Abstract
We describe a sensitive, robust, high-throughput method for quantifying the formation of micronuclei, markers of genome instability, in mouse erythrocytes. Micronuclei are whole chromosomes or chromosome segments that have been separated from the nucleus. Other methods of detection rely on labour-intensive, microscopy-based techniques. Here, we describe a 2-d, 96-well plate-based flow cytometric method of micronucleus scoring that is simple enough for a research technician experienced in flow cytometry to perform. The assay detects low levels of genome instability that cannot be readily identified by classic phenotyping, using 25 μl of blood. By using this assay, we have screened >10,000 blood samples and discovered novel genes that contribute to vertebrate genome maintenance, as well as novel disease models and mechanisms of genome instability disorders. We discuss experimental design considerations, including statistical power calculation, we provide troubleshooting tips, and we discuss factors that contribute to a false-positive increase in the number of micronucleated red blood cells and to experimental variability.
Keywords: Genome instability, DNA damage, micronucleus, mouse, in vivo, assay
INTRODUCTION
Micronuclei are extra nuclear segments of chromatin that can arise as a result of DNA double-strand breaks or mitotic spindle apparatus dysfunction. Micronuclei form infrequently during mitosis of healthy cells, and they may either be acentric chromosomal fragments or whole chromosomes that have failed to be incorporated into the nucleus at the completion of mitosis1 (Fig. 1a, 1b). The use of micronuclei as a means to quantify cytogenetic damage was originally proposed by Evans et al.2. Cells derived from people with hereditary genome-instability disorders, such as ataxia telangiectasia and Fanconi anemia, have a high frequency of micronucleus formation, and micronucleus assays can be used to aid the diagnosis of these disorders 3,4. In addition, many genome instability disorders predispose people to cancer, and genome instability is a hallmark of spontaneously arising cancers5.
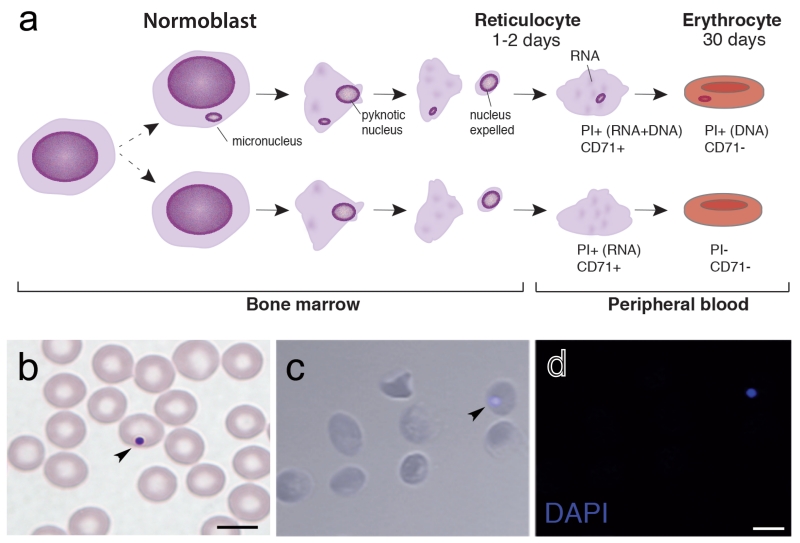
Figure 1. Micronucleus induction during erythropoiesis.
a) Erythropoiesis is characterized by maturation of the multipotent hematopoietic stem cell (not shown) to a smaller, mitotically inactive normoblast. Within the normoblast, chromatin begins to condense, and eventually the pyknotic nucleus is expelled, yielding a cell that is known as a reticulocyte (RET). The RETs migrate from the bone marrow to the peripheral circulation, where, 1 or 2d later, it matures into an NCE, which is characterized by degradation of the reticular RNA mesh and loss of transferrin receptor (CD71) expression. Micronuclei can form spontaneously during the mitotic stages of erythropoiesis, for example, within the multipotent hematopoietic stem cell or differentiated precursor cells (normoblasts); however, in normal cells this is an extremely rare event. Although the nucleus is expelled during red blood cell maturation, the micronucleus is retained. If cells are genomically unstable or mice have been treated with a genotoxin, the frequency of micronucleus formation increases. Micronuclei are PI positive, and they can be differentially identified in NCEs or RETs by co-staining with CD71. b) May-Grünwald-Giemsa staining of fresh blood sample from a wild-type mouse shows a Giemsa positive DNA fragment (micronucleus) in an erythrocyte. (c,d) DAPI-stained micronucleated erythrocyte from a blood sample prepared according to the methods indicated in this protocol (heparin anticoagulation followed by fixation in −80°C methanol). The methanol-fixed blood was cytospun onto a slide and DAPI-stained (c shows staining with overlay phase-contrast/DAPI, whereas d shows staining with DAPI alone). Scale bars, 5 μm. Arrows indicate micronuclei.
Recent genomic analyses have identified a novel form of genome instability in cancer cells, which is a phenomenon termed ‘chromothripsis’6. Chromothripsis involves up to thousands of clustered chromosomal rearrangements, thought to occur as a single event and confined to genomic regions in one or a few chromosomes. This phenomenon has sparked further interest in micronuclei, which have been shown to undergo highly error-prone DNA replication and DNA breakage7,8. It is thought that DNA fragments could be reassembled in the wrong order by the process of nonhomologous end joining, leading to massive chromosomal rearrangements in mitosis. Micronuclei have been shown to re-incorporate into the same nuclear compartment as the main chromosomes during mitosis7. This raises the possibility that genomic re-integration of a genomically rearranged micronucleus might occur in cancers, and it could be selected for if the rearrangement causes an oncogenic event such as loss or disruption of a tumour suppressor gene7,9,10.
Elevated levels of micronuclei have been associated with tumor predisposition in genetically modified mice11-13, and micronucleus assays have been used to assess the carcinogenic potential of compounds in toxicity studies, as well as to biomonitor mice and humans who have been exposed to mutagens such as radiation14-17.
Development and applications of the method
Fliedner et al. first proposed the idea of scoring micronuclei as a biological dosimeter of radiation exposure in humans18. Measuring numbers of micronuclei in mammalian red blood cells is relatively straightforward because normoblasts (erythrocyte precursor cells that reside in the bone marrow) expel their nuclei during red blood cell development, leaving behind any micronuclei that have formed (Fig. 1). In the blood, micronucleated erythrocytes were first identified by microscopy using a basophilic stain, and they were called ‘Howell-Jolly bodies’ after the two scientists who first discovered them19-22. Although micronucleated red blood cells are extremely rare in humans, partly owing to their efficient clearance by the spleen, mice are less efficient at clearing micronucleated red blood cells from the peripheral circulation, and thus they are more readily detected in this model organism23.
Hutter and Stohr24 were the first to describe a flow cytometry–based method of scoring micronucleated erythrocytes from bone marrow, which they compared with standard microscopy-based scoring. Their method involved the fixation of samples followed by the addition of the fluorescent DNA stain DAPI and the protein fluorochrome sulforhodamine 101 to discriminate between normal anuclear erythrocytes and micronucleated erythrocytes. Their method was later refined and applied to the analysis of peripheral erythrocytes using a variety of DNA stains, including DAPI, Hoechst and propidium iodide (PI)25,26. In mice, 1-3% of red blood cells are reticulocytes (RETs), immature erythrocytes that are characterized by their cytoplasmic reticular mesh of RNA and their large size, whereas the remaining 93-94% are mature normochromatic erythrocytes (NCEs), which are characterized by their ‘donut’ shape and the absence of a nucleus (Fig. 1a). In 1992, Grawé et al. described a method that distinguished between micronucleated RETs, which are thiazole orange positive, and NCEs using a dual-laser flow cytometer configuration25,27. The distinction between RETs and NCEs is important for assessing the effects of genotoxic challenge, because micronucleated RETs are indicative of recent damage, whereas micronucleated NCEs are indicative of damage caused >72 h earlier. RETs can be distinguished from NCEs by studying the expression of the mouse transferrin receptor (CD71), which is present on the surface of most dividing hematopoietic progenitor cells28. In 1996, Dertinger et al. refined the method using a fluorescein-conjugated monoclonal antibody to the CD71 receptor, which labels the RET population29. The use of PI to stain DNA (after removal of RNA from RETs by incubation with RNase) simplified the assay by enabling the use of a single-laser flow cytometer for enumeration of cells (patent nos. US6100038A and US5858667A)30.
