding the data collection efforts to incorporate multiple tissues, cell types and environmental factors, and even signals from wearable consumer health monitoring devices will be critical for creating comprehensive models that best capture the entire system and produce more accurate estimates of the key drivers underlying disease. These efforts will require new statistical models to merge data appropriately and test the relationships to offer estimates of uncertainty that can assist clinical decisions. Additionally, collecting these data across multiple time points, toward real-time monitoring, will provide better forecasts of tumor evolution and treatment response, allowing clinicians to make finer adjustments to alter the disease course in ways that could result in better long-term outcomes.
Enlarging the therapeutic options for oncology depends on knowing what components within a biological network to target and having the proper feedback and testing procedures to evaluate the consequence of a given intervention. Multiscale models offer a framework for determining key drivers of disease and performing in silico screening of perturbations to identify better therapeutics for a specific tumor. This type of screening would not be limited to compounds that have been approved for oncology, but could systematically consider the effect of any type of intervention such as off-label drugs (e.g., repurposing existing therapies), experimental compounds, natural products, diet or exercise. More radically, these mathematical models and analyses will suggest drivers for where no current compound exists, thus suggesting targets for novel therapeutics.
Monitoring devices and systems for sharing data offer exciting opportunities for expanding collaborations among the broader team of researchers, healthcare providers, hospitals and patients. Such collaborations will require substantial upgrades in computing power and storage, as well as, enhancements to systems for standardizing and transferring information so data can be processed and operated on in near real-time to make reports available in a timely manner. Concerns about data sharing with regards to privacy will need to be addressed [93], but the solutions will ultimately advance our understanding of cancer, improve our models of biological systems and result in better treatments for individuals.
Executive summary.
Background
High-throughput `omics' technologies possess tremendous power to characterize the molecular and cellular aberrations driving cancer.
A major challenge for systems medicine is how to extract relevant and actionable information from vast data that can be collected on individual patients and convert this information into clinically meaningful results.
Challenges & opportunities for personalized oncology
Tumor heterogeneity and evolution make cancer a moving molecular target.
Integrative modeling that captures multiple data sources provides a more precise lock on cancer.
Applying new technological developments to personalized oncology
New technologies are providing unprecedented information about the state of the biological system and how it changes over time or in response to treatment.
Oncology is at the forefront of adopting new technologies, which is resulting in tangible benefits for patients.
Impact of targeted therapeutics on personal cancer medicine
Targeted therapies provide precise manipulation of cellular states.
New technologies for better molecular diagnosis offer improved guidance for therapy selection (single or combination).
Computational & collaborative approach to personalized medicine
Multiscale network models provide a framework for organizing the information contained in large-scale data sets to determine key drivers of disease and identify optimal therapies for personalized medicine.
Multidisciplinary teams made up of clinical geneticists, clinical oncologists, bioinformaticians and genomic scientists are critical for providing care for personalized oncology.
Matching drug & disease profiles in personalized oncology
The main objective for personalized oncology is to identify the optimal molecular match between a patient's tumor profile and a drug response profile.
Optimal matches are achieved through systematic comparisons of the drug space with a comprehensive characterization of the patient's physiological and molecular state.
Case study in multiple myeloma
Computational drug matching identified azacitidine as the best therapy for the patient's status.
Paraprotein levels dropped greater than 50% when treated with a combination of azacitidine, dexamethasone and pomalidomide.
Lessons learned thus far
Initiating genomics testing early and tracking tumor evolution provides better predictive modeling and understanding of the disease course to optimize treatment direction.
The critical care and monitoring of cancer patients provides a “successive learning pipeline” in which the patient-specific network model can improve with each visit.
How this approach can be applied at other institutions
We propose a multi-institutional pilot study to share resources so the community can learn and refine best practices of personalized oncology.
Future perspective
The next series of advances in personalized oncology will be driven by improved mathematical models that link together more data – from both physiological and environmental sources – with better predictive algorithms to classify tumor types and identify drug matches.
