Summary
Dosage compensation mechanisms provide a paradigm to study the contribution of chromosomal conformation towards targeting and spreading of epigenetic regulators over a specific chromosome. By using Hi-C and 4C analyses we show that high-affinity sites (HAS), landing platforms of the male-specific lethal (MSL) complex, are enriched around topologically associating domain (TAD) boundaries on the X chromosome and harbor more long-range contacts in a sex-independent manner. Ectopically expressed roX1 and roX2 RNA target HAS on the X chromosome in trans and, via spatial proximity, induce spreading of the MSL complex in cis, leading to increased expression of neighboring autosomal genes. We show that the MSL complex regulates nucleosome positioning at HAS, thus acting locally rather than influencing the overall chromosomal architecture. We propose that sex-independent three-dimensional conformation of the X chromosome poises it for exploitation by the MSL complex, thereby facilitating spreading in males.
Introduction
The organization of chromosomes within the nucleus and the spatial arrangement of genes within a chromosome territory are gaining fundamental importance during epigenetic control of gene expression (Quinodoz and Guttman, 2014). Notably, the regulatory mechanisms of sex chromosomes offer ideal paradigms to understand how expression of an entire chromosome, and therefore thousands of genes at once, can be controlled by epigenetic mechanisms (Brockdorff and Turner, 2015).
Dimorphic sex chromosomes genetically determine sex in many organisms. In the XX/XY sex determination system, males are heterogametic (XY) and females are homogametic (XX). To overcome the risk of an unequal transcriptional output, different organisms have evolved independent strategies (termed “dosage compensation”) to balance the X chromosomal gene dose between the sexes (Vicoso and Charlesworth, 2006): in mammals, expression of the long non-coding RNA (lncRNA) Xist from only one of the two female X chromosomes leads to recruitment of silencing complexes in cis, through which this chromosome becomes compacted and heterochromatinized (Heard and Disteche, 2006). In Drosophila melanogaster, dosage compensation happens on the single male X chromosome by formation of the male-specific lethal (MSL) complex that promotes approximately 2-fold transcriptional upregulation (Conrad and Akhtar, 2012). The MSL complex consists of four core proteins (MSL1, MSL2, MSL3 and males absent on the first (MOF)) that together form a heterooctameric complex which is further stabilized by integration of two lncRNAs, called RNA on the X (roX) 1 and 2 by the ATP-dependent RNA helicase maleless (MLE) (Keller and Akhtar, 2015). The formation of this ribonucleoprotein complex is believed to occur at the roX gene loci as roX RNAs are the only components of the complex being produced within the nucleus.
Based on both genetic and genomic analyses, the complex is thought to first target genomic regions called high-affinity sites (HAS), which include the roX genes, and then to spread to lower affinity sites. During this process, MOF acetylates histone H4 lysine 16 across the entire X chromosome, which ultimately upregulates transcription (Conrad and Akhtar, 2012). Yet, how HAS are organized to allow the complex to reach the whole X chromosome continues to be an enigma. The MSL complex preferentially binds to an active chromatin environment containing a consensus sequence motif, termed MSL recognition element (MRE), that is flanked by sequences of elevated GC content (Alekseyenko et al., 2012; Conrad and Akhtar, 2012). However, these features, although moderately enriched on the X, are also found on autosomes and thus it has not been possible to fully characterize HAS. Current models are based on linear genomic analysis or DNA fluorescent in situ hybridization (DNA FISH) on few individual loci (Grimaud and Becker, 2009) without accounting for the potential influence of global chromosome conformation.
In this study, we used Hi-C, a technique that enables the study of all chromosomal interactions within a genome at once (Lieberman-Aiden et al., 2009). This was further complemented by 4C-seq (Splinter et al., 2012) and three-dimensional double-label DNA FISH (3D DNA FISH) analysis on single cells to study the interaction patterns of individual loci on the X. Our data highlight a distinct mechanism in flies in which specific features at TAD boundaries on the X provide an advantageous location for the MSL complex to spread to spatially close regions and induce dosage compensation. Moreover, we show that rather than modifying global chromosomal domain organization, the MSL complex acts locally by inducing chromatin remodeling at HAS.
Results
High-affinity sites preferentially occur at boundaries of topologically associating domains
Previously published Hi-C studies in fruit flies could not address male-specific dosage compensation as either sex-mixed embryos (Sexton et al., 2012) or the female Kc cell line (Hou et al., 2012) alone were used. Therefore, we generated wild type Hi-C contact maps using the restriction enzyme HindIII for two widely used male model cell lines, CME W1 cl.8+ (Currie et al., 1988) (clone8) and Schneider’s line 2 (Schneider, 1972) (S2) in biological duplicates. To ensure consistent comparisons, we also reprocessed previously published Hi-C contact maps for mixed-sex embryos and for Kc cell lines using the same mapping and normalization procedures (Figure S1A-B, Table S1). The correlation within replicates was very high for raw and corrected Hi-C counts (0.96 Pearson correlation in both cases, Figure S1C-F) as well as the correlation between different cell types, when raw and corrected Hi-C counts were considered. For the corrected counts we see expected power-law decay of interaction frequencies with increasing genomic distance (Hou et al., 2012; Lieberman-Aiden et al., 2009; Sexton et al., 2012) and a similar decay for all chromosomes in the different Hi-C datasets (Figure S1G-I).
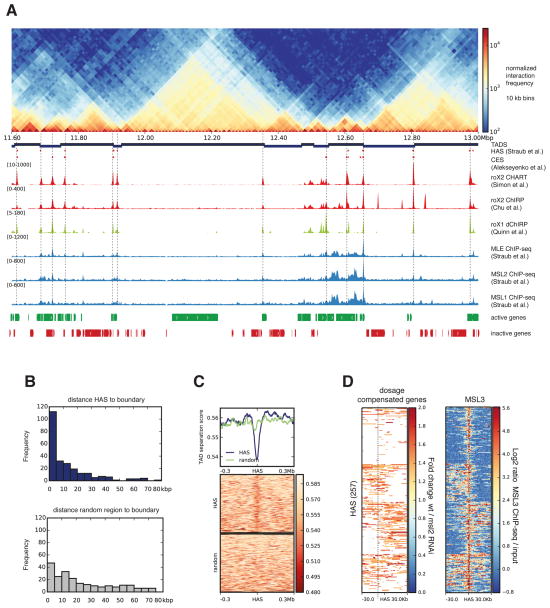
When combining our Hi-C data for S2 cells with published high resolution roX occupancy sites (indicative of HAS) from CHART (Simon et al., 2011) and ChIRP (Chu et al., 2011) for this cell type, we observed that HAS have a tendency to localize at or near topologically associating domains (TADs) (Dixon et al., 2012; Hou et al., 2012; Nora et al., 2012; Sexton et al., 2012) boundaries on the X chromosome. This can be readily seen by simple visual inspection of the data (Figure 1A, Figure S2A-B). To corroborate this finding, we used a domain caller (see Experimental Procedures, Figures S3A-C) to define TAD boundaries in S2 and clone-8 cells. Since the TAD structure is highly conserved between the cell types studied (Figures S3D), the boundaries obtained for S2 and clone-8 are very similar to the published domain partitions for Kc (Hou et al., 2012) and Drosophila embryos (Sexton et al., 2012) (Figures S3E, F). For the X, we identified a total of 257 HAS using the genome-wide mapping of roX2 (Simon et al., 2011) and MSL2 (Straub et al., 2013)(see also Table S2 and Extended Experimental Procedures). This association to TAD boundaries is significantly different from a random distribution (p-value = 1.9 ×10−11 Fisher’s exact test on the number of overlaps between boundaries and HAS, see Extended Experimental Procedures). We found that 68% of HAS lie within 20 kilobase pair (kb) distance to the nearest boundary. When calculated within a 5 kb distance, 45% of the HAS are located near a boundary in contrast to 9% expected by chance (Figure 1B, Figure S3G). To further validate this finding, we computed the TAD separation score at each HAS (see Extended Experimental Procedures, Figure S3A, B) and verified that it tends to have a minimum, which is indicative of boundary regions, at HAS (Figure 1C). For comparison, similarly low values are obtained when the TAD separation score is evaluated at architectural protein binding sites (APBS) (Van Bortle et al., 2014), that are thought to localize at TAD boundaries (Dixon et al., 2012; Hou et al., 2012; Sexton et al., 2012; Sofueva et al., 2013; Van Bortle et al., 2014) (Figure S3H). When studying dosage compensated genes (measured by downregulation of expression upon MSL2 depletion (Zhang et al., 2010)), we found that they appear frequently either upstream or downstream of HAS (Figure 1D) in a pattern similar to the one observed for TAD boundaries that separate active and inactive chromatin (Figure S3I). Taken together, we conclude that the apparent linear arrangement of HAS along the genome follows a particular pattern dictated by the three dimensional TAD organization.