In 2003, Shima et al. used the assay developed by Dertinger et al. to genetically screen mice for novel genes required for vertebrate genome maintenance31,32. Here we have scaled down Dertinger’s method to perform the assay in 96-well plates (Fig. 2), which we have used to assess genome stability in >10,000 mice as part of a large-scale reverse genetic screen in association with the Mouse Genetics Project at the Wellcome Trust Sanger Institute33. The program screens genetically modified mice as part of an international effort to associate gene function with developmental abnormalities and disease. By combining the assay with high-throughput phenotyping of mice, we have identified several novel mouse models of rare genome-instability disorders, novel genome-maintenance-related genes, including Cenpj and Mysm1, and cancer predisposition-related genes including Slx411,34-38. If such a screen were to be performed for the human population, albeit with a slightly modified method that enriches for RETs before assay processing owing to the efficient clearance of micronucleated erythrocytes by the human spleen39, the assay could aid the diagnosis of novel genome instability syndromes or identify individuals who may be predisposed to cancer owing to elevated basal levels of genome instability. Furthermore, by incorporating a challenge with ionizing radiation into the mouse phenotyping pipeline, we have identified radiation-sensitive mutants, which exhibit increased levels of micronucleated RETs after irradiation but not before irradiation. Here we provide additional details on data analysis, troubleshooting and potential sources of data variability and false-positive increases in numbers of micronucleated red blood cells.
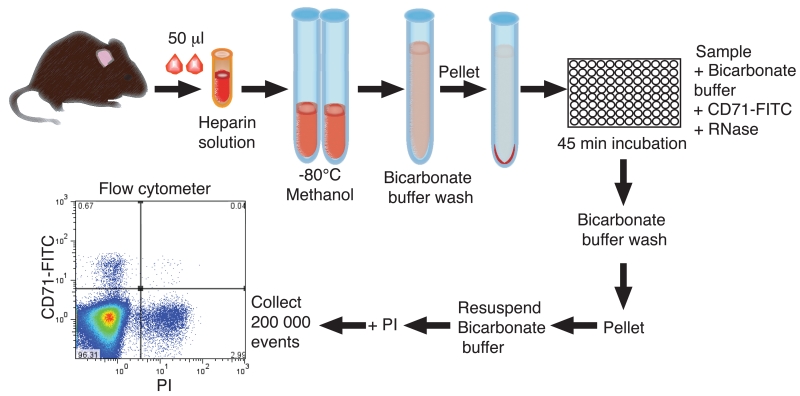
Figure 2. Workflow of the micronucleus assay.
Mouse blood samples are collected into liquid heparin solution and fixed in cold methanol. Samples are washed in bicarbonate buffer and cells are pelleted and resuspended in buffer, before incubation in bicarbonate buffer containing FITC-conjugated anti-transferrin receptor (CD71) antibody and RNase. Samples are washed out of the incubation solution with bicarbonate buffer, and they are resuspended in buffer before adding PI and collecting 200,000 events by flow cytometry.
Our high-throughput flow cytometric micronucleus assay is relatively quick, inexpensive, and simple to perform. It uses equipment that is available in most research organizations, and it is reliable if performed according to the instructions provided herein. It therefore makes an ideal test for the detection of genome instability in vivo that is minimally invasive and does not require the death of the animal, meaning that it can easily be incorporated into a phenotyping screen. We provide step by-step technical advice, as well as a troubleshooting guide with tips gleaned from our use of the method over the past few years.
Limitations of the method
The high-throughput micronucleus assay gives an indication of genome stability as measured by the frequency of micronucleated erythrocytes in a population. The assay does not provide information on the structure of the micronucleus, and therefore it does not distinguish between the different mechanisms of action of inducers, such as clastogens or aneugens, nor does the assay inform on whether a cell has a single micronucleus or multiple micronuclei, as could be determined by conventional microscopy. Therefore, further experiments are needed to determine the structure of the micronucleus and mechanism of its formation. For example, we have previously shown that fibroblasts derived from Cenpj-deficient mice (1.5-fold increase in the percentage of micronucleated NCEs) are prone to tetraploidy, as shown by multiplex metaphase fluorescent in situ hybridization and indicated by cell cycle analysis35. This suggested that micronuclei were likely to be the result of chromosome mis-segregation; fluorescence microscopy revealed that fibroblasts had an increased frequency of supernumerary centrosomes and multi polar spindles, which have been shown to cause aneuploidy35.
Alternatively, micronuclei can also be detected in vitro using the cytokinesis block micronucleus cytome assay40. In addition to quantifying micronuclei, the cytokinesis block micronucleus cytome assay also detects cytostasis, cytotoxicity and provides information on the structure of the micronucleus. This added information is useful for determining the likely mechanism of micronucleus formation. For example, nucleoplasmic bridges are a biomarker of misrepair and/or telomere end fusions, whereas nuclear buds are a biomarker of elimination of amplified DNA and/or DNA repair complexes40. However, the cytokinesis block micronucleus cytome assay is time-consuming and it relies on a complicated scoring system. In summary, the high-throughput micronucleus assay is a fast and sensitive method for the detection of micronuclei, but further experiments are needed to ascertain the mechanism of its formation.
Through screening for micronucleus induction in parallel with a large-scale, unbiased mouse phenotyping pipeline, we have identified several factors that may influence the detection of true, micronucleated red blood cells with the assay. Regenerative anemia causes increased production of red blood cells (reticulocytosis) and activation of extramedullary hematopoiesis41. In extreme cases of reticulocytosis, such as that observed in 16-20-week-old C57BL/6J Apcmin/+ mice (Suppl. Fig. 1a-b), the main nucleus may not be expelled during erythropoiesis, resulting in erythroblastosis, which is the presence of nucleated red blood cells in the peripheral circulation. The assay does not distinguish between micronucleated and nucleated red blood cells, and thus this causes a false-positive increase in the percentage of micronucleated NCEs (Suppl. Fig. 1a-b). With the high-throughput micronucleus assay, there is a positive correlation between the percentage of RETs and the percentage of micronucleated NCEs (Suppl. Fig. 1b). The use of an anti-CD71 transferrin receptor antibody enables the researcher to quantify RETs to ensure that numbers are within normal limits (e.g. for healthy C57BL/6 mice, RETs should constitute 1-2% of the total red blood cell population; Suppl. Fig. 1c). Furthermore, the assay relies on the distinction of blood cells by size and granularity, and therefore any changes to these parameters could result in a false-positive increase in the number of micronucleated red blood cells. As a rule of thumb, if other changes in other hematopoietic parameters are observed, such as but not limited to red blood cell distribution width, we would confirm the micronucleated cell type using microscopy. A simple method for confirming a change in micronucleated NCEs would be to perform cell counts on fresh blood smears stained with May-Grünwald-Giemsa (Fig. 1b) or on methanol-fixed blood samples co-stained with DAPI on slides (Fig. 1c, 1d). For the quantification of micronuclei from slides stained with May-Grünwald-Giemsa, we recommend counting >6000 red blood cells or cells from 15 fields of view (40× magnification) per sample, for at least 3 mice per test group. For a wild-type mouse, we would expect to see 1 or 2 micronucleated cells per 1000 erythrocytes. For a positive-control mouse, we would expect to see at least 2-4 micronucleated erythrocytes per 1000 erythrocytes.