Mobile devices and real-time monitoring provide promising opportunities for better detection and follow-up.
Acknowledgements
We would like to thank M Rosi-Schumacher for her help on the “Applying new technological developments to personalized oncology” section.
The preparation of this manuscript was funded in part by grants to JT Dudley from NIH/NIDDK R01DK098242, PhRMA Foundation Research Starter Grant and NCI U54CA189201. JT Dudley has served as a consultant to Janssen Pharmaceuticals and holds equity in NuMedii, Inc., Ayasdi Inc. and LifeMap Inc.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest; •• of considerable interest
- 1.Hood L, Friend SH. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat. Rev. Clin. Oncol. 2011;8(3):184–187. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- 2.Schadt E. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461(7261):218–223. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- 3.Phan JH, Quo CF, Cheng C, Wang MD. Multiscale integration of -omic, imaging, and clinical data in biomedical informatics. IEEE Rev. Biomed. Eng. 2012;5:74–87. doi: 10.1109/RBME.2012.2212427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, Sova P, Xu Q, et al. Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate Cell regulation. PLoS Biol. 2012;10(4):e1001301. doi: 10.1371/journal.pbio.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Large-scale data integration of six different data types into a probabilistic causal multiscale network.
- 5.Chen R, Mias GI, Li-Pook-Than J, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Comprehensive monitoring of multiple `omics' data in a single individual over a 14-month period, capturing system changes during healthy and diseased states.
- 6.Ashley E, Butte A, Wheeler M, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375(9725):1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The first integrated analysis of a human genome along with clinical guidelines and actions, providing a framework for personal genomics and medicine.
- 7.Hamburg M, Collins F. The path to personalized medicine. N. Engl. J. Med. 2010;363(4):301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 8.West M, Ginsburg GS, Huang AT, Nevins JR. Embracing the complexity of genomic data for personalized medicine. Genome Res. 2006;16(5):559–566. doi: 10.1101/gr.3851306. [DOI] [PubMed] [Google Scholar]
- 9.Zhang B, Gaiteri C, Bodea L-G, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153(3):707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang IM, Zhang B, Yang X, et al. Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Mol. Syst. Biol. 2012;8:594. doi: 10.1038/msb.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kidd BA, Peters L, Schadt EE, Dudley JT. Unifying immunology with informatics and multiscale biology. Nat. Immunol. 2014;15(2):118–127. doi: 10.1038/ni.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilal E, Dutkowski J, Guinney J, et al. Improving breast cancer survival analysis through competition-based multidimensional modeling. PLoS Comput. Biol. 2013;9(5):e1003047. doi: 10.1371/journal.pcbi.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence M, Stojanov P, Mermel C, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meacham C, Morrison S. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schadt E, Björkegren J. Network-Enabled Wisdom in biology, medicine, and health care. Sci. Transl. Med. 2012;4(115):115rv111. doi: 10.1126/scitranslmed.3002132. [DOI] [PubMed] [Google Scholar]
- 16.Califano A, Butte A, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat. Genet. 2012;44(8):841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clayton TA, Lindon JC, Cloarec O, et al. Pharmacometabonomic phenotyping and personalized drug treatment. Nature. 2006;440(7087):1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 18.Mansour J, Schwarz R. Molecular mechanisms for individualized cancer care. J. Am. Coll. Surg. 2008;207(2):250–258. doi: 10.1016/j.jamcollsurg.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Van't Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452(7187):564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 20.Wu D, Sherwood A, Fromm J, et al. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci. Transl. Med. 2012;4(134):134ra163. doi: 10.1126/scitranslmed.3003656. [DOI] [PubMed] [Google Scholar]
- 21.Robins H, Srivastava S, Campregher P, et al. Overlap and effective size of the human CD8+ T Cell receptor repertoire. Sci. Transl. Med. 2010;2(47):47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd S, Marshall E, Merker J, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci. Transl. Med. 2009;1(12):12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortazavi A, Williams B, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Gerstein M, Snyder M. RNA-Seq. a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bendall S, Simonds E, Qiu P, et al. Single-Cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roychowdhury S, Iyer M, Robinson D, et al. Personalized oncology through integrative high-throughput sequencing. a pilot study. Sci. Transl. Med. 2011;3(111):111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The authors established a clinical protocol and feasibility for an integrative sequencing approach to personalized oncology.