Figure 1. High affinity sites (HAS) are enriched at TAD boundaries.
A. Normalized Hi-C counts at 10 kb resolution for the region 11.6-13.0 Mb in chromosome X of S2 cells. From top, the tracks are as follows: partitioning of the genome into topologically associating domains (TADs); HAS as defined by Straub et al. (Straub et al., 2008); HAS reported by (Alekseyenko et al., 2008) (originally called chromosome entry sites (CES)); roX2 CHART (Simon et al., 2011); roX2 ChIRP (Chu et al., 2011); roX1 dChIRP (Quinn et al., 2014); MLE-, MSL1-, MSL2- ChIP-seq (Straub et al., 2013); active and inactive genes in S2 cells (Cherbas et al., 2011). Vertical lines are high resolution HAS based on roX2 and MSL2 binding. B. Distribution of distances from the boundaries to HAS (top, in blue) and from boundaries to the same number of shuffled random regions (bottom, in grey) within chromosome X. X-axis represents the distance in kb from HAS to boundary in bins of 5 kb, y-axis contains the number of HAS per bin. C. TAD separation score (see Extended Experimental Procedures) around HAS shows the tendency of the MSL complex to land at boundaries. Lower scores indicate better TAD separation. D. Dosage compensated genes (based on RNA-seq data from wild type and MSL2 RNAi treated S2 cells (Zhang et al., 2010)) and MSL3 ChIP-seq (Straub et al., 2013) up to 30 kb away from HAS. For the dosage compensation panel, only genes showing activity are colored. The heatmap rows were divided into four clusters based on the kmeans algorithm using deepTools (Ramírez et al., 2014). See also Table S1 and S2 and Figure S1, S2 and S3.
HAS show more enriched Hi-C contacts in a sex-independent manner
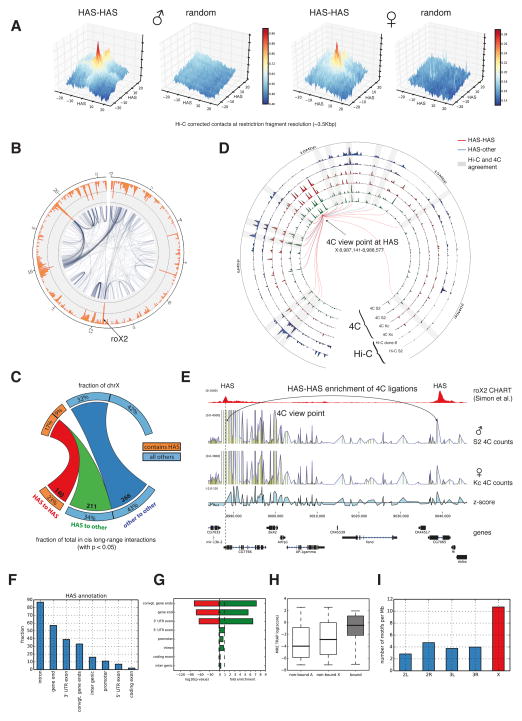
Previous analyses of Hi-C data have shown that active chromatin regions tend to interact together (Lieberman-Aiden et al., 2009; Sexton et al., 2012). Moreover, boundary regions have been shown to be enriched in Hi-C contacts (Hou et al., 2012; Rao et al., 2014) and also long-range contacts between architectural proteins have been suggested (Liang et al., 2014). Since HAS are frequently associated with active chromatin (Alekseyenko et al., 2012) and appear at boundaries, we explored the possibility that the spreading in cis of the MSL complex could be mediated by long-range associations of HAS. Using a method similar to the PE-scan (de Wit et al., 2013) (Figure 2A) we find that HAS, and in general TAD boundaries on all chromosomes, are frequently enriched for Hi-C contacts in the different cell lines, including female Kc cells (Figure 2A and Figure S4A,B). We also investigated the differences of cis long-range contacts on the X chromosome for each cell type and compared them to each other using a rigorous estimation of significant long-range Hi-C contacts (Figure 2B, Figure S4C,D, see Extended Experimental Procedures). To gain statistical confidence and to avoid spurious results, we segmented the genome into 25 kb bins for this analysis. Our results reveal that HAS tend to exhibit more long-range contacts than other regions. HAS are found in 26% of the 25 kb bins on the X and these regions contribute to 61% of all significant intra-X chromosomal long-range HiC contacts (Figure 2C). The spatial association between different HAS is highly significant as only 77 (11%) interactions are expected by chance (p-value 5 ×10−16Fisher’s exact test, one-tailed). Similar results were found for all cell types studied, when using different p-value thresholds to call long-range contacts, and for boundaries on other chromosomes (Table S3). The same analysis using 10 kb bins from a merge of all Hi-C samples for S2 cells (see Extended Experimental Procedures) show very comparable results (Figure S4E, see also Figure S4F for examples of enriched HAS-HAS contacts). As the Hi-C results represent population averages, where each cell may contain a unique chromosomal conformation (Nagano et al., 2013), we do not expect that all enriched HAS contacts are found in each cell but rather that HAS form a dynamic interaction network on the X. We analyzed long-range contacts for bins containing either a HAS and a boundary, a HAS only, or a boundary only. We find that bins which contain a HAS (either alone or together with a boundary) have on average more long-range contacts than the bins that only contain a boundary (Figure S5A). We additionally compared bins containing a boundary on the chromosome X versus those on autosomes and found the average number of enriched contacts to be similar. While the majority of HAS are in the same bins as boundaries, all bins that contain a HAS have on average more long-range contacts as the bins that only contained a boundary but no HAS (Figure S5A). This result suggests that HAS tend to be associated to boundaries with more long-range contacts.
Figure 2. HAS show more Hi-C and 4C ligations with other HAS.