Experimental design
Test and control mice should be matched by sex, age and genetic background, since all of these factors influence micronucleus induction in mice (Box 1 and Fig. 3a,b). The assay is highly sensitive and reproducible if screening is performed with mice of the same sex that are bred on a pure genetic background (Fig. 3c). It is possible to detect minimal changes with statistical significance using the appropriate sample number (Box 1 and Fig. 3d). The simplest way to generate positive control animals for the assay is to expose wild-type mice to genotoxic agents, including treatment with 1 mg/kg mitomycin C (MMC) or 0.75 gray (Gy) irradiation (Box 2; Fig. 4a,b, respectively). Alternatively, several mouse models with elevated micronucleus levels, such as the Slx4-null mouse (Fig. 4c-e), have previously been characterized, and mice, or their blood samples, could be requested from the corresponding author of the study (see review by Balmus & McIntyre42). Notably, the experimenter should ensure that the experiment is performed in accordance with relevant animal research regulations and ARRIVE guidelines43.
Box 1. Sources of variation in micronucleus frequency.
Below we highlight significant sources of variation in micronucleus frequency that should be taken into consideration during experimental design.
Sex
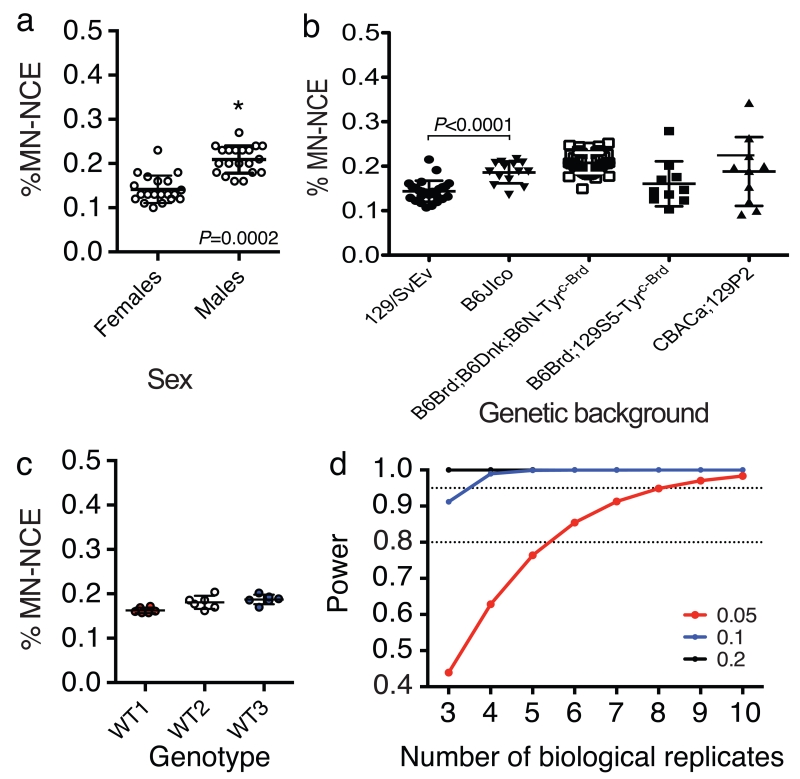
For all mouse strains that we have tested, the frequency of micronucleated NCEs for males was significantly increased when compared with female mice. For example, female C57BL/6N mice have a mean of 0.14% micronucleated NCEs, whereas the mean for male mice is 0.21% (P=0.0002, mixed model analysis where sex is a fixed effect and assay date is a random effect; Fig. 3a).
Genetic background
The assay is sensitive enough to detect significant differences between inbred strains of mice (Fig. 3b). Compared with 129SvEv mice (n = 27), C57BL/6JIco mice (n = 15) have a higher basal level of %micronucleated NCEs (0.186% compared with 0.143%; P=0.002; mixed model analysis for which core strain is a fixed effect and assay date a random effect). Relative to the variance of 129SvEv (i.e. variance of 1), the variance of C57BL/6JIco is similar (relative variance=1.18), whereas the variance of mixed strains is more than three times greater (relative variance of B6Brd;129S5-Tyrc-Brd = 3.29, CBACa;129S5 = 3.63; P = 0.0065 mixed model analysis, Fig. 3b), demonstrating that data variability is greater for mice with mixed genetic backgrounds than for those with pure genetic backgrounds. This finding should be considered when determining statistical power. It should be noted that, when comparing mixed genetic backgrounds comprising substrains of laboratory mice the variance is low (Fig. 3b). For example, if we compare the variance of a mixed strain of mice, where all strains are historically derived from BL/6 mice, then the variance is similar to that of a pure inbred strain (variance of B6Brd;B6Dnk;B6N-Tyrc-Brd =0.92 relative to the pure, inbred strain 129SvEv). In summary, assay sensitivity is highest when mice are of the same sex and are bred on a pure genetic background (Fig. 3c).
Age
DNA damage is thought to accumulate with age and therefore older mice may have higher frequencies of spontaneous micronucleus formation than young mice45. We recommend that mice are age-matched where possible.
Assay date
By performing the micronucleus assay as part of a large-scale phenotyping pipeline, we have found that, as with other phenotyping variables, assay date is a significant source of variation46. The variation with date arises from multiple factors including researcher, reagent lot, cage, and mother and litter size. This temporal variation has a significant impact on data analysis, as readings taken on the same day are more similar than readings on different days. The simplest approach to manage this source of variation is to process all samples at the same time, which will enable the use of standard statistical methods such as the Student’s t-test. When it is not possible to collect samples and process them on the same day, such as with a large-scale phenotyping pipeline, micronucleus data can be analyzed with a mixed model approach, in which genotype is treated as a fixed effect and assay date is treated as a random effect. This separates the genotype effect from the temporal variation by assuming that temporal variation is normally distributed with a mean of zero and with a defined variance. We refer the reader to Karp et al. 2012 for further statistical design considerations with regard to assay date46.
Statistical power
Data from 200 blood samples obtained from male C57BL/6N wild-type mice, which were screened over seven assay dates, were used to determine the appropriate statistical method and associated statistical power. Across the seven datasets, the percentage of micronucleated NCE data were normally distributed, as assessed by graphical inspection (normal Q-Q plot) and Shapiro-Wilk normality test (data not shown). These data characteristics indicate that a standard Student’s t-test is appropriate to detect changes in the mean percentage of micronucleated NCEs between the control group and the test group, subject to the experimental design implemented. The average s.d. was estimated to be 0.026%.
By using the variance estimate, we assessed the power to detect changes of ±0.05%, 0.1% and 0.2% micronucleated NCEs for different samples sizes when using a two-tailed, two-sampled Student’s t-test47. For hypothesis testing of age-, sex- and genetic background-matched test and control mice, we recommend a target power of >0.8. For confirmation of genomic instability mutants, we suggest a power ≥ 0.95 (Fig. 3d). The power calculations showed that the assay is very sensitive and that n = 3 mice per genotype is enough to test for differences >0.1% micronucleated NCEs (~1.5-fold change for C57BL/6N mice) in a confirmation experiment, and that n = 6 mice per genotype are required to test differences as low as 0.05% micronucleated NCEs (~1.25-fold change for C57BL/6N mice) in a screening experiment. The variance of mixed genetic backgrounds is greater than for pure genetic backgrounds and this will reduce the statistical power (Fig. 3b).
Figure 3. Effect of sex and genetic background on micronucleus induction and determination of statistical power.