- 27.Merker J, Valouev A, Gotlib J. Next-generation sequencing in hematologic malignancies. what will be the dividends? Ther. Adv. Hematol. 2012;3(6):333–339. doi: 10.1177/2040620712458948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon R, Roychowdhury S. Implementing personalized cancer genomics in clinical trials. Nat. Rev. Drug Discov. 2013;12(5):358–369. doi: 10.1038/nrd3979. [DOI] [PubMed] [Google Scholar]
- 31.Walter MJ, Shen D, Ding L, et al. Clonal architecture of secondary acute myeloid leukemia. N. Engl. J. Med. 2012;366(12):1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan G, Kaiser M. How to use new biology to guide therapy in multiple myeloma. Hematology Am. Soc. Hematol. Educ. Program. 2012;2012(1):342–349. doi: 10.1182/asheducation-2012.1.342. [DOI] [PubMed] [Google Scholar]
- 34.Hagemann I, Govindan R, Javidan-Nejad C, Pfeifer J, Cottrell C. Stabilization of disease after targeted therapy in a thymic carcinoma with KIT mutation detected by clinical next-generation sequencing. J. Thorac. Oncol. 2014;9(2):e12–e16. doi: 10.1097/JTO.0b013e3182a7d22e. [DOI] [PubMed] [Google Scholar]
- 35.Irish J, Czerwinski D, Nolan G, Levy R. Altered B-Cell receptor signaling kinetics distinguish human follicular lymphoma B Cells from tumor-infiltrating nonmalignant B Cells. Blood. 2006;108(9):3135–3142. doi: 10.1182/blood-2006-02-003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bendall S, Nolan G. From single cells to deep phenotypes in cancer. Nat. Biotechnol. 2012;30(7):639–647. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- 37.Giesen C, Wang HA, Schapiro D, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods. 2014;11(4):417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 38.Angelo M, Bendall SC, Finck R, et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 2014;20(4):436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y-M, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3(6):636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med. 2009;361(10):958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 41.Mok T, Wu Y-L, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 42.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405). An open label, randomised Phase 3 trial. Lancet Oncol. 2010;11(2):121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 43.Hyman D, Zhou Q, Iasonos A, et al. Improved survival for BRCA2-associated serous ovarian cancer compared with both BRCA-negative and BRCA1-associated serous ovarian cancer. Cancer. 2012;118(15):3703–3709. doi: 10.1002/cncr.26655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellis M, Ding L, Shen D, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasche B, Absher D. Whole-genome sequencing. a step closer to personalized medicine. JAMA. 2011;305(15):1596–1597. doi: 10.1001/jama.2011.484. [DOI] [PubMed] [Google Scholar]
- 46.Bolton K, Chenevix-Trench G, Goh C, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307(4):382–390. doi: 10.1001/jama.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faham M, Zheng J, Moorhead M, et al. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2012;120(26):5173–5180. doi: 10.1182/blood-2012-07-444042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng WK, Armstrong R, Arai S, Desmarais C, Hoppe R, Kim YH. Minimal residual disease monitoring with high-throughput sequencing of T cell receptors in cutaneous T cell lymphoma. Sci. Transl. Med. 2013;5(214):214ra171. doi: 10.1126/scitranslmed.3007420. [DOI] [PubMed] [Google Scholar]
- 49.Hirsch H, Iliopoulos D, Joshi A, et al. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17(4):348–361. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhodes D, Yu J, Shanker K, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl Acad. Sci. USA. 2004;101(25):9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waldman Y, Geiger T, Ruppin E. A genome-wide systematic analysis reveals different and predictive proliferation expression signatures of cancerous vs. non-cancerous cells. PLoS Genet. 2013;9(9):e1003806. doi: 10.1371/journal.pgen.1003806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin G, Fu C, Zhao H, Cui K, Chang J, Wong S. A novel method of transcriptional response analysis to facilitate drug repositioning for cancer therapy. Cancer Res. 2012;72(1):33–44. doi: 10.1158/0008-5472.CAN-11-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The authors developed a Bayesian network-based screening tool to identify potential off-target effects that might guide drug repositioning efforts in cancer.