A. Enrichment of Hi-C contacts between HAS loci in male (S2) and female (Kc) data (see Experimental Procedures). The x- and y-axes show distance from HAS in restriction fragment resolution. The z-axis contains the mean value of all pooled normalized sub-matrices corresponding to a HAS-HAS intersection. B. Representation of long-range Hi-C contacts from S2 cells as a network (25 kb bins, p-value<0.05, see Experimental Procedures). The orange outer rim shows the total number of long-range Hi-C contacts per bin. C. Quantification of long-range interactions on chromosome X from HAS to HAS (red), HAS to other regions (green) and within other regions (dark blue). Multiple HAS found in same bins were considered once. D. Comparison of enriched 4C contacts in males and females and Hi-C contacts in two cell lines for the view point at HAS position X:8,987,141-8,988,577. Tracks displayed from outside towards the inside: Hi-C S2 and Hi-C clone-8 adjusted –log(p-values) for a 25 kb binning; 4C contact enrichment –log(p-values) for Kc replicate1, Kc replicate2, S2 replicate1 and S2 replicate2 at 25 kb binning. Lines represent enriched contacts detected in one of the 4C S2 replicates. Red lines indicate interactions between HAS and the blue line indicates interactions from HAS to other regions. Peripheral numbering indicates genomic position in Mb. Grey rectangles highlight the agreement between Hi-C and 4C at the enriched 4C contacts at the connecting lines. Close to the view point the detection of enrichments that are close to TAD size is limited when using Hi-C data. E. 4C data for a viewpoint at the end of gene CG7766 confirms the interaction between two HAS that are 50 kb apart. Further 4C examples are in Figure S5C. F. Graph representing the location of HAS on different regions in and around genes. G. Enrichment of MREs bound by the MSL complex in contrast to the genome-wide background occurrence of MREs. H. TRAP score for MREs on autosomes (non-bound A), MREs not bound by the MSL complex on chromosome X (non-bound X), and MREs bound to the MSL complex (bound). H. Occurrence of MREs at different chromosomes filtered by the following criteria: the MRE has to be on active chromatin determined using H3K36me3 (GEO accession GSM685608), the MRE has to be located near the gene end, the MRE needs to have a TRAP score of at least -2 and the MRE should be located within 25 kb of the nearest boundary. MREs on chromosome X are enriched for these features. See also Table S3 and S4 and Figure S4 and S5.
We validated our Hi-C results using 4C-seq on male S2 and female Kc cell lines (for 4C viewpoints see also Table S4). Figure 2D and Figure S5B show comparisons of adjusted p-values where the high correspondence between Hi-C and 4C can be seen. These comparisons show that our estimations of long-range contacts are reliable and can be reproduced by a different experimental and processing method as the one used for Hi-C. Furthermore, the 4C data allowed us to explore the enriched contacts at short genomic distances (5-10 kb) that are not possible with Hi-C. At this resolution, we consistently detect HAS-HAS enriched contacts that remain unchanged between the male and female cell lines (Figures 2E and further on Figure S5C).
Depletion of MSL2 or MSL3 does not change global TAD organization
The similarity between male and female results from Hi-C and 4C experiments suggested that, contrary to expectation (Grimaud and Becker, 2009), the MSL complex may not alter the conformation of the X chromosome. To directly investigate this, we generated Hi-C data in male S2 cells depleted of the MSL complex members (via RNAi-mediated knockdown of either MSL2 or MSL3, Figure S6A) and compared it with control knockdown (EGFP RNAi) or wild type Hi-C samples. The resulting Hi-C counts showed a high correlation between all samples (Figure S6B), while the HAS-HAS enriched contacts of the knockdown samples did not differ from those of the wild type or EGFP RNAi control (Figure S6C, Table S3) and the TAD structure remained virtually identical (Figure S6D). These data indicate that the dosage compensation machinery in Drosophila does not broadly alter chromosomal topology but rather acts over a pre-existing chromosome conformation, independent of the sex.
The X chromosome harbors stronger MREs at boundaries compared to autosomes
Since TAD boundaries are present on all chromosomes and are frequently enriched for long-range contacts (Figure S4B), we next addressed the relationship between TADs and HAS located on the X. Previous lower resolution approaches showed that HAS tend to be located at gene bodies or at the end of genes (Alekseyenko et al., 2008; Gilfillan et al., 2006). Using the high resolution HAS derived from CHART (Simon et al., 2011) and ChIRP (Chu et al., 2011) methods, we observe that HAS often appear on intronic regions (35%) and in the proximity of gene ends (51%) which include the 3’ UTR exon and convergent gene ends (Figure 2F). Moreover, HAS are virtually never found at coding exons. The genomic distribution of the MRE associated to HAS over the different gene annotations revealed that gene ends are significantly enriched for MREs that are bound by the MSL complex (i.e. HAS) (Figure 2G). Also, HAS are always associated with active genes decorated with H3K36me3 histone mark, and tend to have a DNA sequence with higher binding energy (TRAP score (Thomas-Chollier et al., 2011)) containing usually several copies of the MRE. When we considered only MREs that are at gene ends, are in active chromatin, have a log (TRAP score) higher than -2, and are within 20 kb from the nearest boundary, we found that such combination of features is enriched on chromosome X (Figures 2H-I). These data suggest that a combination of MREs, chromatin state and gene architecture are required for the specificity of the MSL complex towards the X chromosome.
Differential positioning of active regions within the X chromosomal territory
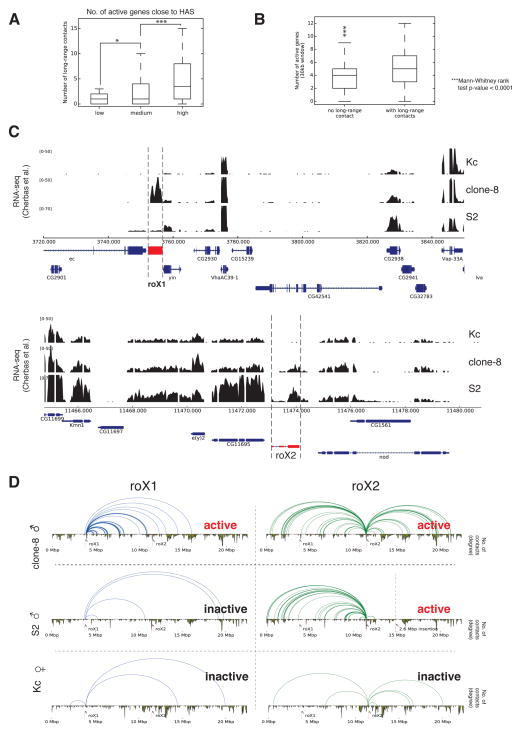
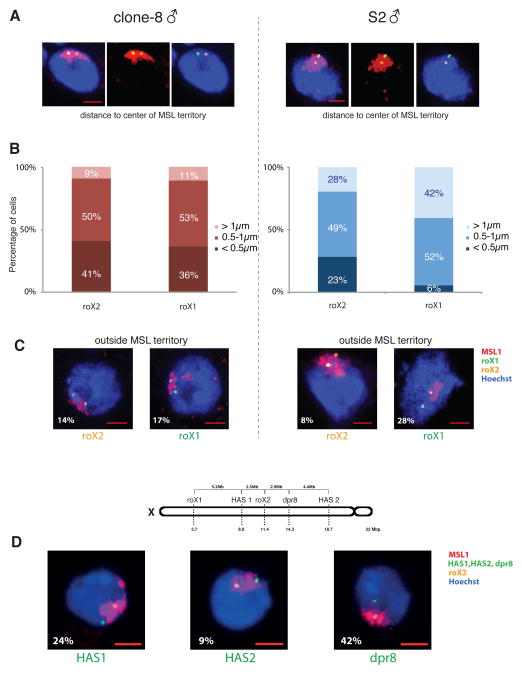
Correlation of expression and long-range contacts revealed that transcriptionally active HAS show more contacts compared to HAS located within inactive genes (Figure 3A-B). Analysis of two prominent HAS, roX1 and roX2, allowed us to explore this finding in more detail: in clone8 cells, both roX RNAs are actively transcribed (Cherbas et al., 2011), while >99.5% of S2 cells do not show roX1 expression (Johansson et al., 2011) and female Kc cells do not express any of the roX genes (Cherbas et al., 2011). In contrast, genes surrounding roX1 and roX2 are similarly expressed in the three cell lines (Figure 3C). We observed a greater number of long-range contacts when the roX genes are active (Figures 3D and Figure S6E-F), although no changes were seen on the TAD structure. Next, we explored whether this difference between the two roX loci in the number of long-range contacts is reflected by their nuclear positioning in individual cells. For this, we performed 3D DNA FISH for roX1 and roX2, in clone8 and S2R+ cells (a derivative of the original S2 cells with similar transcriptional profiles), and measured radial distances of the probes to the center of mass of the MSL1 immunostained region (MSL territory) (Figure 4A and Table S5). In clone8 cells, both expressed roX genes showed almost equal distance distributions, while in S2R+ cells the non expressed roX1 appeared farther away compared to the expressed roX2 (p-value=1.9×10−4, t-test one-tailed) (Figure 4B). In clone8 cells, both roX probes can be found almost equally often outside of the MSL territory (17% of roX1 and 14% of roX2 signals), whereas in S2R+ roX1 is found outside over three times as often as roX2 (28% vs 8%, Figure 4C). Taken together, the differences in the relative locations of roX1 and roX2 with respect to the MSL territory in clone8 and S2R+ cells suggest that such positioning is related to their transcriptional activity. Consistent with this hypothesis, we observed that a FISH probe over a non-HAS (dpr8, inactive gene) was more frequently found outside of the MSL territory in comparison to two different HAS (HAS1 and HAS2, active genes) with respect to roX2 (Figure 4D). These data suggest that differential positioning of active regions within chromosomes could serve as an elegant mechanism for tissue-specific fine-tuning without changes in TAD structure, thus providing plasticity for gene regulation, while maintaining stability of the overall chromosome shape.