(a) Wild-type female mice have a significantly decreased percentage of micronucleated normochromatic erythrocytes (%MN-NCEs; mean ± s.d.: 0.141 ± 0.031; n = 20) relative to wild-type male mice (mean ± s.d.: 0.209 ± 0.03; n = 20; P = 0.0002, mixed model analysis with assay date as a random effect and sex as a fixed effect). (b) Graph shows %MN-NCEs for 16-week-old male mice bred on different genetic backgrounds; the assay is sensitive enough to detect significant differences between strains. Data represent mean (bar) and s.d. (error bar): 0.144 ± 0.024, n = 26 129SvEv versus 0.186 ± 0.025 n = 14 BL6/JIco, P=0.002, mixed model analysis with assay date as a random effect and core strain as a fixed effect; 0.208 ± 0.023, n = 46 B6Brd;B6Dnk;B6N-Tyrc-Brd; 0.160 ± 0.051, n=10 B6Brd;129S5-Tyrc-Brd; 0.188 ± 0.077, n = 10 CBACa;129P2. Note the increased variance for mixed genetic backgrounds e.g. 129P2 × CBACa, compared with mixed genetic substrains, e.g. B6Brd;B6Dnk;B6N-Tyrc-Brd. (c) Graph shows data for five technical replicates for blood samples obtained from three C57BL/6 wild-type male mice. Data demonstrate that there is good reproducibility when using mice of the same sex and genetic background. Data show mean (bar) and error bars represent s.d. (d) Power analysis shows the estimated power as the number of biological replicates increases. The relationship was studied for three effect sizes changes (absolute difference of 0.05, 0.1 or 0.2% MN-NCEs). The dotted lines highlight the target power of 0.8 for a hypothesis-generating experiment or 0.95 for a hypothesis-testing experiment.
Box 2. DNA damage response challenge.
TIMING 3d
DNA damage response (DDR) defects can be revealed by treating mice that show normal spontaneous micronucleus frequencies with genotoxic agents. Upon treatment, micronuclei form in mitotically dividing erythroid progenitor cells, and they can be detected 24-72 h later in peripheral RETs (Fig. 1 and Fig. 4a,b). This procedure can be used to create positive control samples from wild-type control animals or for the identification of deficiencies that are only apparent, or amplified, in response to DDR challenge. This enables the researcher to detect genes that are not necessary for the response to endogenous DDR stress, thus their deregulation does not produce elevated spontaneous micronucleus levels, but which are crucial for the response to DNA damage.
PROCEDURE
-
1 |
Before genotoxic challenge, collect and fix 50 μl of blood, as detailed in the spontaneous micronucleus procedure (Steps 1-3); include an untreated control cohort to ensure that the procedure does not cause marked reticulocytosis (see ‘Limitations of the method’ and TROUBLESHOOTING).
-
2 |
Challenge the animals with the specific genotoxic agent; the recommended dose of irradiation is 0.75 Gy whole-body irradiation, and the recommended dose of MMC is 1 mg/kg body weight via i.p. injection.
-
3 |
Collect and fix 50 μl of blood 24, 48 and 72 h post treatment, as detailed in the spontaneous micronucleus procedure (Steps 1-3).
-
4 |
Continue as detailed in the spontaneous micronucleus procedure (Steps 4-24);
ANTICIPATED RESULTS
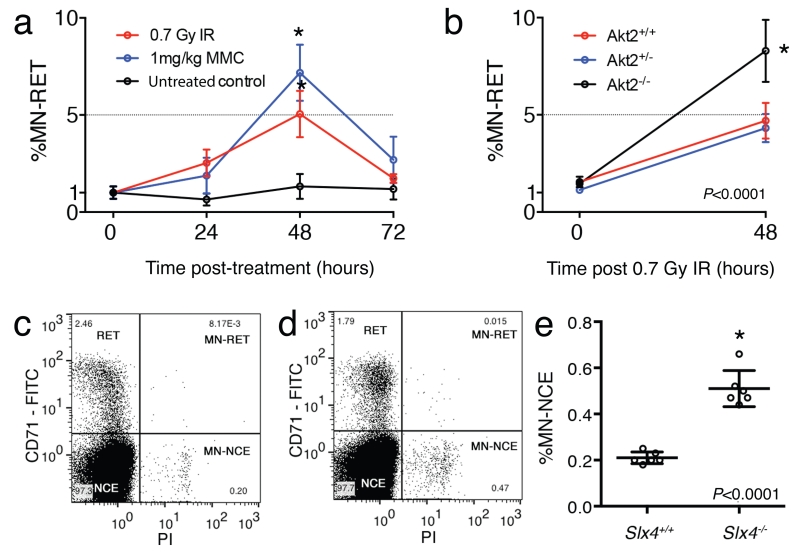
Wild-type mice treated with these genotoxins show a significant increase in the percentage of micronucleated RETs 24 and 48 h after treatment, and recovery is observed 72 h after treatment (Fig. 4a,4b). For example, 48 h after 0.75 Gy, wild-type mice have a mean fivefold increase in the percentage of micronucleated RETs (baseline mean = 0.61% for n = 10; 48 h after 0.75 Gy mean = 3.05% for n = 3), whereas 48 h after administration of 1 mg/kg MMC wild-type mice have a sevenfold increase (baseline mean = 0.61% for n = 10; 48 h after treatment mean = 4.23% for n = 3). Genotoxin sensitivity is characterized by a normal perentage of micronucleated RET frequency before treatment, but a marked increase in the percentage of micronucleated RETs versus control, 48 h after treatment. Fig. 4b demonstrates the radiosensitivity of Akt2−/− (tm1a[EUCOMM]WTSI) mice. Treatment must be carried out in accordance with the local regulations for the use of animals in research.
Figure 4. Micronucleus induction after genotoxic treatment or genetic modification.
a) Graph shows mean and s.d. for induction of micronucleus (MN) formation in reticulocytes (RETs) of wild-type mice treated with the DNA cross-linking agent mitomycin C (MMC; 1 mg/kg; n=3) or 0.75 Gy ionizing radiation (Gy IR; n = 4), relative to untreated mice (n = 4). Mice were bled at the indicated time points, and % MN-RETs were calculated as a percentage of total RETs. Forty-eight hours after MMC or IR treatment, %MN-RET is significantly increased relative to untreated control mice (P<0.01, Student’s two-tailed t-test). (b) Graph shows mean and s.d. for n≥5 Akt2+/+; Akt2+/− and Akt2−/− male mice after 0.75 Gy IR. Akt2−/− mice have a higher frequency of %MN-RET after IR, when compared to littermate controls (*P<0.0001; two-tailed Student’s t-test). (c,d) Increased %MN-normochromatic erythrocytes (NCEs) in a mouse model of Fanconi anaemia. Blood was collected from Slx4+/+ (c) and Slx4−/− (d) mice at 16 weeks of age and processed with the high-throughput micronucleus assay protocol. Representative plots are shown. Note the increased frequency of MN-NCEs in blood obtained from Slx4−/− mice (d) when compared with that of control Slx4+/+ mice (c). (e) Graph shows MN-NCEs as a percentage of total NCEs. Slx4−/− mice have significantly higher levels of spontaneous micronucleus induction than Slx4+/+ mice (mean ± standard deviation: 0.21 ± 0.025; Slx4+/+ versus 0.51 ± 0.078; Slx4−/−; n = 6, B6Brd;B6Dnk;B6N-Tyrc-Brd males for each genotype; *P<0.0001, Student’s two-tailed t-test). Data are duplicated from Crossan et al.11 to provide an example of a genetic positive control.