- 53.Jahchan N, Dudley J, Mazur P, et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3(12):1364–1377. doi: 10.1158/2159-8290.CD-13-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meldrum C, Doyle M, Tothill R. Next-generation sequencing for cancer diagnostics. a practical perspective. Clin. Biochem. Rev. 2011;32(4):177–195. [PMC free article] [PubMed] [Google Scholar]
- 55.Soverini S, De Benedittis C, Machova Polakova K, et al. Unraveling the complexity of tyrosine kinase inhibitor-resistant populations by ultra-deep sequencing of the BCR-ABL kinase domain. Blood. 2013;122(9):1634–1648. doi: 10.1182/blood-2013-03-487728. [DOI] [PubMed] [Google Scholar]
- 56.Martignetti J, Camacho-Vanegas O, Priedigkeit N, et al. Personalized ovarian cancer disease surveillance and detection of candidate therapeutic drug target in circulating tumor DNA. Neoplasia. 2014;16(1):97–103. doi: 10.1593/neo.131900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dawson S-J, Tsui D, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 58.Dudley JT, Sirota M, Shenoy M, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl. Med. 2011;3(96):96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The authors developed a molecular matching algorithm to connect drug response signatures with disease state signatures, providing a tool for drug selection based on a patient's transcriptional profile.
- 59.National Cancer Institute Targetted Cancer Therapies. www.cancer.gov/cancertopics/factsheet/Therapy/targeted.
- 60.Kobayashi S, Boggon T, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl.J. Med. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 61.Mahadevan D, Cooke L, Riley C, et al. A novel tyrosine kinase switch is a mechanism of imatinib resistance in gastrointestinal stromal tumors. Oncogene. 2007;26(27):3909–3919. doi: 10.1038/sj.onc.1210173. [DOI] [PubMed] [Google Scholar]
- 62.Gorre M, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 63.Bozic I, Reiter JG, Allen B, et al. Evolutionary dynamics of cancer in response to targeted combination therapy. Elife. 2013;2:e00747. doi: 10.7554/eLife.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The authors developed a mathematical model to evaluate the effectiveness of combination therapy in various cancers.
- 64.Chao MP, Alizadeh AA, Tang C, et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell. 2010;142(5):699–713. doi: 10.1016/j.cell.2010.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitsui J, Nishikawa H, Muraoka D, et al. wo distinct mechanisms of augmented antitumor activity by modulation of immunostimulatory/inhibitory signals. Clin. Cancer Res. 2010;16(10):2781–2791. doi: 10.1158/1078-0432.CCR-09-3243. [DOI] [PubMed] [Google Scholar]
- 66.Zhu J, Lum P, Lamb J, et al. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet. Genome Res. 2004;105(2–4):363–374. doi: 10.1159/000078209. [DOI] [PubMed] [Google Scholar]
- 67.Dobrin R, Zhu J, Molony C, et al. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol. 2009;10(5):R55. doi: 10.1186/gb-2009-10-5-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sieberts S, Schadt E. Moving toward a system genetics view of disease. Mamm. Genome. 2007;18(6–7):389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schadt E, Lamb J, Yang X, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 2005;37(7):710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu J, Zhang B, Smith E, et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet. 2008;40(7):854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nat. Rev. Drug Discov. 2009;8(4):286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]; •• The authors outlined a roadmap for using multiscale networks for drug discovery.