Figure 3. Comparison of gene expression activity and long-range contacts.
A. Comparison of the number of active genes associated to HAS and the number of long-range contacts. We stratified the number of active genes within a 15 kb window up- and downstream of HAS into low (< 4 active genes), medium (between 4 and 7) and high (8 or more). B. Number of active genes in vicinity (±15 kb) to HAS with our without long-range contacts. C. Gene expression from (Cherbas et al., 2011) RNA-seq for genes close to roX1 and roX2. In clone-8 roX1 and roX2 are active while in S2 only roX2 is active. D. Schematic representation of long-range Hi-C contacts originating either from the roX1 (± 25 kb) or the roX2 locus (± 25 kb) in clone8, S2 and Kc cells. The activity status for each of the roX genes is shown in the respective panels. Complementary images showing 4C contacts are found in Figure S6E and S6F. See also Figure S6 and Table S4.
Figure 4. Differential positioning of HAS relative to the MSL territory.
A. Representative maximum intensity projections of confocal image stacks of 3D FISH experiments with roX1 (green) and roX2 (orange) probes in S2R+ and clone8 male D. melanogaster cells. MSL1 immunostaining is shown in red, DNA is counterstained with Hoechst (blue). B. 3D distances from the center of mass of probes to the center of mass of the MSL territory. Graphs show the distribution of roX1 and roX2 probes in the different bins, within the two cell lines; clone8 (left) and S2R+ cells (right). C. Percentage of cells in which either roX1 or roX2 are localized outside the MSL territory, as demarcated by MSL1 immunostaining, in clone8 (left) and S2R+ cells (right). Scale Bars represent 2 μm. n=64 for clone8 cells and n=71 for S2R+. D. Three additional DNA FISH probes (dpr8, HAS1, HAS2) were used in S2 cells, paired with roX2, to study their positioning with respect to the MSL territory. Representative pictures of each probe pair are shown, with the percentage of probes escaping the MSL territory indicated at the bottom left of each panel. Above is a schematic representation of the genomic location of all the probes used in this study on the X chromosome of D. melanogaster. Scale Bars represent 2 μm. n=45 for HAS1, n=23 for HAS2, n=30 for dpr8. See also Table S5.
The MSL complex spreads from HAS to spatially proximal regions
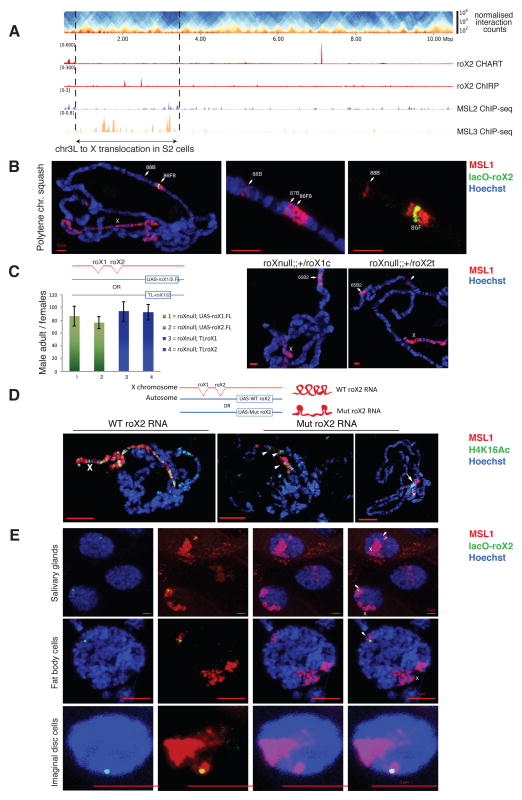
Knowing that HAS are located at regions often engaging in long-range contacts, we tested whether this is a sufficient condition for targeting of the MSL complex to loci in 3D-proximity to the roX genes by studying a previously unnoticed large (~2.67 Mega base pair (Mb)) insertion of chromosome 3L (chr3L: ~796,745-3,468,912) into the X (chrX: ~14,809,484) that was revealed by our Hi-C analysis of S2 cells (Figure S7A) and is most likely present in one of the two X chromosomes in the tetraploid S2 cells. This large region appears enriched for the MSL3 protein –a hallmark of MSL complex spreading in ChIP-seq data– although we do not detect typical HAS (in Figure 5A we only see seemingly spurious roX peaks that are not consistent between the two methods used to determine roX-DNA contacts (CHART (Simon et al., 2011) and ChIRP (Chu et al., 2011)) and do not observe other MSL proteins). For comparison, an X-linked region of similar size as the insertion contains on average 30 HAS. Using a 4C viewpoint on the translocated region we observed long-range contacts with the X chromosome. The same viewpoint in Kc cells, lacking this translocation, does not show any contacts (Figure S7B). These data demonstrate that by being physically associated to the X, the MSL complex can spread via long-range contact over a region lacking HAS.
Figure 5. Insertion of an autosomal fragment into the X chromosome and of the roX2 HAS into an ectopic autosomal location.