MATERIALS
REAGENTS
Laboratory mice (adult C57BL/6J mice of same sex and similar age [+/− 2 months] from The Jackson Laboratory, cat. no.000664)
Methanol (CH3OH; Sigma-Aldrich, #494437)
Heparin (Sigma-Aldrich, #H3393; ≥180 U.S.P. (U.S. Pharmacopeia) U/mg)
Sodium chloride (NaCl; e.g. Sigma-Aldrich, #S3014)
Sodium phosphate dibasic (HNa2O4P; e.g. Sigma-Aldrich, #S5136)
Sodium bicarbonate (CHNaO3; Sigma-Aldrich, #S6297)
Hydrochloric acid (HCl; Sigma-Aldrich, #84415)
RNase A from bovine pancreas (RNase A; Sigma-Aldrich, #R4642; activity ≥ 70 Kunitz units/mg protein)
FITC-conjugated rat anti-mouse CD71 transferrin receptor antibody (Southern Biotech, #1720-02; 0.5 mg/ml)
Mitomycin C from Streptomyces caespitosus (MMC; Sigma-Aldrich, #P4864) CRITICAL MMC is only required if you are using MMC to produce a positive control.
Propidium iodide 1.0 mg/ml solution (PI; Sigma-Aldrich, # P4864)
EQUIPMENT
Flow cytometer capable of handling multi-titer-well plates (96-well plates) and equipped with 488-nm blue and 561-nm green lasers (e.g. LSR Fortessa with high-throughput sampler option, Becton Dickinson) or, alternatively, with only a 488-nm laser.
Centrifuge with 4°C setting (Eppendorf 5810R) or centrifuge located in a cold room
−80°C freezer
pH meter (Meter Toledo MP220)
Biological safety cabinet (Fisher Scientific, #12372419)
Reciprocating shaker (Stuart SSL2)
Aluminum foil (Terinex)
Single-use 0.22 μm filters (Corning, #430521)
10 ml serological pipets (Corning - Product #4101)
15 ml polypropylene conical centrifuge tubes (Corning, #352097)
5 ml round bottom polystyrene tubes for flow cytometry (Corning, #352054)
96-well deep-well plates with 0.8 ml capacity (ABgene, #AB-0859)
96-well clear flat-bottom plate for flow cytometry assay (Corning, #353075)
1.5 ml microcentrifuge tubes (Eppendorf, #0030120191)
Flow cytometry data analysis software (FlowJo, Tree Star, Inc.) and statistical analysis software (Prism, GraphPad)
Optional: live animal irradiator (CIS-IBL 437 C; C137)
Optional: sterile lithium-heparin-coated Vacutainer blood collection tubes (Microtainer, Beckton Dickinson, #365953)
REAGENT SETUP
Mitomycin C (MMC)
Prepare MMC solution in a sterile laminar flow cabinet (Class II). Prepare 0.5 mg/ml stock solution with sterile water. Store at 4°C for up to two weeks. CAUTION: MMC is potentially mutagenic or carcinogenic; wear gloves, safety glasses and a laboratory coat when handling it.
Heparin
Prepare a 500 U.S.P. U/ml (2.35 mg/ml) solution in sterile PBS. Filter (0.22 μm filter) and divide the solution into 300 μl aliquots in 1.5 ml tubes and store at −20°C for up to 1 year. Do not freeze-thaw more than three times. The solution may be stored at 4°C for several weeks in 1.5 ml tubes, but evaporation occurs over longer periods.
Fixative
More than 12 h before blood collection, divide methanol into 2 ml aliquots in 15 ml conical tubes in a chemical safety cabinet and cap them tightly. It is recommended to prepare at least two tubes per blood sample from each mouse, but to process one sample and keep the second sample for technical repetition or for back-up. Place tubes in −80°C freezer for >12 h and < one month. We do not recommend long-term storage of methanol aliquots in the freezer; rather, methanol should be stored in a suitable ‘flammables’ cupboard away from sources of ignition. CAUTION: methanol is flammable, corrosive and toxic, and thus it should be handled in a chemical safety hood away from sources of ignition, and wear gloves, safety glasses and a laboratory coat when handling it.
Bicarbonate buffer
Prepare 0.9% (wt/vol) NaCl, 5.3 mM sodium bicarbonate (84 g/mole), and bring the pH to 7.5 by adding HCl. Autoclave and store the buffer at 4°C for up to 1 year. A 10× stock solution can be prepared, autoclaved and stored at 4°C for up to a year, but the pH must be determined for the 1× solution again when diluted for use. CAUTION: HCl is corrosive, so wear gloves, safety glasses and a laboratory coat when handling the reagent, and pipette it in a chemical safety cabinet. CRITICAL: Bicarbonate buffer must be cold before use, so allow plenty of time for cooling. If fixed blood samples are washed with warm buffer, they will turn brown and will be unsuitable for analysis.
CD71/RNase solution
This solution should be freshly prepared. While working in dimmed lighting, prepare enough bicarbonate buffer/RNase/CD71-FITC antibody mix for the required number of samples plus an extra 10% (‘dead’ volume). To each sample, add 0.2 μl anti-CD71-FITC (0.5 mg/ml) and 7 μl RNase A to bicarbonate buffer (72.8 μl) to make a total volume of 80 μl. To test the activity of new batches of RNase, we recommend performing a dilution series and comparing profiles after incubation with, for example, 0, 4, 8, 12 μl of RNase. CRITICAL: Sufficient RNase A activity is required to digest the RNA present in RETs; low-quality RNase A may lead to inaccurate results, as PI binds to RNA and DNA (see Suppl. Fig. 2a,b for profiles associated with incomplete digestion of RNA).
Animals
We obtain our mice from The Jackson Laboratory. The care and use of all mice that are used to generate data for this protocol was carried out in accordance with UK Home Office regulations, UK Animals (Scientific Procedures) Act of 2013. Our mice are maintained in a specific pathogen-free unit on a 12-h light: 12-h dark cycle with lights off at 19:30 and no twilight period. The ambient temperature is 21±2°C and the humidity is 55±10%. Mice are housed using a stocking density of 3–5 mice per cage (overall dimensions of caging: (L×W×H) 365×207×140 mm, floor area 530 cm2) in individually ventilated caging (Tecniplast Seal Safe1284L) receiving 60 air changes per hour. In addition to Aspen bedding substrate, standard environmental enrichment of two Nestlets, a cardboard fun tunnel and three wooden chew blocks are provided. Mice are given water and diet ad libitum.
EQUIPMENT SETUP
Flow cytometer
Warm up the lasers for 30 min and prepare the instrument for high-throughput sample analysis using a 96-well plate (refer to the manufacturer’s instrument setup guide). The fluorescence emitted from FITC-conjugated anti-CD71 and PI is collected through a 530/30-nm and a 610/20-nm band-pass filter after excitation by the blue 488-nm and the green 561-nm laser, respectively. Data resolution is improved and compensation is not required when the fluorochromes are excited and detected independently of each other using a 488 nm and 561 nm laser configuration. When a single 488 nm blue laser is used in this experiment, compensation is required during data acquisition, as FITC and PI have a slight spectral overlap (please refer to reagent supplier’s technical sheet), the same excitation wavelength and the same detection light path (please refer to your instrument’s technical specification).
Set the high-throughput sampler to mix each sample 2-4 times before acquiring 200,000 events per sample at a rate of 1000-2000 events per second. The preparation of single fluorochrome and unstained controls from surplus sample to setup the regional gates for the analysis of the erythrocyte cell population is included in the main protocol. CRITICAL STEP: Maintain an acquisition rate of < 2,500 events per second on a volumetric flow rate of 0.5 μl-1 μl/s. A higher volumetric flow rate can result in an increased rate of false positive events for the CD71-FITC-positive, PI-positive population (Suppl. Fig. 2a and 2c).
PROCEDURE
Blood collection, fixation and storage
TIMING: ~ 10 min per mouse.