- 72.Lamb J, Crawford ED, Peck D, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 73.Yang W, Soares J, Greninger P, et al. Genomics of drug sensitivity in cancer (GDSC). A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:d955–d961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Basu A, Bodycombe NE, Cheah JH, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154(5):1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Welter D, Macarthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rustici G, Kolesnikov N, Brandizi M, et al. ArrayExpress update—trends in database growth and links to data analysis tools. Nucleic Acids Res. 2013;41:D987–D990. doi: 10.1093/nar/gks1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus. NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 81.Kumar SK, Dispenzieri A, Lacy MQ, et al. Continued improvement in survival in multiple myeloma. changes in early mortality and outcomes in older patients. Leukemia. 2013;28(5):1122–1128. doi: 10.1038/leu.2013.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Munshi NC, Anderson KC. New strategies in the treatment of multiple myeloma. Clin. Cancer Res. 2013;19(13):3337–3344. doi: 10.1158/1078-0432.CCR-12-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mccarthy PL, Hahn T. Strategies for induction, autologous hematopoietic stem cell transplantation, consolidation, and maintenance for transplantation-eligible multiple myeloma patients. Hematology Am. Soc. Hematol. Educ. Program. 2013;2013(1):496–503. doi: 10.1182/asheducation-2013.1.496. [DOI] [PubMed] [Google Scholar]
- 84.Richardson PG, Barlogie B, Berenson J, et al. A Phase 2 study of bortezomib in relapsed, refractory myeloma. N. Engl.J. Med. 2003;348(26):2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 85.Lohr J, Stojanov P, Carter S, et al. Widespread genetic heterogeneity in multiple myeloma. Implications for targeted therapy. Cancer Cell. 2014;25(1):91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Anderson KC. Oncogenomics to target myeloma in the bone marrow microenvironment. Clin. Cancer Res. 2011;17(6):1225–1233. doi: 10.1158/1078-0432.CCR-10-3366. [DOI] [PubMed] [Google Scholar]
- 87.Relating Clinical Outcomes in Multiple Myeloma to Personal Assessment of Genetic Profile (CoMMpass) http://clinicaltrials.gov/ct2/show/NCT01454297.
- 88.Sirota M, Dudley JT, Kim J, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci. Transl. Med. 2011;3(96):96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee YG, Kim I, Yoon SS, et al. Comparative analysis between azacitidine and decitabine for the treatment of myelodysplastic syndromes. Br. J. Haematol. 2013;161(3):339–347. doi: 10.1111/bjh.12256. [DOI] [PubMed] [Google Scholar]
- 90.Khong T, Sharkey J, Spencer A. The effect of azacitidine on interleukin-6 signaling and nuclear factor-kappaB activation and its in vitro and in vivo activity against multiple myeloma. Haematologica. 2008;93(6):860–869. doi: 10.3324/haematol.12261. [DOI] [PubMed] [Google Scholar]
- 91.Reu FJ, Khan SN, Mahfouz R, et al. Azacitidine with lenalidomide and low-dose dexamethasone (Rd) in relapsed or refractory myeloma (RRMM). Phase I results. Presented at: 14th International Myeloma Workshop; Kyoto, Japan. Apr 3–7, 2013. [Google Scholar]
- 92.Dudley JT, Tibshirani R, Deshpande T, Butte AJ. Disease signatures are robust across tissues and experiments. Mol. Syst. Biol. 2009;5:307. doi: 10.1038/msb.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schadt EE. The changing privacy landscape in the era of big data. Mol. Syst. Biol. 2012;8:612. doi: 10.1038/msb.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Durie BG, Harousseau JL, Miguel JS, et al. International uniform response criteria for multiple myeloma. Leukemia. 2006;20(9):1467–1473. doi: 10.1038/sj.leu.2404284. [DOI] [PubMed] [Google Scholar]