A. Hi-C heatmap of chromosome 3L highlighting a 2.67 Mb translocation (chr3L: ~796,745-3,468,912) into chromosome X. Figure S7B shows 4C long-range contacts from the translocation to the chromosome X. B. The autosomal insertion of LacO-roX2 (green) into 86F8 on chromosome arm 3R efficiently targets the MSL complex (marked by MSL1, red) to the X chromosome in males and causes local spreading from the insertion site in polytene chromosomal immunostainings of salivary glands from male third instar larvae. DNA is stained with Hoechst 33342 (blue). Figure S7C contains the Hi-C counts from the insertion point on band 86F8 in S2 cells. C. (Left) Rescue of roX1SMC17A, roX2Δ double mutant (roXnull) male-specific lethality by daGal4-induced expression from a full-length (FL) UAS-roX1 transgene (#1), UAS-roX2 transgene (#2) or by expression driven by endogenous promoters upon translocation (TL) of roX1 (#3) or roX2 (#4) to the 3rd chromosome. All transgenes are inserted in the same autosomal location VK33. Transgenic males surviving to adulthood were counted and normalized to female siblings (100% viable) (~1100 flies/genotype). Columns show averages of n≥3 separate crosses ±SD. See Experimental Procedures for details of the genetic crosses. Below the graph, a schematic representation of the transgenes used. (Right) Two representative polytene chromosome squashes are shown for roXnull; UAS-roX1/2. D. A mutant form of roX RNA lacking important stem loop structures is expressed from the VK33 autosomal insertion. This causes inefficient targeting of the MSL complex (marked by MSL1, red) to the X chromosome and mislocalization of H4K16 acetylation (in green) as visualized on polytene chromosomal immunostainings of salivary glands from male third instar larvae (See Extended Experimental Procedures and Figure S7D for further details). E. As C. but intact nuclei of whole salivary glands (top), fat body cells (middle) and imaginal disc diploid cells (bottom). The label “X” indicates the X chromosome and arrowheads point at the region around the lacO-roX2 autosomal insertion. See also Figure S7.
The MSL complex can target a HAS independent of the proximity of roX RNA production site
As a testable prediction following the above observations we expected that autosomes would also display MSL spreading if a roX gene is placed in the appropriate region. In an adaptation of a classical rescue experiment (Meller, 2002) we next generated transgenic flies carrying as a sole source of roX lncRNA a roX2-insertion on the right arm of the third chromosome (3R) at a precise position (86F8) in a roX1/2 double mutant background. The inserted roX2 gene was complemented with an array of lac operon sequences and an EGFP-fused lacI as a reporter to visualize the transgene without the need of DNA FISH (see Extended Experimental Procedures). Consistent with previous reports (Kelley et al., 1999), roX RNA produced from the autosomal roX insertion properly targeted the X, enabling functional dosage compensation and restoring male viability. Moreover, the roX transgene was able to recruit the MSL complex to the ectopic insertion site where, in addition to local spreading of the MSL complex into direct flanking regions on the autosome (seen in salivary gland polytene chromosomes), we repeatedly detected MSL binding at band 88B on the third chromosome (Figure 5B). Interestingly, this cytological position resides ~2.6 Mb away from the insertion site, with no detectable binding within a ~1.7 Mb region in between targeted bands, supporting an involvement of the 3D conformation. Although S2 and clone-8 cells are not expected to share the 3D structure of polytene chromosomes, it is quite remarkable that, out of many other possibilities, we detect an enrichment of Hi-C contacts in S2 (Figure S7C) and clone-8 cells (not shown) between 86F and 88B. This suggests that HAS (such as roX loci) enable spreading of the MSL complex to interaction sites that can be far away on a linear scale and that do not necessarily need to be HAS themselves. We further confirmed this observation by two independent approaches involving the ectopic expression of roX1 and roX2 genes from an autosomal location (VK33 on chromosome 3L) in the roX1/2 double null background: insertion of UAS-driven roX1 or roX2 genes, and translocation of an X-chromosomal segment containing either roX1 or roX2 genes to the ectopic site. In all cases, the autosomally expressed roX fully rescued male lethality and induced MSL targeting to the X chromosome, as seen in polytene chromosome spreads (Figure 5C). A nonfunctional roX RNA, lacking important stem loop structures and expressed from VK33 under its native promoter, on the other hand, was not able to rescue male lethality and led to diminished or mis-targeting to the chromocenter (Ilik et al., 2013) (Figure 5D and Figure S7D). These experiments verify that the properties of the ectopic roX1/2 are independent of the promoter or location of the roX genes but require intact roX structure.
It has been proposed that the X chromosomal territory forms in a self-organizing process around the roX genes by attracting HAS (Grimaud and Becker, 2010). However, such a model seems incompatible with the targeting of the X chromosome observed for polytene chromosomes in the previous experiments, unless the autosomal roX2 was looping into the X chromosomal territory in order to physically reach the X chromosome. To test this, we analyzed intact nuclei from the LacO-roX2 line where we observed the formation of a distinct additional MSL territory surrounding the ectopic insertion of roX2, frequently distant to the main MSL territory in three different tissues (salivary glands and fat body (examples of polytenic tissues) as well as imaginal discs (diploid tissue)) (Figure 5E). These results suggest that physical proximity is not necessary for the transfer of roX RNA from its production site to its targets on the X, and can occur efficiently despite possibly distinct 3D chromosome conformation of different tissues. Still, we also show that spreading to spatially proximal regions, situated at a long genetic distance, is possible, as the extra MSL territory surrounding the insertion site appears discontinuous on polytene chromosome squashes (Figure 5B).
Ectopic insertion of roX-HAS leads to transcriptional up-regulation of neighboring autosomal genes in vivo
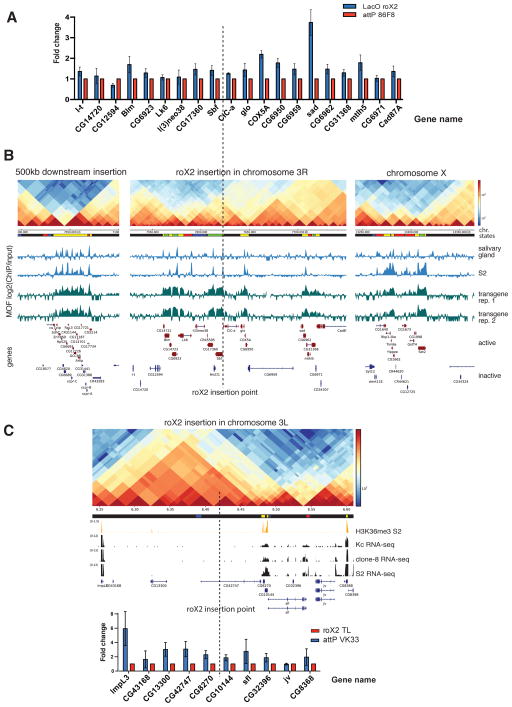
To further investigate the spreading of the MSL complex from ectopic roX2 HAS to nearby autosomal locations, we isolated RNA from flies with or without ectopic roX2 HAS and measured the expression of 10 genes upstream and 10 genes downstream of the insertion site. Active genes surrounding the ectopic roX2 HAS showed a consistent upregulation of expression in comparison to control flies (Figure 6A). To complement this analysis, we performed ChIP-seq with an antibody against MOF from the same transgenic larvae. On autosomes, MOF binds only to gene promoters, while on the X, it also binds to gene bodies ((Kind et al., 2008), Figure 6B right panel). In transgenic flies carrying an ectopic roX2, MOF binds not only to the bodies of active genes surrounding the autosomal insertion point but also at a distance from it (Figure 6B left and central panels). Importantly, we could detect MOF binding at the body of active genes up to 0.5 Mb upstream (Figure 6B) and 2 Mb downstream of the insertion site (data not shown). This suggests that the presence of a HAS on an ectopic autosomal location enables MOF to spread to proximal and distal regions on autosomes. To further validate the influence of ectopic roX insertion on the expression of neighboring genes, we also determined the expression of genes neighboring a different transgene used in this study, i.e. the X chromosomal translocation to 65B2 on chromosome arm 3L (VK33), where roX2 is expressed under its native promoter and also observed enhanced expression of 10 genes surrounding this insertion site (Figure 6C). Taken together, these observations corroborate that the presence of a roX2 HAS enables the MSL complex to spread distally from the site of insertion and that it has activating potential on gene expression in vivo.