-
1 |
Collect 50 μl of blood (e.g. two or three drops from a tail vein or retro-orbital bleed) into 300 μl of heparin solution in a 1.5 ml tube44. Alternatively, blood can be collected into a heparinized Vacutainer; however, it must then be transferred to the heparin solution to ensure proper single-cell dissociation of the cells before fixation. CAUTION: blood must be obtained from mice using a technique that is approved by local regulatory bodies and/or ethics committees44.
CRITICAL STEP: Ensure that the blood sample is fixed within 12 h of collection to minimize the risk of sample degradation, which is one of the most common sources of false positives (Suppl. Fig. 3).
TROUBLESHOOTING.
-
2 |
Rapidly expel 150 μl of heparin-blood mix directly over the 2-ml aliquot of cold methanol (taken straight out of the −80°C freezer) using a pipette and cap the 15 ml tube. Repeat this step for the duplicate tube. CRITICAL STEP: Do not put the pipette tip in the methanol, and do not let the blood touch the side of the tube, or aggregation may occur. CRITICAL STEP: The temperature of the methanol must be −80°C before fixation, as warmer methanol causes the blood to turn red-brown (Suppl Fig. 2d) and it alters the size and granularity of the blood cells, leading to a false-positive increase in micronucleated erythrocytes (Suppl. Fig. 3). The blood sample should remain bright red in fixative. TROUBLESHOOTING.
-
3 |
Invert the sample 4-5 times to prevent aggregation, and store the tubes at −80°C for at least 12 h before continuing. We recommend that the tubes be returned to their original polystyrene packaging, as this provides added protection should the freezer fail. We do not recommend transferring samples to a −20°C freezer after fixation, as warmer methanol temperatures change the size and granularity of blood cells, which can result in a slight elevation of the micronucleated NCE population (Suppl. Fig. 3). PAUSE POINT: When methanol-fixed samples are stored at −80°C, they remain stable for up to 1 year (Suppl. Fig. 2e and 4). At this stage, the 15-ml tubes of methanol-fixed samples can be placed in a sealed bag, shipped on dry ice and stored in a −80°C freezer upon arrival at the new destination.
Sample preparation
TIMING 3-4 h for 96 samples
-
4 |
Working on ice, add 12 ml of cold (4°C) bicarbonate buffer, and cap and invert the 15-ml tubes to mix the contents. Centrifuge the tubes at 500 g for 5 min at 4 °C. CRITICAL STEP: Samples must remain cold, especially while the cells are in undiluted methanol, if samples become warm at this stage, they may turn red-brown (Suppl. Fig. 2f) the size and granularity of blood cells can change and the data may be uninterpretable (Suppl. Fig. 3). TROUBLE SHOOTING.
-
5 |
Pour off the supernatant, add 200 μl of cold bicarbonate buffer and resuspend the cell pellet by pipetting gently up and down.
-
6 |
Transfer 20 μl of sample to each well in a 96-deep-well plate (800 μl per well capacity). Add 20 μl of any sample to three 1.5 ml microcentrifuge tubes for preparation of unstained control, PI-only control and CD71-FITC-only control, which are required for the flow cytometry setup.
-
7 |
Add 80 μl of CD71-FITC/RNase solution to test samples in the plate and to the CD71-FITC-single-stain control tube. Add 7 μl of RNase and 73 μl of buffer to the unstained control and PI-only control tubes. Incomplete RNase digestion will lead to a false increase in the MN-RET population, because PI also binds the RNA present in RET.
TROUBLESHOOTING
-
8 |
Cover plate with foil and incubate it for 45-60 min at 4°C with gentle agitation (~ 60 r.p.m) on a shaker.
-
9 |
Wash the samples (in wells and Eppendorf tubes) by addition of 600 μl of cold bicarbonate buffer (4°C) and centrifuge them at 500 g for 5 min at 4°C.
-
10 |
Pour off the supernatant by quickly inverting the plate over a waste container. The cells will remain in the wells. Pipette off the supernatant from the control tubes. Resuspend the pellets in 500 μl of bicarbonate buffer by pipetting gently up and down.
-
11 |
Transfer 200 μl of sample to a clear, flat-bottomed 96-well plate. The remaining sample can be stored at 4°C and used for technical replication if required.
Flow cytometric analysis
TIMING: 2-3 hours for 96 samples
-
12 |
Transfer 500 μl of each control sample into 5 ml round-bottom polystyrene tubes.
-
13 |
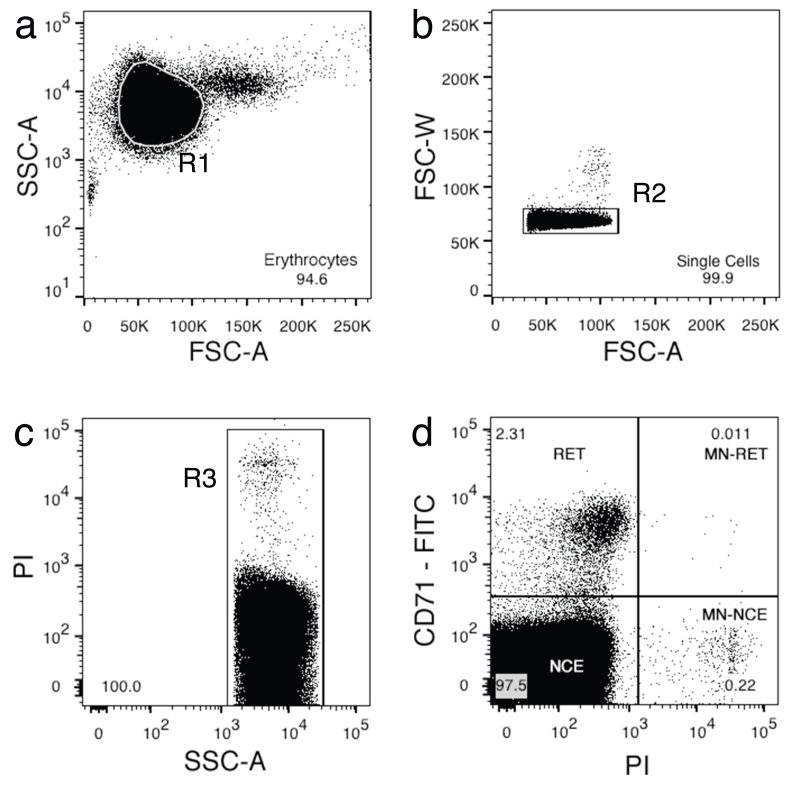
By using the unstained control, acquire events with logarithmic side scatter (SSC-A-log) versus linear forward scatter (FSC-A-Lin) axes. The SSC-versus-FSC plot should reveal two or three overlapping populations of red blood cells with reasonably well-defined edges. Centre the main population (erythrocytes) using the voltage and gain settings, and create the first gate (R1) just inside the erythrocyte population (Fig. 5a).
-
14 |
Isolate doublets by plotting FSC-W-Lin versus FSC-A-Lin and set gate R2 around single cells (Fig. 5b).
-
15 |
Add 2.5 μl of PI to the PI-only control tube, leave a few minutes and acquire the events. Adjust the voltage and gain to ensure that all events are within the axes, and draw a quadrant gate to separate the positive and negative populations. Repeat this step for the CD71-FITC-only control.
-
16 |
Use the PI single-stain control to identify events caused by autofluorescence (Suppl. Fig. 5) and remove by setting gate R3 on the PI biexponential scale versus SSC biexponential scale axes (Fig. 5c and Suppl. Fig. 5d).
-
17 |
On CD71-FITC-biexponential versus PI-biexponential axes, gate the four different populations (RET, Q1; MN-RET, Q2; MN-NCE, Q3; NCE, Q4; Fig. 5d).
-
18 |
Save the experiment settings on the flow cytometer.
-
19 |
Switch the flow cytometer from tube mode to multititer-well plate mode.
-
20 |
Set the volume to 70 μl, mixes to 4 and volumetric flow rate to 0.5 μl or 1 μl/s (to achieve ~ 1000-2000 events per second).
-
21 |
Add 1 μl PI to each well of the plate using a multipipettor, loading 24 wells at a time. Loading more than 24 wells at a time may increase variability (Suppl. Fig. 4b). Although pipetting 1 μl is not as accurate as pipetting larger volumes, our data suggest high assay reproducibility (Box 1). We routinely use an eight-channel Biopette (1-10 μl) or an Eppendorf Multipette plus. Mix by pipetting up and down with a multi-channel pipette (20-200 μl), and then incubate the plate for a few minutes at room temperature.
TROUBLESHOOTING
-
22 |
Load the saved protocol, and then acquire and save data.
TROUBLESHOOTING
-
23 |
Export the data; repeat the analysis (create gate R1 around erythrocytes, isolate doublets and set gate R2, remove autofluorescence by setting gate R3, gate the four populations) using appropriate software (e.g. FlowJo, TreeStar).
-
24 |To calculate the % micronucleus (MN) formation use the absolute event counts for each of the indicated quadrants:
Figure 5. Analysis of micronucleated red blood cells by flow cytometry.
(a) The erythrocyte cell population is resolved from the rest of the cell population by region gate R1, created on side scatter logarithmic scale (SSC-A-log) versus forward scatter linear scale (FSC-A-lin). (b) Single cells are isolated by region gate R2 on forward scatter width (FSC-W) versus FSC-A-lin. (c) Autofluorescent cells are excluded by gating on propidium iodide (PI) versus SSC-A (see also Suppl. Fig. 5). (d) The gated cells (R1, R2 and R3) are analyzed according to the intensity of fluorescence in the FITC channel (CD71-FITC) and the PI channel, and a quadrant gate is drawn to separate the four cell populations: normochromatic erythrocytes (NCEs) are CD71−/PI−; reticulocytes (RETs) are CD71+/PI−; micronucleated RETs are CD71+/PI+ (MN-RETs); while micronucleated normochromatic erythrocytes (MN-NCEs) are CD71−/PI+.
TIMING
For 96 samples
Day 1 (Reagent Setup): Prepare bicarbonate buffer (10 min), autoclave it and place it at 4°C. Label 192 15-ml tubes (96 × 2), transfer 2 ml of methanol into each and place at −80°C (~2h).
Day 2 (Steps 1-3): collect 96 blood samples into heparin solution. If you are experienced in the tail vein bleed technique, this should take ~ 4h. Fix blood in methanol (−80°C; ~1.5h).
Day 3 (Steps 4-24): Wash the samples in buffer, centrifuge them and resuspend them in buffer (~2.5h), incubate the samples for 45-60 min in CD71-FITC/RNase solution, wash them in buffer and centrifuge them (15 min), add PI and analyze the samples by flow cytometry (~2h).
TROUBLESHOOTING
See Table 1 for troubleshooting guidance.
Table 1.
Troubleshooting
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 1, 13, 22, 23 | Debris in the bottom left hand corner of the SSC-versus-FSC plot. (Suppl. Fig. 3b) |
|
|
|
| |||
| 2-4 |
|
|
|
|
| |||
| 7, 17, 22, 23 | CD71-FITC-positive-population predominates in Q2 rather than Q1 (See Suppl. Fig. 2b) |
|
|
|
| |||
| 20 | Spikes of events in Q2 (FITC-versus-PI plot) (Fig. 3c) |
|
|
|
| |||
| 21, 24 | Large variability in micronucleated NCEs (Q3) for mice of same genotype |
|
|
|
| |||
| 22, 23 | Excessive number of events (>50-fold increase in total reticulocytes) in Q1 versus control mouse (RET; see Suppl. Fig. 1a) | Reticulocytosis | Extreme reticulocytosis is likely to cause a false-positive increase in the number of FITC−, PI+ events (MN-NCE). Therefore it would be preferable to score the cells for the presence of a nucleus or MN with a microscope after smearing methanol-fixed blood onto slides and co-staining with DAPI (Fig 1c,d)13. |
| Mice are of a mixed genetic background. | Increase sample number. | ||
| Genotyping error. | Re-genotype. | ||
The flow cytometric quadrants (FITC-versus-PI plot ) are defined as Q1, RET; Q2, MN-RET; Q3, MN-NCE; Q4, NCE.
ANTICIPATED RESULTS
The anticipated results with the flow cytometric micronucleus assay depend on the mouse genetic background, sex, age and level of exposure to genotoxic agents (Box 1; Figs. 3 and 4). Any substantial increase over the wild-type mouse baseline can be identified with high sensitivity using simple statistics that are appropriate for the experimental design (Box 1). The successful procedure is dependent on proficient sample collection, storage, preparation and acquisition on the flow cytometer (see TROUBLESHOOTING). Upon sample collection, the blood should not be allowed to coagulate, and the fixation should be done in timely manner in −80°C methanol. Upon fixation, the solution should have a distinctive bright red color that will preserve throughout storage; brown coloration will indicate defective fixation. During data acquisition on the flow cytometer, the quality of the blood preparation can be easily observed on the graph generated on side scatter logarithmic scale (SSC-log) versus forward scatter linear scale (FSC-lin) where the total blood has a distinctive pattern that is easily distinguishable (Fig. 5a); deviations from this pattern might indicate sample degradation or defective fixation (see TROUBLESHOOTING). The comparison between the unstained, CD71-only, PI-only and the CD71/PI-stained samples will demonstrate successful sample staining. The addition of wild-type and positive controls to each plate (see Experimental design) will confirm that the assay has been run successfully.
Supplementary Material
Editorial summary: This protocol describes a sensitive, robust, high-throughput method for quantifying the formation of micronuclei in mouse erythrocytes. It detects low levels of genome instability and can be used to identify novel genome maintenance genes and models.