Figure 6. Ectopic insertion of a HAS leads to spreading of MOF to and enhanced expression of neighboring autosomal genes.
A. HAS on a autosomal location causes an up-regulation of autosomal genes. Expression of autosomal genes neighboring roX2 insertion was analyzed by qRT-PCR. Flies analyzed were roX double mutant (roXnull) combined with either a transgene carrying LacO-roX2 (under the endogenous promoter) (LacO-roX2, blue bars) or the attP transposon only in the same insertion site (AttP 86Fb, red bars). Relative gene expression levels of LacO-roX2 flies were calculated over that of AttP 86Fb flies. Data are represented as mean of triplicates ± SEM. B. MOF spreads to distal regions from the roX insertion site on chromosome 3R. The tracks shown from top to bottom are: Hi-C contacts from S2; chromatin states (Filion et al., 2010); input-normalized MOF ChIP-seq signals for third instar larva salivary gland (Conrad and Akhtar, 2012); S2 cells (Straub et al., 2013) and LacO-roX2 flies (2 replicates). The Hi-C heat map contains corrected counts from single restriction fragments. In the chromatin states track the yellow and red regions represent active chromatin, the black corresponds to repressive chromatin, and blue and green are heterochromatin. The tracks are shown for a 100 kb region, located 500 kb downstream of the roX2 insertion site (left), a 250 kb region around the insertion site (middle) and a 100 kb region on the X (right). Dotted line depicts autosomal insertion site of roX2 (chromosome 3R, 86F8). C. Expression of autosomal genes neighboring the roX2 insertion was analyzed by qRT-PCR. Flies analyzed were roX double mutant (roXnull) combined with either a translocation of roX2 (roX 2 TL) to an insertion site on chromosome 3L (VK33) or the attP transposon alone in the same VK33 insertion site. Relative gene expression levels (± SEM) of roX2 TL flies were calculated over that of AttP VK33 flies. Data are represented as mean of triplicates ± SEM. Genes in repressive or heterochromatin have low or no up-regulation (e.g. CG14720, CG15594 in A. and CG43168, jv in C.) H3K36me3 and RNA-seq tracks are used to indicate gene transcription activities in different cell lines.
Nucleosome positioning at HAS is dependent on the presence of MSL2
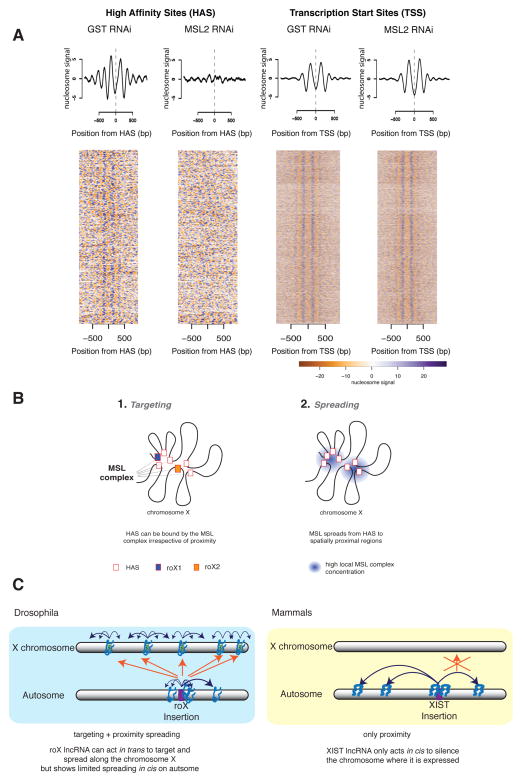
Investigating functional HAS more closely, we observed that they tend to display increased DNaseI hypersensitivity (Figure S7E). We therefore asked whether the local chromatin structure of HAS can be influenced by the MSL complex. To this end, we mapped the positioning of nucleosomes in S2 cells by treating the chromatin with micrococcal nuclease followed by deep sequencing (MNase-seq) (Mavrich et al., 2008). We found nucleosome depleted regions on HAS, flanked by well-positioned nucleosomes (Figure 7A). To test if the MSL complex has any roles in maintaining the nucleosome configuration, we performed MNase-seq analysis in S2 cells depleted of MSL2. Strikingly, the nucleosome pattern around HAS was lost upon depletion of MSL2 (p-values < 2.2e-16, see Extended Experimental Procedures), while it was preserved at TSS. We conclude that the MSL complex is crucial for the maintenance of the local nucleosome arrangement specifically at HAS.
Figure 7. Depletion of MSL complex severely affects nucleosome positioning at HAS but not at TSS.
A. Summary plots (top) and heat maps (bottom) showing normalized nucleosome occupancy for regions centered around HAS (left, n= 257) and active TSS (right, n= 5985) in control (GST RNAi) and MSL2-depleted cells (MSL2 RNAi). Scale bar representing nucleosome signal is shown below. B. Conformation-based affinity model: we propose a model in which (1.) chromosome X is targeted via HAS by the MSL complex independently of spatial proximity. Then in (2.) the complex spreads from the HAS to spatially proximal regions. In (1.), the MSL complex binds specific regions of chromosome X containing a sequence motif, (the MRE), if this appears at the end of active genes that are at TAD boundaries. In (2.) the complex spreads (probably by diffusion) from HAS to spatially close regions. Because TAD boundaries appear enriched in contacts within each other as well as to other regions it is expected that they are physically close, thus offering an optimal location from which to reach all active genes on chromosome X. C. Comparison of ectopic expression of roX RNA versus Xist RNA on autosomes. Blue arrows denote spreading of the lncRNA in 3D proximity; orange arrows denote targeting of the lncRNA in trans. (Blue box) In Drosophila, ectopically expressed roX displays restricted spreading on the autosome indicating that spatial proximity is beneficial but not sufficient for roX RNA spreading. Importantly, roX RNA expressed from an autosomal location specifically targets the X in trans, leading to coating of the entire X chromosome by roX/MSL complex independent of spatial proximity. (Yellow box) In mammals, ectopically expressed Xist RNA can spread and coat the autosome using spatial proximity (Engreitz et al., 2013; Jiang et al., 2013; Simon et al., 2013) without targeting the X in trans. See also Figure S7.
Discussion
This study provides a first step towards understanding the role of chromosome conformation in dosage compensation in Drosophila melanogaster. We observe that HAS, the landing regions of the MSL complex on the X chromosome, frequently reside in proximity to TAD boundaries. We demonstrate that HAS are enriched in Hi-C contacts to each other and to other X chromosomal regions and that this organization remains comparable between male and female cells.
“Conformation-based affinity” model explains MSL complex targeting and spreading on the X chromosome
Our analysis revealed that HAS are characterized by a combination of DNA sequence (MREs), chromatin state (active), and gene architecture which drive the specificity of the MSL complex towards the X chromosome (targeting) (Figure 7B, (1)). Our data suggest that once the MSL complex binds to HAS, it then spreads (either via an active mechanism or via diffusion) to spatially close regions to place the H4K16ac mark on active genes (Figure 7B, (2)). We propose a “conformation-based affinity” model based on the strategic location of HAS at highly interconnected regions of the D. melanogaster X chromosome that efficiently distribute the MSL complex over the X by attracting the MSL complex to cis-interacting HAS on the X, this system ensures that only this chromosome is specifically and globally targeted. By spreading from those HAS over short (3D) distances, all active genes on the X chromosome are then reached and acetylated without influencing the autosomes. We suggest that this system is resilient to major perturbations exemplified by the large autosomal insertion from chromosome 3L and the ectopic expression of the roX genes that produce viable cells and flies, respectively (Figure 5).
MSL complex is crucial for maintaining nucleosome free regions at HAS
Our MNase-seq analysis shows for the first time a direct effect of the MSL complex on nucleosome organization specifically on HAS (Figure 7A) and not on TSS, albeit prominent binding of MSL1/2 to promoter regions. The MSL complex may act similar to a pioneer DNA binding protein (Magnani et al., 2011) to establish nucleosome patterns at HAS and may act on neighboring active regions rather than modifying TAD boundaries. This system may be unique to flies as the Drosophila dosage compensation evolved a fine-tuning transcription activation mechanism rather than a complete shutdown of gene transcription as seen in mammalian X inactivation. It would be very interesting to see how nucleosome positioning is affected upon Xist binding in mammals.
HAS locate at regions with abundant long-range contacts to facilitate spreading
Although many factors, including CTCF as well as tRNA and housekeeping genes have been shown to be enriched at boundaries (Dixon et al., 2012; Hou et al., 2012; Sexton et al., 2012), by dissecting the targeting and spreading activity of the MSL complex for the X chromosome, we offer a plausible explanation behind the advantages of HAS localization: HAS are enriched at the X chromosomal boundaries and not at autosomal boundaries where all other boundary factors will bind indiscriminately. Furthermore, we find that the few HAS that are not near a boundary also occupy locations of elevated number of long-range contacts (Figure S5A) indicating that HAS form interaction hubs for the spreading of the MSL complex.
Activity of roX genes is associated with a higher abundance of long-range contacts
Hi-C as well as in vivo immunofluorescence show that active roX genes have more contacts and are closer to each other than inactive regions (Figure 3). These observations are in line with previous reports showing that active chromatin compartments interact more often with each other (Lieberman-Aiden et al., 2009; Sexton et al., 2012) and that active chromatin localizes to the interface of the chromosomal territory (Nagano et al., 2013). Our results imply that different transcriptional programs in each cell line or tissue are likely to be associated with a particular arrangement of long-range contacts, suggesting that the dosage compensation must be flexible to act over such diverse conformations without disturbing them. This idea is consistent with the observation that the chromosome conformation remains unchanged after knockdown of the MSL complex (Figure S6A-D), and stays in contrast to mammalian X inactivation, which involves chromatin condensation, gene inactivation, and alterations in chromosome conformation (Nora et al., 2012).
lncRNAs work differently in coordinating fly and mammalian dosage compensation
Dosage compensation mechanisms in flies and mammals lead to opposite outcomes, namely gene activation versus gene repression, yet both systems use lncRNAs transcribed from the dosage compensated X chromosome: roX1 and roX2 RNA are expressed from the male hyperactivated X chromosome in D. melanogaster while Xist is expressed from the inactivated X in mammalian females (Brockdorff and Turner, 2015; Grimaud and Becker, 2010). Recent work has shown that Xist spreads to distal sites on the X chromosome. Interestingly, this spreading is dependent on the spatial proximity of sites distal to the Xist gene (Engreitz et al., 2013; Simon et al., 2013). This is further exemplified by ectopic expression of Xist from chromosome 21, where Xist spread only in cis on this chromosome (Jiang et al., 2013). In our study, ectopic insertion of roX transgenes on autosomes demonstrated that the roX/MSL complex can reach the X and rescue male lethality (Figure 5B-D). Thus, acting in trans is a special feature of roX RNAs (in conjunction with the MSL complex), not observed for Xist, indicating that the two systems utilize the respective lncRNAs differently. In both systems though, the lncRNAs need to be functional, as the stem loop structures of the roX RNAs are required for dosage compensation in D. melanogaster (Ilik et al., 2013) (Figure S7D), while Xist needs the “A-repeat domain” to induce mammalian X inactivation (Engreitz et al., 2013). The distinct mechanisms utilized by the Xist and roX RNAs exemplify the great versatility by which lncRNAs can be involved in the global regulation of single chromosomes and might reflect important differences between the two systems: in mammals, only one of the two X chromosomes needs to be inactivated. Therefore, a trans action of Xist RNA on the sister X would be detrimental to the organism. In contrast, the dosage compensated X chromosome is present singularly in males in Drosophila. However, since the roX RNAs can act in trans it may be disadvantageous to target the activating MSL complex to active genes on autosomes; hence the need for specific target regions (the HAS) unique to the X.
Faster evolution of the X chromosome may favor positioning of HAS to interaction hubs
To fully understand the occurrence of HAS at sites with extensive long-range interactions on the X chromosomes, it could be helpful to consider evolutionary models proposing that X chromosomes tend to evolve faster than autosomes (faster-X effect) (Vicoso and Charlesworth, 2006): under the faster-X effect, traits only beneficial for the males can introduce significant changes specific to the X chromosome on a short evolutionary time scale (Parsch and Ellegren, 2013; Vicoso and Charlesworth, 2006). Based on these and other observations suggesting that the X chromosome in flies is different to autosomes (Alekseyenko et al., 2012; Gallach, 2014; Meisel and Connallon, 2013; Zhang and Oliver, 2010), we assume that selective pressures on males favored the occurrence of HAS at regions of increased interactions like TAD boundaries. Future analyses of different Drosophila species will open exciting opportunities to study the evolutionary changes of HAS in the context of X chromosomal architecture. Moreover, conformation-based affinity could be a generic mechanism for other regulatory elements to exert their functions. It has to be seen in which contexts in cis versus in trans action of different lncRNAs is essential for their function and how chromosome conformation, long-range contacts, HAS and regulation of transcription have co-evolved for dosage compensation.
Experimental Procedures
Hi-C experimental procedure and analysis
Hi-C in S2 or clone8 cells using HindIII as restriction enzyme was carried out as described by Belton et al. (Belton et al., 2012) with the following minor modifications: Starting material for all samples was 50 million insect cells per sample. After lysis, samples were taken up in 125μl and split into two aliquots of 50μl (Hi-C samples) and the remaining 25μl (3C control) were used to adjust for the smaller size of the Drosophila genome as compared to mammalian cells. Accordingly, for each 3C control only half the volumes per tube compared to the original protocol were used. For details see Extended Experimental Procedures.
4C-seq experimental procedure and analysis
4C-seq in S2 and Kc cells was carried out as described by Splinter et al. (Splinter et al., 2012) with minor modifications as follows: 50-100 million S2 cells or Kc cells, fixed as described above, were used for two biological replicates. DpnII (NEB) was used as the primary and Csp6I (ThermoScientific) as the secondary restriction enzyme. For each viewpoint, two 160ng PCR reactions (8 cycles with 55°C annealing temperature, followed by 18 cycles with 63°C) were prepared and cleaned up. All different viewpoint libraries for one biological replicate were mixed equimolarly and sequenced on separate lanes on an Illumina HiSeq2500 DNA sequencer. Primer sequences and coordinates for the experiments are found in Table S4.