Acknowledgments
We thank Marie Hitcham and Nick Harman for assistance with blood collections, William Cheng for assistance with flow cytometry during high-throughput screening and Kate Dry for comments on the manuscript. R.E.M. is supported by Cancer Research UK (CRUK; Project Grant C20510/A12401). D.J.A is supported by CRUK. D.J.A and B.L.N are supported by the Wellcome Trust. Research in the Jackson laboratory is funded by CRUK programme grant C6/A11224, the European Research Council, and the European Community Seventh Framework Programme grant agreement no. HEALTH-F2-2010-259893 (DDResponse). Core funding is provided by CRUK (C6946/A14492) and the Wellcome Trust (WT092096). S.P.J receives his salary from the University of Cambridge, UK, supplemented by CRUK. G.B. is funded by CRUK programme grant C6/A11224.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Fenech M, et al. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011;26:125–32. doi: 10.1093/mutage/geq052. [DOI] [PubMed] [Google Scholar]
- 2.Evans HJ, Neary GJ, Williamson FS. The relative biological efficiency of single doses of fast neutrons and gamma-rays on Vicia faba roots and the effect of oxygen. Part II. Chromosone damage: the production of micronuclei. Int J Radiat Biol. 1959;1:216–29. doi: 10.1080/09553005914550311. [DOI] [PubMed] [Google Scholar]
- 3.Maluf SW, Erdtmann B. Genomic instability in Down syndrome and Fanconi anemia assessed by micronucleus analysis and single-cell gel electrophoresis. Cancer Genet Cytogenet. 2001;124:71–5. doi: 10.1016/s0165-4608(00)00322-8. [DOI] [PubMed] [Google Scholar]
- 4.Holland N, et al. The micronucleus assay in human buccal cells as a tool for biomonitoring DNA damage: the HUMN project perspective on current status and knowledge gaps. Mutat Res. 2008;659:93–108. doi: 10.1016/j.mrrev.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–8. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang CZ, Leibowitz ML, Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27:2513–30. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–70. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- 11.Crossan GP, et al. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet. 2011;43:147–52. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shima N, et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–8. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- 13.Hodskinson MR, et al. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair. Mol Cell. 2014;54:472–84. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakachi K, Hayashi T, Hamatani K, Eguchi H, Kusunoki Y. Sixty years of follow-up of Hiroshima and Nagasaki survivors: current progress in molecular epidemiology studies. Mutat Res. 2008;659:109–17. doi: 10.1016/j.mrrev.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Doherty AT. The in vitro micronucleus assay. Methods Mol Biol. 2012;817:121–41. doi: 10.1007/978-1-61779-421-6_7. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Liu Y, Ni G, Liu S. Flow cytometric scoring of micronucleated reticulocytes as a possible high-throughput radiation biodosimeter. Environ Mol Mutagen. 2010;51:215–21. doi: 10.1002/em.20523. [DOI] [PubMed] [Google Scholar]
- 17.Maluf SW, Passos DF, Bacelar A, Speit G, Erdtmann B. Assessment of DNA damage in lymphocytes of workers exposed to X-radiation using the micronucleus test and the comet assay. Environ Mol Mutagen. 2001;38:311–5. doi: 10.1002/em.10029. [DOI] [PubMed] [Google Scholar]
- 18.Fliedner TM, Andrews GA, Cronkite EP, Bond VP. Early and Late Cytologic Effects of Whole Body Irradiation on Human Marrow. Blood. 1964;23:471–87. [PubMed] [Google Scholar]
- 19.Howell WH. The life-history of the formed elements of the blood, especially the red blood corpuscles. Journal of Morphology. 1891;4:57–116. [Google Scholar]
- 20.Jolly JMJ. Sur la formation des globules rouges des mammiféres. Comptes rendus de la Société de Biologie. 1905;58:528–531. [Google Scholar]
- 21.Countryman PI, Heddle JA. The production of micronuclei from chromosome aberrations in irradiated cultures of human lymphocytes. Mutat Res. 1976;41:321–32. doi: 10.1016/0027-5107(76)90105-6. [DOI] [PubMed] [Google Scholar]
- 22.Sears DA, Udden MM. Howell-Jolly bodies: a brief historical review. Am J Med Sci. 2012;343:407–9. doi: 10.1097/MAJ.0b013e31823020d1. [DOI] [PubMed] [Google Scholar]
- 23.MacGregor JT, Wehr CM, Gould DH. Clastogen-induced micronuclei in peripheral blood erythrocytes: the basis of an improved micronucleus test. Environ Mutagen. 1980;2:509–14. doi: 10.1002/em.2860020408. [DOI] [PubMed] [Google Scholar]
- 24.Hutter KJ, Stohr M. Rapid detection of mutagen induced micronucleated erythrocytes by flow cytometry. Histochemistry. 1982;75:353–62. doi: 10.1007/BF00496738. [DOI] [PubMed] [Google Scholar]
- 25.Grawe J, Zetterberg G, Amneus H. Flow-cytometric enumeration of micronucleated polychromatic erythrocytes in mouse peripheral blood. Cytometry. 1992;13:750–8. doi: 10.1002/cyto.990130711. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi M, Norppa H, Sofuni T, Ishidate M., Jr. Flow cytometric micronucleus test with mouse peripheral erythrocytes. Mutagenesis. 1992;7:257–64. doi: 10.1093/mutage/7.4.257. [DOI] [PubMed] [Google Scholar]
- 27.Lee LG, Chen CH, Chiu LA. Thiazole orange: a new dye for reticulocyte analysis. Cytometry. 1986;7:508–17. doi: 10.1002/cyto.990070603. [DOI] [PubMed] [Google Scholar]
- 28.Bender JG, et al. Identification and comparison of CD34-positive cells and their subpopulations from normal peripheral blood and bone marrow using multicolor flow cytometry. Blood. 1991;77:2591–6. [PubMed] [Google Scholar]
- 29.Dertinger SD, Torous DK, Tometsko KR. Simple and reliable enumeration of micronucleated reticulocytes with a single-laser flow cytometer. Mutat Res. 1996;371:283–92. doi: 10.1016/s0165-1218(96)90117-2. [DOI] [PubMed] [Google Scholar]
- 30.Dertinger SD, Torous DK, Hayashi M, MacGregor JT. Flow cytometric scoring of micronucleated erythrocytes: an efficient platform for assessing in vivo cytogenetic damage. Mutagenesis. 2011;26:139–45. doi: 10.1093/mutage/geq055. [DOI] [PubMed] [Google Scholar]
- 31.Reinholdt L, Ashley T, Schimenti J, Shima N. Forward genetic screens for meiotic and mitotic recombination-defective mutants in mice. Methods Mol Biol. 2004;262:87–107. doi: 10.1385/1-59259-761-0:087. [DOI] [PubMed] [Google Scholar]
- 32.Shima N, et al. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–40. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White JK, et al. Genome-wide Generation and Systematic Phenotyping of Knockout Mice Reveals New Roles for Many Genes. Cell. 2013;154:452–64. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, et al. Mcph1-Deficient Mice Reveal a Role for MCPH1 in Otitis Media. PLoS One. 2013;8:e58156. doi: 10.1371/journal.pone.0058156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mclntyre RE, et al. Disruption of mouse Cenpj, a regulator of centriole biogenesis, phenocopies Seckel syndrome. PLoS Genet. 2012;8:e1003022. doi: 10.1371/journal.pgen.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nijnik A, et al. The critical role of histone H2A-deubiquitinase Mysm1 in hematopoiesis and lymphocyte differentiation. Blood. 2012;119:1370–9. doi: 10.1182/blood-2011-05-352666. [DOI] [PubMed] [Google Scholar]
- 37.Balmus G, et al. Disease severity in a mouse model of ataxia telangiectasia is modulated by the DNA damage checkpoint gene Hus1. Hum Mol Genet. 2012;21:3408–20. doi: 10.1093/hmg/dds173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holloway JK, et al. Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genet. 2011;7:e1002094. doi: 10.1371/journal.pgen.1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dertinger SD, et al. Enumeration of micronucleated CD71-positive human reticulocytes with a single-laser flow cytometer. Mutat Res. 2002;515:3–14. doi: 10.1016/s1383-5718(02)00009-8. [DOI] [PubMed] [Google Scholar]
- 40.Fenech M. Cytokinesis-block micronucleus cytome assay. Nat Protoc. 2007;2:1084–104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- 41.Danise P, et al. Evaluation of nucleated red blood cells in the peripheral blood of hematological diseases. Clin Chem Lab Med. 2012;50:357–60. doi: 10.1515/CCLM.2011.766. [DOI] [PubMed] [Google Scholar]
- 42.Balmus G, Mclntyre RE. Genetic screens in mice for genome integrity maintenance and cancer predisposition. Curr Opin Genet Dev. 2014;24:1–7. doi: 10.1016/j.gde.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 43.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parasuraman S, Raveendran R, Kesavan R. Blood sample collection in small laboratory animals. J Pharmacol Pharmacother. 2010;1:87–93. doi: 10.4103/0976-500X.72350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–74. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 46.Karp NA, Melvin D, Sanger Mouse Genetics, P. Mott RF. Robust and sensitive analysis of mouse knockout phenotypes. PLoS One. 2012;7:e52410. doi: 10.1371/journal.pone.0052410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenth RV. Statistical power calculations. J Anim Sci. 2007;85:E24–9. doi: 10.2527/jas.2006-449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.