Fly culture and genetics
Details of fly culture conditions and genetics is explained in Extended Experimental Procedures.
Fluorescence in situ hybridization (FISH)
Fluorescence in situ hybridization procedures were performed as described previously (Vaquerizas et al., 2010). To perform DNA FISH, approximately 10-5 kb regions were chosen in the genome and amplified by PCR from genomic DNA with 5–10 primers pairs, each covering around 0.5–3 kb. Primer sequences are available on request. roX1 probe sequence was taken from (Vaquerizas et al., 2010). roX2 and dpr8 probe sequences was taken from Grimaud and Becker (Grimaud and Becker, 2009). HAS1 and HAS2 probes were designed and generated in this study.
Nucleosome positioning analysis in Drosophila
Nucleosome positioning analysis using MNase-seq was performed essentially as described in (Mavrich et al., 2008). For details of the analysis see Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We thank all members of the Akhtar laboratory for discussions and especially F. Dündar, A. Gaub, T. Aktas, T. Khanam, T. Chelmicki, I. Ilik and N. Iovino for critical reading of the manuscript. We thank G. Arib, I. Ilik for generating the lacO-roX2 and UAS-roX1/2 fly lines, respectively, M. Shvedunova for initial help with imaging, A. Panhale for culturing clone8 cells and N. Gutierrez for support on the generation of FISH probes. We thank U. Bönisch and E. Betancourt for help with deep sequencing. This work was supported by EU-funded “EpiGeneSys”, DFG:CRC992 and DFG:CRC746 awarded to AA. AA is also part of the DFG-funded BIOSSII excellence cluster.
Footnotes
Accession numbers
Hi-C, 4C-seq and ChIP-seq raw and processed data are available at the Gene Expression Omnibus (GEO) site under accession number GSE58821.
Supplemental information includes seven supplemental figures, five supplemental tables, and Extended Experimental Procedures.
Author contribution
Conceptualization, F.R., T.L., T.M., A.A.; Methodology, F.R., T.L., S.T., K.L.; Software, F.R.; Formal Analysis, F.R., T.L., S.T., K.L., H.C.; Investigation, F.R., T.L., S.T., K.L., P.G.; Resources, P.G., B.L., Y.Z., J.D., E.d.W., W.d.L.; Data Curation, F.R. and B.L.; Writing – Original Draft, F.R., T.L., S.T., and A.A.; Writing – Review & Editing, F.R., T.L., S.T., K.L., E.d.W., W.d.L., T.M. and A.A.; Visualization, F.R., T.L., S.T., K.L. and A.A.; Supervision, W.d.L., J.D., T.M. and A.A.; Funding Acquisition, A.A.
References
- Alekseyenko AA, Ho JWK, Peng S, Gelbart M, Tolstorukov MY, Plachetka A, Kharchenko PV, Jung YL, Gorchakov AA, Larschan E, et al. Sequence-Specific Targeting of Dosage Compensation in Drosophila Favors an Active Chromatin Context. PLoS Genet. 2012;8:e1002646. doi: 10.1371/journal.pgen.1002646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee O-K, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. A Sequence Motif within Chromatin Entry Sites Directs MSL Establishment on the Drosophila X Chromosome. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi–C: A comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Turner BM. Dosage Compensation in Mammals. Cold Spring Harbor Perspectives in Biology. 2015;7:a019406. doi: 10.1101/cshperspect.a019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, Carlson JW, Landolin JM, Kapranov P, Dumais J, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Research. 2011;21:301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nature Publishing Group. 2012;13:123–134. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- Currie DA, Milner MJ, Evans CW. The growth and differentiation in vitro of leg and wing imaginal disc cells from Drosophila melanogaster. Development. 1988;102:805. [Google Scholar]
- de Wit E, Bouwman BAM, Zhu Y, Klous P, Splinter E, Verstegen MJAM, Krijger PHL, Festuccia N, Nora EP, Welling M, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973–1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic Protein Location Mapping Reveals Five Principal Chromatin Types in Drosophila. Cells. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M. Recurrent turnover of chromosome-specific satellites in Drosophila. Genome Biol Evol. 2014;6:1279–1286. doi: 10.1093/gbe/evu104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, Becker PB. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. 2006;20:858–870. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Becker PB. The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. 2009;23:2490–2495. doi: 10.1101/gad.539509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Becker PB. Form and function of dosage-compensated chromosomes - a chicken-and-egg relationship. BioEssays. 2010;32:709–717. doi: 10.1002/bies.201000029. [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. Gene Density, Transcription, and Insulators Contribute to the Partition of the Drosophila Genome into Physical Domains. Mol Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, Toscano S, Wan Y, Spitale RC, Luscombe N, Backofen R, et al. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol Cell. 2013;51:156–173. doi: 10.1016/j.molcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A-M, Allgardsson A, Stenberg P, Larsson J. msl2 mRNA is bound by free nuclear MSL complex in Drosophila melanogaster. 2011;39:6428–6439. doi: 10.1093/nar/gkr236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CI, Akhtar A. The MSL complex: juggling RNA-protein interactions for dosage compensation and beyond. Curr Opin Genet Dev. 2015;31:1–11. doi: 10.1016/j.gde.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic Spreading of the Drosophila Dosage Compensation Complex from roX RNA Genes into Flanking Chromatin. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, Akhtar A. Genome-wide Analysis Reveals MOF as a Key Regulator of Dosage Compensation and Gene Expression in Drosophila. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- Liang J, Lacroix L, Gamot A, Cuddapah S, Queille S, Lhoumaud P, Lepetit P, Martin PGP, Vogelmann J, Court F, et al. Chromatin immunoprecipitation indirect peaks highlight long-range interactions of insulator proteins and Pol II pausing. Mol Cell. 2014;53:672–681. doi: 10.1016/j.molcel.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani L, Eeckhoute J, Lupien M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet. 2011;27:465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Connallon T. The faster-X effect: integrating theory and data. Trends Genet. 2013;29:537–544. doi: 10.1016/j.tig.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. Embo J. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, Ellegren H. The evolutionary causes and consequences of sex-biased gene expression. Nat Rev Genet. 2013;14:83–87. doi: 10.1038/nrg3376. [DOI] [PubMed] [Google Scholar]
- Quinn JJ, Ilik IA, Qu K, Georgiev P, Chu C, Akhtar A, Chang HY. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32:933–940. doi: 10.1038/nbt.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651–663. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USa. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. Embo J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Wit E, van de Werken HJG, Klous P, de Laat W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods. 2012;58:221–230. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The Chromosomal High-Affinity Binding Sites for the Drosophila Dosage Compensation Complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Zabel A, Gilfillan GD, Feller C, Becker PB. Different chromatin interfaces of the Drosophila dosage compensation complex revealed by high-shear ChIP-seq. Genome Research. 2013;23:473–485. doi: 10.1101/gr.146407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, Hufton A, Heinig M, O'Keeffe S, Masri NE, Roider HG, Manke T, Vingron M. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat Protoc. 2011;6:1860–1869. doi: 10.1038/nprot.2011.409. [DOI] [PubMed] [Google Scholar]
- Van Bortle K, Nichols MH, Li L, Ong CT, Takenaka N, Qin ZS, Corces VG. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Drosophila Genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Oliver B. An evolutionary consequence of dosage compensation on Drosophila melanogaster female X-chromatin structure? BMC Genomics. 2010;11:6. doi: 10.1186/1471-2164-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, Macalpine DM, Oliver B. Expression in Aneuploid Drosophila S2 Cells. PLoS Biol. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.